Abstract
Nanoparticles (NPs) are paving the way for improved treatments for difficult to treat diseases diseases; however, much is unknown about their fate in the body. One important factor is the interaction between NPs and blood proteins leading to the formation known as the “protein corona” (PC). The PC, consisting of the Hard (HC) and Soft Corona (SC), varies greatly based on the NP composition, size, and surface properties. This highlights the need for specific studies to differentiate the PC formation for each individual NP system. This work focused on comparing the HC and SC of three NPs with different matrix compositions: a) polymeric NPs based on poly(lactic-co-glycolic) acid (PLGA), b) hybrid NPs consisting of PLGA and Cholesterol, and c) lipidic NPs made only of Cholesterol. NPs were formulated and characterized for their physico-chemical characteristics and composition, and then were incubated in human plasma. In-depth purification, identification, and statistical analysis were then performed to identify the HC and SC components. Finally, similar investigations demonstrated whether the presence of a targeting ligand on the NP surface would affect the PC makeup. These results highlighted the different PC fingerprints of these NPs, which will be critical to better understand the biological influences of the PC and improve future NP designs.
Keywords: Nanomedicine, Polymeric PLGA nanoparticles, Lipidic nanoparticles, Protein corona, Cholesterol, Targeted nanomedicines
Graphical abstract
Highlights
-
•
NPs with different matrices were formulated: PLGA, Cholesterol, or mixed PLGA-Chol hybrids.
-
•
The hard and soft corona of each formulation was quantified and compared.
-
•
The PC seems to be more strongly affected by the polymer rather than the lipid in mixed NPs.
-
•
The soft corona depends more on the hard corona composition than on the matrix.
-
•
Surface modification with a targeting ligand did not influence PC composition.
1. Introduction
Nanoparticles (NPs) represent one of the most innovative tools in the medical field. They can offer a wide range of potential advantages to drug delivery, such as the ability to a) incorporate a broad range of characteristically diverse active substances (Duskey et al., 2017; Sigg et al., 2016; Zu et al., 2021); b) stabilize and protect molecules from degradation in different biological environments (Duskey et al., 2021; Duskey et al., 2020a; Fornaguera and García-Celma, 2017); c) be surface-modified for specific targeted delivery (i.e. to tissue, cell types, or intra-cellular receptors) (Amini et al., 2021; Hoyos-Ceballos et al., 2020; Pederzoli et al., 2019); d) selectively modulate drug release (i.e. prolonged or retarded release) (Bagheri et al., 2021; Dinu et al., 2016; Warsi, 2021). NPs used for medical applications should be non-toxic, cause a limited immune response activation, and be site-specific to limit negative off-target effects (Tewabe et al., 2021). Nevertheless, once intravenously dosed, their biological effects can be very difficult to predict due to the various interactions of the NPs with proteins in the blood. These interactions lead to the formation of various layers around the NP surface, termed “the protein corona” (PC), made up of numerous blood proteins that can affect the fate of the NPs; moreover, the composition of this PC is usually hard to predict, necessitating thorough analyses for each NP system (Tosi, 2017).
The PC absorbed on the surface of NPs is very complex, but can generally be broken down into two distinct layers: 1) the more stable and slowly exchanging Hard Corona (HC) which interacts more intimately with the surface of the NPs, and 2) the more dynamic and weakly associated Soft Corona (SC) (García-Álvarez and Vallet-Regí, 2021; Lundqvist and Cedervall, 2020; Milani et al., 2012; Tenzer et al., 2013). The composition of the two PCs has been shown to influence circulation and biodistribution (Bertrand et al., 2017; Chinen et al., 2017; Falahati et al., 2019; Ke et al., 2017; Shanwar et al., 2021; Tekie et al., 2020), drug targeting (Dai et al., 2015; Mahmoudi et al., 2015; Prapainop et al., 2012), cellular uptake (De Paoli et al., 2014; Francia et al., 2019; Lesniak et al., 2012; Yan et al., 2013), and toxicity (Bélteky et al., 2019; Dobrovolskaia et al., 2014; Nyström and Fadeel, 2012; Shannahan et al., 2015) of both organic and inorganic NPs. These differences can be further complicated by biological parameters such as protein concentration (Caracciolo et al., 2011; Monopoli et al., 2011; Partikel et al., 2019a), exposure time (Barrán-Berdón et al., 2013; Natte et al., 2013; Ren et al., 2022), and temperature (Mahmoudi et al., 2014, Mahmoudi et al., 2013), which could vary from patient to patient or be due to a pathological state. For these reasons, the study of the PC is considered an urgent issue in the field of NP characterization as a necessary requirement to better predict the biological identity, biological consequences, and therapeutic outcomes of NPs (Ahsan et al., 2018; Kopac, 2021; Monopoli et al., 2012; Nguyen and Lee, 2017).
The formation of the PC, both the HC and SC, is influenced not only by the characteristics of the blood composition, but also by the NP characteristics. In particular, the composition (Caracciolo et al., 2015), hydrophobicity (Gessner et al., 2000; Lindman et al., 2007; Moyano et al., 2012), physical characteristics such as size and surface charge (Dobrovolskaia et al., 2009; Lacerda et al., 2010; Lundqvist et al., 2008; Tenzer et al., 2011), shape (Chakraborty et al., 2011; García-Álvarez et al., 2018), and surface modifications with targeting ligands or coatings (Almalik et al., 2017; Ashby et al., 2014; Pederzoli et al., 2018; Saha et al., 2016). All of these factors are interconnected and can influence critical variations in the PC fingerprint between delivery systems. This leads to the conclusion that there is no “universal” PC. Each individual factor ranging from the matrix, surfactant, size, and morphology can have important effects on the PC composition. Therefore, it is crucial to analyse the PC for each NP to determine which factors play the most important roles and which proteins lead to the strongest effects on the delivery of the NPs in vivo.
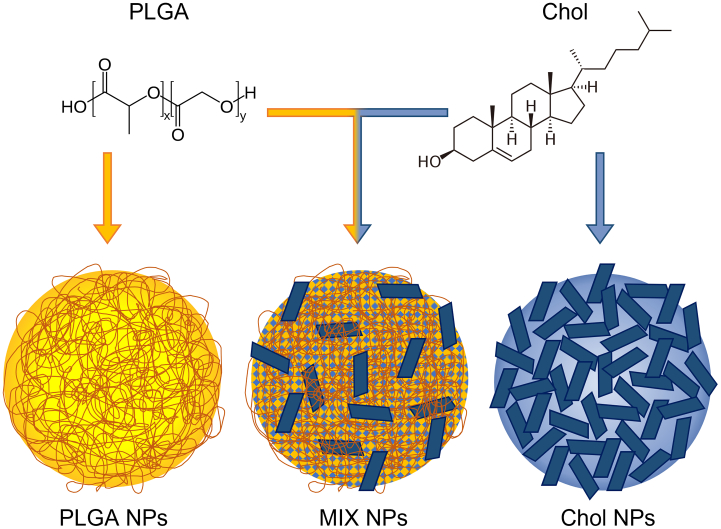
The present work aims to characterize the entire PC of three NPs with different compositions: 1) polymeric NPs that form solid matrix cores composed of poly-lactic co-glycolic acid (PLGA NPs), 2) hybrid NPs composed of a homogeneous matrix of PLGA and cholesterol (MIX NPs), and 3) solid-core lipidic NPs composed of cholesterol (Chol NPs) (Fig. 1). PLGA is a well-known FDA approved polymer that has been extensively used for the encapsulation and delivery of pharmaceutics. Cholesterol is a natural, ubiquitous lipid that has been widely used to formulate liposomes and hybrid NPs (Belletti et al., 2016; Ottonelli et al., 2021; Zhang et al., 2022b) and has recently been adapted to form lipidic NPs without any helper component (patent pending). Finally, hybrid NPs which have gained promise in the literature with the capacity to combine the advantages of both polymeric and lipidic systems as improved delivery tools (Ghitman et al., 2020). Therefore, these three NPs represent highly studied delivery systems that bridge the gap between the two extremes of polymeric and lipidic NPs.
Fig. 1.
The chemical structure of the polymer PLGA and Cholesterol and a schematic repre-sentation of the structure of PLGA, hybrid PLGA-Cholesterol (MIX), and Cholesterol NPs.
To evaluate the impact of the NP core composition on the formation of the HC and SC, NPs were incubated in plasma and the proteins absorbed were identified, evaluated, quantified, and compared. HPLC ms-ms analysis was used to identify the major proteins present and supported with statistical analysis (volcano plots) to determine the congruency between each NP type based on the proteins present in the HC or SC. A second set of NPs with the same composition was then produced and surfaced modified with the g7 peptide, known to promote BBB crossing in vivo and the HC and SC were analysed using identical methodologies to determine if the core composition or the surface engineering led to greater disparities in the PC layers. With this in-depth characterization of the PC of these 3 NPs (PLGA, MIX, and Chol), and determining the effect of surface modifications on the NPs, we hope to supplement the vast amount of similar research being performed on other types of NPs (Bing et al., 2021; Nguyen and Lee, 2017; Zhang et al., 2022a). This will help to create a field-wide understanding of how the PC affects the in vivo behaviour of NPs, allowing for the intelligent design of NP systems based on the identity of their PCs and their potential biological effects and reduce unanticipated biological responses during translation to in vivo testing.
