Summary
Low monocyte (m)HLA-DR expression is associated with mortality in sepsis. G-286A∗rs3087456 polymorphism in promoter III of HLA class II transactivator (CIITA), the master regulator of HLA, has been associated with autoimmune diseases but its role in sepsis has never been demonstrated. In 203 patients in septic shock, GG genotype was associated with 28-day mortality and mHLA-DR remained low whereas it increased in patients with AA or AG genotype. In ex vivo cells, mHLA-DR failed to augment in GG in comparison with AG or AA genotype on exposure to IFN-γ. Promoter III transcript levels were similar in control monocytes regardless of genotype and exposure to IFN-γ. Promoter III activity was decreased in GG genotype in monocyte cell line but restored after stimulation with IFN-γ. Hereby, we demonstrated that G-286A∗rs3087456 significantly impact mHLA-DR expression in patients with septic shock in part through CIITA promoter III activity, that can be rescued using IFN-γ.
Subject areas: Health sciences, Genetics, Immunology
Graphical abstract
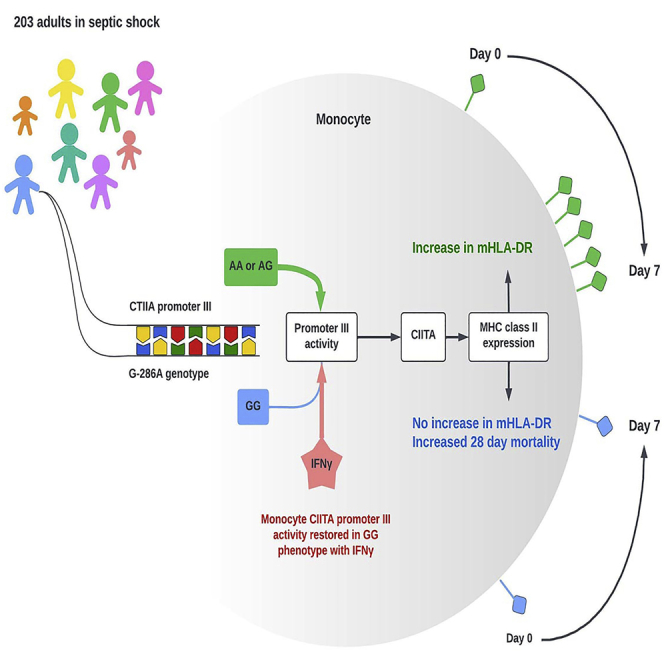
Highlights
-
•
CIITA G-286A∗rs3087456 polymorphism impact mortality in septic shock
-
•
CIITA G-286A∗rs3087456 reduces promotor activity and influences mHLA-DR expression
-
•
Downregulatory role of this SNP on mHLA-DR can be reverse by IFN-γ in septic patients
Health sciences; Genetics; Immunology.
Introduction
Sepsis is a life-threatening condition because of a dysregulated immune response to infection (Singer et al., 2016), and leads to more than 6 million deaths per year (Fleischmann et al., 2016; Reinhart et al., 2017). Dysregulated immune response is currently considered as a paradigm in pathogenesis of sepsis and recognized as an important component of sepsis prognosis (Venet and Monneret, 2018). Although the management of sepsis is well standardized (Evans et al., 2021), specific immunomodulatory therapies, such as high dose steroids (Minneci et al., 2009), anti-endotoxin (Ziegler et al., 1991), inhibitors of pro-inflammatory cytokines IL-1, TNF-α, IL-6 (Eichacker et al., 2002; Shakoory et al., 2016), and anticoagulants such as activated protein C (Abraham et al., 2005; Warren et al., 2002) have shown equivocal results in patients with sepsis (Fang et al., 2019; Suffredini and Munford, 2011). Despite this, post-hoc analyses have shown specific benefits (or harms) in groups of patients when stratified by immunological phenotype or transcriptomic profile (Antcliffe et al., 2019; Kernan et al., 2019; Shakoory et al., 2016), suggesting a role for personalized immunomodulation in sepsis. Recently, therapies targeting the restoration of monocyte (m)HLA-DR level with interferon (IFN)-γ as gain interest to reverse sepsis-induced immunodepression (Nguyen et al., 2021; Payen et al., 2019) (ClinicalTrials.gov Identifier: NCT01649921, NCT01374711).
Decreased expression of HLA-DR – an MHC class II molecule required for antigen presentation to T cells – on the cell surface of circulating monocytes is widely recognized as a marker of sepsis-induced immunodepression and is strongly associated with both sepsis-induced mortality and increased risk of secondary infections (Conway Morris et al., 2018; Hotchkiss et al., 2013; Lukaszewicz et al., 2009). MHC class II transcript expression is controlled by the class II transactivator (CIITA), a highly conserved regulatory module found in the genes encoding the α-chain and β-chain of all classical human MHC class II molecules (HLA-DP,-DQ, and-DR) (Ting and Trowsdale, 2002). In humans, control of CIITA activity is mediated by three distinct promoters, pI (expressed in cells of myeloid origin), pIII (expressed in cells of lymphoid origin) and pIV (expressed in cells of non-hematopoietic origin when stimulated by IFN-γ), although the cell-specific expression and response to IFN-γ increasingly appears interchangeable (Reith et al., 2005; Zinzow-Kramer et al., 2012). Absence of CIITA promoter activity results in a failure to express HLA-DR molecules and severe immunodeficiency (bare lymphocyte syndrome). (Mach et al., 1994; Steimle et al., 1993). In addition, G-286A∗rs3087456 single nucleotide polymorphism (SNP) in the CIITA promoter pIII has been associated with rheumatological disease and cancer in some cohorts (Bronson et al., 2008; Eike et al., 2012; Harrison et al., 2007; Swanberg et al., 2005; Xue et al., 2020). Moreover, Vasseur et al. provided some evidence of a positive selection of the 5′ genomic region of CIITA (Vasseur et al., 2012). The positive selection, detected by LD-based tests and Fst tests, identified as highly differentiated in Europe (27% have a G variant, 73% an A). In addition, SNP G-712A∗rs12596540 is shown to be in high linkage disequilibrium (r2>0.6) with SNP G-286A∗rs3087456 (Vasseur et al., 2012). A systematic investigation of regulatory variants on monocyte gene expression showed that differential regulation between individuals was dependent on innate immune activity, known as context-dependent expression quantitative loci (eQTLs) (Fairfax et al., 2014). Considering the role of CIITA in the regulation of HLA-DR, and its association with immune-inflammatory diseases, we therefore investigated the relationship between genetic variation in the CIITA promoter, HLA-DR expression and outcome. We hypothesized that linked SNPs in the CIITA promoter pIII (G-286A∗rs3087456, characterized by the presence of G>An SNP at chr16:10877045, GRCh38.p13 or G-712A∗rs12596540, characterized by A>G at chr16:10876619, GRCh38.p13 (Vasseur et al., 2012)) would affect mHLA-DR expression kinetics in patients with sepsis. Conversely, we hypothesized that eQTLs may be modulated by IFN-γ treatment in septic patients and therefore investigated the molecular and cellular regulatory mechanisms.
Results
Severity and mortality in septic shock in association to CIITA G-286A∗rs3087456 and G-712A∗rs12596540 genotypes
Of 221 patients with septic shock recruited to the study, daily data from day of ICU admission to ≥7 days and genomic DNA was available from 203 patients (Figure S1). The median age of the study cohort was 61 years (range: 18-94), with a majority of men (65%) (Table 1). The cohort had a high degree of disease on admission, as indicated by a median SOFA score of 7 (range: 1-16) (Table 1). Almost a quarter of the patients were surgical patients (23.7%) and the majority of sepsis (53.7%) was of pulmonary origin (Table 1).
Table 1.
