Abstract
Non-coding RNAs are emerging targets for drug development because they are involved in various cellular processes. However, there are a few reliable design strategies for small molecules that can target RNAs. This paper reports a simple and efficient method to comprehensively analyze RNA motifs that can be bound by a specific small molecule. The method involves Dicer-mediated pre-miRNA cleavage and subsequent analysis of the reaction products by high-throughput sequencing. A pre-miRNA mutant library containing a randomized region at the Dicer cleavage site was used as the substrate for the reaction. Sequencing analysis of the products of the reaction carried out in the presence or absence of a synthetic small molecule identified the pre-miRNA mutants whose Dicer-mediated cleavage was significantly altered by the addition of the small molecule. The binding of the small molecule to the identified pre-miRNA mutants was confirmed by surface plasmon resonance, demonstrating the feasibility of our method.
Introduction
The Encyclopedia of DNA Elements (ENCODE) project1 has revealed that >70% of the human genome is transcribed into RNA, whereas only ∼3% of the genome encodes proteins. The RNAs that are not translated to proteins are called non-coding RNAs (ncRNAs), and they have been found to exhibit diverse functions in various biological processes.2−4 With an increasing number of reports describing the important roles of ncRNAs in cellular functions and diseases,5 strategies that target specific ncRNAs and modulate their functions using small molecules have attracted the interest of researchers from various research fields, including cell biology, medicine, biotechnology, and chemistry.6−9
In the past decade, methods have been developed to identify small molecules and drugs that target RNAs. They are in vitro(10−17) or in silico(18,19) screening of a chemical library, structural modification of natural products,20−22 and more recently, design of small molecules using databases of RNA-small molecule interactions.23,24 Although these methods successfully discovered small molecules that target various RNAs including microRNA (miRNA) precursors17,18,24 and long ncRNAs (lncRNAs),25 identification of small molecules that bind target RNAs with high affinity and selectivity remains a challenge. This is partly due to our limited understanding of the small molecule–RNA interactions. To understand the nature of small molecule–RNA interactions, collecting and analyzing RNA binding pairs of a small molecule comprehensively is of crucial importance.
Next-generation sequencing (NGS) technology, also known as high-throughput sequencing technology, enables simultaneous sequencing of millions of DNA fragments,26,27 and has been applied in various investigations, including genomic analysis,28−30 transcriptome profiling,31,32 and epigenomic analysis.33 Using NGS technology, synthetic RNA libraries have been screened to identify RNA aptamers34,35 and catalytic RNAs.35,36 RNA sequencing by NGS has been applied to collect massive data sets for small molecule–RNA interactions, which should advance the development of small molecules that target RNAs. In particular, by combining NGS with other detection methods and bioinformatics analysis, target RNA motifs of small molecules have been identified from RNA libraries.11
Herein, we describe a simple and efficient method to identify binding motifs in a pre-miRNA-like hairpin for a given small molecule. We have previously shown that a small molecule can inhibit the cleavage of precursor miRNAs (pre-miRNAs) by binding to the Dicer cleavage site.37,38 Dicer recognizes the 3′ end of a pre-miRNA and cuts it at a fixed distance from the end with little sequence preference. This mode of cleavage by the Dicer and our previous results inspired us to design a new method for the identification of RNA motifs that can be bound by small molecules in the pre-miRNA structural context. We use a pre-miRNA mutant library containing a randomized sequence at the Dicer cleavage site that can serve as the binding site for a small molecule as the substrate for the Dicer. We hypothesized that the binding of a small molecule to the randomized region would affect the Dicer-mediated cleavage of particular pre-miRNA mutants and this can be assessed by analyzing the reaction products by NGS. To demonstrate our strategy, we analyzed the effect of a small molecule, naphthyridine carbamate dimer (NCD), on Dicer-mediated cleavage of the pre-miRNA mutant library and identified pre-miRNA mutants whose cleavage was affected by NCD. The identified pre-miRNA mutants were evaluated for their binding to NCD by surface plasmon resonance (SPR) and the effect of NCD binding on the Dicer-mediated cleavage by gel electrophoresis to confirm the accuracy and effectiveness of our proposed method.
Results and Discussion
Sequencing Analysis of Dicer-Cleaved Pre-miRNA Mutant Library
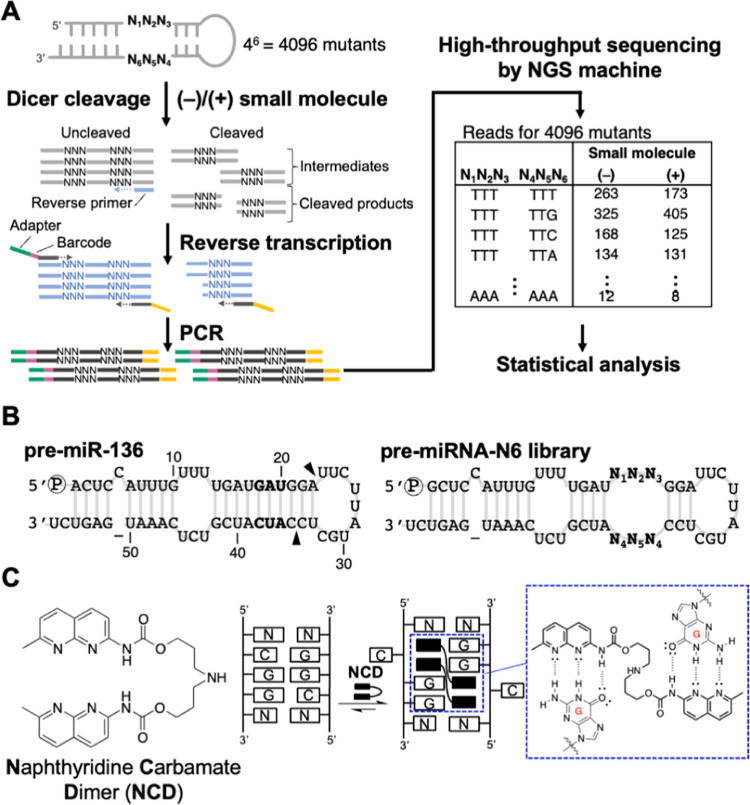
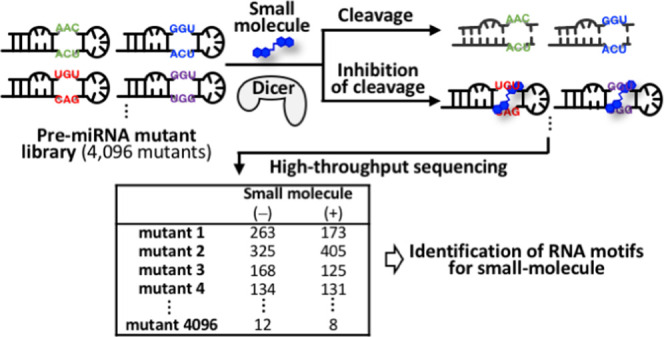
Our method involves the enzymatic digestion of target RNA substrates in a pre-miRNA mutant library followed by high-throughput sequencing of uncleaved substrates by the NGS platform (Figure 1A). The pre-miRNA mutant library containing a randomized sequence adjacent to the putative Dicer cleavage site was designed and used. Following the addition of the small molecule and Dicer treatment, the uncleaved substrates were converted into a DNA library for high-throughput sequencing. The sequencing data were analyzed to count the number of reads for each pre-miRNA mutant. The difference in the frequency of each mutant between the library with or without the small molecule was statistically analyzed.