2. Materials and Methods
2.1. Materials
Poly-(D,L-lactic-co-glycolic) acid RG503H (PLGA) 50:50, MW 11,000 Da, (Evonik, Essen, Germany), was used as received from the manufacturer. The g7 peptide was purchased from Mimotopes, Mulgrave, Australia: sequence GFtGFLS(O-β-Glc)-CONH2, MW 888.97. Cholesterol and Pluronic® F68 were purchased from Sigma Aldrich, Milan, Italy. Sterile filtered human K2EDTA plasma from a healthy donor (protein concentration estimated 6.4 g/dL; Gentaur, Bergamo, Italy), pre-stained Protein Sharp Mass V Plus (Euroclone, Milan, Italy), and Coomassie Brilliant Blue R-250 (Bio-Rad, Milan, Italy) were used as received from the manufacturers. All other solvents (analytical grade), products, and reagents were purchased from Sigma Aldrich (Milan, Italy) and used without any further purification (unless otherwise specified). High-purity water was supplied by a MilliQ water system (Millipore, MA, USA).
To ensure that samples were free of erroneous proteins, all multi-use glassware, plasticware, spatulas, and stir bars were pre-washed with 70% ethanol and then heated at 200 °C for at least 2 h. Mono-use sterile, protein-free, DNAse free, RNAse free materials were used for all other experiments. Finally, to further avoid sample contamination, all preparation methods were performed under a sterile vertical laminar flow hood (Asalair Vertical 700, Asalair, Firenze, Italy).
2.2. Formulation of the NPs
The g7 peptide was conjugated to PLGA or Cholesterol prior to NP formulation via a peptide coupling reaction as previously described (Costantino et al., 2005; Duskey et al., 2020b). PLGA, hybrid PLGA-Cholesterol (MIX), and Cholesterol (Chol) NPs were prepared using the nanoprecipitation procedure as previously reported (Belletti et al., 2016; Birolini et al., 2021; Pederzoli et al., 2019). Briefly, polymers and/or lipids were dissolved in the organic solvent and added dropwise to water containing Pluronic® F68 and under magnetic stirring (1300 rpms). The organic solvent was then removed at 30 °C under reduced pressure (R 114 Rotavapor, Buchi, Cornaredo, Italy). The recovered NP suspensions were then ultracentrifuged (J21, Beckman, CA, USA; 16,000 rpms) for 10 min at 4 °C and the supernatant was discarded to remove unformed polymer and or lipid and free surfactant. The NPs were resuspended in water, stored at 4 °C, and used within a week.
2.3. Chemico-physical characterization
2.3.1. Weight yield
A measured amount (200 μL) of the purified NP suspension was transferred to a pre-weighed Eppendorf tube and freeze-dried (−60 °C, 3 mm/Hg, for 48 h; LyoLab 3000, Heto-Holten, Allerod, Denmark) to calculate the Yield%:
| (1) |
2.3.2. Size and zeta potential analysis
Each NP sample was diluted in MilliQ water to a final concentration of 0.1 mg/mL to measure the size (Z-Average) and the polydispersity index (PDI) of the samples determined at room temperature by photon correlation spectroscopy using a Zetasizer Nano ZS (Malvern, Malvern, UK) equipped with a 4 mW He—Ne Laser at 633 nm, automatic Laser attenuator, Avalanche photodiode Detector, with a quantum efficiency (Q.E.) >50% at 633 nm.
The ζ potential (ζ-pot) was calculated starting from the electrophoretic mobility measured using the same sample and instrument equipped with a combination of laser Doppler velocimetry and phase analysis light scattering. For each preparation, the Z-Average, PDI, and ζ-pot were calculated as the mean of triplicate measurements from 3 distinct NP formulations.
2.3.3. The NP morphology
The NP morphology was investigated using a scanning transmission electron microscopy (STEM). Samples were prepared by immersing a 200-mesh Cu grid (TABB Laboratories Equipment, Berks, UK) into the NP suspension (∼ 0.1 mg/mL) and letting it air dry at room temperature. Images were then achieved using a Nova Nano SEM 450 (FEI Co., OR, USA) (acceleration voltage 30 KV, Spot 1.5) with a scanning transmission electron microscope II detector.
2.3.4. Residual Pluronic® F68
The residual Pluronic® F68 associated with the NPs was quantified following a previously published colorimetric reaction (Childs, 1975). Briefly, the surfactant was extracted from 10 mg of lyophilized NPs by solubilisation in 2 mL of dichloromethane followed by the addition of 10 mL of MilliQ water. After the dichloromethane was completely evaporated, the solution was filtered through a 0.2 μm cellulose acetate filter to remove any insoluble components. An aliquot of the filtrate was treated with 1 mL of a 0.005% (w/v) BaCl2 solution in 0.1 M HCl and 1 mL of an I2/KI solution (0.05 M / 0.15 M). After a 15 min incubation at room temperature, the absorbance was measured at 540 nm using a UV/Vis Spectrophotometer V-530 (Jasco Europe, Cremella, Italy) and the amount of Pluronic® F68 was calculated using a standard curve created that day under the same conditions.
2.4. PC purification and characterization
Characterization of the PC was performed following previously published methods (thoroughly described in the Supplementary material) (Pederzoli et al., 2018). Briefly, NP samples were first incubated in human plasma. To isolate NPs with only the HC, samples were centrifuged to remove the unbound material and SC from the NP-HC complex. In order to collect the NPs with the SC still intact, size exclusion chromatographic purification was used as a more gentle alternative (Cedervall et al., 2007b). Eluted fractions were monitored by SDS-PAGE to distinguish the free proteins in solution from the samples containing NP-PC complexes. Both samples, with only the HC or with the HC and SC, were then characterized in terms of total amount of proteins using the bicinchoninic acid (BCA) assay following the manufacturer's instructions. To identify the proteins present, these samples were processed with reducing and alkylating agents, and eventually trypsin, before being analysed by mass spectrometry. Plasma samples without NPs were treated similarly as controls.
2.5. Software and statistical analysis
Statistical analysis regarding the chemico-physical properties of NPs was performed using the Student's t-Test, where * p < 0.05 and ** p < 0.01, included in the software GraphPad Prism 6 (GraphPad Holdings, San Diego, CA, USA). All results are the mean with standard deviation (SD) measuring at least 3 different samples: n > 3.
Protein identification was achieved by searching the protein databases Swiss-Prot (2018_05, Homo Sapiens 557,491 entries) for peptides, and c-RAP for contaminants (ftp://ftp.thegpm.org/fasta/cRAP, 116 entries), using the MASCOT protein identification software (Version 2.4, Matrix Science, London, UK). Once identified, proteins were semi-quantified by normalizing their presence to the total amount of proteins and expressed using the “exponentially modified Protein Abundance Index” (emPAI) (Arike and Peil, 2014).
Eventually, results were statistically evaluated with the MSStat tool (Choi et al., 2014) and graphed as volcano plots with the binary logarithm of the fold change between two samples in the x-axis and the –log2 of the adjusted p on the y-axis. Proteins present in the PC with a log fold change < −2 or > 2 and an adjusted p < 0.05 were considered statistically differently between samples.
3. Results
3.1. NP chemico-physical characterization
The physico-chemical characteristics of NPs have been demonstrated to directly influence the PC formation. Therefore, a complete chemico-physical analysis of each different formulation was performed (Table 1, Fig. S1). Independently of the NP matrix all three formulations formed homogenous and reproducible structures as evidenced by a low PDI (< 0.3) (Shao et al., 2015). As expected from previous literature (Duskey et al., 2021; Hoyos-Ceballos et al., 2020), PLGA NPs exhibited the smallest size compared to the other formulations (∼ 150 nm), while MIX and Chol NPs displayed slightly larger structures of around 250 nm (Belletti et al., 2018, Belletti et al., 2016; Birolini et al., 2021). Regarding the Zeta potential, all formulations were statistically similar, ranging from −20 to −35 mV, with a trend towards more negative values correlating to the number of carboxylic moieties of PLGA present in the matrix (PLGA < MIX < Chol NPs). While all the NPs were recovered with high weight yields of almost 80%, differences were found in the amount of residual Pluronic® F68 between each NP type. PLGA NPs were found to have a lower percentage of surfactant compared to MIX and Chol NPs, in accordance with previous literature (14 vs 26%) (Belletti et al., 2018, 2016; Tosi et al., 2007). This increase of residual Pluronic® F68 could be explained by its high affinity with cholesterol, leading to higher non-specific interactions and more being retained in the matrix (Maskarinec et al., 2002; Owens and Peppas, 2006). This is important because contradicting literature suggests in some cases that the amount or location of the surfactant (in the matrix or absorbed on the surface) does not lead to interference of protein absorption and PC formation. Other sources claim that while it does not block PC formation, surfactants can influence its composition: this must be taken into consideration before conclusions are made about the importance of NP matrix on the PC proteins (Akhter et al., 2021; Neagu et al., 2017; Petry et al., 2019).
Table 1.