Baseline characteristics of patients in septic shock
| G-286A∗rs3087456 |
G-712A∗rs12596540 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA (n = 92) | AG (n = 84) | GG (n = 27) | All (n = 203) | P Value ∗ | AA (n = 113) | AG (n = 76) | GG (n = 14) | All (n = 203) | P Value ∗ | |
| Age, years | 62 (18-91) | 60 (19-94) | 63 (30-88) | 61 (18-94) | 0.25 | 62 (18-91) | 59 (19-94) | 58 (33-88) | 61 (18-94) | 0.73 |
| Gender, % male | 68.5 | 59.5 | 70.4 | 65 | 0.38 | 66.4 | 63.2 | 64.3 | 65 | 0.90 |
| SAPS II | 49 (22-120) | 49 (15-96) | 45 (19-93) | 49 (15-120) | 0.44 | 48 (22-120) | 50 (15-96) | 44 (19-65) | 49 (15-120) | 0.58 |
| SOFA | 7 (2-15) | 7 (2-16) | 7 (1-16) | 7 (1-16) | 0.87 | 7 (2-15) | 7 (2-16) | 7 (1-11) | 7 (1-16) | 0.91 |
| Surgical, % | 22.8 | 25 | 22.2 | 23.6 | 0.93 | 21.2 | 26.3 | 28.6 | 23.6 | 0.65 |
| Sepsis origin, % | ||||||||||
| Lung | 54.3 | 50 | 59.3 | 53.7 | 0.67 | 56.7 | 47.4 | 64.3 | 53.7 | 0.36 |
| Abdomen | 20.7 | 22.6 | 14.8 | 20.7 | 0.68 | 20.4 | 21.1 | 21.4 | 20.7 | 0.99 |
| Urinary | 4.3 | 4.8 | 3.7 | 4.4 | 0.97 | 3.5 | 5.3 | 7.1 | 4.4 | 0.75 |
| Laboratory variables | ||||||||||
| Leukocyte count, x 103/mm3 | 11.4 (0.1-55) | 12.4 (0.1-92.5) | 9.1 (0.4-32.7) | 11.4 (0.1-92.5) | 0.76 | 10.7 (0.1-55) | 12.3 (0.1-92.5) | 15.8 (1.8-32.7) | 11.4 (0.1-92.5) | 0.42 |
| Neutrophil count, x 103/mm3 | 10.1 (0.1-46.2) | 10.4 (0.1-74.9) | 8.4 (0.5-29.3) | 9.1 (0.1-74.9) | 0.56 | 9.3 (0.1-46.2) | 10.4 (0.1-74.9) | 9.7 (1-28.4) | 9.1 (0.1-74.9) | 0.84 |
| Lymphocyte count, x 103/mm3 | 0.7 (0.1-11.9) | 0.6 (0.1-5.8) | 0.9 (0.2-2.5) | 0.7 (0.1-11.9) | 0.17 | 0.6 (0.1-11.9) | 0.7 (0.1-3.7) | 1.7 (0.2-2.5) | 0.7 (0.1-11.9) | 0.005 |
| Monocyte count, x 103/mm3 | 0.5 (0.1-2.7) | 0.6 (0.1-4.6) | 0.5 (0.1-7.9) | 0.6 (0.1-7.9) | 0.87 | 0.5 (0.1-2.9) | 0.6 (0.1-4.6) | 0.9 (0.1-7.9) | 0.6 (0.1-7.9) | 0.24 |
SAPS II: Simplified Acute Physiology Score II; SOFA = Sequential Organ Failure Assessment. Data are median (interquartile range) for continuous variable. ∗p values were calculated with the use of chi-square test and Kruskal-Wallis test.
Allele frequency and genotype distribution were G-286A∗rs3087456: AA 45%, AG 41%, GG 13%; G-712A∗rs12596540: AA 56%, AG 37%, GG 7%. Both polymorphisms met the Hardy Weinberg law in our cohort. No significant association was found for both SNPs with clinical severity scores (SAPS II and SOFA scores) at day 1 of ICU admission, although patients who had the CIITA G-712A∗rs12596540 GG genotype had a higher lymphocyte count at day 1 (p=0.005, Table 1). In an A-dominant model, patients homozygous for CIITA pIII G-286A∗rs3087456 GG genotype had a significant increased risk of 28-day mortality (55.6% for GG genotype vs. 35.2% for AA and AG genotypes, OR: 2.298, CI 95% [1.013-5.217], p = 0.043) (Table 2). There was also a non-significant trend toward an increase in 7-day mortality for GG genotype for G-712A∗rs12596540 (AA or AG: 25.9% vs. GG: 50%, OR: 2.857, CI 95% [0.954-8,558], p = 0.05) (Table 2). No association was found for secondary infections between the different genotypes of CIITA pIII gene in both SNPs (Table 2). We further evaluated the impact of both SNPs on mHLA-DR expression in septic shock patients, using the A-dominant model.
Table 2.
Genotyped related mortality
|
A-dominant model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G-286Aars3087456 |
G-712Aars12596540 |
|||||||||
| AA or AG (n = 176) | GG (n = 27) | OR | 95% CI | P Valuea | AA or AG (n = 189) | GG (n = 14) | OR | 95% CI | P Valuea | |
| 28-daymortality, n (%) | 62 (35.2) | 15 (55.6) | 2.298 | 1.013-5.217 | 0.043 | 69 (36.5) | 8 (57.1) | 2.319 | 0.773-6.960 | 0.13 |
| 7-day mortality, n (%) | 46 (50) | 10 (37) | 1.662 | 0.710-3.891 | 0.24 | 49 (25.9) | 7 (50) | 2.857 | 0.954-8.558 | 0.05 |
| Secondary infections, n (%) | 39 (20.7) | 4 (14.8) | 0.611 | 0.199-1.872 | 0.38 | 41 (21.7) | 2 (14.3) | 0.602 | 0.129-2.796 | 0.49 |
|
Co-dominant model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G-286Aars3087456 |
G-712Aars12596540 |
|||||||||
| AA (n = 92) | AG (n = 84) | GG (n = 27) | All (n = 203) | P Valuea | AA (n = 113) | AG (n = 76) | GG (n = 14) | All (n = 203) | P Valuea | |
| 28-day mortality, n (%) | 35 (38) | 27 (32.1) | 15 (55.6) | 77 (37.9) | 0.10 | 45 (39.8) | 24 (31.6) | 8 (57.1) | 77 (37.9) | 0.16 |
| 7-day mortality, n (%) | 25 (27.2) | 21 (25) | 10 (37) | 56 (27.6) | 0.47 | 31 (27.2) | 18 (23.7) | 7 (50) | 56 (27.6) | 0.13 |
| Secondary infections, n (%) | 19 (20.7) | 20 (23.8) | 4 (14.8) | 43 (21.2) | 0.61 | 23 (20.3) | 18 (23.7) | 2 (14.3) | 43 (21.2) | 0.70 |
p values were calculated with the use of chi-square test and Kruskal-Wallis test.
Monocyte HLA-DR surface expression in septic shock in association with CIITA G-286A∗rs3087456 and G-712A∗rs12596540 genotypes
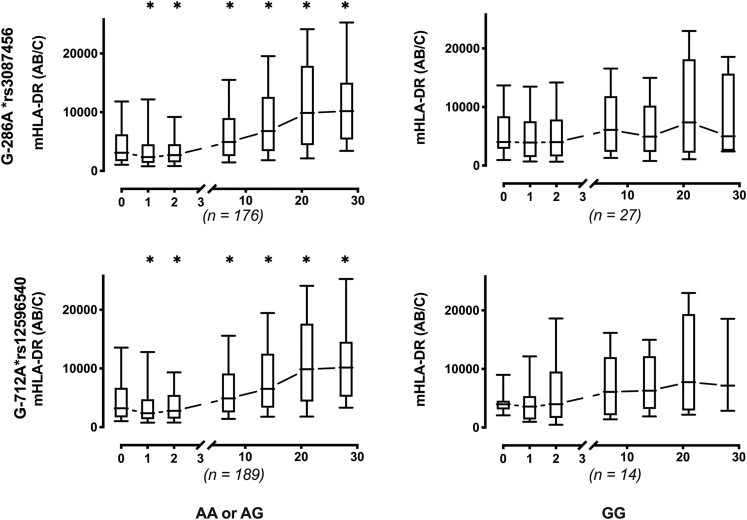
Longitudinal measurements of circulating mHLA-DR expression were measured during the ICU stay. For G-712A∗rs12596540 and G-286A∗rs3087456, septic patients with AA or AG genotype showed a significant early increase in mHLA-DR level, whereas patients with GG genotype displayed a persistent low mHLA-DR values (Wilcoxon signed rank test, p< 0.03 vs. basal) (Figure 1).