Figure 1.
(A) Outline of the method in this study. (B) Sequences and predicted secondary structures of pre-miR-136 and the pre-miRNA-N6 library. The arrowheads indicate the putative Dicer cleavage site. (C) Chemical structure of NCD and binding to 5′-CGG-3′/5′-CGG-3′ in dsDNA. The dashed blue box shows the hydrogen bonding between the N-methoxycarbonyl-1,8-naphthyridine unit and guanine.
We first constructed a pre-miRNA library for the Dicer-mediated cleavage reaction. We chose pre-miR-136 as a structural template (Figure 1B). Pre-miR-136 was demonstrated to be a suitable substrate for the Dicer with high cleavage efficiency in our previous study.39 Pre-miR-136 has a cytosine bulge near the 5′ end, a 5′-UUU-3′/5′-UCU-3′ internal loop in the middle of the stem, and a 9-nt terminal loop. Six nucleotides, 5′-GAU-3′/5′-AUC-3′ (shown in bold in Figure 1B), close to the putative cleavage site of the Dicer were randomized as 5′-N1N2N3-3′/5′-N4N5N6-3′ to generate the pre-miRNA-N6 library that consists of 4,096 pre-miRNA mutants. In this study, we used the compound NCD (Figure 1C) from our chemical library to test this method. NCD is a synthetic compound that contains two naphthyridine rings connected by a flexible methylene linker. NCD binds to the 5′-CGG-3′/5′-CGG-3′ sequence motif in double-stranded DNA by forming hydrogen bonds with the unpaired guanines in the motif.40 However, NCD exhibits only weak binding to the same sequence motif in a double-stranded RNA (unpublished data). Recently, we coincidentally found a high-affinity interaction between NCD and RNA.41
The in vitro Dicer reaction of the pre-miRNA-N6 library was performed in the absence or presence of NCD. Cleaved and uncleaved RNAs in the reaction were reverse-transcribed into cDNA using a primer partially complementary to the 3′ arm sequence of the pre-miRNA-N6 library. The cDNA was amplified using PCR. Adapter sequences and a barcode sequence required for sequencing analysis were incorporated during the PCR (Figure 1A). The reverse-transcription primer binds to uncleaved and cleaved RNAs that contain the 3′-arm sequence of the library but not to cleaved RNAs that contain only the 5′-arm sequence. During the PCR amplification, the forward primer binds only to cDNAs that contain the sequence partially complementary to the 5′ arm sequence of the pre-miRNA-N6 library. This results in selective amplification of cDNAs corresponding to the uncleaved RNA mutants. The obtained dsDNA library was sequenced on the ion PGM platform. We conducted three independent experiments (experiments #1, #2, and #3) and investigated how the addition of NCD affects Dicer-mediated cleavage of each pre-miRNA mutant in the library.
The sequencing results are summarized in Table 1. We identified reads that perfectly matched the primer regions and internal fixed regions using an in-house Perl script. In experiment #1, the number of matched reads was 584 738 for the NCD (−) sample and 484 051 for the NCD (+) sample. On average from three independent experiments, the number of matched reads was 510 942 for the NCD (−) sample and 442 556 for the NCD (+) sample. The matched reads were counted for all 4096 mutants, and the obtained counts were transformed into log2RP100K (reads per 100 000 total reads) values. First, we generated histograms to compare the frequency distribution of log-fold-change (log FC) values for 4096 mutants between NCD (−) and (+) samples of each experiment (Figure 2). The histograms obtained from experiments #1 and #3 showed a similar frequency distribution of log FC, where the majority of the mutants were within the range of −1 < log FC < +1 (Figure 2A,C,D).
Table 1. Summary Statistics for the High-Throughput Sequencing Dataa.
| experiment
no. 1 |
experiment no. 2 |
experiment no. 3 |
average |
|||||
|---|---|---|---|---|---|---|---|---|
| NCD | – | + | – | + | – | + | – | + |
| matched reads | 584 738 (52.1%) | 484 051 (45.6%) | 485 458 (54.5%) | 387 013 (49.5%) | 462 631 (53.0%) | 456 634 (54.4%) | 510 942 | 442 566 |
| non-matched reads | 537 127 (47.9%) | 576 637 (54.4%) | 405 952 (45.5%) | 395 258 (50.5%) | 410 202 (47.0%) | 383 587 (45.6%) | 451 094 | 451 827 |
| total reads | 1 121 865 | 1 060 688 | 891 410 | 782 271 | 872 833 | 840 221 | 962 036 | 894 393 |
Values in parentheses are percentages of the matched or nonmatched reads to total reads.
Figure 2.

Frequency distribution of log-fold-change (log FC) values for 4096 mutants. The log FC values were calculated by subtracting the log2RP100K values in the NCD (−) sample from the log2RP100K values in the NCD (+) sample in (A) experiment #1, (B) experiment #2, and (C) experiment #3. (D) Box-plot representation of the frequency distribution of log FC values.
In contrast, the frequency of the log FC values in experiment #2 had a broader distribution with a peak shift compared with that of the other experiments (Figure 2B). Most of the mutants showed a decreased frequency of reads in the NCD (+) sample, indicated by negative log FC values (Figure 2B,D). The difference in the frequency distribution of log FC among the three experiments leads to false identification of mutants if analyzed by the same statistical procedures. Therefore, we performed a linear regression analysis on the log2RP100K values between NCD (−) and (+) samples using the middle 50% mutants of the log FC distribution, and normalized the experimental data by the obtained equations (Figure S1). The normalized log2RP100K values of the mutants from the NCD (−) and (+) samples were plotted and are shown in Figure 3A–C. The normalized log2RP100K values were converted back to the read counts and a statistical analysis of the difference in frequency of each mutant obtained for NCD (−) and (+) samples was performed using Pearson’s chi-square test with Bonferroni correction.42 Details of the chi-square test are described in the Methods section. We identified 70, 110, and 14 mutants that exhibited significant differences (q <0.05) between NCD (−) and (+) samples in Experiment #1, #2, and #3, respectively. Among these mutants identified, 67, 105, and 11 mutants (obtained from experiments #1, #2, and #3, respectively) yielded more reads in the NCD (+) sample, showing that Dicer-mediated cleavage of these mutants proceeded less efficiently in the presence of NCD than in the absence of NCD. We refer to these mutants as inhibited mutants (Figure 3A–C, red dots). In contrast, 3, 5, and 3 mutants (obtained from experiments #1, #2, and #3, respectively) yielded more reads in the NCD (−) sample, showing that Dicer-mediated cleavage of these mutants proceeded more efficiently in the presence than in the absence of NCD. We refer to these mutants as the promoted mutants (Figure 3A–C, light blue dots). The numbers of the inhibited mutants and promoted mutants obtained from each of the three experiments are shown in Table 2. We found 11 inhibited mutants and two promoted mutants that were identified in all three experiments (Table 3).