Chemico-physical characteristics of the three NP formulations.
| NP sample | Size (SD), nm |
PDI (SD) | Electrophoretic Mobility (SD), μmcm/Vs |
ζ-pot (SD), mV |
Weight Yield % w/w |
Pluronic® F68% w/w |
|---|---|---|---|---|---|---|
| PLGA-NPs | 152 (13) | 0.12 (0.02) | −2.52 (0.12) | −34.4 (6.2) | 74.9 (6.6) | 14 (4) |
| MIX-NPs | 237 (17) | 0.26 (0.01) | −2.00 (0.44) | −25.8 (5.3) | 76.7 (6.6) | 27 (3) |
| Chol-NPs | 257 (14) | 0.18 (0.05) | −1.55 (0.05) | −19.1 (6.8) | 78.7 (6.0) | 26 (9) |
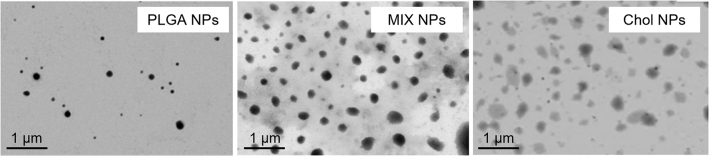
The physical characteristics of the NPs recorded by light scattering were also confirmed by STEM microscopy. Images acquired evidenced the presence of homogeneous populations of NPs in each sample while defining differences previously described, such as PLGA yielding smaller, more dense NPs, while Chol NPs being slightly larger and less electron dense. MIX NPs displayed intermediate characteristics between the two, as MIX NPs are formed by the homogeneous mixture in the NP matrix of both PLGA and Chol (Belletti et al., 2018)(Fig. 2).
Fig. 2.
STEM images of PLGA, MIX, and Chol NPs.
3.2. Hard corona studies
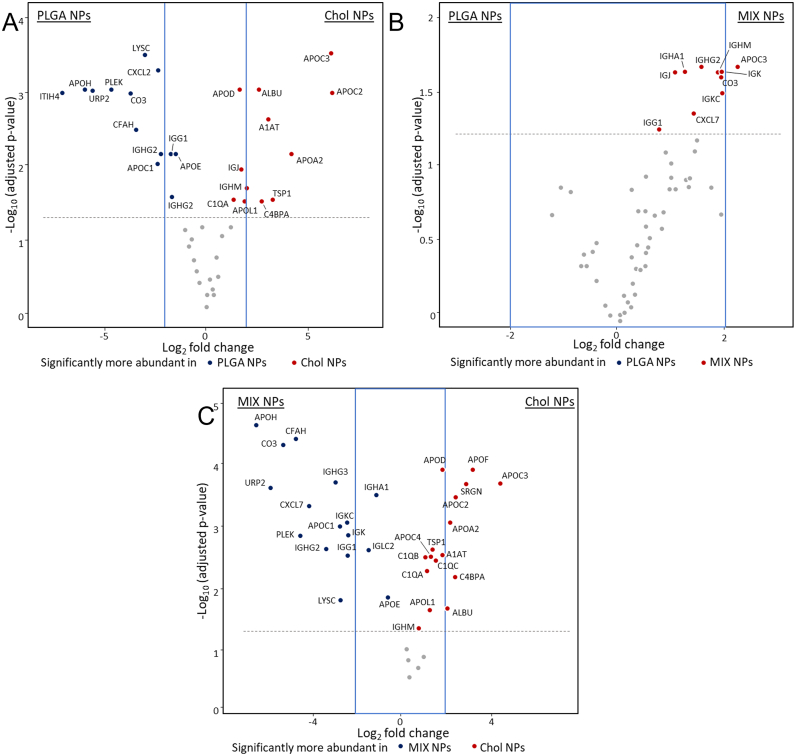
To study the HC, quantitative analysis of the major detectable proteins was compared between each NP matrix type (expressed as % normalized emPAI) while using pure plasma as a standard (Fig. 3). Statistical analysis in the relative protein abundance was determined by group comparisons of each matrix using volcano plots: dots above the grey line have a p value <0.05. To increase the confidence of the results, only proteins outside the range of −2 < Log2 fold change <2 (represented by a blue rectangle) were considered as representing significant differences not linked to possible measurement variations (Fig. 4).
Fig. 3.
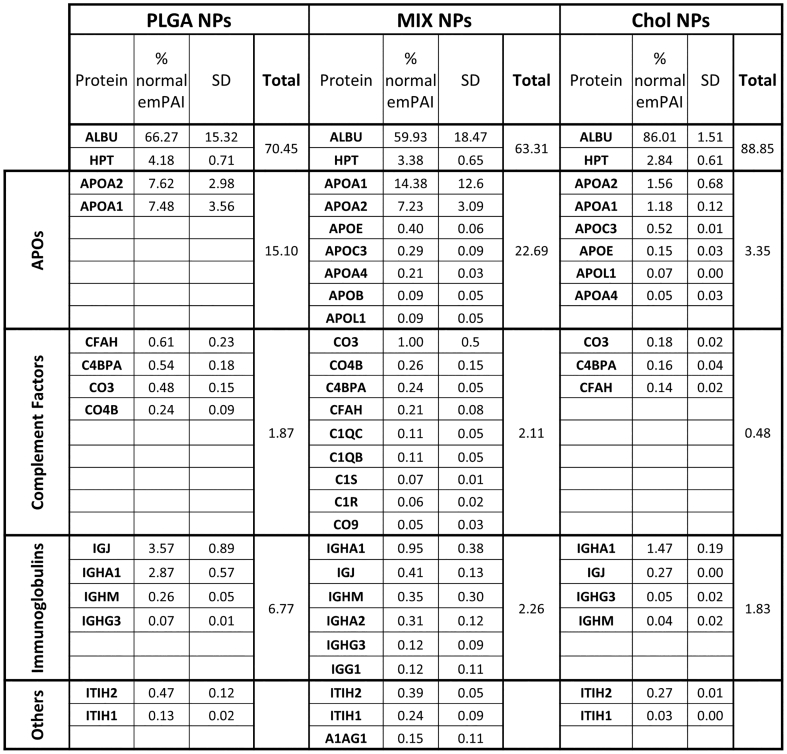
Qualitative and quantitative analysis of the HC. HC protein analysis of PLGA, MIX, and Chol NPs divided into the major subgroups and ordered by emPAI score. The standard devi-ation was determined by the analysis of three different NP formulations. The complete com-position analysis of the HC is reported in Fig. S2.
Fig. 4.
Volcano plots representing relative abundance of the proteins detected in the HC comparing NPs of different matrices. A) PLGA vs MIX NPs. B) PLGA vs Chol NPs. C) MIX vs Chol NPs. The dotted line represents the limit of statistical significance; the blue square represents a secondary selection for those proteins that are at least 2-fold more present in one sample than its counterpart. The complete statistical analysis is reported in Fig. S3. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
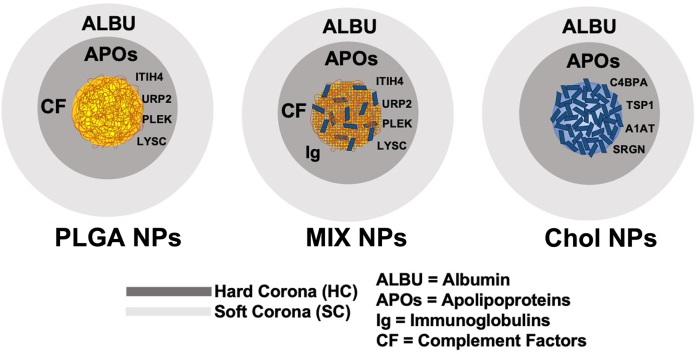
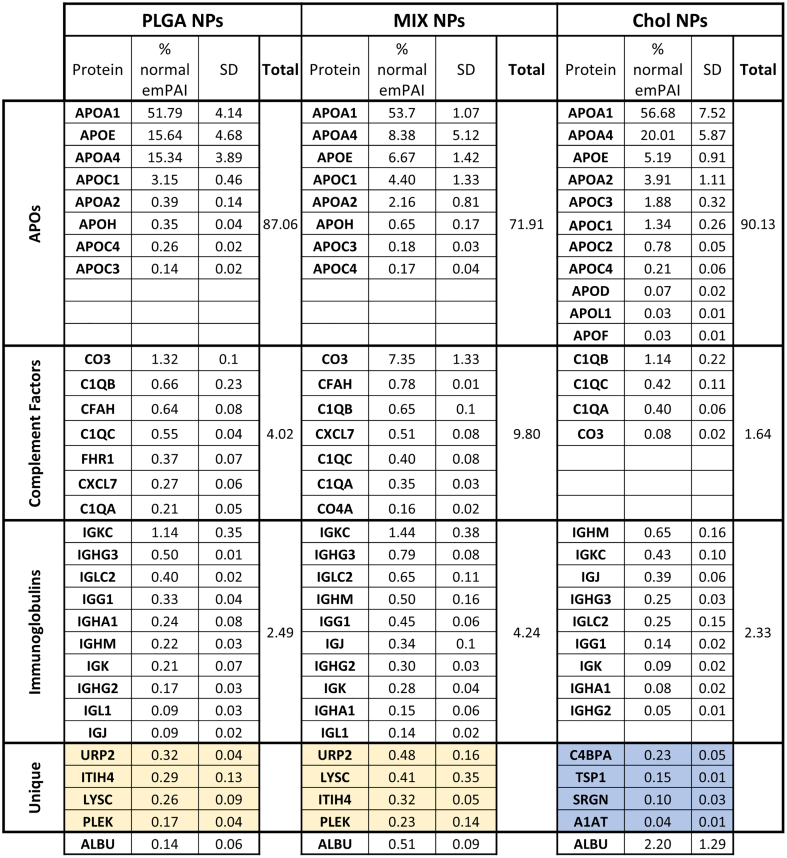
Quantitative analysis of the plasma control sample showed albumin (HSA) as the most abundant protein, with the next highest proteins being transferrin (0.86%), Immunoglobulin H (0.17%), Haptoglobin (0.14%), and Vitamin D binding protein (0.10%), with a long list of below 0.10% (Fig. S2A). Of these most prominent proteins in the plasma, only albumin was consistently detectable in the HC of the different NPs; however, while being the most abundant plasma protein it was always <3% of the NP HC composition. Interestingly, the albumin amount appeared to be directly related to the amount of Cholesterol in the NPs: Chol NPs (2.20%), MIX NPs (0.51%) NPs, and in a much minor fraction for PLGA NPs (0.14%) (Fig. 3, Fig. S2B) (Meierhofer et al., 2010; Peng et al., 2008; Teir et al., 2012). These results demonstrate that the plasma abundance is not the critical determining factor for which proteins form the HC. Instead, the most abundant HC proteins, representing 95–99% of the total HC proteins according to emPAI calculations, could be separated into three distinct categories: apolipoproteins complement factors, and immunoglobulins (Fig. 3). This categorization of the most abundant HC proteins is in accordance with studies for other NPs where similar observations were seen (García-Álvarez and Vallet-Regí, 2021; Kopac, 2021; Walkey and Chan, 2012).