Figure 1.
Monocyte HLA-DR expression in 203 patients with septic shock
Circulating monocyte HLA-DR expression is expressed as number of antibodies bound per cell (AB/C). Results are presented as median with 95% CI and by genotype of CIITA rs12596540 and G-286A∗rs3087456 in a dominant model. ∗p< 0.03 vs. basal (Wilcoxon signed rank test).
Ex vivo mHLA-DR surface expression in healthy controls following IFN-γ stimulation in association with G-286A∗rs3087456 and G-712A∗12596540
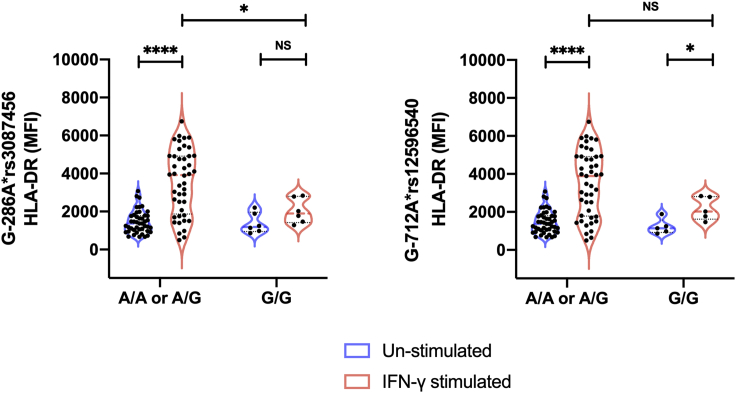
To investigate the functional role of CIITA pIII polymorphisms, we measured basal HLA-DR expression in monocytes from 50 healthy controls and evaluated the response of monocytes to IFN-γ exposure. Of these 50 healthy controls, 26 had the AA genotype, 18 had the AG genotype and 6 had the GG genotype for G-286A∗rs3087456, respectively; and 29 had the AA genotype, 15 had the AG genotype and 6 had the GG genotype for G-712A∗rs12596540, respectively. At baseline (no IFN-γ) there was no difference between different genotypes in both polymorphisms (G-286A∗rs3087456: AA or AG = 1371 [95%CI, 1154 to 1625] MFI vs. GG = 1192 [95% CI, 864 to 2207] MFI, p = 0.87; G-712A∗rs12596540: AA or AG = 1374 [95%CI, 1154 to 1642] MFI vs. GG = 1137 [95%CI, 864 to 1889] MFI, p =0.45; Figure 2). Following IFN-γ stimulation, mHLA-DR significantly increased in healthy controls with AA or AG genotype for both alleles (G-286A∗rs3087456: 1371 vs. 3919 MFI [CI 95%, 1521 to 2934], p< 0.0001; G-712∗rs12596540: 1374 vs. 3889 MFI [CI 95%, 1400 to 2843], p< 0.0001; Figure 2). In contrast, there was a non-significant trend to augment mHLA-DR expression from healthy controls with GG genotype for G-286A∗rs3087456 (1192 vs. 1898 MFI [CI 95%,−186 to 1652], p =0.09; Figure 2). Healthy controls with GG genotype for G-712A∗rs12596540 showed a significant ability to augment mHLA-DR expression in response to IFN-γ stimulation (G-712A∗rs12596540: 1137 vs. 2021 MFI [CI 95%, 132 to 1854], p< 0.05; Figure 2).
Figure 2.
Ex vivo HLA-DR expression in monocytes of healthy controls
Results are presented as violin plot with median and quartiles lines, and by genotype of CIITA G-712A∗rs12596540 and G-286A∗rs3087456 in an A-dominant model (results for the co-dominant model presented in Figure S2). p values were calculated using Mann-Whitney Test. ∗∗∗∗p< 0.0001 and ∗p< 0.05.
Those results need to be interpreted in regard to the effect of SNP G-286A∗rs3087456 on CIITA expression.
Ex vivo CIITA promoter isoforms expression in association with G-286A∗rs3087456
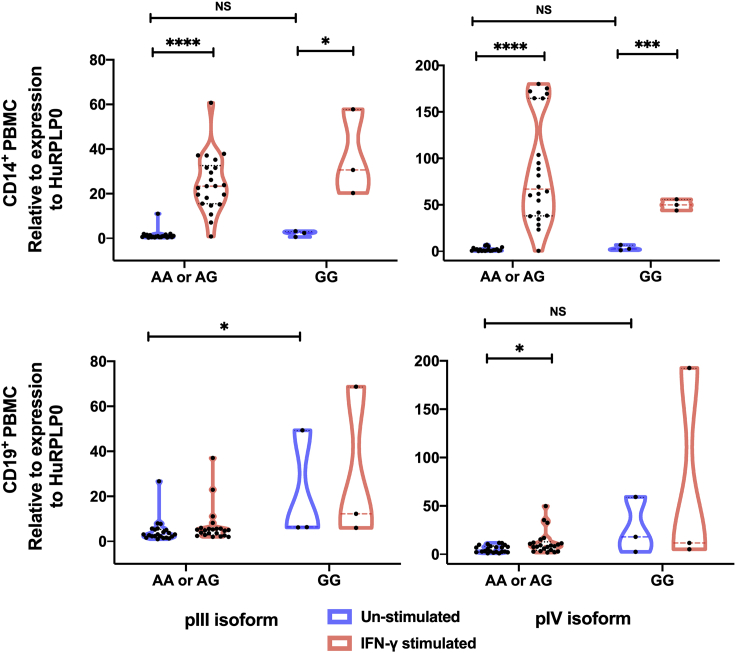
To determine whether the G-286A∗rs3087456 polymorphism was associated with different expression of isoforms of the CIITA promoter pIII and pIV, we quantified mRNA from ex vivo healthy control purified monocytes (CD14+) and B cells (CD19+) by Q-PCR. In monocytes, there was no difference in basal expression level of pIII (p = 0.18) and pIV transcripts (p = 0.24) between AA or AG and GG genotypes (Figure 3). Following 6 h of ex vivo cells stimulation with IFN-γ, pIII and pIV transcripts were significantly up-regulated in monocytes for AA or AG as well as GG genotype (p< 0.0001, Figure 3). In B cells, no difference was observed in pIII or pIV transcripts expression in unstimulated cells.
Figure 3.
Ex vivo mRNA levels for promoter III and IV isoforms
mRNA level in positively selected CD14+ (monocytes) and CD19+ (lymphocytes) from healthy controls. Cells were stimulated or not with IFN-γ during 6h before qPCR. Results are presented as truncated violin plot with median and quartiles lines and by genotype of CIITA G-286A∗rs3087456 in an A-dominant model. p values were calculated using Mann-Whitney Test. ∗∗∗∗p< 0.0001, ∗∗∗p< 0.001 and ∗p< 0.05.
Therefore, G-286A∗rs3087456 genotype did not impact ex vivo basal expression of CIITA promoter isoforms, and all three genotypes (AA, AG, GG) were similarly inducible by IFN-γ in monocytes from healthy controls, but not in B cells. We therefore evaluated the effect of G-286A∗rs3087456 on the transcriptional activity of the promoter CIITA pIII activity in different leukemoid cell lines.
