Abstract
Fracture healing is a complex process with multiple overlapping metabolic and differentiation phases. Small non-coding RNAs are involved in the regulation of fracture healing and their presence in circulation is under current interest due to their obvious value as potential biomarkers. Circulating microRNAs (miRNAs) have been characterized to some extent but the current knowledge on tRNA-derived small RNA fragments (tsRNAs) is relatively scarce, especially in circulation.
In this study, the spectrum of circulating miRNAs and tsRNAs was analysed by next generation sequencing to show their differential expression during fracture healing in vivo. Analysed tsRNA fragments included stress-induced translation interfering tRNA fragments (tiRNAs or tRNA halves) and internal tRNA fragments (i-tRF), within the size range of 28–36 bp. To unveil the expression of these non-coding RNAs, genome-wide analysis was performed on two months old C57BL/6 mice on days 1, 5, 7, 10, and 14 (D1, D5, D7, D10, and D14) after a closed tibial fracture.
Valine isoacceptor tRNA-derived Val-AAC 5′end and Val-CAC 5′end fragments were the major types of 5′end tiRNAs in circulation, comprising about 65 % of the total counts. Their expression was not affected by fracture. After a fracture, the levels of two 5′end tiRNAs Lys-TTT 5′ and Lys-CTT 5′ were decreased and His-GTG 5′ was increased through D1-D14. The level of miR-451a was decreased on the first post-fracture day (D1), whereas miR-328-3p, miR-133a-3p, miR-375-3p, miR-423-5p, and miR-150-5p were increased post-fracture. These data provide evidence on how fracture healing could provoke systemic metabolic effects and further pinpoint the potential of small non-coding RNAs as biomarkers for tissue regeneration.
Keywords: Fracture healing, Circulating small non-coding RNA, tsRNA, tiRNA, miRNA, Biomarker
Abbreviations: tsRNA, transfer RNA derived small non-coding RNA fragment; i-tRF, internal tRNA fragment; tiRNA, stress-induced tRNA fragment, tRNA halves
1. Introduction
Healing of a long-bone fracture is a complex process involving a succession of several overlapping events such as hematoma formation, inflammation, migration of the mesenchymal stromal cells (MSCs) and their differentiation at the fracture site, bone formation, angiogenesis, and finally bone remodelling (Einhorn and Gerstenfeld, 2015; Gerstenfeld et al., 2003). Bone formation through endochondral or intramembranous ossification requires differentiation of mesenchymal stromal cells or transdifferentiation of hypertrophic chondrocytes to osteoblasts (Hu et al., 2017).
Small non-coding RNAs, including microRNAs (miRNAs) and tRNA fragments, have emerged as key regulators of cellular homeostasis. Mature miRNAs are small single-stranded non-coding RNA molecules with a size range of 19 to 23 nucleotides. They regulate many biological functions post-transcriptionally by binding mostly to the 3′ UTR of their target mRNA molecule (Bartel, 2004). Transfer-RNA derived small RNAs (tsRNA) are a group of functional non-coding RNAs which in recent years have been described to have diverse biological functions rather than being only breakdown by-products of tRNA molecules (Kim, 2019, Kumar et al., 2016, Krishna et al., 2019). Endonucleases involved in the biogenesis of tsRNA are still under debate, but several types of tsRNAs, including tiRNAs (stress-induced translation interfering tRNA fragments, tRNA halves), tRFs (short tsRNA between 15 and 30 nucleotides), and internal tRNA fragments (i-tRFs varying in size most often 36 nt long) are produced from tRNA molecules. Upon cellular or physiological stress, tiRNAs are processed from tRNA molecules by angiogenin ribonuclease but probably also by angiogenin-independent endonucleases (Krishna et al., 2019; Saikia and Hatzoglou, 2015; Su et al., 2019). This cleavage results in two 28 to 36 nucleotides long 5′end and 3′end fragments (Saikia and Hatzoglou, 2015).
Non-coding RNAs are gaining value as physiological circulating biomarkers for the clinical prognosis of most cancers (Anfossi et al., 2018). Earlier studies have indicated the association of tsRNAs with multiple functions such as regulation of mRNA expression, protection of cells from apoptosis, and interfering with virus replication (S. Li et al., 2018). Further, an increasing number of 5′end tiRNAs have been associated with cell differentiation processes (Krishna et al., 2019). More recently, we reported tsRNA expression in bone and callus tissues during fracture healing (Bourgery et al., 2021). Currently, there is only limited data available concerning tsRNAs in circulation and no studies regarding fracture healing.
In this study, we demonstrated differential expression of circulating miRNAs during fracture healing in vivo and also observed association of tsRNAs (including 5′end tiRNAs and i-tRFs) with the fracture healing processes (Fig. 1A).
Fig. 1.
The overall distribution of cellular tsRNA reads after closed bone fracture in circulation in mice. (A) Schematic structures of tRNA derived fragments analysed in this study, produced by angiogenin (ANG) for 5′end tiRNAs or unknown ribonucleases for i-tRFs. Data analysis revealed isoacceptor tRNA fragments between 28 and 36 nucleotides, including stress-induced 5′end tiRNA fragments and internal (i-tRF) with variable size, as listed in the panel. Isoacceptor tRNAs carry the same amino acid although they have different anticodons. (B) Circulating cellular tsRNA reads originating from 5′end tiRNAs or i-tRFs are presented by bars as mean ± SD of normalized tsRNA reads (FPM, fragment per million) in D0 (control), D1, D5, D7, D10, and D14 serum samples. Kruskal-Wallis statistical analysis indicated significant differences in the abundance of tiRNAs (p = 0.013) and i-tRFs (p = 0.048) between sample groups. Pairwise comparison using the Conover post-hoc test detected significance between D0 and D7 (p = 0.0024) and D0 and D10 (p = 0.0024) in tiRNAs, and D0 and D14 (p = 0.033) in i-tRFs. SD = sqrt(sum variances). D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). (C) Radar plots demonstrate the relative expression of the four most prevalent isoacceptor-derived tsRNAs identified in the serum tissue after fracture on D0, D1, D5, D7, D10, and D14, based on normalized counts calculated by DESeq2. The whole list of serum tsRNAs with their expression levels is presented in Supplemental file 1.
The overall distribution of cellular tsRNA reads after closed bone fracture in circulation in mice. (A) Schematic structures of tRNA derived fragments analysed in this study, produced by angiogenin (ANG) for 5′end tiRNAs or unknown ribonucleases for i-tRFs. Data analysis revealed isoacceptor tRNA fragments between 28 and 36 nucleotides, including stress-induced 5′end tiRNA fragments and internal (i-tRF) with variable size, as listed in the panel. Isoacceptor tRNAs carry the same amino acid although they have different anticodons. (B) Circulating cellular tsRNA reads originating from 5′end tiRNAs or i-tRFs are presented by bars as mean ± SD of normalized tsRNA reads (FPM, fragment per million) in D0 (control), D1, D5, D7, D10, and D14 serum samples. Kruskal-Wallis statistical analysis indicated significant differences in the abundance of tiRNAs (p = 0.013) and i-tRFs (p = 0.048) between sample groups. Pairwise comparison using the Conover post-hoc test detected significance between D0 and D7 (p = 0.0024) and D0 and D10 (p = 0.0024) in tiRNAs, and D0 and D14 (p = 0.033) in i-tRFs. SD = sqrt(sum variances). D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). (C) Radar plots demonstrate the relative expression of the four most prevalent isoacceptor-derived tsRNAs identified in the serum tissue after fracture on D0, D1, D5, D7, D10, and D14, based on normalized counts calculated by DESeq2. The whole list of serum tsRNAs with their expression levels is presented in Supplemental file 1.
2. Material and methods
2.1. Animals and experimental setup
The study plan and the use of animal material were approved by the National Animal Experiment Board ELLA (project license ESAVI/6129/04.10.03/2011). C57BL/6NHsd mice were supplied by Envigo (The Netherlands), and animals were maintained in the Central Animal Laboratory of the University of Turku, and they received a soya-free diet as previously reported (Bourgery et al., 2021). Animal care was in accordance with the international guidelines following the 3Rs principles.
2.2. Fracture generation and collection of samples
Standard closed fracture (age 70–74 days) was produced in mouse tibia by an impact device as previously described (Bourgery et al., 2021; Hiltunen et al., 1993; Puolakkainen et al., 2017).
Changes in small RNA profiles during fracture healing were followed by collecting callus tissue samples (published in Bourgery et al., 2021) and blood samples at D0 (intact control tibial diaphysis) and at D1, D5, D7, D10, and D14 after fracture.