Figure 3.
(A–C) Scatter plots representing the normalized log2-transformed read counts for 4096 pre-miRNA mutants obtained from the reaction in the absence (x-axis) and presence (y-axis) of NCD. Red dots represent the inhibited mutants, and light blue dots represent the promoted mutants.
Table 2. Summary of the Number of Mutants That Showed Significant Difference in the Frequency Obtained for the NCD (−) Sample and NCD (+) Sample.
| experiment no. | inhibited mutants | promoted mutants | total |
|---|---|---|---|
| 1 | 67 | 3 | 70 |
| 2 | 105 | 5 | 110 |
| 3 | 11 | 3 | 14 |
| identified in 1, 2, and 3 | 11 | 2 | 13 |
Table 3. List of the Inhibited Mutants and the Promoted Mutants.
| experiment no. 1 |
experiment no. 2 |
experiment no. 3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| log2RP100K |
log2RP100K |
log2RP100K |
|||||||
| sequence: 5′-N1N2N3-3′/5′-N4N5N6-3′ | NCD (−) | NCD (+) | log FC | NCD (−) | NCD (+) | log FC | NCD (−) | NCD (+) | log FC |
| Inhibited Mutants | |||||||||
| GUG GGU | 6.71 | 8.68 | 1.97 | 5.93 | 9.64 | 3.71 | 6.87 | 7.73 | 0.86 |
| GGU GUG | 7.11 | 8.78 | 1.67 | 6.21 | 9.91 | 3.70 | 7.17 | 7.94 | 0.77 |
| GGU GUA | 6.64 | 8.78 | 2.14 | 6.13 | 10.02 | 3.88 | 6.91 | 7.78 | 0.88 |
| GGU GGU | 7.33 | 9.14 | 1.81 | 6.56 | 10.45 | 3.89 | 7.56 | 8.19 | 0.63 |
| GGG GUU | 7.34 | 9.07 | 1.73 | 6.92 | 10.12 | 3.20 | 7.68 | 8.28 | 0.60 |
| GGG GUG | 6.51 | 8.14 | 1.64 | 5.94 | 9.07 | 3.13 | 6.69 | 7.50 | 0.81 |
| GGG GGU | 7.18 | 8.73 | 1.55 | 6.54 | 9.83 | 3.29 | 7.28 | 8.11 | 0.82 |
| GGG GGG | 6.20 | 7.55 | 1.35 | 5.27 | 8.68 | 3.42 | 6.35 | 7.31 | 0.96 |
| GGC GGU | 6.77 | 9.13 | 2.36 | 6.26 | 10.57 | 4.30 | 6.99 | 7.93 | 0.94 |
| GCA GGU | 5.93 | 8.25 | 2.31 | 5.46 | 9.25 | 3.80 | 6.17 | 7.16 | 1.00 |
| AGG GGU | 5.96 | 8.36 | 2.40 | 5.70 | 9.62 | 3.93 | 6.28 | 7.33 | 1.06 |
| Promoted Mutants | |||||||||
| CGG AGU | 7.53 | 5.62 | –1.91 | 6.95 | 6.10 | –0.85 | 7.63 | 5.24 | –2.40 |
| CGG AGC | 6.12 | 4.20 | –1.92 | 5.39 | 4.51 | –0.88 | 6.13 | 4.08 | –2.06 |
The 11 inhibited mutants exhibited a clear preference for the guanine base in the randomized region (Figure 4A). The overall content of guanines in the randomized region was >70%, which is considerably higher than that of the other bases. Significantly high frequencies of guanine were observed at positions N1 (10 out of 11 mutants, 91%), N2 (9 out of 11 mutants, 82%), and N4 (100%). On the other hand, uracil was preferred at position N6 (7 out of 11 mutants, 64%). The frequencies of adenine and cytosine bases in the randomized region were as low as 5% and 3%, respectively. The two promoted mutants share a similar sequence (5′-CGG-3′/5′-AGU-3′ and 5′-CGG-3′/5′-AGC-3′) in the randomized region, where the only difference is in the nucleobase at the N6 position (U or C) (Figure 4B). To confirm that the observed changes in the frequency of mutants are indeed due to the effect of NCD addition to the Dicer reaction and not because of the effect of NCD on other steps such as reverse transcription and PCR, we performed sequencing analysis on the pre-miRNA-N6 library without subjecting it to Dicer cleavage but treating with NCD (Table S1 and Figure S2). We identified one promoted mutant that exhibited significant differences (q < 0.05) between the NCD (−) and (+) samples in experiment #3, but this mutant was not identical to any of the mutants identified above in the samples with Dicer cleavage.
Figure 4.

Sequence logo of the randomized region for (A) 11 inhibited mutants and (B) two promoted mutants identified in all of the three experiments.
Validation of the Identified RNA Motifs
To validate the sequencing data, the inhibited mutants and the promoted mutants (Table 3) were separately prepared and cleaved by the Dicer in the presence or absence of NCD. We also selected 7 mutants that exhibited a high q value (non-responder) in all three experiments and used them as controls for the Dicer cleavage reaction. The reaction products were analyzed by denaturing PAGE, and RNA bands were visualized by staining the gel with SYBR Gold (Figure 5A–C). Densitometric analyses of the visualized bands were performed to determine whether the addition of NCD to the Dicer reaction affects the relative abundance of cleaved RNAs and the uncleaved substrate (Figure 5D–F). In all of the inhibited mutants, the intensity of the band and the fraction of the uncleaved substrate significantly increased in the presence of NCD (Figure 5A,D). For the 5′-GGU-3′/5′-GUG-3′ mutant, the fraction of the uncleaved substrate increased by 12.3-fold from 7.2 ± 3.2% in the absence to 88.4 ± 3.0% in the presence of NCD. A similar increase of 11.8-fold was observed for the 5′-GUG-3′/5′-GGU-3′ mutant, where the fraction of the uncleaved substrate was 6.6 ± 2.1% in the absence and 78.6 ± 3.0% in the presence of NCD. The inhibitory effect of NCD on the cleavage of the 5′-GGU-3′/5′-GUG-3′ mutant and 5′-GUG-3′/5′-GGU-3′ mutant was concentration-dependent and the IC50 values of NCD were estimated as 4.5 ± 0.1 and 5.2 ± 0.4 μM, respectively (Figure S3). Among the inhibited mutants, relatively small changes in the fraction of the uncleaved substrate were observed for 5′-GGG-3′/5′-GUU-3′, 5′-GGG-3′/5′-GUG-3′, and 5′-GGG-3′/5′-GGG-3′ mutants (1.5-, 1.4-, and 1.5-fold increase, respectively), probably because of the moderate cleavage of these mutants by the Dicer in the absence of NCD. These data obtained for the inhibited mutants demonstrate the inhibitory effect of NCD on Dicer-mediated cleavage of these mutants.