In vitro CIITA type III promoter activity in association with rs3087456 genotype
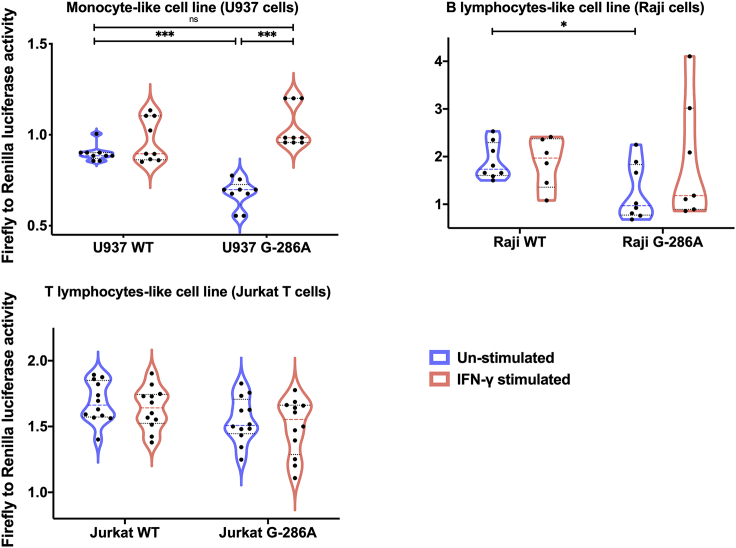
Dual luciferase reporter gene assays were used to detect specific CIITA promoter activity. Unexpectedly, after transfection into a monocyte-like cell line (U937), G-286A∗rs3087456 pIII activity was significantly lower than in wild-type (WT) construct (p< 0.001) (Figure 4). Following 6 h stimulation with IFN-γ, activity of G-286A∗rs3087456, pIII recovered a similar activity than WT promoter (p< 0.001) (Figure 4). As alternative isoforms of pIII are specific to B and activated-T cells, we transfected a B lymphocyte-like cell line (Raji) and a T lymphocyte-like cell line (Jurkat T). In B cells, luciferase activity of G-286A∗rs3087456 pIII was significantly lower in comparison with WT promoter (p< 0.05), and IFN-γ did not restore activity of mutated pIII (p = 0.23) (Figure 4). For T cells, transfection of G-286A∗rs3087456 pIII did not affect baseline promoter activity, nor was pIII activity increased in response to IFN-γ stimulation for transfected or WT promoter (Figure 4). Thus, although CIITA pIII basal activity was reduced in monocytes transfected with G-286A∗rs3087456 than WT, CIITA pIII activity was fully restored in G-286A∗rs3087456 following IFN-γ stimulation.
Figure 4.
In vitro transient transfection of U937, Raji and Jurkat T cells with CIITA pIII luciferase reporting plasmid
Transfected cells were stimulated during 6h with IFN-γ. CIITA promoter III activity is represented by Firefly to Renilla luciferase activity over empty vector. Results are presented as violin plot, truncated for Raji cells, with median and quartiles lines. WT, WildType. p values were calculated using Mann-Whitney Test. ∗∗∗p< 0.001 and ∗p< 0.05.
Discussion
In this study, we showed that G-286A∗rs3087456 CIITA pIII polymorphism is associated with an increase 28-day mortality in septic shock and a decreased ability to increase mHLA-DR expression across the trajectory of ICU admission. We showed that G-286A∗rs3087456 CIITA polymorphism impairs basal activity of pIII promoter without altering CIITA pIII and pIV isoforms expression, but this reduced activity is fully rescuable by IFN-γ. Difference in the induction profiles between in vitro promoter activity and ex vivo transcripts expression suggests that IFN-γ may act as a transcriptional inducer in G-286A∗rs3087456 but not in WT, where effect appears at a post-transcriptional level (e.g., transcript stabilization).
Genetic polymorphisms are widely recognized as important determinants of the host response to pathogens and have been strongly selected in human evolution (Quintana-Murci, 2019). Such polymorphisms may be pathogen-specific immunodeficiencies (Casanova and Abel, 2007), and/or only quantifiable following immune challenge, i.e. eQTLs, that are dependent on innate immune context (Fairfax et al., 2014). Here, we provide data supporting a functionally important variant of the CIITA promoter III on mHLA-DR expression in patients and ex vivo experiments, which in turn is associated with poor outcome in sepsis and may be a realistic target for personalized immunomodulation.
The effect of the G-286A∗rs3087456 polymorphism on the regulation of HLA-DR expression has not been previously studied. To our knowledge, this is the first study to investigate the regulatory effects of this promoter polymorphism. Our results suggest that GG genotype alters pIII activity in monocytes and B cells but not in T cells, in a cell-type specific manner. Unexpectedly, on basal condition (e.g., monocytes and B lymphocytes from healthy volunteers), pIII related transcripts induction is not significantly different between all haplotypes. Following ex vivo stimulation with IFN-γ, pIII transcripts expression was increased only in monocytes in both GG and AA or AG genotypes. Similarly, pIV transcripts expression is not different in the basal condition, but expression increases after IFN-γ only in monocytes. These suggest that, in healthy condition, transcriptional induction counteracts altered promoter activity of GG or repressor inhibit transcription in AA or AG haplotypes, both hypotheses encompassing complex transcription regulatory mechanisms.
This study was unable to identify the precise mechanisms by which CIITA pIII polymorphisms are associated with decreased mHLA-DR expression in individuals homozygous for G-286A∗rs3087456. This is unsurprising given the heterogeneity of transcriptional response during sepsis and complexity of HLA-DR gene regulation. Recently, immune-enhancing CpG and non-CpG oligodeoxynucleotides (ODNs) have been described as having a role in regulating expression of MHC class II and costimulatory molecules. Wang et al. (2007) showed that non-CpG ODNs (rODNs) downregulate HLA-DR expression in monocytes and block promoter III-directed transcription of CIITA in these cells. The precise inhibitory molecular effects of rODN on pIII activity are not understood, and more specifically, its precise promoter interaction not studied. Lohsen et al. (2014) described a distal regulatory element, known as hypersensitive site 1 - HSS1 (located ∼3kb upstream of pI) with PU.1, NF-κB, and Sp1 interacting with pIII through a looping interaction. Of interest, proximal regulatory elements were described, and include inducers (e.g., AP-1, Sp1, CREB, NF-Y, ARE-1, ARE-2), and repressors (e.g., PRDI-BF1/BLIMP-1, ZBTB32). Other mechanisms are involved in CIITA expression regulation. Among these, in B cells, restricted access of pI because of extensive DNA methylation induces a switch to pIII activation (Lohsen et al., 2014). The −300 region of CIITA has been well identified as an important proximal regulatory region (Ghosh et al., 1999). It is plausible that pIII G-286A∗rs3087456 polymorphism may interfere with transcription in monocyte during sepsis and CIITA expression be rescued through IFN-γ stimulation. Whether this may be directly related to a cis- or trans-regulatory mechanism is unknown. Ni et al. (2008) identified a dependent distal enhancer regulating IFN-γ-induce expression of CIITA through pIV. This IFN-γ induce promoter activation is dependent on BRG1, an ATP-dependent remodeling factor, which promotes the formation of a dynamic three-dimensional chromatin loop and demonstrates the importance of distant enhancer/silencer regions in CIITA expression regulation. In our study we showed that expression in monocyte of pIV isoforms following IFN-γ is ∼2.7-fold higher than pIII, which confirms the preferential activation of pIV after IFN-γ induction. Altogether, these facts support the evidence that IFN-γ is a preferential inducer of CIITA expression that may overcome monocytic MHC class II phenotype triggered by G-286A∗rs3087456 polymorphism in septic patients. The detailed transcriptional mechanism is unknown but the complexity of the regulation of CIITA expression renders difficult the precise determination out of high-throughput promoter analysis.
CIITA has been shown to regulate the expression of several non-MHC class II immune related genes such as interleukin (IL)-4, collagen α2, Fas ligand, and plexin A1 (Ting and Trowsdale, 2002). CIITA was able to inhibit MMP-9 expression, a matrix metalloproteinase involved in the degradation of extracellular matrix in lung injury associated with sepsis (Nozell et al., 2004). This effect was mediated by the binding and sequestration of CREB binding protein by CIITA, and therefore a reduction in promoter activity for MMP-9 – illustrating the complexity of CIITA-mediated signaling through transcriptional, and non-transcriptional pathways. Promoters of CIITA have previously been implicated in autoimmune diseases, highlighting their role in immunoregulation more broadly (Swanberg et al., 2005; Vasseur et al., 2012). Indeed, the greatest challenge to a true understanding of gene regulation in monocytes is likely to be the integration of proximal (cis) and distal (trans) regulatory effects on genetic loci in complexes disease, rather than in vitro (Fairfax et al., 2014).