Even though apolipoproteins account for only 4.8% of the plasma proteins (when excluding albumin), they were the most abundant proteins in the HC for all of the NPs and accounted for approximately 70–90% of the HC. Comparing the three formulations, slight variations were evident. While APO_A1 accounts for approximately 53% of proteins across all of the formulations, APO_E, APO_A4, and other APOs are more variable, being most prevalent in PLGA and Chol NPs compared to MIX NPs. This high abundance of APOs in the HC is in accordance with numerous other studies of different nanomedicines and are probably linked to the hydrophobic nature of these structures, but is interesting as to why they were less abundant in the MIX NPs (Pozzi et al., 2015, Pozzi et al., 2014; Sempf et al., 2013).
The other most abundant classes of proteins found in the HC consisted of a mix of complement factors and immunoglobulins. In the HC of all three NPs, CO3 was the most abundant complement factor, ranging from 1.3%, 0.1%, and 7.4% for PLGA, Chol, and MIX NPs respectively. This is followed prominently by C1QB, C1QC, and CFAH with only a small trace of numerous others. Immunoglobulins present in the HC represented 2–4% of the total proteins. In all three matrix types, IGKC was the major immunoglobulin protein followed by IGHG3, IGLC2, and various others. Altogether, a major difference can be seen in the amount of these proteins in the HC of the NP types. The HC of MIX NPs had the highest amount of complement factors and IG levels compared to PLGA and Chol NPs (14% vs 6.5% and 3% respectively).
Altogether, APOs, complement factors, and IGs added up to >90% of the total composition of the HC for each NP matrix type. Nevertheless, it is also important to consider other potentially important candidates that are unique to the HC of each matrix type whether they were quantifiable in the plasma control or not. In fact, several proteins that were too low to be detected in the plasma were detectable in the HC of the PLGA and MIX NPs, including URP2, LYSC, ITIH4, PLEK, and CYTC (Fig. 3, yellow), but were not detected in the Chol NP HC. At the same time, numerous unique proteins were found in the HC of Chol NP but not in PLGA or MIX NPs, namely C4BPA, A1AT, SRGN, and TSP1 (Fig. 3, blue).
After identifying the main proteins that compose the HC, a statistical analysis was performed to evaluate the significant differences in the protein abundance linked to each matrix type. Fig. 4A represents the comparison between the two most different NPs in terms of characteristics and nature: PLGA and Chol NPs. The classes of proteins that were the most prominent in both the plasma and HC, discussed above, showed no significant difference in abundance between PLGA NPs and Chol NPs with a few exceptions. In particular, the HC of the PLGA NPs had significantly higher amounts of the complement factors (CO3, and CFAH), while Chol NPs had significantly higher levels of APO_C2, APO_C3, and albumin. This statistical analysis also supported previous observations regarding the proteins uniquely found in either the HC of Chol or PLGA NPs.
The comparison of PLGA NPs and MIX NPs highlighted little to no differences in the composition of the HC with the only difference being that APO_C3 was statistically more abundant in the HC of the MIX NPs (Fig. 4B). On the contrary, a comparison between Chol and MIX NPs exhibited a surprisingly similar trend to the Chol vs PLGA NPs. MIX NPs showed statistically more proteins that were also seen in the polymeric PLGA NPs, such as URP2, PLEK, and LYSC, as well as the complement factors CO3 and CFAH. Interestingly, they also showed a significant predominance of immunoglobulins compared to Chol NPs, with IGKC, IGHG3, and IGG1(Fig. 4C). Both MIX and Chol NPs showed an abundance of a different variety of APOs.
These data could have important implications on the fate of these NPs. APOs were found to be more present in the HC of Chol NPs compared to the other two types. This effect could be expected, as this class of proteins is involved in the transport of lipidic and hydrophobic material in the bloodstream generally represented by high-density and low-density lipoproteins rich in cholesterol (HDL and LDL) (Bolanos-Garcia and Miguel, 2003; Fielding and Fielding, 1995; Lai et al., 2020; Mahley, 1988; Reschly et al., 2002). At the same time, the total amount of complement factors and immunoglobulins is an important factor to be considered. Unlike APOs, complement factors have been linked to more negative biological repercussions, inciting both an inflammatory and immunogenic effect (Goldberg and Ackerman, 2020; Moghimi et al., 2011; Xiao and Gao, 2018).
3.3. Soft corona
While the HC is made up of proteins intimately associated with the surface of the NPs, the soft corona (SC) is much more dynamic and less intimately bound. This does not negate the need to characterize this entity as an equilibrium will still be reached in the plasma environment leading to effects on the biological fate of the NPs (Fig. 5, Fig. S4) (Caracciolo et al., 2017). One major difference seen in the emPAI scores is that the SC of both the PLGA and Chol NPs consisted of fewer proteins compared to the HC (30 vs 50 proteins). Of these, the most abundant proteins were albumin and haptoglobin, which ranged from 60 to 86%, with no statistical difference between the three matrix types (Fig. 6). This was followed by low amounts of the three major categories previously seen in the HC: APOs, complement factors, and immunoglobulins. Chol and MIX NPs shared the highest variety of APOs compared to the few found in the PLGA NPs. This variety could be linked to the biological role of these proteins in transporting Chol as described previously in the section regarding the HC; however, it is also important to consider the amount of APOs present in the SC. In Chol NPs, APOs represented only 3% of the SC proteins while in MIX NPs they represented over 20%. Similarly, the SC of MIX NPs consisted of a notably larger variety of complement factors compared to the other two NPs even though the total emPAI percentage was no different from that of PLGA NPs (2.11% and 1.87% respectively). On the other hand, Chol NPs showed the smallest amount of complement factors with a total of 0.48% of the total composition of the SC. Unlike what was reported for the HC, the SC of the different NPs did not contain “unique” proteins, but instead consisted of variations in the amount and number of the major proteins, suggesting the difference of the soft equilibrium formed.
Fig. 5.
Qualitative analysis of the SC. SC protein analysis of PLGA, MIX and Chol NPs di-vided into the major subgroups and ordered by emPAI score. The standard deviation was de-termined by the analysis of three different NP formulations. The complete composition analysis of the SC is reported in Fig. S4.
Fig. 6.
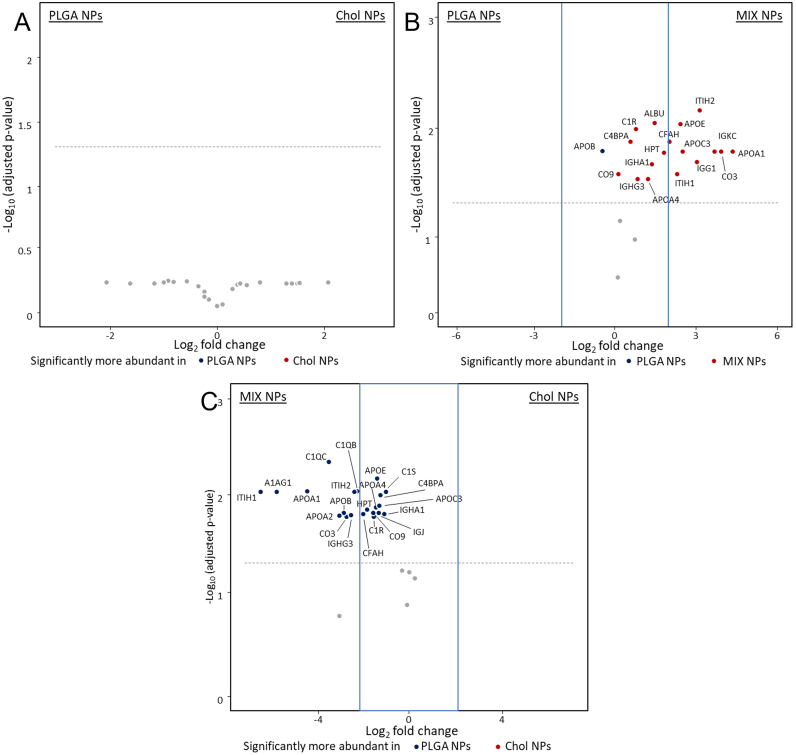
Volcano plots representing relative abundance of the proteins detected in the SC com-paring NPs of different matrices. A) PLGA vs MIX NPs. B) PLGA vs Chol NPs. C) MIX vs Chol NPs. The dotted line represents the limit of statistical significance; the blue square rep-resents a secondary selection for those proteins that are present at least 2-fold higher in one sample compared to its counterpart. The complete statistical analysis is reported in Fig. S5. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Statistical analysis with volcano plots supported these results. PLGA and Chol NPs showed no statistical differences in the identity of the SC proteins (Fig. 6A). On the contrary, MIX NPs showed statistically higher amounts of apolipoproteins (such as APO_A, APO_C, and APO_E), complement factors (CO5, CO3, C1QC), and immunoglobulins (IGKC, IGHM) compared to both other NP matrix types (Fig. 6B and C).