Figure 5.
Denaturing PAGE analysis of Dicer-mediated cleavage in the presence or absence of NCD of (A) the inhibited mutants, (B) the promoted mutants, and (C) the non-responders. Densitometric analyses were performed on SYBR Gold-stained gels and the fractions of uncleaved substrate (%) in the absence (light gray bars) and presence (dark gray bars) of NCD are shown as bar graphs in panels (D) the inhibited mutant, (E) the promoted mutants, and (F) the non-responders. NCD concentration: 100 μM, S: Substrate, I: Intermediate, P: Product. Data are represented as mean ± SD of three independent experiments. *p < 0.05 and **p < 0.01 versus the sample without NCD addition (paired t-test).
In contrast to the inhibited mutants, the effect of NCD addition was not evident for Dicer-mediated cleavage of the promoted mutants (Figure 5B,E). The fractions of the uncleaved substrate for 5′-CGG-3′/5′-AGU-3′ and 5′-CGG-3′/5′-AGC-3′ mutants did not change significantly in the presence of NCD, 1.1- and 1.0-fold changes, respectively. NCD had different effects on the Dicer-mediated cleavage of the non-responders (Figure 5C,F). In 4 out of 7 non-responders that were tested, NCD addition did not significantly change the fraction of the uncleaved substrate, suggesting that NCD had little effect on Dicer-mediated cleavage of those mutants. On the other hand, NCD affected Dicer-mediated cleavage of the other three mutants with statistical significance. Densitometric analysis showed that the fraction of the the uncleaved substrate increased for 5′-GGC-3′/5′-GUG-3′ and 5′-ACA-3′/5′-GCG-3′ mutants in the presence of NCD (5.1- and 1.9-fold increase, respectively). The addition of NCD decreased the fraction of the uncleaved substrate for the 5′-CAC-3′/5′-GCG-3′ mutant (0.4-fold). The discrepancy observed between the sequencing and PAGE results for the promoted mutants and non-responders is discussed later in this paper.
Confirmation of NCD Binding to the Identified RNA Motifs
Next, we assessed whether NCD binds to these selected mutants by surface plasmon resonance (SPR) assay. NCD was immobilized onto a sensor chip surface (Scheme S1) and the association and dissociation kinetics of the mutants to the sensor surface were recorded at five concentrations ranging from 0.01 to 1 μM (Figure 6). Among the 11 inhibited mutants, eight mutants produced a significant increase in the SPR signal for the NCD-immobilized sensor surface, whereas three mutants (5′-GGG-3′/5′-GUU-3′, 5′-GGC-3′/5′-GGU-3′, and 5′-GCA-3′/5′-GGU-3′ mutants) showed weaker responses (Figure 6A). The apparent dissociation constant (KDapp) values calculated from rate constants, assuming a 1:1 binding stoichiometry, revealed that these inhibited mutants have an affinity for NCD in the nanomolar range (15.5–110 nM) (Table S2). The 2 promoted mutants showed weak responses to the NCD-immobilized sensor surface (Figure 6B), and they exhibited higher KDapp values than the inhibited mutants (Table S2). The SPR responses obtained for the non-responders were smaller than those obtained for the inhibited mutants, except for one mutant 5′-GGC-3′/5′-GUG-3′ (Figure S3). The KDapp values calculated for the non-responders varied, ranging from 65.5 nM to 23.1 μM (Table S2). Overall, the SPR responses obtained for the inhibited mutants were higher than those obtained for the promoted mutants and non-responders. However, the KDapp values obtained for the binding of NCD to the promoted mutants or non-responders are within 4-times the highest KDapp value of 110 nM obtained for the inhibited mutant 5′-GCA-3′/5′-GGU-3′, except for two non-responders (5′-GCC-3′/5′-GGC-3′ and 5′-CAC-3′/5′-GCG-3′) that showed extremely high KDapp values.
Figure 6.
SPR analysis of the binding of NCD to (A) the inhibited mutants and (B) the promoted mutants. RNA was added stepwise at 0.01 μM (yellow), 0.03 μM (green), 0.1 μM (blue), 0.3 (purple), and 1.0 μM (red) to the NCD-immobilized sensor surface. Insets are higher magnification views.
Search for the Identified RNA Motifs in Endogenous Pre-miRNA Hairpin
MiRNAs regulate gene expression at the post-transcriptional level. They are produced by sequential cleavage of hairpin-structured precursors by two RNase III enzymes, Drosha and Dicer.43,44 Mature miRNAs are loaded onto argonaute 2 protein (AGO2) to form an RNA-induced silencing complex, where they bind to their target sequence in mRNA and lead mRNA cleavage or translational repression. Because miRNAs are involved in many cellular processes including carcinogenesis, targeting the biogenesis of a specific miRNA by small molecules can be a therapeutic strategy for the treatment of diseases associated with miRNA dysregulation.
We, therefore, looked at whether the identified NCD-binding RNA motifs are present in the endogenous pre-miRNA hairpins. To date, 34 675 sequences of pre-miRNA hairpins have been deposited in the miRBase database.
The sequences of pre-miRNA hairpins in the database were downloaded and their secondary structures were predicted using the ViennaRNA package.45 The RNA motifs identified in the 11 inhibited mutants were searched in the stem region, and the pre-miRNA hairpins that was found to contain the RNA motifs were counted. We found that the 11 RNA motifs are present in 738 positions in the pre-miRNA hairpins in the database (Figure S5A,B), and 49 positions in 41 human pre-miRNA hairpins (Figure 7), respectively. Among the 11 RNA motifs, the 5′-GGU-3′/5′-GGU-3′ motif was the most frequently found in the human pre-miRNA hairpins, and 5′-GCA-3′/5′-GGU-3′, and 5′-GGC-3′/5′-GGU-3′ were found to be the second and third most frequently observed. We selected three miRNA precursors, pre-miR-216a, pre-miR-3689c, and pre-miR-6073, which contain one or more of the 11 motifs found in the inhibited mutants, and investigated the effect of NCD on Dicer-mediated cleavage of these miRNA precursors (Figure S6). Addition of NCD decreased the fraction of cleaved products for pre-miR-3689c and pre-miR-6073. On the other hand, the fraction of cleaved products and uncleaved substrates did not change significantly upon addition of NCD in the reaction for pre-miR-216a. This difference in the effect of NCD may be related to the position of a possible NCD-binding site in the miRNA precursor, which is consistent with the result observed in the previous study.38
Figure 7.