Hereby, we have showed that homozygosity for G-286A∗rs3087456 was associated with low mHLA-DR expression in patients with sepsis, and that IFN-γ was able to restore CIITA promoter III activity. In the seminal article by Döcke et al. (1997), authors showed that IFN-γ was able to restore monocyte functions in 8 patients with sepsis. More recently, in a cohort of 20 pediatrics and adults’ patients with sepsis, we showed that immunotherapy with IFN-γ was safe to improve the immune host defense, particularly the monocyte HLA-DR expression in patients with prolonged downregulation of mHLA-DR, and was associated with a very high survival rate (Nguyen et al., 2021; Payen et al., 2019). These results may suggest the use of IFN-γ as a targeted immune therapy on G-286A∗rs3087456 homozygotes GG to rescue mHLA-DR expression. Leentjens et al. (2012) suggested in healthy volunteers with LPS-induced immunoparalysis that the modulatory effect of IFN-γ is variable, despite the significant increase in mHLA-DR expression. Of interest, GM-CSF did not induce a significant increase in mHLA-DR in this study (Leentjens et al., 2012). This is in agreement with the study of Hornell et al. (2003) showing that GM-CSF increase CIITA type I and III transcript expression but not of type IV. The use of mHLA-DR to assess the risk of secondary infection after septic shock is well admitted (Lukaszewicz et al., 2010; Wu et al., 2011). The use of a threshold of 8,000 AB/C to define a reduction in mHLA-DR expression to diagnose an acquired immunosuppression gets more credence, in particular if it is lasting after day 3 post admission (Monneret et al., 2006; Payen et al., 2019). Hereby, we showed that rs3087456 GG genotype was significantly associated with unresolving low mHLA-DR kinetics, predicting a persistent immunodepression phenotype in patients with septic shock.
This study raised number of questions. Among these, the specific effect of CIITA G-286A∗rs3087456 on CIITA transcripts expression in septic patients is unknown. It is probable that regulatory mechanism (e.g., acute phase response, cytokines, steroids, adrenergic drugs) encountered during sepsis may significantly modified both transcriptional and post-transcriptional regulation of CIITA, but regarding the rarity of this SNP, the necessity of immediate ex vivo experiments, such study may be difficult to realize.
This study provides two important insights: First, a genetic susceptibility affecting MHC class II master regulator CIITA has been shown to be associated with defective mHLA-DR expression in patients with septic shock and dismal outcome. This allows us to anticipate the onset of sepsis-induced immunosuppression. Second, the demonstration of the effect of IFN-γ in restoring the functional consequence of this genotype, suggests the use of this genetic biomarker to identify patients who may benefit of IFN-γ-based immunomodulatory therapy. In conclusion, we report that CIITA G-286A∗rs3087456 GG genotype is associated with impaired mHLA-DR expression in septic shock, predictive of development of persistent immunosuppression, and demonstrate the effect of IFN-γ to rescue altered MHC class II expression.
Limitations of the study
There are some limitations to be discussed. First, to validate that CIITA G-286A∗rs3087456 polymorphism can be considered as a biomarker of susceptibility to worsening prognosis in patients with septic shock, a study on a larger cohort of patients would be necessary. Second, because of inclusion before the release of the new definition of septic shock (Singer et al., 2016), patients were included on the criteria of sepsis with organ failure and hypotension despite adequate volume resuscitation (Levy et al., 2003). This definition remains close to the more recent one, and they both target the most critically ill patients in septic shock. In addition, recent data have shown that immunosuppression in ICU, also known as delayed-injury acquired immunodeficiency, is present in subgroups of severely injured patients, in both septic and non-septic patients (Venet et al., 2022). Therefore, this study provides an opportunity to discuss an example of the use of genetic biomarker through the understanding of the underlying immune mechanism, which can be used in all severe ICU patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Quantibrite Anti-Human HLA-DR PE/Monocyte PerCP-Cy™5.5 | BD Biosciences | Cat#340827; RRID: AB_400137 |
| PE Phycoerythrin Fluorescence Quantitation Kit | BD Biosciences | Cat#340495; RRID: AB_2868736 |
| CD20 Antibody, anti-human, FITC | Miltenyi Biotec | Cat#130-113-373; RRID: AB_2726142 |
| PE Mouse Anti-Human CD14 | BD Biosciences | Cat#347497; RRID: AB_400312 |
| Fc Receptor Binding Inhibitor Polyclonal Antibody, eBioscienceTM | Thermo Fisher Scientific | Cat#14-9161-73; RRID: AB_468582 |
| PE Mouse Anti-Human CD14 | BD Biosciences | Cat#555398; RRID: AB_395799 |
| PE-Cy™7 Mouse Anti-Human HLA-DR | BD Biosciences | Cat#560651; RRID: AB_1727528 |
| Biological samples | ||
| Blood sample from adult patient | Critical Care Unit of the Department of Anesthesiology, LariboisièreUniversity Hospital, Assistance Publique Hôpitaux deParis, France | Cochin Hospital Ethics Committee (# CCPPRB 2061) |
| Blood sample from healthy blood donors | French national blood collection center (Etablissement Français du Sang, EFS) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Human Interferon Gamma (rh IFN-gamma) | ImmunoTools | Cat#11343537 |
| Critical commercial assays | ||
| iScript™ cDNA Synthesis Kit | Bio-Rad | Cat#1708891 |
| Dual-Glo® Luciferase Assay System | Promega | E2940 |
| SNP Genotyping Analysis Using TaqMan Assays | ThermoFisher Scientific | N/A |
| TurboFect Transfection Reagent | ThermoFisher Scientific | Cat#R0531 |
| Deposited data | ||
| FHU Sepsis: personalized interventions | This paper | https://www.fhu-sepsis.uvsq.fr |
| Experimental models: Cell lines | ||
| U-937 | ATCC | CRL-15932 |
| Raji | ATCC | CCL-86 |
| Oligonucleotides | ||
| SNP genotyping; CIITA-P3 SNP-712 Taqman probe: ATGGGAGTCAGTATTATTTAGCATC[A/G]CTTTGG CGGGTCACCCCAAACCATC |
This paper | N/A |
| SNP genotyping; CIITA-P3 SNP-286 Taqman probe: GAAGTGAAATTAATTTCAGAGGTGT[A/G]GGGAG GGCTTAAGGGAGTGTGGTAA |
This paper | N/A |
| QPCR CIITA isoforms; Promoter1 f: CATGGTGGCAGCTCAC | This paper | N/A |
| QPCR CIITA isoforms; Promoter3 f: CCCAAGGCAGCTCACA | This paper | N/A |
| QPCR CIITA isoforms; Promoter4 f: GAACAGCGGCAGCTCA | This paper | N/A |
| QPCR CIITA isoforms; Exon2 r: GTAGCCACCTTCTAGGG | This paper | N/A |
| HuRPLP03 f: AGGCTTTAGGTATCACCACTAA | This paper | N/A |
| HuRPLP03 r: ACATCACTCAGGATTTCAATGG | This paper | N/A |
| Recombinant DNA | ||
| pGL3 Luciferase Reporter Vectors | Promega | E1761 |
| pRL Renilla Luciferase Control Reporter Vectors TK | Promega | E2241 |
| pRL Renilla Luciferase Control Reporter Vectors no promoter | Promega | E2271 |
| Software and algorithms | ||
| Arlequin ver 3.11 | Excoffier and Schneider, 2005 | http://cmpg.unibe.ch/software/arlequin3/ |
| PLINK | Purcell et al., 2007 | https://zzz.bwh.harvard.edu/plink/ |
| Stata | Stata Corp, 2016 | https://www.stata.com |
| GraphPad Prism version 9.0 | GraphPad Software | www.graphpad.com |
| FlowJo | BD Life Sciences | https://www.flowjo.com/solutions/flowjo |
| Beacon Designer 7.0 | PREMIER Biosoft | http://www.premierbiosoft.com/molecular_beacons/ |
| Other | ||
| One-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol | Liu and Naismith, 2008 | https://pubmed.ncbi.nlm.nih.gov/19055817/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pierre Tissieres (pierre.tissieres@i2bc.paris-saclay.fr).