2.3. Handling of blood samples
Blood samples were collected with a cardiac puncture, allowed to clot for 60 min at room temperature, and then centrifuged for 10 min at 3000 ×g. The supernatant was collected and further centrifuged for 2 min at 3000 ×g to remove any blood cell traces. Samples with hemolysis were discarded and only pale yellow serum samples were accepted for the NGS analysis. Serum samples were snap frozen in liquid nitrogen in 500 μl aliquots and stored at −80 °C until RNA isolation. ExoQuick was used for isolation of an exosome-containing fraction from 250 μl of serum collected at D0 (n = 3, intact control animal), and at D1, D5, D7, D10 and D14 (n = 4 in each group) following the manufacturer's instructions (System Biosciences, ref# EXOQ5A-1).
2.4. Processing of samples for NGS analysis
Total RNA was isolated from exosome isolate using miRNeasy Mini Kit (Qiagen, cat# 217004). Total RNA concentration was measured using Nanodrop spectrometry (Thermo Fisher Scientific).
Small RNA libraries were prepared using TruSeq Small RNA Sample Preparation Kit (Illumina, USA) with multiplexing adapters, following the kit user guide (Rev. E), and sequenced by MiSeq V3 flow cell using Illumina MiSeq reagent kit V3 for 150 cycles and 36 bp reads with single-end chemistry as previously described (Bourgery et al., 2021).
2.5. Bioinformatics analysis
Raw reads were subjected to quality control using FastQC. MicroRNA reads were mapped to the UCSC mouse genome (mm10) using TopHat version 2.0.9 and aligned using miRDeep2 (Friedlander et al., 2012).
TsRNA reads were extracted using the same method as previously described (Bourgery et al., 2021). In brief, tsRNA reads were mapped and aligned using SPORTS1.1 (Shi et al., 2018) against mm10 UCSC genome and fasta files from GtRNAdb using SPORTS default settings. The reads mapping to tsRNA sequences (28 to 36 nt long) were extracted from SPORTS output text file using R. Then, the data were analysed using R version 4.1.1. Differential expression for miRNA and tsRNA reads was calculated in R using DESeq2 version 1.32.0 (Love et al., 2014). Data were normalized against control serum samples. FPM (fragment per million) was calculated after raw counts normalization using SPORTS output in R.
Raw counts >10 reads across all samples were selected for differential expression analysis. p-Values were corrected using Benjamini-Hochberg multiple testing adjustment procedure (Benjamini and Hochberg, 1995). Taking to account the temporality of the data, genes were tested using a likelihood ratio test (LRT).
Non-coding RNAs were regarded as differentially expressed when fitting the criteria with baseMean > 10, absolute value of log2 fold change (FC) > 1.5 and p-adjusted value < 0.1.
2.6. Statistical analyses
Statistical analyses were performed in R. All statistical analyses for differential expression analysis were performed using DESeq2 based on negative binomial generalized linear models. The overall distribution of tsRNA reads analysis was performed using a nonparametric Kruskal-Wallis test, followed by a post-hoc Conover test or using a Wilcoxon rank sum test after violation of normality assumption.
3. Results and discussion
3.1. Expression of tiRNAs in circulation and in callus tissue
The expression profiles of tsRNAs were investigated by NGS in small RNA fractions at D0–D14. A total of 76 tRNA fragments were annotated, including 29 5′end tiRNAs, and 35 i-tRFs (Fig. 1, Supplemental file 1). The distribution during fracture healing of the total normalized reads of circulating cellular 5′end tiRNAs and i-tRFs were analysed (Fig. 1A and B).
The majority of the tiRNAs originated from the 5′end, forming about 94 % of the total tiRNA reads (Fig. 1B, Supplemental file 1). In fracture healing groups, a 1.4 to 1.8 fold decrease was observed in their amount. About 6 % of the normalized reads corresponded to the i-tRFs. Full data, including cellular tRNA fragments derived from the 3′end as well as mitochondrial tRNA fragments, are presented in Supplemental file 1. Fragments from the 3′end composed <1 % of the total tiRNAs, with a read number below 10 in the analysis, and were therefore filtered out of the data presented in Fig. 1. Traditional sequencing technologies, as was also used in the present study, are not adapted to the analysis of tsRNA biogenesis with their multiple modifications, especially found at the 3′end of the tRNA fragments (Podnar et al., 2014). Recent improvements in library processing technologies and workflows provide possibilities to overcome these problems (van Dijk et al., 2019; Maguire et al., 2020; Shi et al., 2021).
Relative expression of the four most prevalent isoacceptor tRNA-derived tsRNA fragments in serum were analysed at D0 - D14 (Fig. 1C, Supplemental file 1). Isoacceptors are tRNAs that carry the same amino acid although they have different anticodons.
The major proportion of parental tRNA-derived fragments in samples originated from Val-tRNAs (from 61 to 68 %) and the remaining from Lys-tRNA (from 33 to 11 %), His-tRNA (from 2 to 14 %), and Glu-tRNA (from 2 to 7 %) as illustrated by radar plots for the four most prevailing isoacceptors (Fig. 1C, Supplemental file 1).
3.2. Differential expression of tsRNAs
Valine isoacceptor-derived tiRNAs were the most prevalent tiRNAs in serum, but their expression remained relatively stable during fracture healing (Table 1). Expression levels in serum of four other 5′end tiRNAs Glu-CTC-5′, Glu-TTC-5′, iMet-CAT-5′, and Cys-GCA-5′, and two i-tRFs Val-AAC, and Val-CAC were not affected by the fracture. Val-AAC-5′end and Val-CAC-5′end tiRNAs were also highly expressed in callus tissue and upregulated by 1.5 to 1.9 log2FC during fracture healing in comparison to the control diaphyseal bone (Bourgery et al., 2021). Functions of tiRNA-Val family members associate, e.g., with angiogenesis and, in the mouse retinal tissue with diabetic retinopathy 5′end tiRNA-Val was demonstrated to directly target the 3′UTR of Sirt1 and repress its expression (Xu et al., 2022). This finding is interesting as angiogenesis is a crucial process also during fracture healing (Einhorn and Gerstenfeld, 2015; Gerstenfeld et al., 2003).
Table 1.
Expression of mature 5′end tiRNA and internal tRNA fragments in circulation after closed tibial bone fracture in mice. Green and blue shadings indicate at D1, D5, D7, D10 and D14 after fracture the statistically significant increased and decreased expression, respectively. Change was considered statistically significant if the absolute value of log2FC was above 1.5 and the adjusted p-value below 0.1. These values are highlighted by red fonts. Fourteen most prevalent tsRNAs with baseMean > 10 are presented in this table. A full list of the tsRNA data is presented in Supplemental file 1.
Expression of mature 5′end tiRNA and internal tRNA fragments in circulation after closed tibial bone fracture in mice. Green and blue shadings indicate at D1, D5, D7, D10 and D14 after fracture the statistically significant increased and decreased expression, respectively. Change was considered statistically significant if the absolute value of log2FC was above 1.5 and the adjusted p-value below 0.1. These values are highlighted by red fonts. Fourteen most prevalent tsRNAs with baseMean > 10 are presented in this table. A full list of the tsRNA data is presented in Supplemental file 1.
Small non-coding RNAs in circulation can be found in extracellular vesicles (including exosomes) and associated with lipids and RNA binding proteins. These carriers play important role in transmitting systemic effects by transporting their cargo (including tsRNAs) to the target tissues and organs. Current understanding of the biological function of the circulating tsRNAs or other tRFs is at a very early age and the major interest has been on their potential as biomarkers and therapeutic targets (Barile and Vassalli, 2017; Liu et al., 2021).
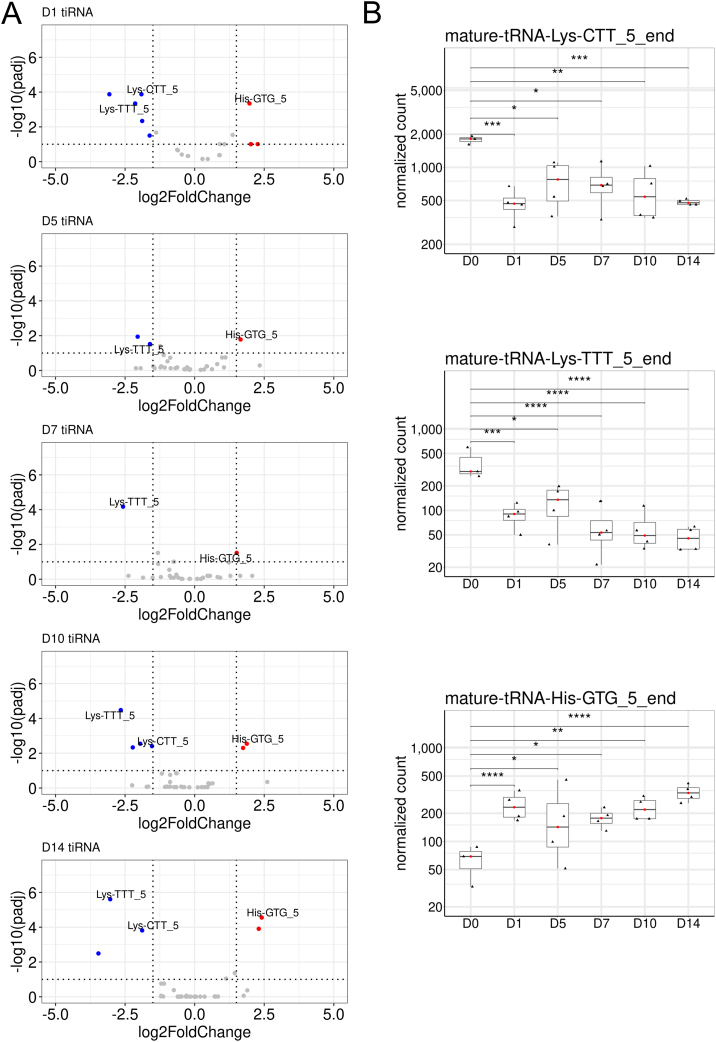
Three 5′end tiRNA fragments and three i-tRFs were differentially expressed during fracture healing in circulation (Fig. 2, Table 1). Levels of Lys-tRNA-derived 5′end tiRNAs Lys-CTT-5′, and Lys-TTT-5′, and i-tRFs Lys-TTT and Lys-CTT were reduced in serum, although their expression in callus tissue was either stable or increased (Bourgery et al., 2021).