Motif search in the human pre-miRNA hairpins. The RNA motifs in the 11 inhibited mutants were searched in the sequences of human pre-miRNA hairpins in the miRbase. The table of counts for each motif that were found in the human pre-miRNA hairpins (left), and the bar-graph representation (right).
To see the effect of NCD treatment on miRNA expression in cells, we performed small RNA sequencing on NCD-untreated and NCD-treated HeLa cells with each condition run in triplicate. The sequencing reads were mapped to the human reference genome sequence (hg19) and the miRNA loci were annotated with the miRBase database.
We obtained 1422 unique miRNAs in 1013 pre-miRNA hairpins that were detected, more than one read in either of the six samples. Among these unique miRNAs detected, differentially expressed miRNAs between the NCD-untreated and NCD-treated samples were identified using DESeq.2 (Figure 8A). We set a stringent filter of base mean ≥16 to obtain a reliable set of differentially expressed miRNAs. With this filter, 476 miRNAs corresponding to 357 pre-miRNA hairpins were obtained, and 48 upregulated and 31 downregulated miRNAs (belonging to 44 and 28 pre-miRNA hairpins, respectively) by NCD treatment were identified (fold change ≥3 or ≤1/3, Benjamini and Hochberg method-adjusted p-value < 0.05).
Figure 8.
(A) MA plot representing the miRNAs that differentially expressed upon treatment of HeLa cells with NCD. Black dots: upregulated miRNAs. Red dots: downregulated miRNAs. (B) Venn diagram showing the overlap between the pre-miRNA hairpin carrying the inhibited motif and the pre-miRNA hairpins whose miRNA expressions were downregulated by NCD treatment. (C) The pre-miRNA hairpins containing the inhibited motifs in their wobble base pairs. (D) Bar-graph representation of the normalized raw read counts for the hsa-miR-671-3p/-5p and hsa-miR-3605-3p. FC: fold change. *p-value < 0.05.
We examined whether there is a relationship between the motif-containing pre-miRNA hairpins and miRNAs downregulated by NCD treatment. Among 41 human pre-miRNA hairpins that were found to contain at least one of the 11 inhibited RNA motifs, five pre-miRNA hairpins (has-mir-192, hsa-mir-671, hsa-mir-1306, hsa-mir-3605, and hsa-mir-3661) were included in the 357 pre-miRNA hairpins obtained above (Table S3). We found two overlapping pre-miRNA hairpins (hsa-mir-671 and hsa-mir-3605) between these five pre-miRNA hairpins and the 28 pre-miRNA hairpins whose miRNA expressions were downregulated by NCD treatment (Figure 8B). We performed the hypergeometric-distribution test to evaluate the significance of this overlap. Although this overlapping was 5.1-fold enriched than the expected value, the calculated p-value did not meet the p <0.05 criterion, which means the relationship between the motif-containing pre-miRNA hairpins and the downregulated miRNAs was not statistically significant. Looking more carefully at the secondary structure of these overlapping pre-miRNA hairpins, we noticed that both hairpins have the inhibited motif in their wobble base pairs (Figure 8C), while the other three pre-miRNA hairpins have the motif in their stem regions (Figure S5C). When we focused on the two overlapping pre-miRNA hairpins, which were defined with the NCD-binding motifs on the wobble base pairs, the comparisons with the 28 pre-miRNA hairpins deriving downregulated miRNAs provided 12.7-fold enrichment than expected and a p-value < 0.05, and we observed a clear decrease in the expression level of their corresponding miRNAs upon NCD treatment (Figure 8D). These results suggest a correlation with the occurrence of the inhibited RNA motifs in the wobble base pairs, which implies the effect of NCD treatment on the expression of some miRNAs.
Discussion
In this study, we employed a high-throughput sequencing of a pre-miRNA mutant library to investigate the effect of a small molecule on the cleavage of pre-miRNA mutants by a Dicer and to comprehensively identify sequence motifs functioning as a binding pocket for the small molecule. The high-throughput sequencing analysis identified pre-miRNA mutants whose Dicer-mediated cleavage can be either promoted or inhibited by the addition of the small molecule NCD. These identified pre-miRNA mutants were further evaluated by PAGE and densitometric analysis of the Dicer cleavage reaction and by SPR analysis of their binding ability to NCD. Finally, we performed a small RNA sequencing to study the effect of NCD treatment on miRNA expression in cells. For the 11 inhibited mutants identified from the pre-miRNA library by high-throughput sequencing analysis, Dicer-mediated cleavage was demonstrated to be inhibited by the addition of NCD. SPR analysis revealed that these inhibited mutants bind to NCD with affinities in the nanomolar range. These results indicate that the high-throughput sequencing of the Dicer-cleaved pre-miRNA library with and without the addition of NCD successfully identified RNA sequence motifs whose cleavage was inhibited by NCD binding.
The secondary-structure models for the inhibited mutants help predict consensus motifs required for NCD binding (Figure S7). Among 11 inhibited mutants, 4 mutants have a GG mismatch in the randomized region, which was found only in one non-responder (5′-GGC-3′/5′-GUG-3′) and not found in the other mutants (Figure S8). The position of the GG mismatch relative to the putative Dicer cleavage site differs from each other among the inhibited mutants, which could cause a different degree of effect of NCD binding on the Dicer-mediated cleavage of these mutants.
In contrast, the Dicer cleavage of the promoted mutants was shown not to be significantly affected by the addition of NCD when analyzed by PAGE. The discrepancy between the sequencing result and the PAGE analysis for the promoted mutants may be related to a moderate number of reads obtained from both NCD (−) and (+) samples (Table S4). These promoted mutants exhibited binding to the NCD-immobilized SPR sensor surface, but their affinities were lower than those obtained for the inhibited mutants. This might also explain why there was no clear effect of NCD in promoting the cleavage of those promoted mutants in the PAGE analysis. To most of the non-responders tested, NCD did not affect their Dicer-mediated cleavage. However, the cleavage of a few non-responders was affected by the addition of NCD, which is inconsistent with the sequencing result. The numbers of reads obtained for the non-responders were all small regardless of the addition of NCD (Table S4), and this is why no statistical significance was found in the frequency of these mutants in sequencing results. The discrepancies found between the sequencing results and the PAGE analysis of the Dicer cleavage reaction for the promoted mutants and non-responders suggest the need for a cut-off threshold on the number of reads for selecting mutants in addition to a threshold of q value. Overall, our method successfully identified pre-miRNA mutants whose cleavage by the Dicer was inhibited by the addition of a small molecule with no false positive cases.