3.4. The PC of NPs with targeting ligands
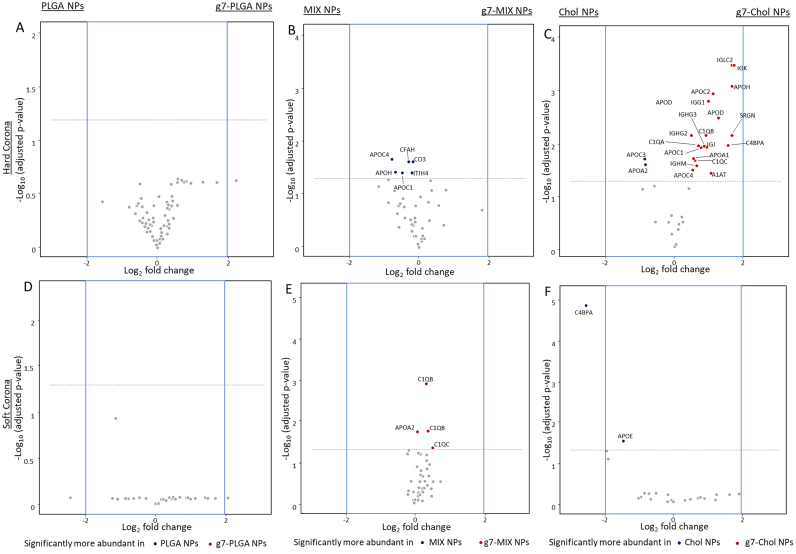
The surface modification of NPs has been demonstrated to be critical for the composition of the PC. Therefore, the PC of NPs surface modified with a targeting ligand was analysed to test if the differences in PC linked to the matrix composition would be modified by the presence of a surface ligand. For this purpose, the g7 peptide was chosen: this is a well-known BBB targeting ligand proven to promote CNS targeting when conjugated onto the surface of NPs (Birolini et al., 2021; Rigon et al., 2019; Tosi et al., 2007). Overall, the emPAI scores for the HC and SC of each matrix were very similar for each NP type with or without the ligand present (Fig. S6 and S7). Statistical analysis confirmed that all proteins forming the HC and SC remained under the significance threshold when comparing the g7-modified NPs to their unmodified counterparts (Fig. 7). The only exception was an increased amount of C4BPA in the SC for unmodified Chol NPs. In opposition to many literature sources, these data suggest that the presence of the g7 ligand on the surface of these NPs did not significantly alter the HC or SC for any of the different matrix types of NPs and that the differences seen in previous sections are derived solely due to the different matrix compositions.
Fig. 7.
Volcano plots representing relative abundance of the proteins detected in the HC and SC comparing NPs with or without the g7 peptide. A) HC and D) SC of PLGA vs g7-PLGA NPs. B) HC and E) SC of MIX vs g7-MIX NPs. C) HC and F) SC of Chol vs g7-Chol NPs. The dotted line represents the limit of statistical significance; the blue square represents a secondary selection for those proteins that are at least 2-fold more present in one sample than its counterpart. The complete statistical analysis is reported in Fig. S8. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Analysis of the PC is crucial to understand the interactions between NPs and the biological environment (Bai et al., 2021; Mohammad-Beigi et al., 2020). In fact, the PC is known to affect the fate of NPs after administration by influencing their biodistribution, circulation time, surface properties, cell uptake dynamics, toxicity, and drug release profile (Behzadi et al., 2014; Godara et al., 2020; Rampado et al., 2020). At the same time, several chemico-physical features of the NPs, including their composition, size, charge, and hydrophobicity have been reported to strongly influence the composition of the PC, suggesting that PC should be investigated for each individual NP formulation (Rahman et al., 2013; Singh et al., 2021). Here we reported the investigation of the PC of three different NPs: polymeric PLGA NPs, fully lipidic Cholesterol NPs, and hybrid NPs (MIX) composed of PLGA and Chol. In-depth knowledge of the PC of different prominent NPs is critical for understanding their potential biological and pharmacological effects, in order to improve the specific design of NPs by taking into account the composition of their PC (Rezaei et al., 2019; Wang et al., 2018).
As reported in the literature, apolipoproteins are a major component of the PC independent of the matrix type for all NPs (Cedervall et al., 2007a; Charbgoo et al., 2018; Huang et al., 2021; Lundqvist et al., 2008; Tenzer et al., 2011; Yu et al., 2020). Previous literature has demonstrated that APOs can have a positive effect on the fate of NPs (Ahsan et al., 2018; Kreuter et al., 2002; Martínez-Negro et al., 2021; Wagner et al., 2012). In fact, APOs can change the surface properties of NPs minimizing toxicity of the matrix, increasing blood circulation, and improving biodistribution. Moreover, many APOs have even been demonstrated to aid in BBB crossing, a crucial effect to consider when designing NPs (Ju et al., 2016; Li et al., 2021; Michaelis et al., 2006). On the other hand, complement factors and immunoglobulins represent a fundamental part of the immune system, and their presence in the HC of NPs must be taken into great consideration due to the negative effects they could have on early clearance and immunogenicity (Ahsan et al., 2018; Barbero et al., 2017; Buchman et al., 2019; Chen et al., 2017; Ehrenstein and Notley, 2010; Sobczynski and Eniola-Adefeso, 2017; Wang et al., 2016). Quantification of these proteins with emPAI calculations showed their presence was 2-fold higher in MIX NPs compared to both PLGA and Chol NPs. This was also confirmed by statistical analysis, where several complement factors and immunoglobulins were found to be significantly more expressed in the HC of MIX NPs compared to the other two formulations.
When considering the potential biological effects of the PC, it is also important to look beyond the proteins found in abundance in both the plasma and on the NP surface: in fact, other potentially important candidates might be in such low concentrations in the plasma that they are unquantifiable, but could still play crucial roles when incorporated in the HC formation. This is what was observed for a series of “unique” proteins whose presence appeared to be completely dependent on the NP matrix. In fact, several proteins were too low to be detected in the plasma but were present in the HC of both PLGA and MIX NPs while being absent in the HC of the Chol NPs. This could suggest that the PLGA in both NP types is the cause of these proteins being present in the HC. At the same time, numerous proteins were found in the HC of Chol NPs but not in PLGA NPs. These considerations were supported also by statistical analysis, highlighting the significant difference in the composition of the HC for these NPs which would create unique protein profiles directly linked to the matrix composition.
It could be hypothesised that the lipidic and polymeric NPs had different PC characteristics due to their different chemical properties; however, statistical comparisons demonstrated near identical PCs between PLGA and MIX NPs, while variations were observed with Chol NPs. This difference could suggest that the polymeric portion of the MIX NPs plays a larger role in the composition of the minor proteins in the HC. Interestingly, a unique profile for MIX NPs was also reported in the composition of the SC. More specifically, MIX NPs showed higher variability in the type and total amount of complement factors forming the SC compared to the other NPs. This result could be linked to the composition of the HC in which the higher presence of immunoglobulins could lead to higher complement factor recruitment in the outer SC layer of the PC (Fischinger et al., 2019).
It is interesting to note that the proteins associated with the HC of PLGA and Chol NPs play very different biological roles. Proteins uniquely found in the polymeric and hybrid NPs, such as complement factors and immunoglobulins, were mainly linked to the immune response, and are involved in inflammatory and stress response pathways. On the other hand, the HC of Chol NPs had higher levels of APOs which are involved in intracellular transport and storage and a smaller amount of immune response linked proteins. The HC profile could be easily explained through the high biocompatibility of cholesterol, which is a physiologic molecule normally present in the blood with high affinity to APOs. Altogether, these data suggest that the use of Chol NPs as drug delivery systems could be promising due to their potentially low immunogenicity, improved biodistribution and reduced toxicity compared to PLGA-based NPs.