Materials availability
-
•
This study did not generate new unique reagents.
-
•
Plasmids generated in this study have not been deposited.
Experimental model and subject details
Study design and setting
This was a single center observational study with sequential recruitment of patients admitted in the Critical Care Unit of the Department of Anesthesiology, Lariboisière University Hospital, Paris, France, between February 2004 and November 2005. The study was approved by the Cochin Hospital Ethics Committee (# CCPPRB, 2061), Assistance Publique Hôpitaux deParis. Patients were screened at admission by attending intensive care physicians for the inclusion criteria: presence of septic shock (Levy et al., 2003) and age ≥18 years. There were no exclusion criteria. Eligible patients (or their representatives) gave informed written consent for the study. Clinical and biological characteristics were prospectively collected from recruited patients, including demographic characteristics, severity scores (SAPS II: Simplified Acute Physiology Score II and SOFA: Sequential Organ Failure Assessment), infection foci, baseline health status and medications and routinely measured laboratory markers. Peripheral blood was sampled into EDTA anticoagulants tubes (Vacutainer, Becton Dickinson, USA) at days 1, 2, 7, 14, 21 and 28 following ICU admission for mHLA-DR assays, and upon recruitment to the study for genotyping. Of 221 patients with septic shock recruited to the study, daily data from day of ICU admission to ≥7 days and genomic DNA was available from 203 patients (Figure S1). The median age of the study cohort was 61 years (range: 18-94), with a majority of men (65%) (Table 1).
Healthy human control samples
For ex vivo and in vitro experiments, peripheral blood from healthy blood donors were collected at the French national blood collection center (Etablissement Français du Sang, EFS) following written informed consent was obtained from healthy volunteers.
Cell lines
Human leukemic monocyte lymphoma cell line U937 and human Burkitt’s lymphoma cells (Raji) were cultured in RPMI 1640 containing 10% heat-inactivated fetal bovine serum (FBS), 1.0 mM sodium pyruvate, amino acids, 20 mM HEPES, 100 U/ml penicillin, 100 μg/mL streptomycin at 37°C in 5% CO2.
Method details
Genotyping
Genotyping was performed blinded to the clinical data. Genomic DNA (gDNA) was extracted from 200 μL of peripheral whole blood using the NucleoSpin Blood kit (Macherey-Nagel, Düren, Deutschland) following manufacturer’s instruction. Presence of two SNPs: G-286A∗rs3087456 and G-712A∗rs12596540 located on the CIITA pIII was assessed using commercially available TaqMan assays (Life Technologies, La Jolla, CA) to discriminate between the two SNPs (Table S1). Real time allelic discrimination assays were performed using SsoFast Probes Supermix (Bio-Rad, Hercules, CA) and a CFX-96 real-time PCR system (Bio-Rad, Hercules, CA). Genotyping was performed blinded to the clinical data. Genomic DNA concentration was measured (NanoDrop, 2000 spectrophotometer, ThermoFisher, Waltham, MA) and quality assessed (260/280 ratio between 1.7 and 1.9).
Flow cytometry to assay mHLA-DR
Quantification of mHLA-DR on circulating monocytes from patients with sepsis was performed using a standardized flow cytometry assay (Demaret et al., 2013). Anticoagulated whole blood was stained with antibodies to HLA-DR and CD14 mixture (QuantiBrite anti-HLA-DR PE/anti-monocyte CD14 PerCP-Cy5.5, Becton Dickinson, CA), prior to lysis, washing, and acquiring of HLA-DR expression on monocytes using BD FACSCanto (BD Biosciences, Franklin Lakes, NJ). To quantify, the median fluorescence intensity of the entire monocyte population was then transformed to number of antibodies per cell (AB/C) using calibrated PE beads (BD QuantiBRITE PE BEADS, Becton-Dickinson San Jose, CA). Results were analyzed using FlowJo (TreeStar Inc, Ashland, OR). and mixed with calibrated PE beads (BD Quantibrite PE BEADS, Becton Dickinson, CA).
mHLA-DR expression upon exposure to IFN-γ
Peripheral blood mononuclear cells (PBMCs) from healthy volunteers were isolated by Ficoll density-gradient centrifugation. B cells and monocytes from blood donors were isolated using positive selection with MS columns and the CD19 and CD14 Microbeads kits, respectively (Miltenyi Biotec, Bergisch Gladbach, Germany) following manufacturer’s protocols. Purity was assessed by flow cytometry, using anti-CD20-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany), and anti-CD14-PE (BD Biosciences, Le Pont deClaix, France). Isolated cells had an average purity of 90% (±8%).
For analysis of healthy donors’ blood, isolated peripheral blood mononuclear cells (PBMCs), monocytes and B cells were washed once in PBS. After being washed once in cold PBS, cells were incubated at 4°C for 15′ with the Human Fc Receptor Binding Inhibitor (eBioscience, San Diego, CA) to avoid unspecific binding of labeled antibodies. Cells were then incubated with fluorescently labeled antibodies were added for 15minat 4°C. The antibodies mix contained: Fluorescein isothiocyanate (FITC)-conjugated anti-CD20 (mouse monoclonal IgG1κ, clone LT20, Miltenyi Biotec, Bergisch Gladbach, Germany), phycoerythrin (PE)-conjugated anti-CD14 (mouse monoclonal IgG2α,κ, clone M5E2, BD Biosciences, Franklin Lakes NJ) and PE-cyanine 7 (PE-Cy7)-conjugated anti-HLADR (mouse monoclonal IgG2α,κ, clone G46-6, BD Biosciences, Franklin Lakes, NJ). After two PBS washes, cells were suspended in 1% paraformaldehyde. Flow cytometry was performed on a BD FACSCanto (BD Biosciences, Franklin Lakes, NJ) and results analyzed by the software FlowJo (TreeStar Inc, Ashland, OR).
PBMCs were cultured in 24-well plates at 2 × 106 cells/ml in RPMI with 10% FBS at 37°C, 5% CO2. Isolated CD14+ cells were cultured at 2.5 × 105 in 24-well plates in DMEM with 5% FBS at 37°C, 5% CO2. Isolated CD19+ cells were cultured at 105 cells in 96-well plates in RPMI 10% FBS at 37°C, 5% CO2. Cultured cells were incubated in the presence or absence of 50 UI/ml of human recombinant IFN-γ (ImmunoTools, Friesoythe, Germany). After 6 h, CD14+ and CD19+ cells were harvested for RNA extraction, whilst after 24 h, PBMCs were collected for flow cytometry analysis.
Total RNA isolation and quantitative PCR analysis
Total RNA was isolated from monocytes and B cells using the NucleoSpin® RNA XS (Macherey-Nagel, Düren, Deutschland), following manufacturer’s instruction. cDNA was generated from 200 ng of total RNA using iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Primers for each CIITA isoform and for the large subunit of ribosomal protein (RPLPO), were designed using the software Beacon Designer 7.0 (PREMIER Biosoft, Palo Alto, CA) and are reported in together with working concentrations and amplification efficiencies (Table S1). RPLPO was used as a reference gene and the relative quantity of the transcript was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001). PCR reactions were performed in a CFX96 real time PCR system (Bio-Rad Hercules, CA) using the SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA).
Transfection and plasmid and dual-luciferase reporter assays
Four x 105 cells (Human leukemic monocyte lymphoma cell line U937 and human Burkitt’s lymphoma cells Raji) were transfected with 1 μg of reporter plasmid and 10 ηg of pRL Renilla luciferase vector (Promega, Madison, WI) using Turbofect™ Transfection Reagent (Thermo Fisher Scientific, Fermentas, Waltham, MA). After transfection, the cells were incubated for 30 h and stimulated by 50 UI/ml rIFN-γ (ImmunoTools, Friesoythe, Germany) for 6h or left untreated.