The results of this study provide strong evidence that our method is effective at identifying RNA sequence motifs serving as targets for a small molecule in the pre-miRNA structural context. Our method does not require any labeling and immobilization of either RNAs or small molecules, which enables easy application of the method to different target molecules. Although we examined one small molecule against 4096 pre-miRNA mutants in this study, a more significant number of small molecules can be evaluated simultaneously using barcoding and pooling of all samples. Binding assays such as SPR can be used to identify small-molecule binders for RNAs; however, not all such molecules are functionally active against target RNAs. Our method can identify small molecule–RNA interactions that can modulate the function of the target RNAs. Although there needs to be further refinement of criteria for identifying target pre-miRNA mutants from high-throughput sequencing data, we believe that our method can contribute to the identification of new RNA targets of small molecules.
Methods
Preparation of the Pre-miRNA Mutant Library
The double-stranded DNA (dsDNA) library was generated by PCR using a synthesized oligonucleotide containing 6-nt random nucleotides (5′-GCT CCA TTT GTT TTG ATN NNG GAT TCT TAT GCT CCN NNA TCG TCT CAA ATG AGTCT-3′, where N represents any nucleotide) as a template with Platinum Pfx DNA polymerase (Invitrogen), followed by purification with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel). The primers used for the PCR amplification were: forward primer (5′-TAA TAC GAC TCA CTA TAG CTC CAT TTG TTT TGA-3′ [T7 promoter sequence is underlined]) and reverse primer (5′- A*G*A CTC ATT TGA GAC GA-3′ [*2′-OMe-modified nucleosides]). The dsDNA library was transcribed in vitro with T7 RNA polymerase (MEGAshortscript T7 Transcription Kit, Invitrogen), followed by DNase I treatment. The resulting mixture was separated on an 8% native polyacrylamide gel, and the RNA band similar in size to the wild-type pre-miR-136 was excised and eluted from the gel. The gel-purified RNA was treated with 5′ RNA polyphosphatase (Epicentre) to convert triphosphate to monophosphate at the 5′ end, and purified by denaturing polyacrylamide electrophoresis.
In Vitro Dicer Reaction
RNA was incubated with the in-house-prepared recombinant human Dicer46 in the presence or absence of the compound (NCD, 100 μM) at 37 °C. The incubation time varies with different RNA substrates (10 h for the pre-miRNA mutant library and 3 h for the individual mutants). A typical reaction mixture contained 2 μM RNA, 74 nM recombinant human Dicer, 1 mM ATP, 10 mM MgCl2, 0.5 mg/mL BSA, and 1× Dicer Reaction Buffer (Genlantis) in a 10 μL-reaction. The reaction was stopped by adding Dicer stop solution (Genlantis), and was analyzed by 15% denaturing polyacrylamide gel electrophoresis (acrylamide/bis-acrylamide = 19:1, 6 M urea). The gel was stained with SYBR Gold and the RNA bands were visualized using a LAS 4010 system and quantified using ImageQuant (GE Healthcare).
Preparation of DNA Libraries for Sequencing
After the in vitro Dicer reaction, the reaction mixture was reverse-transcribed with PrimeScript II Reverse Transcriptase (TAKARA) at 42 °C for 1 h using the primer (5′-AGA CTC ATT TGA GAC GA-3′). The cDNA was amplified by PCR with the following primers: forward primer (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG XXX XXX XXX XGA TGC TCC ATT TGT TTT GA -3′ [XXXXXXXXXX denotes a barcode sequence for multiplexing]) and reverse primer (5′-CCT CTC TAT GGG CAG TCG GTG ATA GAC TCA TTT GAG ACG A-3′). The PCR products were cleaned with NucleoSpin Gel and PCR Clean-up kit and quantified using the Qubit dsDNA HS Assay Kit (Invitrogen). The DNA libraries prepared above were pooled and diluted in nuclease-free water to obtain a final concentration of 8 pM. The library solution was subjected to emulsion PCR with the Ion OneTouch 2 system using the Ion PGM Hi-Q View OT2 200 Kit (Thermo Fisher Scientific). After recovery and enrichment of ion sphere particles, sequencing was performed using the Ion PGM Hi-Q View Sequencing Kit on an Ion 318 Chip V2 (Thermo Fisher Scientific). The primer sequences used for the library preparation are shown in Table S5.
Analysis of Sequencing Data
Sequence reads obtained from each sample were first filtered to remove reads that do not have the 5′-side (GCT CCA TTT GTT TTG AT), 3′-side (ATC GTC TCA AAT GAG TCT), and internal (GGA TTC TTA TGC TCC) fixed sequence. The reads that perfectly matched these fixed sequences were sorted to count the number of reads for each pre-miRNA mutant. A chi-square test of independence was performed to determine whether there is a significant difference in the frequency of each mutant between the library obtained from NCD (−) and NCD (+) samples. The chi-square statistics (χ2) was calculated for each mutant using the chi-square function in R (version 3.6.3) as follows:
| NCD (−) | NCD (+) | total | |
| mutant 1 | N1 | N2 | N1 + N2 |
| nonmutant 1 | n1 | n2 | n1 + n2 |
| total | N1 + n1 | N2 + n2 | N1 + N2 + n1 + n2 |
χ2 = (N1 + N2 + n1 + n2) (N1n2 – N2n1)2/(N1 + N2) (N2 + n2) (n2 + n1) (n1 + N1), where N1 and N2 are the number of reads for mutant 1 obtained from NCD (−) and NCD (+) samples, whereas n1 and n2 are the number of reads for nonmutant 1 obtained from NCD (−) and NCD (+) samples. The probability of the chi-square test (p-value) was calculated by applying the R function p-value with one degree of freedom. The resulting p-value was adjusted using Bonferroni’s method to obtain the q value by multiplying the p-value by 4096 (the number of pre-miRNA mutant sequences).
Binding Analysis by SPR
Interaction between NCD and RNA was evaluated by surface plasmon resonance (SPR) with a Biacore T200 SPR system. Chemically synthesized RNAs were diluted to 1 μM in HBS-EP+ buffer (GE Healthcare), heated at 80 °C for 3 min, and then slowly cooled to room temperature for annealing. The resulting RNA solutions were injected at increasing concentrations (0, 0.01, 0.03, 0.1, 0.3, and 1 μM) to the NCD-immobilized sensor surface with the regeneration step between each concentration. Injections were done at 30 μL/min with a contact time of 180 s, followed by dissociation with running buffer for 180 s. All sensorgrams were corrected by subtraction of blank flow cell response and buffer injection response. The obtained sensorgrams were fitted to a 1:1 binding model to determine the association and dissociation rate constants for RNA-NCD complex using BIAevaluation software (GE Healthcare).
Motif Search in Endogenous Pre-miRNA Hairpins
The sequences of pre-miRNA hairpins (34 675 sequences) were downloaded in FASTA format from the miRBase miRNA database (https://www.mirbase.org/). The secondary structure of each input sequence with the lowest free energy was predicted using ViennaRNA (ver.2.4.14). The dot-bracket notation of the predicted secondary structure was used to determine the stem region and the terminal loop in each sequence. The RNA motifs identified in the 11 inhibited mutants were searched in the stem region determined above. The hairpin miRNA precursors that were found to contain the RNA motifs were counted and extracted using an in-house script.