As previously mentioned, the size of NPs as well as their surface composition are important factors that can affect the composition of the PC (Akhter et al., 2021; Dobrovolskaia et al., 2009; Lacerda et al., 2010; Neagu et al., 2017; Petry et al., 2019; Tenzer et al., 2011); however, other studies report that a difference in size ranging 150–250 nm might not be enough to produce differences in terms of PC composition (Lundqvist et al., 2008; Partikel et al., 2019b; Zhang et al., 2011). Our data support this literature precedence: in our study, MIX NPs (237 nm and 27% residual Pluronic® F68) were shown to have a HC with a more similar composition to PLGA NPs (152 nm and 14% residual Pluronic® F68) than Chol NPs (257 nm and 26% residual Pluronic® F68). Our data suggest that, for polymeric or lipidic NPs in this size range, the NP matrix composition had a larger impact on the formation and identity of the PC proteins respective of both the size or the amount of Pluronic® F68 that was either internalized or on the surface of the NPs. This highlights an important gap in the literature and emphasizes the need of thorough analysis of all the factors that affect the formation of the PC. In fact, most studies on the PC are now focused on evaluating the impact of single NP features, often leading to contradictory findings. Thus, an extensive and comprehensive study is still missing to understand which parameters play a predominant role in affecting the PC composition.
One of the most fascinating applications of NPs is the possibility to achieve targeted delivery to specific cells. Current approaches mainly depend on the modification of the NP surface with targeting ligands (Yoo et al., 2019). Because NP surface modifications are considered a major factor that can affect the formation of the PC, peptide targeted NPs of each matrix composition were analysed. Comparing the ligand modified with the unmodified NPs, no significant differences were observed in the composition of both the HC and SC. This result is in contrast to what is found in the literature, however, it must be considered that the ligand density on the surface of these NPs is often so low that their presence do not change the physical characteristics, thus not affecting the PC (Segets et al., 2011). It is important to note that several studies have linked the formation of the PC to a loss of targeting potential of NPs in vitro (Li et al., 2021; Salvati et al., 2013; Su et al., 2018; Varnamkhasti et al., 2015). Nevertheless, recent findings evidence how the PC in vivo has different dynamics compared to in vitro studies which is supported by the successful targeted delivery results in vivo already published (Hadjidemetriou et al., 2015; Su et al., 2018; Zhang et al., 2018).
Research on the PC is crucial to better understand the biological implications of NPs. Although much research is being done to progress in this direction, some points still need to be addressed. While identifying and quantifying the proteins in the PC is important, it will be critical in future experiments to assess the effective concentration needed for these proteins to elicit a biological effect. This will help determine if the difference in the amount or variety of proteins facilitate a difference in biological effects. Another factor to be considered is the conformational state of the bound proteins. Literature precedence highlights that protein interaction with the surface of the NP could provoke a conformational change in the structure of adsorbed proteins resulting in a change or loss of activity. This calls for the need to not only quantify the proteins in the PC, but also to analyse their conformation after binding (Linse et al., 2007; Park, 2020; Roach et al., 2005; Smith et al., 2009; Tsai et al., 2011). Such studies will further allow researchers to determine which key proteins need to be specifically evaluated and which can be considered innocent bystanders in the fate of the NPs. Intricate experiments both in vitro and in vivo will be required, but this information could lead to fascinating discoveries for the future of nanomedicine.
5. Conclusions
Research on the PC is rapidly growing despite the difficulty in accurately isolating, identifying, and quantifying the proteins in the HC and SC. While there are some general consistencies across the proteins found in the HC and SC of all NPs, each individual NP shows individual differences in the PC that will lead to drastic effects on the biodistribution, toxicity, immunogenicity, and general therapeutic potential making these individual studies critical for each NP system. Strong examples in this work include the seemingly small effect of both size and surfactant amount on the PC as stated in other literature works compared to the major differences in matrix composition, as well as the increase of immunoglobulins in the SC due to the higher presence of complement factors in the HC of MIX NPs. This work advanced this research by analysing the HC and SC composition of three types of NPs with different matrices that have recently shown great promise in the literature and evaluated if the matrix or the presence of a targeting ligand on the surface would have a more critical impact on the proteins present. Overall, this work represents a “piece of the puzzle” in the world of PC studies, where continuous research for different NPs would lead to more controlled and safe tools, thus paving the way for a brighter future for Nanomedicine.
Patents
PATENT: Italian patent application number 102020000025126 registered October 23rd 2020. PCT application PCT/IB2021/059750 registered October 21st 2021.
Author contributions
Conceptualization J.T.D., F.P., I.O., B.R. Investigation J.T.D., F.P., F.G., I.O., R.C. and G.T. Writing — original draft preparation J.T.D., F.P., F.G., I.O., R.C., and B.R. Writing — review and editing I.O., J.T.D., M.V., and R.C. Supervision; J.T.D., G.T., M.A.V., and B.R. Project administration B.R., M.A.V., G.T. and J.T.D. Funding acquisition J.T.D, G.T, M.V., M.A.V., and B.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by an IMI EU Grant “Investigating Mechanisms and Models predictive of accessibility of therapeutics (IM2PACT) Into the Brain” IMI2 - Call 12, GA n.807015(im2pact.org); FAR Unimore Fondazione di Modena “Nano-immuno targeting per il trattamento del glioblastoma multiforme” NIT Project; GLIOSILK Project (Reference Number: EURONANOMED2019–075); a Ministero degli Esteri e della Cooperazione Internazionale MAECI grant, Progetti di ricerca scientifica e tecnologica di grande rilevanza by Ministero degli Esteri, Progetti Italy-USA, “Nanomedicine for Blood Brain Barrier (BBB)-crossing in CNS oncologic pathologies”, Prot. nr. MAE00691612020–06-26; Creutzfeldt–Jakob disease Foundation (CJDF); Telethon GGP19113 project: “Pigment Epithelium-derived Factor (PEDF) peptide as therapeutic agents for inherited retinal degeneration”.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
The authors acknowledge the Fondazione Cassa di Risparmio di Modena for funding the HPLC ESI-QToF system at the Centro Interdipartimentale Grandi Strumenti (CIGS).
Footnotes
Methods for PC analysis, Fig. S1: Correlogram and Intensity function of the PCS analysis on PLGA, MIX, and Chol NPs, Fig. S2: A) Total protein composition of plasma. B) Total composition of the HC of PLGA, MIX and Chol NPs, Fig. S3: Volcano plots representing relative abundance of the proteins detected in the HC comparing NPs of different matrices, Fig. S4: Total composition of the SC of PLGA, MIX and Chol NPs, Fig. S5: Volcano plots representing relative abundance of the proteins detected in the SC comparing NPs of different matrices, Fig. S6: Total composition of the HC of g7-modified NPs, Fig. S7: Total composition of the SC of g7-modified NPs, Fig. S8: Volcano plots representing relative abundance of the proteins detected in the HC and SC comparing NPs with or without the g7 peptide. Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpx.2022.100136
Contributor Information
Ilaria Ottonelli, Email: ilaria.ottonelli@unimore.it.
Jason Thomas Duskey, Email: jasonthomas.duskey@unimore.it.
Filippo Genovese, Email: filippo.genovese@unimore.it.
Riccardo Caraffi, Email: riccardo.caraffi@unimore.it.
Marta Valenza, Email: marta.valenza@unimi.it.
Giovanni Tosi, Email: gtosi@unimore.it.
Maria Angela Vandelli, Email: mariaangela.vandelli@unimore.it.