Fig. 2.
Differential expression of cellular 5′tiRNAs in circulation after closed tibial bone fracture in mice. (A) Volcano plots visualize the differential expression of 5′ end tiRNAs at D5, D7, D10 and D14 after fracture. Blue dots visualize the tiRNAs with a log2FC < −1.5 and p-adjusted value < 0.1. Red dots visualize the tiRNAs with a log2FC > 1.5 and p-adjusted value < 0.1. tiRNAs with baseMean > 100 are annotated. (B) Box plot graphs show the expression levels of the circulating 5′end tiRNAs Lys-CTT-5′, Lys-TTT-5′, and His-GTG-5′ in D0 control and experimental samples on D1-D14 after fracture. D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). p-Adjusted value was calculated by DESeq2 during differential expression analysis: ns, non-significant; *: <0.1; **: <0.01; ***: <0.001; ****: <0.0001. Full data is presented in Supplemental file 1.
Differential expression of cellular 5′tiRNAs in circulation after closed tibial bone fracture in mice. (A) Volcano plots visualize the differential expression of 5′ end tiRNAs at D5, D7, D10 and D14 after fracture. Blue dots visualize the tiRNAs with a log2FC < −1.5 and p-adjusted value < 0.1. Red dots visualize the tiRNAs with a log2FC > 1.5 and p-adjusted value < 0.1. tiRNAs with baseMean > 100 are annotated. (B) Box plot graphs show the expression levels of the circulating 5′end tiRNAs Lys-CTT-5′, Lys-TTT-5′, and His-GTG-5′ in D0 control and experimental samples on D1-D14 after fracture. D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). p-Adjusted value was calculated by DESeq2 during differential expression analysis: ns, non-significant; *: <0.1; **: <0.01; ***: <0.001; ****: <0.0001. Full data is presented in Supplemental file 1.
Transfer RNA-derived small RNA fragments have diverse biological functions, e.g., they regulate various stages of protein translation. Krishna et al. recently reported on the involvement of 5′end tiRNAs Gln-CTG-5′, Val-CAC-5′ and Lys-TTT-5′ in the regulation of cell proliferation via RNA binding protein Igf2bp1/cMyc expression, thereby affecting pluripotency and maintaining lineage differentiation during rheumatoid arthritis (RA) induced ESC differentiation in vitro (Krishna et al., 2019). Blocking of these 5′end tiRNAs by a pool of antisense oligos during RA-induced differentiation, the stemness indicators (including Lif and Wnt3) were significantly increased. Their data suggest that the primary role of these 5′-tiRNAs is to restrain the expression of stemness-promoting genes, thereby facilitating differentiation.
His-tRNA derived tsRNA fragments tiRNA His-GTG-5′end and i-tRF His-GTG were increased in circulation during fracture healing (Table 1, Fig. 2). They also were upregulated in callus tissue during propagation of the fracture healing process from inflammatory phase (D5) to the advanced anabolic phase (D14) with endochondral ossification in callus samples (Bourgery et al., 2021). Recently, His-GTG-5′ was shown to respond to hypoxic conditions via HIF1α/Ang axis and to promote colorectal cancer by activating LATS2 (Tao et al., 2021). His-GTG-5′ is also essential in cancer cell proliferation and its blocking can induce cell apoptosis. Further, His-GTG-5′ (tiRNA-His-GTG-001) was shown to be elevated in intestinal biopsy samples of patients with an irritable bowel syndrome with diarrhea, targeting GABBR2, TLR4, and GABARAP which are related to, e.g., amino acid (such as glutamate) metabolism, GABAergic synapse, TNF-α signalling pathway, and insulin resistance (Chai et al., 2021). Knockout of TLR4 in mice has been shown to activate Wnt/β-catenin signalling pathway, thereby promoting fracture healing (Zhao et al., 2020). These data suggest that His-GTG-5′ has a role in fracture healing via regulation of macromolecular metabolism.
Recent studies have shown that the differential expression of tRNA genes is involved in the regulation of non-canonical functions by tRNA fragments (Torres et al., 2019; Tosar et al., 2021). Further, it has been shown that the 5′end and not 3′end tiRNAs inhibit translation in vitro (Ivanov et al., 2011). Therefore, differential expression of 5′end tiRNA fragments is expected to affect the translation of its target mRNA similarly to miRNAs although the exact mechanisms are not known on how the circulating tiRNAs are taken up by the recipient cells and how this process is regulated. Based on the current knowledge, it is evident that tsRNA fragments have multiple roles in the regulation of tissue metabolism. In the future, they may turn out to be useful biomarkers for cell differentiation, inflammation and tissue fibrosis and they may also have therapeutic value. Further, trauma caused by bone fracture affected the pool of circulating Lys and His isoacceptor-derived tsRNA fragments. It is not known if their differential expression in circulation has any systemic effects or biological relevance associated with fracture healing but evidently tsRNAs are potential biomarkers of tissue regeneration.
3.3. Differential expression of miRNAs in circulation during fracture healing
Expression profiles of miRNAs in serum were quantified at D1–D14 after fracture against intact control mouse (D0) serum. A total of 93 cellular miRNAs were observed in serum samples with a baseMean >10 (Supplemental file 1). Ten most prevailing miRNAs in serum exosome fraction were miR-486a-5p, miR-486b-5p, miR-22-3p, miR-92a-3p, miR-16-5p, miR-142a-5p, miR-25-3p, miR-451a, miR-215-5p and miR-192-5p.
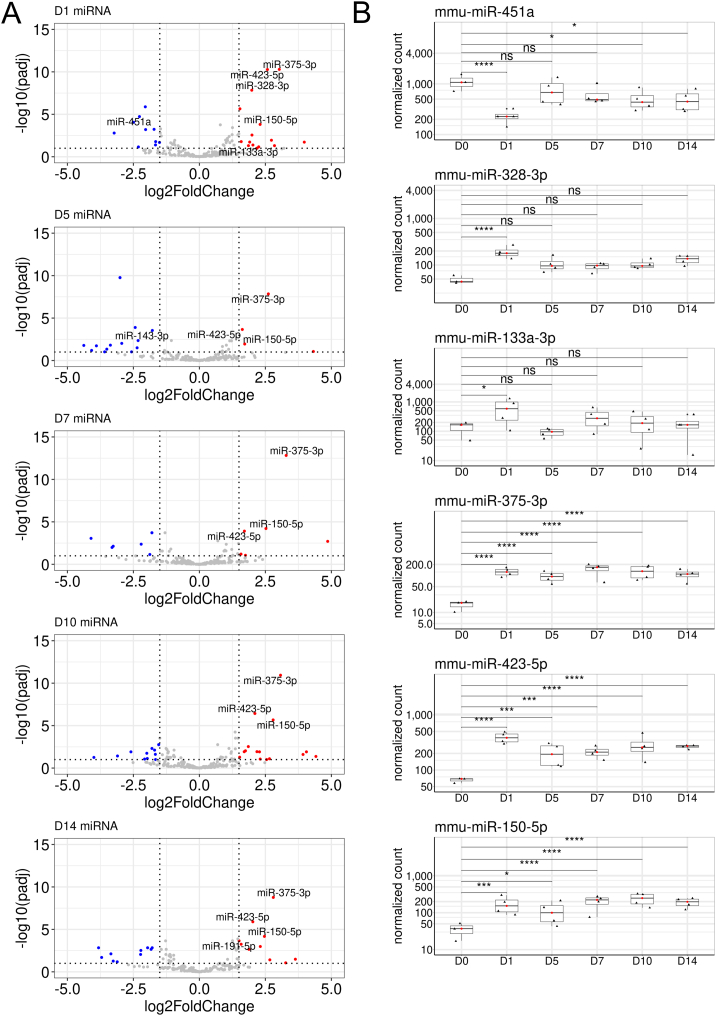
Thirty-four miRNAs were expressed with a baseMean >100 (Fig. 3A). Out of them, six miRNAs were differentially expressed after fracture (Fig. 3B, Table 2). The expression of miR-451a (baseMean 598) was decreased by 2.3 log2FC on D1 post-fracture and increased thereafter but remained at a slightly lower level compared to the control. Two miRNAs miR-328-3p (baseMean 114) and miR-133a-3p (baseMean 270), were increased in serum at D1 but thereafter returned to baseline, thereby also representing early post-fracture response in serum. Levels of three miRNAs, miR-375-3p (baseMean 107), miR-423-5p (baseMean 243), and miR-150-5p (baseMean 164) were increased at D1 and thereafter remained elevated throughout D1-D14.