The 1200 bp of the pIII promoter of MHCIITA was cloned in pGL-3 firefly luciferase reporter vector (Promega, Madison, WI). SNPs G-286A∗rs3087456 and G-712A∗rs12596540 were inserted by using site-specific mutagenesis following published protocols (Stratagene, La Jolla, CA) (Liu and Naismith, 2008). All the inserts were verified by Sanger sequencing. Dual-Luciferase assay was performed with the Dual-Glo luciferase Assay (Promega, Madison, WI), following manufacturer’s instructions. Firefly and Renilla luciferase substrates fluorescence was recorded by a Tri-Star LB 941 (Berthold Technologies, Wilbad, Germany). As an internal control, pRL-TK, expressing the Renilla luciferase under the control of the constitutive TK promoter was co-transfected (Promega, Madison, WI). Results are represented by the mean value and the standard deviation and are expressed as relative luciferase activity (ratio of firefly luciferase activity to renilla luciferase activity). To confirm the results, experiments were performed in triplicates and each one was carried out three times.
Quantification and statistical analysis
Allele and genotype frequencies were calculated by direct counting. Hardy-Weinberg equilibrium for the two SNPs was assessed by the software Arlequin v3.11. Analysis of association between genotype and binomial clinical outcomes using different genetic models was performed by using the software plink (http://pngu.mgh.harvard.edu/∼purcell/plink/).
Results are presented as range and median for continuous variables and as proportion of total for categorical variables. Differences in HLA-DR, SOFA and SAPS II scores for the different genotypes were analyzed by using the Kruskal-Wallis test run in Stata software. The Wilcoxon rank-sum test was applied to analyze continuous variables between different genotypes. Mann-Whitney Test was applied for analysis of luciferase experiments by using the software GraphPad Prism 9.0. p values <0.05 were considered significant.
Acknowledgments
We thank Sergio Crovella (Department of Advanced Diagnostics, Institute for Maternal and Child Health-IRCCS 'Burlo Garofolo', Trieste, Italy and Department of Medical, Surgical and Health Sciences, University of Trieste, Trieste, Italy) for help with paper revisions. The study was supported by a BQR “Bonus Qualité Recherche” grant from the University Paris Sud to P.T.
Author contributions
Investigation, J.M., M.B., S.H., K.Z.M., C.B., V.F., A.C.L., and D.P.; Formal analysis and Visualization, J.M., M.J.C., M.B., and P.T.; Conceptualization and Methodology, J.M., M.B., P.T., D.P. and A.C.L.; Writing – Original Draft, J.M., P.T., and M.B.; Writing – Review and Editing, J.M., P.T., M.J.C., and M.B.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105291.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This article does not report original code. Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Abraham E., Laterre P.-F., Garg R., Levy H., Talwar D., Trzaskoma B.L., François B., Guy J.S., Brückmann M., Rea-Neto A., et al. Administration of drotrecogin alfa (activated) in early stage severe sepsis (ADDRESS) study group Drotrecogin alfa (activated) for adults with severe sepsis and a low risk of death. N. Engl. J. Med. 2005;353:1332–1341. doi: 10.1056/NEJMoa050935. [DOI] [PubMed] [Google Scholar]
- Antcliffe D.B., Burnham K.L., Al-Beidh F., Santhakumaran S., Brett S.J., Hinds C.J., Ashby D., Knight J.C., Gordon A.C. Transcriptomic signatures in sepsis and a differential response to steroids. From the VANISH randomized trial. Am. J. Respir. Crit. Care Med. 2019;199:980–986. doi: 10.1164/rccm.201807-1419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson P.G., Criswell L.A., Barcellos L.F. The MHC2TA -168A/G polymorphism and risk for rheumatoid arthritis: a meta-analysis of 6861 patients and 9270 controls reveals no evidence for association. Ann. Rheum. Dis. 2008;67:933–936. doi: 10.1136/ard.2007.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.-L., Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- Conway Morris A., Datta D., Shankar-Hari M., Stephen J., Weir C.J., Rennie J., Antonelli J., Bateman A., Warner N., Judge K., et al. Cell-surface signatures of immune dysfunction risk-stratify critically ill patients: INFECT study. Intensive Care Med. 2018;44:627–635. doi: 10.1007/s00134-018-5247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaret J., Walencik A., Jacob M.-C., Timsit J.-F., Venet F., Lepape A., Monneret G. Inter-laboratory assessment of flow cytometric monocyte HLA-DR expression in clinical samples. Cytometry B Clin. Cytometry. 2013;84B:59–62. doi: 10.1002/cyto.b.21043. [DOI] [PubMed] [Google Scholar]
- Döcke W.D., Randow F., Syrbe U., Krausch D., Asadullah K., Reinke P., Volk H.D., Kox W. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat. Med. 1997;3:678–681. doi: 10.1038/nm0697-678. [DOI] [PubMed] [Google Scholar]
- Eichacker P.Q., Parent C., Kalil A., Esposito C., Cui X., Banks S.M., Gerstenberger E.P., Fitz Y., Danner R.L., Natanson C. Risk and the efficacy of antiinflammatory agents: retrospective and confirmatory studies of sepsis. Am. J. Respir. Crit. Care Med. 2002;166:1197–1205. doi: 10.1164/rccm.200204-302OC. [DOI] [PubMed] [Google Scholar]
- Eike M.C., Skinningsrud B., Ronninger M., Stormyr A., Kvien T.K., Joner G., Njølstad P.R., Førre O., Flatø B., Alfredsson L., et al. CIITA gene variants are associated with rheumatoid arthritis in Scandinavian populations. Genes Immun. 2012;13:431–436. doi: 10.1038/gene.2012.11. [DOI] [PubMed] [Google Scholar]
- Evans L., Rhodes A., Alhazzani W., Antonelli M., Coopersmith C.M., French C., Machado F.R., Mcintyre L., Ostermann M., Prescott H.C., et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021;47:1181–1247. doi: 10.1007/s00134-021-06506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C., Knight J.C. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Zhang Y., Tang J., Lunsford L.D., Li T., Tang R., He J., Xu P., Faramand A., Xu J., You C. Association of corticosteroid treatment with outcomes in adult patients with sepsis: asystematic Review and meta-analysis. JAMA Intern. Med. 2019;179:213–223. doi: 10.1001/jamainternmed.2018.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann C., Scherag A., Adhikari N.K.J., Hartog C.S., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K., International forum of acute care trialists Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- Ghosh N., Piskurich J.F., Wright G., Hassani K., Ting J.P.-Y., Wright K.L. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J. Biol. Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- Harrison P., Pointon J.J., Farrar C., Harin A., Wordsworth B.P. MHC2TA promoter polymorphism (-168∗G/A, rs3087456) is not associated with susceptibility to rheumatoid arthritis in British Caucasian rheumatoid arthritis patients. Rheumatol. Oxf. Engl. 2007;46:409–411. doi: 10.1093/rheumatology/kel300. [DOI] [PubMed] [Google Scholar]
- Hornell T.M.C., Beresford G.W., Bushey A., Boss J.M., Mellins E.D. Regulation of the class II MHC pathway in primary human monocytes by granulocyte-macrophage colony-stimulating factor. J. Immunol. 2003;171:2374–2383. doi: 10.4049/jimmunol.171.5.2374. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R.S., Monneret G., Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernan K.F., Ghaloul-Gonzalez L., Shakoory B., Kellum J.A., Angus D.C., Carcillo J.A. Adults with septic shock and extreme hyperferritinemia exhibit pathogenic immune variation. Genes Immun. 2019;20:520–526. doi: 10.1038/s41435-018-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leentjens J., Kox M., Koch R.M., Preijers F., Joosten L.A.B., van der Hoeven J.G., Netea M.G., Pickkers P. Reversal of immunoparalysis in humans in vivo: a double-blind, placebo-controlled, randomized pilot study. Am. J. Respir. Crit. Care Med. 2012;186:838–845. doi: 10.1164/rccm.201204-0645OC. [DOI] [PubMed] [Google Scholar]