Barbara Ruozi, Email: barbara.ruozi@unimore.it.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Ahsan S.M., Rao C.M., Ahmad M.F. Nanoparticle-protein interaction: the significance and role of protein corona. Adv. Exp. Med. Biol. 2018;1048:175–198. doi: 10.1007/978-3-319-72041-8_11. [DOI] [PubMed] [Google Scholar]
- Akhter M.H., Khalilullah H., Gupta M., Alfaleh M.A., Alhakamy N.A., Riadi Y., Md S. Impact of protein corona on the biological identity of nanomedicine: understanding the fate of nanomaterials in the biological milieu. Biomedicines. 2021;9:1496. doi: 10.3390/biomedicines9101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almalik A., Benabdelkamel H., Masood A., Alanazi I.O., Alradwan I., Majrashi M.A., Alfadda A.A., Alghamdi W.M., Alrabiah H., Tirelli N., Alhasan A.H. Hyaluronic acid coated chitosan nanoparticles reduced the immunogenicity of the formed protein corona. Sci. Rep. 2017;7:10542. doi: 10.1038/s41598-017-10836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini M.A., Ahmed T., Liu F.-C.F., Abbasi A.Z., Soeandy C.D., Zhang R.X., Prashad P., Cummins C.L., Rauth A.M., Henderson J.T., Wu X.Y. Exploring the transformability of polymer-lipid hybrid nanoparticles and nanomaterial-biology interplay to facilitate tumor penetration, cellular uptake and intracellular targeting of anticancer drugs. Exp. Opinion Drug Deliv. 2021;18:991–1004. doi: 10.1080/17425247.2021.1902984. [DOI] [PubMed] [Google Scholar]
- Arike L., Peil L. Spectral counting label-free proteomics. Methods Mol. Biol. 2014;1156:213–222. doi: 10.1007/978-1-4939-0685-7_14. [DOI] [PubMed] [Google Scholar]
- Ashby J., Pan S., Zhong W. Size and surface functionalization of iron oxide nanoparticles influence the composition and dynamic nature of their protein corona. ACS Appl. Mater. Interfaces. 2014;6:15412–15419. doi: 10.1021/am503909q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri E., Alibolandi M., Abnous K., Taghdisi S.M., Ramezani M. Targeted delivery and controlled release of doxorubicin to cancer cells by smart ATP-responsive Y-shaped DNA structure-capped mesoporous silica nanoparticles. J. Mater. Chem. B. 2021;9:1351–1363. doi: 10.1039/D0TB01960G. [DOI] [PubMed] [Google Scholar]
- Bai X., Wang J., Mu Q., Su G. In vivo protein corona formation: characterizations, effects on engineered nanoparticles’ biobehaviors, and applications. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.646708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero F., Russo L., Vitali M., Piella J., Salvo I., Borrajo M.L., Busquets-Fité M., Grandori R., Bastús N.G., Casals E., Puntes V. Formation of the protein corona: the interface between nanoparticles and the immune system. Semin. Immunol. 2017;34:52–60. doi: 10.1016/j.smim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Barrán-Berdón A.L., Pozzi D., Caracciolo G., Capriotti A.L., Caruso G., Cavaliere C., Riccioli A., Palchetti S., Laganà A. Time evolution of nanoparticle–protein corona in human plasma: relevance for targeted drug delivery. Langmuir. 2013;29:6485–6494. doi: 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- Behzadi S., Serpooshan V., Sakhtianchi R., Müller B., Landfester K., Crespy D., Mahmoudi M. Protein corona change the drug release profile of nanocarriers: the “overlooked” factor at the nanobio interface. Colloids Surf. B: Biointerfaces. 2014;123:143–149. doi: 10.1016/j.colsurfb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Belletti D., Grabrucker A.M., Pederzoli F., Menrath I., Cappello V., Vandelli M.A., Forni F., Tosi G., Ruozi B. Exploiting the versatility of cholesterol in nanoparticles formulation. Int. J. Pharm. 2016;511:331–340. doi: 10.1016/j.ijpharm.2016.07.022. [DOI] [PubMed] [Google Scholar]
- Belletti D., Grabrucker A.M., Pederzoli F., Menrath I., Vandelli M.A., Tosi G., Duskey T.J., Forni F., Ruozi B. Hybrid nanoparticles as a new technological approach to enhance the delivery of cholesterol into the brain. Int. J. Pharm. 2018;543:300–310. doi: 10.1016/j.ijpharm.2018.03.061. [DOI] [PubMed] [Google Scholar]
- Bélteky P., Rónavári A., Igaz N., Szerencsés B., Tóth I.Y., Pfeiffer I., Kiricsi M., Kónya Z. Silver nanoparticles: aggregation behavior in biorelevant conditions and its impact on biological activity. IJN. 2019;14:667–687. doi: 10.2147/IJN.S185965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Grenier P., Mahmoudi M., Lima E.M., Appel E.A., Dormont F., Lim J.-M., Karnik R., Langer R., Farokhzad O.C. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 2017;8:777. doi: 10.1038/s41467-017-00600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing J., Xiao X., McClements D.J., Biao Y., Chongjiang C. Protein corona formation around inorganic nanoparticles: Food plant proteins-TiO2 nanoparticle interactions. Food Hydrocoll. 2021;115 doi: 10.1016/j.foodhyd.2021.106594. [DOI] [Google Scholar]
- Birolini G., Valenza M., Ottonelli I., Passoni A., Favagrossa M., Duskey J.T., Bombaci M., Vandelli M.A., Colombo L., Bagnati R., Caccia C., Leoni V., Taroni F., Forni F., Ruozi B., Salmona M., Tosi G., Cattaneo E. Insights into kinetics, release, and behavioral effects of brain-targeted hybrid nanoparticles for cholesterol delivery in Huntington’s disease. J. Control. Release. 2021;330:587–598. doi: 10.1016/j.jconrel.2020.12.051. [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Miguel R.N. On the structure and function of apolipoproteins: more than a family of lipid-binding proteins. Prog. Biophys. Mol. Biol. 2003;83:47–68. doi: 10.1016/s0079-6107(03)00028-2. [DOI] [PubMed] [Google Scholar]
- Buchman J.T., Hudson-Smith N.V., Landy K.M., Haynes C.L. Understanding nanoparticle toxicity mechanisms to inform redesign strategies to reduce environmental impact. Acc. Chem. Res. 2019;52:1632–1642. doi: 10.1021/acs.accounts.9b00053. [DOI] [PubMed] [Google Scholar]
- Caracciolo G., Pozzi D., Capriotti A.L., Cavaliere C., Foglia P., Amenitsch H., Laganà A. Evolution of the protein corona of lipid gene vectors as a function of plasma concentration. Langmuir. 2011;27:15048–15053. doi: 10.1021/la202912f. [DOI] [PubMed] [Google Scholar]
- Caracciolo G., Pozzi D., Capriotti A.L., Cavaliere C., Piovesana S., Amenitsch H., Laganà A. Lipid composition: a “key factor” for the rational manipulation of the liposome–protein corona by liposome design. RSC Adv. 2015;5:5967–5975. doi: 10.1039/C4RA13335H. [DOI] [Google Scholar]
- Caracciolo G., Farokhzad O.C., Mahmoudi M. Biological identity of nanoparticles in vivo: clinical implications of the protein corona. Trends Biotechnol. 2017;35:257–264. doi: 10.1016/j.tibtech.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Cedervall T., Lynch I., Foy M., Berggård T., Donnelly S.C., Cagney G., Linse S., Dawson K.A. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- Cedervall T., Lynch I., Lindman S., Berggård T., Thulin E., Nilsson H., Dawson K.A., Linse S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Joshi P., Shanker V., Ansari Z.A., Singh S.P., Chakrabarti P. Contrasting effect of gold nanoparticles and nanorods with different surface modifications on the structure and activity of bovine serum albumin. Langmuir. 2011;27:7722–7731. doi: 10.1021/la200787t. [DOI] [PubMed] [Google Scholar]
- Charbgoo F., Nejabat M., Abnous K., Soltani F., Taghdisi S.M., Alibolandi M., Thomas Shier W., Steele T.W.J., Ramezani M. Gold nanoparticle should understand protein corona for being a clinical nanomaterial. J. Control. Release. 2018;272:39–53. doi: 10.1016/j.jconrel.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Chen F., Wang G., Griffin J.I., Brenneman B., Banda N.K., Holers V.M., Backos D.S., Wu L., Moghimi S.M., Simberg D. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotech. 2017;12:387–393. doi: 10.1038/nnano.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs C.E. The determination of polyethylene glycol in gamma globulin solutions. Microchem. J. 1975;20:190–192. doi: 10.1016/0026-265X(75)90038-7. [DOI] [Google Scholar]
- Chinen A.B., Guan C.M., Ko C.H., Mirkin C.A. The impact of protein corona formation on the macrophage cellular uptake and biodistribution of spherical nucleic acids. Small. 2017;13:1603847. doi: 10.1002/smll.201603847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., Vitek O. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- Costantino L., Gandolfi F., Tosi G., Rivasi F., Vandelli M.A., Forni F. Peptide-derivatized biodegradable nanoparticles able to cross the blood-brain barrier. J. Control. Release. 2005;108:84–96. doi: 10.1016/j.jconrel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Dai Q., Yan Y., Ang C.-S., Kempe K., Kamphuis M.M.J., Dodds S.J., Caruso F. Monoclonal antibody-functionalized multilayered particles: targeting cancer cells in the presence of protein coronas. ACS Nano. 2015;9:2876–2885. doi: 10.1021/nn506929e. [DOI] [PubMed] [Google Scholar]
- De Paoli S.H., Diduch L.L., Tegegn T.Z., Orecna M., Strader M.B., Karnaukhova E., Bonevich J.E., Holada K., Simak J. The effect of protein corona composition on the interaction of carbon nanotubes with human blood platelets. Biomaterials. 2014;35:6182–6194. doi: 10.1016/j.biomaterials.2014.04.067. [DOI] [PubMed] [Google Scholar]
- Dinu I.A., Duskey J.T., Car A., Palivan C.G., Meier W. Engineered non-toxic cationic nanocarriers with photo-triggered slow-release properties. Polym. Chem. 2016;7:3451–3464. doi: 10.1039/C6PY00343E. [DOI] [Google Scholar]