Fig. 3.
Differential expression of circulating cellular miRNAs after closed tibial bone fracture in mice. (A) Volcano plots visualize the expression of circulating miRNAs after fracture in serum. Blue dots visualize the tiRNAs with a log2FC < - 1.5 decrease and P-adjusted value < 0.1, and red dots log2FC > 1.5 increase and P-adjusted value < 0.1. Annotated miRNAs have a baseMean > 100. (B) Box plots visualize the expression levels of miR-451a, miR-328-3p, miR-133a-3p, miR-375-3p, miR-423-5p, and mmu-miR-150-5p through D1-D14. D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). P-adjusted value was calculated by DESeq2 during differential expression analysis: ns, non-significant; *: < 0.1; ***: < 0.001; ****: < 0.0001. The whole list of miRNAs with their expression levels is presented in Supplemental file 1.
Differential expression of circulating cellular miRNAs after closed tibial bone fracture in mice. (A) Volcano plots visualize the expression of circulating miRNAs after fracture in serum. Blue dots visualize the tiRNAs with a log2FC < - 1.5 decrease and P-adjusted value < 0.1, and red dots log2FC > 1.5 increase and P-adjusted value < 0.1. Annotated miRNAs have a baseMean > 100. (B) Box plots visualize the expression levels of miR-451a, miR-328-3p, miR-133a-3p, miR-375-3p, miR-423-5p, and mmu-miR-150-5p through D1-D14. D0 (n = 3), D1 (n = 4), D5 (n = 4), D7 (n = 4), D10 (n = 4), D14 (n = 4). P-adjusted value was calculated by DESeq2 during differential expression analysis: ns, non-significant; *: < 0.1; ***: < 0.001; ****: < 0.0001. The whole list of miRNAs with their expression levels is presented in Supplemental file 1.
Table 2.
Top six differentially expressed miRNAs in serum compared to their differential expression in callus after fracture. Callus data is retrieved from Bourgery et al., 2021, Supplemental file 3. Data includes baseMean and adjusted p-value. Log2FC was calculated against intact tibial diaphysis samples. *These miRNAs were among top54 differentially expressed miRNAs in callus (Bourgery et al., 2021, Fig. 5 and Supplemental files 3 and 5). **mRNA typed with bold fonts indicate the significant negative correlation between miRNA and its target mRNA in callus tissue (from Bourgery et al., 2021). Relative baseMean was calculated against the highest baseMean (average normalized reads) value: in serum, baseMean of miR486a-5p was 17,700 and in callus, baseMean of miR-22-3p was 24,666. Statistically significant adjusted p-values (<0.1) are highlighted by red fonts. Green and blue shadings indicate statistically significant increased and decreased absolute value of expression (>1.5 log2FC) after fracture, respectively.
3.3.1. Decreased levels of miR-451a in circulation at D1 after fracture
In serum, miR-451a was one of the ten most prevalent miRNAs (Supplemental file 1). After fracture, miR-451a level initially decreased by log2FC of −2.3 at D1 but returned thereafter to almost a control level (Fig. 3B, Table 2). In callus tissue, miR-451a was downregulated with a − 4.6 to −2.5 log2FC during D5-D14 among the top54 differentially expressed miRNAs of tibial closed fracture in mice (Bourgery et al., 2021). In another study with a femoral closed fracture in mice, miR-451a was also downregulated through D1-D14 by roughly about 55 % where contralateral femoral diaphysis was used as a control (Hadjiargyrou et al., 2016). Conversely, expression of miR-451a was higher on postfracture days 14 and 28 with dynamic expression patterns in a standard fracture healing callus in comparison to fibrous tissue of an unhealing fracture produced by periosteal cauterization in rat femurs (Waki et al., 2016). In mouse calvarial primary osteoblast cultures, miR-451a contributed via a glucose-dependent manner to parathyroid hormone-mediated mineralization and osteoblast differentiation of primary mouse calvarial osteoblasts by suppressing the expression of Osr1, a putative target of miR-451a (Karvande et al., 2018). In callus tissue, miR-451a and Osr1 expression levels negatively correlated with each other, thereby also connecting miR-451a to osteoblast differentiation during fracture healing (Bourgery et al., 2021). In human RA patients, miR-451a was reduced in neutrophils when compared to healthy controls, and in murine neutrophils in vitro, it has been shown to reduce inflammation by targeting p38 MAPK (Murata et al., 2014). Erythrocyte vesicles are rich in miR-451a and may therefore also contribute positively to its amount in circulating exosomes (Thangaraju et al., 2020; Xu et al., 2019). Future studies will show whether this decreased level in circulation after fracture has any physiological role in fracture healing. These studies altogether suggest that miR-451a acts as a fine tuner in callus tissue to regulate dynamically inflammation and osteogenesis during the healing process.
3.3.2. Elevated levels of miR-133a-3p and miR-328-3p in circulation at D1 after fracture
Levels of miR-133a-3p and miR-328-3p increased in serum at D1 but thereafter returned to baseline (Fig. 3B, Table 2). In callus tissue, miR-133a-3p was highly expressed and was upregulated by 2.7 and 1.7 log2FC at D5 and D7 (inflammatory and chondrogenic phases of fracture healing), respectively (Table 2 and Bourgery et al., 2021).
In oral and oesophageal squamous carcinoma cells, miR-133a-3p has been shown to suppress cell proliferation, migration, and invasion via targeting Col1a1 (He et al., 2018; Y. Yin et al., 2019). Further, miR-133a-3p is enriched in muscle cells and hence also called myomiR. Its levels in circulation correlate with myoglobin synthesis and remain high even after 24 h of an 8 km run (Yin et al., 2020). It was also significantly increased in the serum of patients with coronary artery disease (Singh et al., 2020). It has been shown to target MET, a proto-oncogene, and several other growth factor and cytokine receptors, such as receptor tyrosine kinase, EGFR, FGFR1, and IGF1R, thereby inhibiting P13K/AKT signalling in PCa cells and bone metastases (Tang et al., 2018). P13K/AKT signalling is also required in osteogenic differentiation of MSCs, and endochondral ossification via a positive feedback loop with Runx2 to induce osteogenesis (Baker et al., 2015, McGonnell et al., 2012). A chemical inhibitor of C-Met has been shown to promote fracture healing, tissue remodelling and regeneration (R. Wang et al., 2019). In callus tissue, miR-133a-3p levels correlated negatively with Met expression (Bourgery et al., 2021). Recent data indicates that overexpression of miR-133a-3p inhibits osteogenesis of bone marrow mesenchymal stem cells by regulating ankyrin repeat domain 44 (Ankrd44) (Li et al., 2021). Based on these previous studies, we propose that miR-133a-3p functions as a regulator of bone remodelling via targeting P13K/AKT to regulate osteogenic differentiation. Its differential expression in serum may also reflect the muscular trauma at the fracture site.
In the present study, miR-328-3p was increased in serum at D1, whereafter it returned to almost a control level while in callus tissue, miR-328-3p was expressed at a low level, and was mildly downregulated during fracture healing, being lowest at D10 (Table 2 and Bourgery et al., 2021). It is not known whether miR-328-3p in circulation affects osteogenesis but in vitro studies indicate that miR-328-3p inhibits Axin1 and thereby activates Wnt/β-catenin pathway (D. Liu et al., 2018). The level of circulating miR-328-3p was decreased in osteoporotic patients and showed a tight association with Wnt pathway (R. Chen et al., 2018). Further, in vitro studies indicated that miR-328-3p inhibits osteoblast differentiation in human and mouse osteoblastic cell lines via decreased alkaline phosphatase activity and suppress mineral deposition. Also, in another study with osteoporotic fractures, miR-328-3p was reduced in circulation of postmenopausal women, and repression of miR-328-3p reduced significantly alkaline phosphatase activity during osteogenic differentiation of human adipose-derived stem cells but did not affect calcium deposition (Weilner et al., 2015). In humans with fragility fractures, miR-328-3p decreased in serum at 14 and 21 days postfixation (W. Xie et al., 2020). Further, miR-328-3p targets PTEN and its overexpression decreases PTEN/P13K/AKT, increases osteoblast differentiation and decreases apoptotic rate of human osteoblast cells, suggesting that miR-328-3p accelerates fracture healing by promoting osteoblast viability.