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS) [JSPS KAKENHI Grant-in-Aid for Scientific Research (A) (19H00924) to K.N., JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (21H02079) to A.M.] and supported by the JST FOREST Program, Grant Number JPMJFR201W. The authors thank the Osaka University Honors Program for Graduate Schools in Science, Engineering and Informatics for the scholarship for Y.T.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.2c00452.
Additional experimental methods and supplementary figures (sequencing analysis, PAGE analysis of Dicer-mediated cleavage of pre-miRNA mutants and endogenous miRNA precursors, SPR analysis, and predicted secondary structures of the identified pre-miRNA mutants) and tables (kinetic data for SPR analysis, the result of motif search, and supplementary sequencing data) (PDF)
Author Present Address
∥ Department of Material Sciences, Faculty of Engineering Sciences, Kyushu University, 6-1 Kasuga-koen, Kasuga, Fukuoka 816-8580, Japan
Author Present Address
⊥ Department of Life Science, Faculty of Science and Engineering, Kindai University, 3-4-1 Kowakae, Higashi-Osaka, Osaka 577-8502, Japan.
The authors declare no competing financial interest.
Supplementary Material
References
- Pennisi E. ENCODE Project Writes Eulogy for Junk DNA. Science 2012, 337, 1159–1161. 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- Kaikkonen M. U.; Lam M. T. Y.; Glass C. K. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovasc. Res. 2011, 90, 430–440. 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. S.; Ono M. From SnoRNA to MiRNA: Dual Function Regulatory Non-Coding RNAs. Biochimie 2011, 93, 1987–1992. 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Laurent G.; Wahlestedt C.; Kapranov P. The Landscape of Long Noncoding RNA Classification. Trends Genet. 2015, 31, 239–251. 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Sztuba-Solinska J.; Chavez-Calvillo G.; Cline S. E. Unveiling the Druggable RNA Targets and Small Molecule Therapeutics. Bioorg. Med. Chem. 2019, 27, 2149–2165. 10.1016/j.bmc.2019.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi N. F.; Smith G. F. RNA as a Small Molecule Druggable Target. Bioorg. Med. Chem. Lett. 2017, 27, 5083–5088. 10.1016/j.bmcl.2017.10.052. [DOI] [PubMed] [Google Scholar]
- Tor Y. Targeting RNA with Small Molecules. ChemBioChem 2003, 4, 998–1007. 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- Donlic A.; Hargrove A. E. Targeting RNA in Mammalian Systems with Small Molecules. Wiley Interdiscip. Rev. RNA 2018, 9, e1477 10.1002/wrna.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan N. N.; Cai Z.; Newson C. N.; Hargrove A. E. Fluorescent Peptide Displacement as a General Assay for Screening Small Molecule Libraries against RNA. Org. Biomol. Chem. 2019, 17, 1778–1786. 10.1039/c8ob02467g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Seedhouse S. J.; French J.; Disney M. D. Defining the RNA Internal Loops Preferred by Benzimidazole Derivatives via 2D Combinatorial Screening and Computational Analysis. J. Am. Chem. Soc. 2011, 133, 10111–10118. 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.; Disney M. D. Identifying the Preferred RNA Motifs and Chemotypes That Interact by Probing Millions of Combinations. Nat. Commun. 2012, 3, 1125 10.1038/NCOMMS2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi N. F.; Howe J. A.; Nahvi A.; Klein D. J.; Fischmann T. O.; Kim H. Y.; McCoy M. A.; Walker S. S.; Hruza A.; Richards M. P.; Chamberlin C.; Saradjian P.; Butko M. T.; Mercado G.; Burchard J.; Strickland C.; Dandliker P. J.; Smith G. F.; Nickbarg E. B. Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem. Biol. 2018, 13, 820–831. 10.1021/ACSCHEMBIO.7B01013. [DOI] [PubMed] [Google Scholar]
- Howe J. A.; Wang H.; Fischmann T. O.; Balibar C. J.; Xiao L.; Galgoci A. M.; Malinverni J. C.; Mayhood T.; Villafania A.; Nahvi A.; Murgolo N.; Barbieri C. M.; Mann P. A.; Carr D.; Xia E.; Zuck P.; Riley D.; Painter R. E.; Walker S. S.; Sherborne B.; de Jesus R.; Pan W.; Plotkin M. A.; Wu J.; Rindgen D.; Cummings J.; Garlisi C. G.; Zhang R.; Sheth P. R.; Gill C. J.; Tang H.; Roemer T. Selective Small-Molecule Inhibition of an RNA Structural Element. Nature 2015, 526, 672–677. 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- Liu S.; Yang Y.; Li W.; Tian X.; Cui H.; Zhang Q. Identification of Small-Molecule Ligands That Bind to MiR-21 as Potential Therapeutics for Endometriosis by Screening ZINC Database and in-Vitro Assays. Gene 2018, 662, 46–53. 10.1016/j.gene.2018.03.094. [DOI] [PubMed] [Google Scholar]
- Sztuba-Solinska J.; Shenoy S. R.; Gareiss P.; Krumpe L. R. H.; le Grice S. F. J.; O’Keefe B. R.; Schneekloth J. S. Identification of Biologically Active, HIV TAR RNA-Binding Small Molecules Using Small Molecule Microarrays. J. Am. Chem. Soc. 2014, 136, 8402–8410. 10.1021/ja502754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzumi T.; Murata A.; Aikawa H.; Harada Y.; Nakatani K. Exploratory Study on the RNA-Binding Structural Motifs by Library Screening Targeting Pre-MiRNA-29 a. Chem. – Eur. J. 2015, 21, 16859–16867. 10.1002/CHEM.201502913. [DOI] [PubMed] [Google Scholar]
- Periwal V.; Scaria V. Machine Learning Approaches toward Building Predictive Models for Small Molecule Modulators of MiRNA and Its Utility in Virtual Screening of Molecular Databases. Methods Mol. Biol. 2017, 1517, 155–168. 10.1007/978-1-4939-6563-2_11. [DOI] [PubMed] [Google Scholar]
- Ganser L. R.; Lee J.; Rangadurai A.; Merriman D. K.; Kelly M. L.; Kansal A. D.; Sathyamoorthy B.; Al-Hashimi H. M. High-Performance Virtual Screening by Targeting a High-Resolution RNA Dynamic Ensemble. Nat. Struct. Mol. Biol. 2018, 25, 425–434. 10.1038/s41594-018-0062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti M.; Nauwelaerts K.; Herdewijn P. Pre-MicroRNA Binding Aminoglycosides and Antitumor Drugs as Inhibitors of Dicer Catalyzed MicroRNA Processing. Bioorg. Med. Chem. Lett. 2012, 22, 1709–1711. 10.1016/j.bmcl.2011.12.103. [DOI] [PubMed] [Google Scholar]