- Levy M.M., Fink M.P., Marshall J.C., Abraham E., Angus D., Cook D., Cohen J., Opal S.M., Vincent J.-L., Ramsay G. SCCM/ESICM/ACCP/ATS/SIS, 2003. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit. Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lohsen S., Majumder P., Scharer C.D., Barwick B.G., Austin J.W., Zinzow-Kramer W.M., Boss J.M. Common distal elements orchestrate CIITA isoform-specific expression in multiple cell types. Genes Immun. 2014;15:543–555. doi: 10.1038/gene.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A.-C., Faivre V., Payen D. Is monocyte HLA-DR expression monitoring a useful tool to predict the risk of secondary infection? Minerva Anestesiol. 2010;76:737–743. [PubMed] [Google Scholar]
- Lukaszewicz A.-C., Grienay M., Resche-Rigon M., Pirracchio R., Faivre V., Boval B., Payen D. Monocytic HLA-DR expression in intensive care patients: interest for prognosis and secondary infection prediction. Crit. Care Med. 2009;37:2746–2752. doi: 10.1097/CCM.0b013e3181ab858a. [DOI] [PubMed] [Google Scholar]
- Mach B., Steimle V., Reith W. MHC class II-deficient combined immunodeficiency: a disease of gene regulation. Immunol. Rev. 1994;138:207–221. doi: 10.1111/j.1600-065x.1994.tb00853.x. [DOI] [PubMed] [Google Scholar]
- Minneci P.C., Deans K.J., Eichacker P.Q., Natanson C. The effects of steroids during sepsis depend on dose and severity of illness: an updated meta-analysis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2009;15:308–318. doi: 10.1111/j.1469-0691.2009.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneret G., Lepape A., Voirin N., Bohé J., Venet F., Debard A.-L., Thizy H., Bienvenu J., Gueyffier F., Vanhems P. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 2006;32:1175–1183. doi: 10.1007/s00134-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Nguyen L.S., Ait Hamou Z., Gastli N., Chapuis N., Pène F. Potential role for interferon gamma in the treatment of recurrent ventilator-acquired pneumonia in patients with COVID-19: a hypothesis. Intensive Care Med. 2021;47:619–621. doi: 10.1007/s00134-021-06377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Abou El Hassan M., Xu Z., Yu T., Bremner R. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 2008;9:785–793. doi: 10.1038/ni.1619. [DOI] [PubMed] [Google Scholar]
- Nozell S., Ma Z., Wilson C., Shah R., Benveniste E.N. Class II major histocompatibility complex transactivator (CIITA) inhibits matrix metalloproteinase-9 gene expression. J. Biol. Chem. 2004;279:38577–38589. doi: 10.1074/jbc.M403738200. [DOI] [PubMed] [Google Scholar]
- Payen D., Faivre V., Miatello J., Leentjens J., Brumpt C., Tissières P., Dupuis C., Pickkers P., Lukaszewicz A.C. Multicentric experience with interferon gamma therapy in sepsis induced immunosuppression. A case series. BMC Infect. Dis. 2019;19:931. doi: 10.1186/s12879-019-4526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L. Human immunology through the lens of evolutionary genetics. Cell. 2019;177:184–199. doi: 10.1016/j.cell.2019.02.033. [DOI] [PubMed] [Google Scholar]
- Reinhart K., Daniels R., Kissoon N., Machado F.R., Schachter R.D., Finfer S. Recognizing sepsis as a global health priority — a WHO resolution. N. Engl. J. Med. 2017;377:414–417. doi: 10.1056/NEJMp1707170. [DOI] [PubMed] [Google Scholar]
- Reith W., LeibundGut-Landmann S., Waldburger J.-M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- Shakoory B., Carcillo J.A., Chatham W.W., Amdur R.L., Zhao H., Dinarello C.A., Cron R.Q., Opal S.M. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial. Crit. Care Med. 2016;44:275–281. doi: 10.1097/CCM.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle V., Otten L.A., Zufferey M., Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. doi: 10.1016/S0092-8674(05)80090-X. [DOI] [PubMed] [Google Scholar]
- Suffredini A.F., Munford R.S. Novel therapies for septic shock over the past 4 decades. JAMA. 2011;306:194–199. doi: 10.1001/jama.2011.909. [DOI] [PubMed] [Google Scholar]
- Swanberg M., Lidman O., Padyukov L., Eriksson P., Akesson E., Jagodic M., Lobell A., Khademi M., Börjesson O., Lindgren C.M., et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat. Genet. 2005;37:486–494. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- Ting J.P.-Y., Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Vasseur E., Boniotto M., Patin E., Laval G., Quach H., Manry J., Crouau-Roy B., Quintana-Murci L. The evolutionary landscape of cytosolic microbial sensors in humans. Am. J. Hum. Genet. 2012;91:27–37. doi: 10.1016/j.ajhg.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venet F., Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat. Rev. Nephrol. 2018;14:121–137. doi: 10.1038/nrneph.2017.165. [DOI] [PubMed] [Google Scholar]
- Venet F., Textoris J., Blein S., Rol M.-L., Bodinier M., Canard B., Cortez P., Meunier B., Tan L.K., Tipple C., et al. REALISM study group Immune profiling demonstrates a common immune signature of delayed acquired immunodeficiency in patients with various etiologies of severe injury. Crit. Care Med. 2022;50:565–575. doi: 10.1097/CCM.0000000000005270. [DOI] [PubMed] [Google Scholar]
- Wang J., Roderiquez G., Jones T., McPhie P., Norcross M.A. Control of in vitro immune responses by regulatory oligodeoxynucleotides through inhibition of pIII promoter directed expression of MHC class II transactivator in human primary monocytes. J. Immunol. 2007;179:45–52. doi: 10.4049/jimmunol.179.1.45. [DOI] [PubMed] [Google Scholar]
- Warren H.S., Suffredini A.F., Eichacker P.Q., Munford R.S. Risks and benefits of activated protein C treatment for severe sepsis. N. Engl. J. Med. 2002;347:1027–1030. doi: 10.1056/NEJMsb020574. [DOI] [PubMed] [Google Scholar]
- Wu J.-F., Ma J., Chen J., Ou-Yang B., Chen M.-Y., Li L.-F., Liu Y.-J., Lin A.-H., Guan X.-D. Changes of monocyte human leukocyte antigen-DR expression as a reliable predictor of mortality in severe sepsis. Crit. Care. 2011;15:R220. doi: 10.1186/cc10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Tang D., Li S.-J., Zhou J., Hsueh C.-Y., Zhao D.-D., Heng Y., Tao L., Lu L.-M. Link between CIITA rs3087456 polymorphism and the risk of laryngeal squamous cell carcinoma in a Chinese population. Pathol. Res. Pract. 2020;216:152793. doi: 10.1016/j.prp.2019.152793. [DOI] [PubMed] [Google Scholar]
- Ziegler E.J., Fisher C.J., Sprung C.L., Straube R.C., Sadoff J.C., Foulke G.E., Wortel C.H., Fink M.P., Dellinger R.P., Teng N.N. Treatment of gram-negative bacteremia and septic shock with HA-1A human monoclonal antibody against endotoxin. A randomized, double-blind, placebo-controlled trial. The HA-1A Sepsis Study Group. N. Engl. J. Med. 1991;324:429–436. doi: 10.1056/NEJM199102143240701. [DOI] [PubMed] [Google Scholar]
- Zinzow-Kramer W.M., Long A.B., Youngblood B.A., Rosenthal K.M., Butler R., Mohammed A.-U.-R., Skountzou I., Ahmed R., Evavold B.D., Boss J.M. CIITA promoter I CARD-deficient mice express functional MHC class II genes in myeloid and lymphoid compartments. Genes Immun. 2012;13:299–310. doi: 10.1038/gene.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This article does not report original code. Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.