Future studies will show the physiological relevance of miR-133a-3p and miR-328-3p in circulation and how they associate with the regulation of osteogenesis during fracture healing via, e.g., P13K/AKT and Wnt/β-catenin signalling.
3.3.3. Levels of miR-375-3p, miR-423-5p and miR-150-5p were elevated in serum through D1-D14 of fracture healing
In serum, mir-375-3p was increased by 2.6 to 3.3 of log2FC throughout D1-D14 after fracture while in callus its expression was close to the detection limit (Fig. 3B, Table 2 and Bourgery et al., 2021). It has been shown to negatively modulate osteogenesis via decreasing the expression levels of LRP5 and Ctnnb1 and Wnt/β-catenin signalling (Sun et al., 2017). Further, mir-375-3p induces apoptosis of MC3T3-E1 cells in vitro.
A significant increase of circulating miR-375-3p was observed in a CENTRAL Trial study for Lifestyle intervention in participants with prediabetes, which was related to fasting insulin and insulin resistance and reduced visceral and hepatic fat (Heianza et al., 2022). Furthermore, increased serum levels of miR-375-3p were observed with progression of osteoporosis in an experimental rat model, and in postmenopausal women with osteoporotic vertebral fractures, compared to non-fractured controls (Weigl et al., 2021; Zarecki et al., 2020). These studies suggest an association of increased levels of serum miR-375-3p after fracture with the negative regulation of osteogenesis/bone remodelling.
In serum, the miR-423-5p level increased at D1 by log2FC of 2.6 and remained thereafter at a slightly lower but elevated level with log2FC between 1.6 and 2.1, in comparison to intact control mice (Table 2). In a recent study, elevated levels of miR-423-5p were reported in serum of women with osteoporosis and osteoarthritis (Pertusa et al., 2021). Elevated level of miR-423-5p was associated with a lower bone mineral density in an ageing study with healthy adult baboons (Quillen et al., 2022). In a study of two Finnish families with osteoporosis due to the heterozygous p.C218G WNT1 mutation, miR-423-5p was decreased in the circulation of patients with the mutation (Mäkitie et al., 2018). During fracture healing, miR-423-5p was significantly downregulated at D10 and D14 in callus tissue and negatively correlated with the expression of Tnip2, Cdkn1a, and Igf2bp1 (Bourgery et al., 2021) which have been shown by others as verified targets of miR-423-5p (Guo et al., 2018; W. Wang et al., 2017; S. Xie et al., 2020). TNIP2 activates NF-κB signalling, which induces osteoclastogenesis and triggers osteoporosis (Fischer and Haffner-Luntzer, 2022; Wang et al., 2017). Cdkn1a codes for p21, which functions as a regulator of cell cycle progression at G1 and its deletion enhances the osteogenic potential of bone marrow-derived mesenchymal stem cells in vitro (Juran et al., 2021). Igf2bp1 codes for Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) which binds to the mRNA of certain genes, including IGF2 and β-actin, and regulates their transcription (Yakar et al., 2018). Igf2bp1 is also targeted by miR-150-5p (L. Wang et al., 2019). Based on these studies, miR-423-5p is a potential regulator of osteoclastogenesis and bone remodelling during fracture healing. The functional relevance of upregulated levels in serum remains to be solved in future studies but certainly, it has potential as a biomarker.
MiR-150-5p levels were increased in serum and remained high through D1-D14 with log2FC between 1.7 and 2.8 (Table 2). Lower levels of miR-150-5p were found in the serum of RA patients compared to the serum of osteoarthritic patients. Interestingly, studies with miR-150 knockout mice have indicated the importance of miR-150 in regulation of immune system which is also involved in RA (Huang et al., 2015). MiR-150-5p has been previously shown to directly target matrix metalloproteinase 14 (Mmp14) to reduce extracellular matrix modelling (Z. Chen et al., 2018). Furthermore, its overexpression reduces the expression of MMP14 and vascular endothelial growth factor (VEGF) in RA fibroblast-like synoviocyte samples (Z. Chen et al., 2018). In our published callus data, miR-150-5p was downregulated through D5-D14 with lowest expression at D7-D10, and negative correlation was shown between miR-150-5p and its several target mRNAs regulating transcription of multiple cellular functions, e.g., Socs1 (suppression of cytokine signalling), Mmp14 (extracellular matrix remodelling), Rab9 (intracellular protein transport), Slc2a1 (glucose uptake), and Elk1 (binds to promoters of FOS and IER2 and induces target gene transcription by JNK-signalling pathway) (Bourgery et al., 2021). Mature B- and T-cells also express miR-150-5p which downregulates C-MYB, an essential regulator of hematopoiesis (Vasilatou et al., 2010). These studies suggest that its major function in fracture healing is associated with extracellular matrix remodelling and immune system.
3.4. Expression levels in callus and serum are poorly correlated
Profiles of the differentially expressed tsRNAs and miRNAs mainly deviated between serum and callus samples (Table 1, Table 2, and Bourgery et al., 2021). A correlation assay of the six differentially expressed miRNAs in serum revealed no significant correlation between the expression levels in serum and callus tissues (Supplemental file 2). This raises a question on how the serum and callus levels of small non-coding RNAs are coordinated and what is the origin of miRNAs or tsRNA fragments contributing to the altered levels in circulation.
Exosomes contain highly specific RNA cargo and parent cells control the sorting of miRNAs into vesicles and secretion into circulation (Groot and Lee, 2020; O'Brien et al., 2020). Secretion into circulation may be activated as a response to cytokine stimuli or other systemic effects. An interesting mechanism for systemic regulation was presented by Sun and others. They propose that erythrocytes as a response to a certain stimulus release a large number of extracellular vesicles loaded with erythrocyte derived miRNAs which are taken up by recipient cells and modulate their function (Sun et al., 2020).
Circulating miRNA carco functions as cell-cell communication where small RNA is released from the donor cell and taken up by a recipient cell at a distance, resulting in changes of gene expression (Qiu et al., 2021). Selected sorting of a certain small RNA into export with no increase in the endogenous miRNA synthesis might result in reduced levels in callus tissue and increased levels in serum. Immediately after fracture, increased serum levels may result also by the leakage from the fracture site to the circulation. While interpreting the data, it is also important to remember that the comparison is only relative as reliable the comprisons can be made only within serum or callus tissue and at their best they are only relative, notquantitative.
Certain small non-coding RNAs in circulation may originate from specific organs/tissues via different systemic mechanisms. E.g., there is increasing evidence on adipose tissue as a major source of miRNAs in circulation and that miRNAs are a new class of factors mediating endocrine and paracrine functions (Ji and Guo, 2019; Mori et al., 2019; Peng and Wang, 2018; Vienberg et al., 2017). Moreover, adipose-derived miRNAs in circulation can modify gene expression in other tissues (Thomou et al., 2017). Therefore, adipose tissue has a large input into the systemic regulation of body metabolism via miRNA cargo in circulation. An example of miRNAs associated with adipose tissue metabolism and fracture healing is miR-150. It was downregulated in callus and increased in serum during fracture healing. Interestingly, studies with miR-150 knockout mice have shown that miR-150 knockdown significantly decreases the capacity of adipogenic differentiation of adipose-derived stem cells (ADSCs) and its overexpression significantly increases C/EBPα and PPAR-γ expression and lipid formation in ADSCs with adipogenic induction (Li et al., 2019). In addition, in vitro studies suggest that miR-150 suppresses proliferation potential and expression of Nanog in ASDCs and regulates adipogenic differentiation via downregulation of its target gene Notc3. Further, studies with Pld1−/− knockout mice revealed that PLD1 is a novel regulator of bone homeostasis and adipogenic function through Runx2, β-catenin-OPG, PPAR-γ and C/EBPα axis, and it regulates the mesenchymal cell lineage differentiation by increasing osteogenesis while decreasing adipogenesis (Kang et al., 2021). These connections between bone and adipose tissue homeostasis highlight the complexity of the systemic effects in the regulation of body metabolism.
It is tempting to speculate that also other indirect factors, such as reduced exercise and immobilization in association with bone fractures may contribute to the miRNA profile in circulation, e.g., via adipose tissue metabolism, thereby transmitting the systemic effects to the body metabolism.