- Vo D. D.; Tran T. P. A.; Staedel C.; Benhida R.; Darfeuille F.; Di Giorgio A.; Duca M. Oncogenic MicroRNAs Biogenesis as a Drug Target: Structure-Activity Relationship Studies on New Aminoglycoside Conjugates. Chem. – Eur. J. 2016, 22, 5350–5362. 10.1002/chem.201505094. [DOI] [PubMed] [Google Scholar]
- Nahar S.; Ranjan N.; Ray A.; Arya D. P.; Maiti S. Potent Inhibition of MiR-27a by Neomycin-Bisbenzimidazole Conjugates. Chem. Sci. 2015, 6, 5837–5846. 10.1039/c5sc01969a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Gallo S. M.; Disney M. D. Sequence-Based Design of Bioactive Small Molecules That Target Precursor MicroRNAs. Nat. Chem. Biol. 2014, 10, 291–297. 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney J. L.; Disney M. D. Small Molecule Targeting of a MicroRNA Associated with Hepatocellular Carcinoma. ACS Chem. Biol. 2016, 11, 375–380. 10.1021/acschembio.5b00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlic A.; Morgan B. S.; Xu J. L.; Liu A.; Roble C.; Hargrove A. E. Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold. Angew. Chem., Int. Ed. 2018, 57, 13242–13247. 10.1002/ANIE.201808823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. C. Next-Generation Sequencing Transforms Today’s Biology. Nat. Methods 2008, 5, 16–18. 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Churko J. M.; Mantalas G. L.; Snyder M. P.; Wu J. C. Overview of High Throughput Sequencing Technologies to Elucidate Molecular Pathways in Cardiovascular Diseases. Circ. Res. 2013, 112, 1613–1623. 10.1161/CIRCRESAHA.113.300939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. G.; Gianoulis T. A.; Pukatzki S.; Mekalanos J. J.; Ornston L. N.; Gerstein M.; Snyder M. New Insights into Acinetobacter Baumannii Pathogenesis Revealed by High-Density Pyrosequencing and Transposon Mutagenesis. Genes Dev. 2007, 21, 601–614. 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R.; Paul J. S.; Albrechtsen A.; Song Y. S. Genotype and SNP Calling from Next-Generation Sequencing Data. Nat. Rev. Genet. 2011, 12, 443–451. 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R.; Haverkamp T. H. A.; Kumar S.; Lanzén A.; Nederbragt A. J.; Quince C.; Kauserud H. Environmental Microbiology through the Lens of High-Throughput DNA Sequencing: Synopsis of Current Platforms and Bioinformatics Approaches. J. Microbiol. Methods 2012, 91, 106–113. 10.1016/j.mimet.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Tang F.; Barbacioru C.; Wang Y.; Nordman E.; Lee C.; Xu N.; Wang X.; Bodeau J.; Tuch B. B.; Siddiqui A.; Lao K.; Surani M. A. MRNA-Seq Whole-Transcriptome Analysis of a Single Cell. Nat. Methods 2009, 6, 377–382. 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Mutz K. O.; Heilkenbrinker A.; Lönne M.; Walter J. G.; Stahl F. Transcriptome Analysis Using Next-Generation Sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Leblanc V. G.; Marra M. A. Next-Generation Sequencing Approaches in Cancer: Where Have They Brought Us and Wherewill They Take Us?. Cancers 2015, 7, 1925–1958. 10.3390/cancers7030869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I.; Xulvi-Brunet R.; Campbell G. W.; Turk-MacLeod R.; Chen I. A. Comprehensive Experimental Fitness Landscape and Evolutionary Network for Small RNA. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 14984–14989. 10.1073/pnas.1307604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori S.; Takahashi K.; Yokobayashi Y. Deep Sequencing Analysis of Aptazyme Variants Based on a Pistol Ribozyme. ACS Synth. Biol. 2017, 6, 1283–1288. 10.1021/acssynbio.7b00057. [DOI] [PubMed] [Google Scholar]
- Kobori S.; Nomura Y.; Miu A.; Yokobayashi Y. High-Throughput Assay and Engineering of Self-Cleaving Ribozymes by Sequencing. Nucleic Acids Res. 2015, 43, e85. 10.1093/nar/gkv265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata A.; Fukuzumi T.; Umemoto S.; Nakatani K. Xanthone Derivatives as Potential Inhibitors of MiRNA Processing by Human Dicer: Targeting Secondary Structures of Pre-MiRNA by Small Molecules. Bioorg. Med. Chem. Lett. 2013, 23, 252–255. 10.1016/j.bmcl.2012.10.108. [DOI] [PubMed] [Google Scholar]
- Murata A.; Otabe T.; Zhang J.; Nakatani K. BzDANP, a Small-Molecule Modulator of Pre-MiR-29a Maturation by Dicer. ACS Chem. Biol. 2016, 11, 2790–2796. 10.1021/acschembio.6b00214. [DOI] [PubMed] [Google Scholar]
- Otabe T.; Nagano K.; Kawai G.; Murata A.; Nakatani K. Inhibition of Pre-MiRNA-136 Processing by Dicer with Small Molecule BzDANP Suggested the Formation of Ternary Complex of Pre-MiR-136–BzDANP–Dicer. Bioorg. Med. Chem. 2019, 27, 2140–2148. 10.1016/j.bmc.2019.03.031. [DOI] [PubMed] [Google Scholar]
- Peng T.; Nakatani K. Binding of Naphthyridine Carbamate Dimer to the (CGG)n Repeat Results in the Disruption of the G-C Base Pairing. Angew. Chem., Int. Ed. 2005, 44, 7280–7283. 10.1002/anie.200502282. [DOI] [PubMed] [Google Scholar]
- Shibata T.; Nagano K.; Ueyama M.; Ninomiya K.; Hirose T.; Nagai Y.; Ishikawa K.; Kawai G.; Nakatani K. Small Molecule Targeting r(UGGAA)n Disrupts RNA Foci and Alleviates Disease Phenotype in Drosophila Model. Nat. Commun. 2021, 12, 236 10.1038/s41467-020-20487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn O. J. Multiple Comparisons among Means. J. Am. Stat. Assoc. 1961, 56, 52–64. 10.1080/01621459.1961.10482090. [DOI] [Google Scholar]
- Davis-Dusenbery B. N.; Hata A. Mechanisms of Control of MicroRNA Biogenesis. J. Biochem. 2010, 148, 381–392. 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M.; Kim V. N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Lorenz R.; Bernhart S. H.; Höner zu Siederdissen C.; Tafer H.; Flamm C.; Stadler P. F.; Hofacker I. L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata A.; Mori Y.; Di Y.; Sugai A.; Das B.; Takashima Y.; Nakatani K. Small Molecule-Induced Dimerization of Hairpin RNA Interfered with the Dicer Cleavage Reaction. Biochemistry 2021, 60, 245–249. 10.1021/acs.biochem.0c00920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.