Transfer RNA-derived fragments add a whole new level to the regulation of tissue metabolism. Although data on systemic effects is more limited, tRNA fragments are found in circulation, and in the target tissues, they can modulate protein translation and alter the physiology of the recipient cells (Kim, 2019). In addition to the question on the systemic contribution of various organs onto small non-coding RNA profiles in circulation, there are a lot of open questions, e.g., what is a physiologically relevant concentration leading to the changes in recipient cell metabolism or how the circulating RNAs are recognized and taken up in the target tissue and whether, e.g., a specific exported miRNA just ends up as junk in circulation. Interestingly, a recent study revealed specific motifs that determine whether the miRNA is retained in the cell (CELLmotif) or released into exosomes (EXOmotif) (Garcia-Martin et al., 2022). The discovery of this RNA code provides a novel insight into the data analysis on how to link the miRNAs in circulation to the tissue of origin. Yet, it remains largely unknown if all cell types or tissues have their own RNA code and how the target cell is defined for unloading the cargo.
4. Summary and conclusion
Understanding of the functional roles of small non-coding RNAs in circulation is emerging, although the current knowledge particularly on tsRNAs is at a very early age. In this study, we report expression profiles of tsRNAs and miRNAs in circulation in mice and show that closed tibial fracture resulted in differential expression of a pool of tsRNAs and miRNAs during a follow-up period of D1-D14.
Fourteen cellular 5′tiRNAs and i-tRFs were observed in circulation with a baseMean between 10 and 1640, and six of them were differentially expressed after fracture. Val-tsRNAs were the most prevalent tsRNAs in circulation but were not affected by the fracture. Levels of 5′tiRNAs Lys-CTT-5′end, Lys-TTT-5′end and, i-tRFs Lys-TTT and Lys-CTT were decreased while 5′tiRNA His-GTG-5′ and i-tRF His-GTG were increased during fracture healing. Further, 93 miRNAs were observed in circulation with a baseMean over 10 and 34 of them were expressed with a baseMean over 100. Six miRNAs were differentially expressed after fracture. Only the level of miR-451a was decreased after fracture with the major drop at D1, while levels of miR-328-3p and miR-133a-3p were elevated mainly at D1 and returned thereafter close to the control level. These three miRNAs represent an early response to the fracture, while the other three miRNAs miR-375-3p, miR-423-5p and miR-150-5p were expressed at an increased level throughout D1-D14. Based on current published knowledge, these differentially expressed tsRNAs and miRNAs are all capable of targeting pathways to modulate osteogenesis and tissue remodelling during fracture healing, including, e.g., Wnt/β-catenin and P13K/AKT pathways, if taken up by the target tissue. The expression levels in serum did not significantly correlate with the upregulated expression levels in callus tissue, suggesting selected sorting into circulation from the fracture site or release from other tissues due to systemic effects of fracture healing process.
Future characterization on the functional role of these differentially expressed tsRNAs and miRNAs during fracture healing will increase our understanding on the systemic regulation of body metabolism in general.
This study is descriptive in nature, but the purpose of it was to bring out the possibilities of small non-coding RNAs, including tRNA derived fragments as novel tools to monitor fracture healing and tissue regeneration. Functional studies will verify their value in the development of therapeutic applications to target fracture healing process or as biomarker tools which utilize circulating small non-coding RNAs as indicators of faultless fracture healing.
5. Limitation of the study
In this study, ExoQuick precipitation was used to isolate exosomal miRNA fraction after which the total RNA was isolated by miRNeasy (Qiagen). Although precipitation and ultracentrifugation have been considered equally acceptable isolation methods for exosomal miRNA profiling analysis (Rekker et al., 2014), there are studies indicating that freely circulating miRNAs that are associated with lipid/HDL particles and RNA binding proteins in circulation may coprecipitate with the exosome fraction (Karttunen et al., 2019).
As blood cells contain a wide range of RNA molecules, they can also contribute to the small RNA profile in serum. To avoid random contamination, no samples with hemolysis were accepted for the analysis and blood samples were centrifuged twice to remove any cells from the serum before isolation of the exosome fraction. In human plasma, hemolysis-related contamination has been described to contain mir-486-5p, miR-451a, miR-16-5p, miR-106a-5p, miR-17-5p, miR-93-5p, miR-20a-5p, miR-107, and miR-20b-5p (Shkurnikov et al., 2016). Scrutiny of the expression of these miRNAs suggested no systemic input of blood cell contamination in our data. Out of the miRNAs characteristic to hemolysis, miR-451a, miR-16-5p, miR-93-5p, and miR-107 were expressed in our data with a baseMean above 100 while the others were either not detected or their expression level was low with baseMean 5 or below (Supplemental file 1). Erythrocyte enriched (or erythromiR) miR-451a as well as monocyte enriched miRNAs miR-150-5p and miR-423-5p were differentially expressed in our data.
As discussed above in connection with miR-451a, vesicles released from blood cells may contribute to the background of exosome's small RNA profile in circulation. Possible random contamination in a single sample by, e.g., hemolysis would increase variation, and systemic contribution would add background. Differential expression of the six miRNAs in circulation was statistically significant, compared to the control samples, suggesting that the contributing factor was the bone fracture although the source may have been the callus tissue itself or other tissues, e.g., adipose, or blood cells, as a response to the stimulus received from the fracture site (Fig. 3, Table 2).
The following are the supplementary data related to this article.
Full raw data, normalized counts and differential expression of tsRNAs and miRNAs in serum at D1, D5, D7, D10 and D14 after fracture.
Comparison of the expression levels of six differentially expressed miRNAs in circulation to the expression levels in callus tissue.
Funding statement
We are grateful to the Academy of Finland (grant 250671), Turku University Foundation, Institute of Biomedicine, University of Turku, Finland, and Turku Doctoral Programme of Molecular Medicine (TuDMM).
CRediT authorship contribution statement
Matthieu Bourgery: Software, Validation, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft. Erika Ekholm: Investigation, Writing – review & editing. Ari Hiltunen: Investigation, Writing – review & editing. Terhi J. Heino: Investigation, Writing – review & editing. Juha-Pekka Pursiheimo: Investigation, Writing – review & editing. Ameya Bendre: Data curation, Writing – review & editing. Emrah Yatkin: Investigation, Writing – review & editing. Tiina Laitala: Supervision, Funding acquisition, Writing – review & editing. Jorma Määttä: Investigation, Funding acquisition, Writing – review & editing. Anna-Marja Säämänen: Investigation, Supervision, Project administration, Resources, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge Merja Lakkisto and Jukka Karhu for their excellent technical assistance, as well as the Central Animal Laboratory of Turku for taking good care of our animals. Matthieu Bourgery is a member of the University of Turku TUDMM doctoral program.
Data availability
Full raw and normalized data is in Supplemental file 1.
References
- Anfossi S., Babayan A., Pantel K., Calin G.A. Clinical utility of circulating non-coding RNAs - an update. Nat. Rev. Clin. Oncol. 2018;15(9):541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- Baker N., Sohn J., Tuan R.S. Promotion of human mesenchymal stem cell osteogenesis by PI3-kinase/Akt signaling, and the influence of caveolin-1/cholesterol homeostasis. Stem Cell Res Ther. 2015;6:238. doi: 10.1186/s13287-015-0225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile L., Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57(1):289–300. http://www.jstor.org/stable/2346101 [Google Scholar]
- Bourgery M., Ekholm E., Fagerlund K., Hiltunen A., Puolakkainen T., Pursiheimo J.-P., Heino T., Määttä J., Heinonen J., Yatkin E., Laitala T., Säämänen A.-M. Multiple targets identified with genome wide profiling of small RNA and mRNA expression are linked to fracture healing in mice. Bone Rep. 2021;15 doi: 10.1016/j.bonr.2021.101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Lu Y., Yang L., Qiu J., Qin C., Zhang J., Zhang Y., Wang X., Qi G., Liu C., Zhang X., Li D., Zhu H. Identification and potential functions of tRNA-derived small RNAs (tsRNAs) in irritable bowel syndrome with diarrhea. Pharmacol. Res. 2021;173 doi: 10.1016/j.phrs.2021.105881. [DOI] [PubMed] [Google Scholar]
- Chen R., Liao X., Chen F., Wang B., Huang J., Jian G., Huang Z., Yin G., Liu H., Jin D. Circulating microRNAs, miR-10b-5p, miR-328-3p, miR-100 and let-7, are associated with osteoblast differentiation in osteoporosis. Int. J. Clin. Exp. Pathol. 2018;11(3):1383–1390. [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang H., Xia Y., Yan F., Lu Y. Therapeutic potential of mesenchymal cell-derived miRNA-150-5p-expressing exosomes in rheumatoid arthritis mediated by the modulation of MMP14 and VEGF. J. Immunol. 2018;201(8):2472–2482. doi: 10.4049/jimmunol.1800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhorn T.A., Gerstenfeld L.C. Fracture healing: mechanisms and interventions. Nat. Rev. Rheumatol. 2015;11(1):45–54. doi: 10.1038/nrrheum.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer V., Haffner-Luntzer M. Interaction between bone and immune cells: implications for postmenopausal osteoporosis. Semin. Cell Dev. Biol. 2022;123:14–21. doi: 10.1016/j.semcdb.2021.05.014. [DOI] [PubMed] [Google Scholar]
- Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martin R., Wang G., Brandão B.B., Zanotto T.M., Shah S., Kumar Patel S., Schilling B., Kahn C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601(7893):446–451. doi: 10.1038/s41586-021-04234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenfeld L.C., Cullinane D.M., Barnes G.L., Graves D.T., Einhorn T.A. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J. Cell. Biochem. 2003;88(5):873–884. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- Groot M., Lee H. Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells. 2020;9(4) doi: 10.3390/cells9041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Liu Y., Guo Y., Yang Y., Chen B. MicroRNA-423-5p inhibits the progression of trophoblast cells via targeting IGF2BP1. Placenta. 2018;74:1–8. doi: 10.1016/j.placenta.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Hadjiargyrou M., Zhi J., Komatsu D.E. Identification of the microRNA transcriptome during the early phases of mammalian fracture repair. Bone. 2016;87:78–88. doi: 10.1016/j.bone.2016.03.011. [DOI] [PubMed] [Google Scholar]
- He B., Lin X., Tian F., Yu W., Qiao B. MiR-133a-3p inhibits Oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J. Cell. Biochem. 2018;119(1):338–346. doi: 10.1002/jcb.26182. [DOI] [PubMed] [Google Scholar]
- Heianza Y., Krohn K., Yaskolka Meir A., Wang X., Ziesche S., Ceglarek U., Blüher M., Keller M., Kovacs P., Shai I., Qi L. Changes in circulating miR-375-3p and improvements in visceral and hepatic fat contents in response to lifestyle interventions: the CENTRAL trial. Diabetes Care. 2022;45(8):1911–1913. doi: 10.2337/dc21-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen A., Vuorio E., Aro H.T. A standardized experimental fracture in the mouse tibia. J. Orthop. Res. 1993;11(2):305–312. doi: 10.1002/jor.1100110219. [DOI] [PubMed] [Google Scholar]
- Hu D.P., Ferro F., Yang F., Taylor A.J., Chang W., Miclau T., Marcucio R.S., Bahney C.S. Cartilage to bone transformation during fracture healing is coordinated by the invading vasculature and induction of the core pluripotency genes. Development (Cambridge, England) 2017;144(2):221–234. doi: 10.1242/dev.130807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.-L., Zhang L., Li J.-P., Wang Y.-J., Duan Y., Wang J. MicroRNA-150: a potential regulator in pathogens infection and autoimmune diseases. Autoimmunity. 2015;48(8):503–510. doi: 10.3109/08916934.2015.1072518. [DOI] [PubMed] [Google Scholar]
- Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C., Guo X. The clinical potential of circulating microRNAs in obesity. Nat. Rev. Endocrinol. 2019;15(12):731–743. doi: 10.1038/s41574-019-0260-0. [DOI] [PubMed] [Google Scholar]
- Juran C.M., Zvirblyte J., Cheng-Campbell M., Blaber E.A., Almeida E.A.C. Cdkn1a deletion or suppression by cyclic stretch enhance the osteogenic potential of bone marrow mesenchymal stem cell-derived cultures. Stem Cell Res. 2021;56 doi: 10.1016/j.scr.2021.102513. [DOI] [PubMed] [Google Scholar]
- Kang D.W., Hwang W.C., Noh Y.N., Che X., Lee S.-H., Jang Y., Choi K.-Y., Choi J.-Y., Min D.S. Deletion of phospholipase D1 decreases bone mass and increases fat mass via modulation of Runx2, β-catenin-osteoprotegerin, PPAR-γ and C/EBPα signaling axis. Biochim. Biophys. Acta Mol. basis Dis. 2021;1867(5) doi: 10.1016/j.bbadis.2021.166084. [DOI] [PubMed] [Google Scholar]
- Karttunen J., Heiskanen M., Navarro-Ferrandis V., Das Gupta S., Lipponen A., Puhakka N., Rilla K., Koistinen A., Pitkänen A. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J. Extracellular Vesicles. 2019;8(1) doi: 10.1080/20013078.2018.1555410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvande A., Kushwaha P., Ahmad N., Adhikary S., Kothari P., Tripathi A.K., Khedgikar V., Trivedi R. Glucose dependent miR-451a expression contributes to parathyroid hormone mediated osteoblast differentiation. Bone. 2018;117:98–115. doi: 10.1016/j.bone.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Kim H.K. Transfer RNA-derived small non-coding RNA: dual regulator of protein synthesis. Mol. Cells. 2019;42(10):687–692. doi: 10.14348/molcells.2019.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Yim D.G., Lakshmanan V., Tirumalai V., Koh J.L., Park J.E., Cheong J.K., Low J.L., Lim M.J., Sze S.K., Shivaprasad P., Gulyani A., Raghavan S., Palakodeti D., DasGupta R. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019;20(7) doi: 10.15252/embr.201947789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Kuscu C., Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41(8):679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Shen Y.J., Chai S., Bai Y.L., Li Z.H. miR-133a-3p inhibits the osteogenic differentiation of bone marrow mesenchymal stem cells by regulating ankyrin repeat domain 44. Gen. Physiol. Biophys. 2021;40(4):329–339. doi: 10.4149/gpb_2020038. [DOI] [PubMed] [Google Scholar]
- Li S., Xu Z., Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes. 2018;9(5) doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhao Y., Li X., Wang Q., Ao Q., Wang X., Tian X., Tong H., Kong D., Chang S., Bai S., Fan J. MicroRNA-150 modulates adipogenic differentiation of adipose-derived stem cells by targeting Notch3. Stem Cells Int. 2019;2019 doi: 10.1155/2019/2743047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Cao J., Wang X., Guo C., Liu Y., Wang T. Deciphering the tRNA-derived small RNAs: origin, development, and future. Cell Death Dis. 2021;13(1):24. doi: 10.1038/s41419-021-04472-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Kou X., Chen C., Liu S., Liu Y., Yu W., Yu T., Yang R., Wang R., Zhou Y., Shi S. Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Res. 2018;28(9):918–933. doi: 10.1038/s41422-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire S., Lohman G., Guan S. A low-bias and sensitive small RNA library preparation method using randomized splint ligation. Nucleic Acids Res. 2020;48(14) doi: 10.1093/nar/gkaa480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkitie R.E., Hackl M., Niinimäki R., Kakko S., Grillari J., Mäkitie O. Altered MicroRNA profile in osteoporosis caused by impaired WNT signaling. J. Clin. Endocrinol. Metab. 2018;103(5):1985–1996. doi: 10.1210/jc.2017-02585. [DOI] [PubMed] [Google Scholar]
- McGonnell I.M., Grigoriadis A.E., Lam E.W., Price J.S., Sunters A. A specific role for phosphoinositide 3-kinase and AKT in osteoblasts? Front. Endocrinol. 2012;3:88. doi: 10.3389/fendo.2012.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M.A., Ludwig R.G., Garcia-Martin R., Brandão B.B., Kahn C.R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30(4):656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Yoshitomi H., Furu M., Ishikawa M., Shibuya H., Ito H., Matsuda S. MicroRNA-451 down-regulates neutrophil chemotaxis via p38 MAPK. Arthritis Rheumatol. 2014;66(3):549–559. doi: 10.1002/art.38269. [DOI] [PubMed] [Google Scholar]
- O’Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020;21(10):585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Wang Y.-L. Frontiers in Endocrinology. Vol. 9. 2018. Editorial: microRNAs as new players in endocrinology; p. 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertusa C., Tarín J.J., Cano A., García-Pérez M.Á., Mifsut D. Serum microRNAs in osteoporotic fracture and osteoarthritis: a genetic and functional study. Sci. Rep. 2021;11(1):19372. doi: 10.1038/s41598-021-98789-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnar J., Deiderick H., Huerta G., Hunicke-Smith S. Next-generation sequencing RNA-seq library construction. Curr. Protoc. Mol. Biol. 2014;106 doi: 10.1002/0471142727.mb0421s106. 4.21.1–4.21.19. [DOI] [PubMed] [Google Scholar]
- Puolakkainen T., Rummukainen P., Lehto J., Ritvos O., Hiltunen A., Saamanen A.M., Kiviranta R. Soluble activin type IIB receptor improves fracture healing in a closed tibial fracture mouse model. PloS One. 2017;12(7) doi: 10.1371/journal.pone.0180593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Li P., Zhang Z., Wu M. Insights into exosomal non-coding RNAs sorting mechanism and clinical application. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.664904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen E.E., Foster J., Sheldrake A., Stainback M., Glenn J., Cox L.A., Bredbenner T.L. Circulating miRNAs associated with bone mineral density in healthy adult baboons. J. Orthop. Res. 2022;40(8):1827–1833. doi: 10.1002/jor.25215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekker K., Saare M., Roost A.M., Kubo A.-L., Zarovni N., Chiesi A., Salumets A., Peters M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014;47(1–2):135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Saikia M., Hatzoglou M. The many virtues of tRNA-derived stress-induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J. Biol. Chem. 2015;290(50):29761–29768. doi: 10.1074/jbc.R115.694661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Ko E.-A., Sanders K.M., Chen Q., Zhou T. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genomics Proteomics Bioinformatics. 2018;16(2):144–151. doi: 10.1016/j.gpb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhang Y., Tan D., Zhang X., Yan M., Zhang Y., Franklin R., Shahbazi M., Mackinlay K., Liu S., Kuhle B., James E.R., Zhang L., Qu Y., Zhai Q., Zhao W., Zhao L., Zhou C., Gu W., Chen Q. Author correction: PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 2021;23(6):676. doi: 10.1038/s41556-021-00687-w. [DOI] [PubMed] [Google Scholar]
- Shkurnikov M.Y., Knyazev E.N., Fomicheva K.A., Mikhailenko D.S., Nyushko K.M., Saribekyan E.K., Samatov T.R., Alekseev B.Y. Analysis of plasma microRNA associated with hemolysis. Bull. Exp. Biol. Med. 2016;160(6):748–750. doi: 10.1007/s10517-016-3300-y. [DOI] [PubMed] [Google Scholar]
- Singh S., de Ronde M.W.J., Kok M.G.M., Beijk M.A., De Winter R.J., van der Wal A.C., Sondermeijer B.M., Meijers J.C.M., Creemers E.E., Pinto-Sietsma S.-J. MiR-223-3p and miR-122-5p as circulating biomarkers for plaque instability. Open Heart. 2020;7(1) doi: 10.1136/openhrt-2019-001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Kuscu C., Malik A., Shibata E., Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 2019;294(45):16930–16941. doi: 10.1074/jbc.RA119.009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Yu Y., Niu B., Wang D. Red blood cells as potential repositories of MicroRNAs in the circulatory system. Front. Genet. 2020;11:442. doi: 10.3389/fgene.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Li C.-T., Xiong L., Ning Z., Leung F., Peng S., Lu W.W. miR-375-3p negatively regulates osteogenesis by targeting and decreasing the expression levels of LRP5 and β-catenin. PloS One. 2017;12(2) doi: 10.1371/journal.pone.0171281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Pan J., Huang S., Peng X., Zou X., Luo Y., Ren D., Zhang X., Li R., He P., Wa Q. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018;37(1):160. doi: 10.1186/s13046-018-0813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao E.-W., Wang H.-L., Cheng W.Y., Liu Q.-Q., Chen Y.-X., Gao Q.-Y. A specific tRNA half, 5’tiRNA-his-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J. Exp. Clin. Cancer Res. 2021;40(1):67. doi: 10.1186/s13046-021-01836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraju K., Neerukonda S.N., Katneni U., Buehler P.W. Extracellular vesicles from red blood cells and their evolving roles in health, coagulopathy and therapy. Int. J. Mol. Sci. 2020;22(1) doi: 10.3390/ijms22010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., Rao T.N., Winnay J.N., Garcia-Martin R., Grinspoon S.K., Gorden P., Kahn C.R. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A.G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U. S. A. 2019;116(17):8451–8456. doi: 10.1073/pnas.1821120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Ivanov P., Ribas de Pouplana L., Torres A.G. Editorial: understanding the importance of non-canonical tRNA function. Front. Mol. Biosci. 2021;8:769784. doi: 10.3389/fmolb.2021.769784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E.L., Eleftheriou E., Thermes C. Improving small RNA-seq: less bias and better detection of 2'-O-methyl RNAs. J. Vis. Exp. 2019;151 doi: 10.3791/60056. [DOI] [PubMed] [Google Scholar]
- Vasilatou D., Papageorgiou S., Pappa V., Papageorgiou E., Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur. J. Haematol. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- Vienberg S., Geiger J., Madsen S., Dalgaard L.T. MicroRNAs in metabolism. Acta Physiol (Oxf.) 2017;219(2):346–361. doi: 10.1111/apha.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waki T., Lee S.Y., Niikura T., Iwakura T., Dogaki Y., Okumachi E., Oe K., Kuroda R., Kurosaka M. Profiling microRNA expression during fracture healing. BMC Musculoskelet. Disord. 2016;17(1):80–83. doi: 10.1186/s12891-016-0931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Aireti A., Aihaiti A., Li K. Expression of microRNA-150 and its target gene IGF2BP1 in human osteosarcoma and their clinical implications. Pathol. Oncol. Res. 2019;25(2):527–533. doi: 10.1007/s12253-018-0454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xu X., Li Y., Li J., Yao C., Wu R., Jiang Q., Shi D. A C-met chemical inhibitor promotes fracture healing through interacting with osteogenic differentiation via the mTORC1 pathway. Exp. Cell Res. 2019;381(1):50–56. doi: 10.1016/j.yexcr.2019.03.037. [DOI] [PubMed] [Google Scholar]
- Wang W., Gao J., Wang F. MiR-663a/MiR-423-5p are involved in the pathogenesis of lupus nephritis via modulating the activation of NF-κB by targeting TNIP2. Am. J. Transl. Res. 2017;9(8):3796–3803. [PMC free article] [PubMed] [Google Scholar]
- Weigl M., Kocijan R., Ferguson J., Leinfellner G., Heimel P., Feichtinger X., Pietschmann P., Grillari J., Zwerina J., Redl H., Hackl M. Longitudinal changes of circulating miRNAs during bisphosphonate and teriparatide treatment in an animal model of postmenopausal osteoporosis. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2021;36(6):1131–1144. doi: 10.1002/jbmr.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilner S., Skalicky S., Salzer B., Keider V., Wagner M., Hildner F., Gabriel C., Dovjak P., Pietschmann P., Grillari-Voglauer R., Grillari J., Hackl M. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- Xie S., Zhang Q., Zhao J., Hao J., Fu J., Li Y. MiR-423-5p may regulate ovarian response to ovulation induction via CSF1. Reprod. Biol. Endocrinol. 2020;18(1):26. doi: 10.1186/s12958-020-00585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Wang Z., Zhang Y., Zhang Z. Beneficial role of microRNA-328-3p in fracture healing by enhancing osteoblastic viability through the PTEN/PI3K/AKT pathway. Exp. Ther. Med. 2020;20(6):271. doi: 10.3892/etm.2020.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Palmer L.E., Lechauve C., Zhao G., Yao Y., Luan J., Vourekas A., Tan H., Peng J., Schuetz J.D., Mourelatos Z., Wu G., Weiss M.J., Paralkar V.R. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood. 2019;133(23):2518–2528. doi: 10.1182/blood.2018854604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zou H., Ding Q., Zou Y., Tang C., Lu Y., Xu X. tiRNA-val promotes angiogenesis via Sirt1-hif-1α axis in mice with diabetic retinopathy. Biol. Res. 2022;55(1):14. doi: 10.1186/s40659-022-00381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakar S., Werner H., Rosen C.J. 40 years of IGF1: insulin-like growth factors: actions on the skeleton. J. Mol. Endocrinol. 2018;61(1):T115–T137. doi: 10.1530/JME-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Cui S., Li X., Li W., Lu Q.J., Jiang X.H., Wang H., Chen X., Ma J.Z. Regulation of circulatory muscle-specific MicroRNA during 8 km run. Int. J. Sports Med. 2020;41(9):582–588. doi: 10.1055/a-1145-3595. [DOI] [PubMed] [Google Scholar]
- Yin Y., Du L., Li X., Zhang X., Gao Y. miR-133a-3p suppresses cell proliferation, migration, and invasion and promotes apoptosis in esophageal squamous cell carcinoma. J. Cell. Physiol. 2019;234(8):12757–12770. doi: 10.1002/jcp.27896. [DOI] [PubMed] [Google Scholar]
- Zarecki P., Hackl M., Grillari J., Debono M., Eastell R. Serum microRNAs as novel biomarkers for osteoporotic vertebral fractures. Bone. 2020;130 doi: 10.1016/j.bone.2019.115105. [DOI] [PubMed] [Google Scholar]
- Zhao C., Yu T., Dou Q., Guo Y., Yang X., Chen Y. Knockout of TLR4 promotes fracture healing by activating Wnt/β-catenin signaling pathway. Pathol. Res. Pract. 2020;216(2) doi: 10.1016/j.prp.2019.152766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full raw data, normalized counts and differential expression of tsRNAs and miRNAs in serum at D1, D5, D7, D10 and D14 after fracture.
Comparison of the expression levels of six differentially expressed miRNAs in circulation to the expression levels in callus tissue.
Data Availability Statement
Full raw and normalized data is in Supplemental file 1.