- Dobrovolskaia M.A., Patri A.K., Zheng J., Clogston J.D., Ayub N., Aggarwal P., Neun B.W., Hall J.B., McNeil S.E. Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine. 2009;5:106–117. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaia M.A., Neun B.W., Man S., Ye X., Hansen M., Patri A.K., Crist R.M., McNeil S.E. Protein corona composition does not accurately predict hematocompatibility of colloidal gold nanoparticles. Nanomedicine. 2014;10:1453–1463. doi: 10.1016/j.nano.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duskey J.T., Belletti D., Pederzoli F., Vandelli M.A., Forni F., Ruozi B., Tosi G. Current strategies for the delivery of therapeutic proteins and enzymes to treat brain disorders. Int. Rev. Neurobiol. 2017;137:1–28. doi: 10.1016/bs.irn.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Duskey Jason Thomas, da Ros F., Ottonelli I., Zambelli B., Vandelli M.A., Tosi G., Ruozi B. Enzyme stability in nanoparticle preparations Part 1: Bovine serum albumin improves enzyme function. Molecules. 2020;25:4593. doi: 10.3390/molecules25204593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duskey Jason T., Ottonelli I., Da Ros F., Vilella A., Zoli M., Kovachka S., Spyrakis F., Vandelli M.A., Tosi G., Ruozi B. Novel peptide-conjugated nanomedicines for brain targeting: in vivo evidence. Nanomedicine. 2020;28 doi: 10.1016/j.nano.2020.102226. [DOI] [PubMed] [Google Scholar]
- Duskey J.T., Ottonelli I., Rinaldi A., Parmeggiani I., Zambelli B., Wang L.Z., Prud’homme R.K., Vandelli M.A., Tosi G., Ruozi B. Tween® preserves enzyme activity and stability in PLGA nanoparticles. Nanomaterials. 2021;11:2946. doi: 10.3390/nano11112946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein M.R., Notley C.A. The importance of natural IgM: scavenger, protector and regulator. Nat. Rev. Immunol. 2010;10:778–786. doi: 10.1038/nri2849. [DOI] [PubMed] [Google Scholar]
- Falahati M., Attar F., Sharifi M., Haertlé T., Berret J.-F., Khan R.H., Saboury A.A. A health concern regarding the protein corona, aggregation and disaggregation. Biochim. Biophys. Acta Gen. Subj. 2019;1863:971–991. doi: 10.1016/j.bbagen.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding C.J., Fielding P.E. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473 doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornaguera C., García-Celma M.J. Personalized nanomedicine: a revolution at the nanoscale. J. Pers. Med. 2017;7:E12. doi: 10.3390/jpm7040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia V., Yang K., Deville S., Reker-Smit C., Nelissen I., Salvati A. Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano. 2019;13:11107–11121. doi: 10.1021/acsnano.9b03824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Álvarez R., Vallet-Regí M. Hard and soft protein corona of nanomaterials: analysis and relevance. Nanomaterials. 2021;11:888. doi: 10.3390/nano11040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Álvarez R., Hadjidemetriou M., Sánchez-Iglesias A., Liz-Marzán L.M., Kostarelos K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale. 2018;10:1256–1264. doi: 10.1039/C7NR08322J. [DOI] [PubMed] [Google Scholar]
- Gessner A., Waicz R., Lieske A., Paulke B., Mäder K., Müller R.H. Nanoparticles with decreasing surface hydrophobicities: influence on plasma protein adsorption. Int. J. Pharm. 2000;196:245–249. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- Ghitman J., Biru E.I., Stan R., Iovu H. Review of hybrid PLGA nanoparticles: Future of smart drug delivery and theranostics medicine. Mater. Des. 2020;193 doi: 10.1016/j.matdes.2020.108805. [DOI] [Google Scholar]
- Godara S., Lather V., Kirthanashri S.V., Awasthi R., Pandita D. Lipid-PLGA hybrid nanoparticles of paclitaxel: Preparation, characterization, in vitro and in vivo evaluation. Mater. Sci. Eng. C. 2020;109 doi: 10.1016/j.msec.2019.110576. [DOI] [PubMed] [Google Scholar]
- Goldberg B.S., Ackerman M.E. Antibody-mediated complement activation in pathology and protection. Immunol. Cell Biol. 2020;98:305–317. doi: 10.1111/imcb.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou M., Al-Ahmady Z., Mazza M., Collins R.F., Dawson K., Kostarelos K. In vivo biomolecule corona around blood-circulating, clinically used and antibody-targeted lipid bilayer nanoscale vesicles. ACS Nano. 2015;9:8142–8156. doi: 10.1021/acsnano.5b03300. [DOI] [PubMed] [Google Scholar]
- Hoyos-Ceballos G.P., Ruozi B., Ottonelli I., Da Ros F., Vandelli M.A., Forni F., Daini E., Vilella A., Zoli M., Tosi G., Duskey J.T., López-Osorio B.L. PLGA-PEG-ANG-2 nanoparticles for blood–brain barrier crossing: proof-of-concept study. Pharmaceutics. 2020;12:72. doi: 10.3390/pharmaceutics12010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Xiao G., Zhang Y., Min W. Research progress and application opportunities of nanoparticle–protein corona complexes. Biomed. Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111541. [DOI] [PubMed] [Google Scholar]
- Ju Y., Dai Q., Cui J., Dai Y., Suma T., Richardson J.J., Caruso F. Improving targeting of metal–phenolic capsules by the presence of protein coronas. ACS Appl. Mater. Interfaces. 2016;8:22914–22922. doi: 10.1021/acsami.6b07613. [DOI] [PubMed] [Google Scholar]
- Ke P.C., Lin S., Parak W.J., Davis T.P., Caruso F. A decade of the protein corona. ACS Nano. 2017;11:11773–11776. doi: 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Kopac T. Protein corona, understanding the nanoparticle-protein interactions and future perspectives: a critical review. Int. J. Biol. Macromol. 2021;169:290–301. doi: 10.1016/j.ijbiomac.2020.12.108. [DOI] [PubMed] [Google Scholar]
- Kreuter J., Shamenkov D., Petrov V., Ramge P., Cychutek K., Koch-Brandt C., Alyautdin R. Apolipoprotein-mediated transport of nanoparticle-bound drugs across the blood-brain barrier. J. Drug Target. 2002;10:317–325. doi: 10.1080/10611860290031877. [DOI] [PubMed] [Google Scholar]
- Lacerda S.H.D.P., Park J.J., Meuse C., Pristinski D., Becker M.L., Karim A., Douglas J.F. Interaction of gold nanoparticles with common human blood proteins. ACS Nano. 2010;4:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- Lai W., Li D., Wang Q., Nan X., Xiang Z., Ma Y., Liu Y., Chen J., Tian J., Fang Q. A protein corona adsorbed to a bacterial magnetosome affects its cellular uptake. IJN. 2020;15:1481–1498. doi: 10.2147/IJN.S220082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniak A., Fenaroli F., Monopoli M.P., Åberg C., Dawson K.A., Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- Li H., Wang Y., Tang Q., Yin D., Tang C., He E., Zou L., Peng Q. The protein corona and its effects on nanoparticle-based drug delivery systems. Acta Biomater. 2021;129:57–72. doi: 10.1016/j.actbio.2021.05.019. [DOI] [PubMed] [Google Scholar]
- Lindman S., Lynch I., Thulin E., Nilsson H., Dawson K.A., Linse S. Systematic investigation of the thermodynamics of HSA adsorption to n-iso-propylacrylamide/n-tert-butylacrylamide copolymer nanoparticles. effects of particle size and hydrophobicity. Nano Lett. 2007;7:914–920. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- Linse S., Cabaleiro-Lago C., Xue W.-F., Lynch I., Lindman S., Thulin E., Radford S.E., Dawson K.A. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M., Cedervall T. Three decades of research about the corona around nanoparticles: lessons learned and where to go now. Small. 2020;16:2000892. doi: 10.1002/smll.202000892. [DOI] [PubMed] [Google Scholar]
- Lundqvist M., Stigler J., Elia G., Lynch I., Cedervall T., Dawson K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley R.W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Abdelmonem A.M., Behzadi S., Clement J.H., Dutz S., Ejtehadi M.R., Hartmann R., Kantner K., Linne U., Maffre P., Metzler S., Moghadam M.K., Pfeiffer C., Rezaei M., Ruiz-Lozano P., Serpooshan V., Shokrgozar M.A., Nienhaus G.U., Parak W.J. Temperature: the “ignored” factor at the nanobio interface. ACS Nano. 2013;7:6555–6562. doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Lohse S.E., Murphy C.J., Fathizadeh A., Montazeri A., Suslick K.S. Variation of protein corona composition of gold nanoparticles following plasmonic heating. Nano Lett. 2014;14:6–12. doi: 10.1021/nl403419e. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Sheibani S., Milani A.S., Rezaee F., Gauberti M., Dinarvand R., Vali H. Crucial role of the protein corona for the specific targeting of nanoparticles. Nanomedicine. 2015;10:215–226. doi: 10.2217/nnm.14.69. [DOI] [PubMed] [Google Scholar]
- Martínez-Negro M., González-Rubio G., Aicart E., Landfester K., Guerrero-Martínez A., Junquera E. Insights into colloidal nanoparticle-protein corona interactions for nanomedicine applications. Adv. Colloid Interf. Sci. 2021;289 doi: 10.1016/j.cis.2021.102366. [DOI] [PubMed] [Google Scholar]
- Maskarinec S.A., Hannig J., Lee R.C., Lee K.Y.C. Direct observation of poloxamer 188 insertion into lipid monolayers. Biophys. J. 2002;82:1453–1459. doi: 10.1016/S0006-3495(02)75499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhofer T., van den Elsen J.M.H., Cameron P.J., Muñoz-Berbel X., Jenkins A.T.A. The interaction of serum albumin with cholesterol containing lipid vesicles. J. Fluoresc. 2010;20:371–376. doi: 10.1007/s10895-009-0522-7. [DOI] [PubMed] [Google Scholar]
- Michaelis K., Hoffmann M.M., Dreis S., Herbert E., Alyautdin R.N., Michaelis M., Kreuter J., Langer K. Covalent linkage of apolipoprotein E to albumin nanoparticles strongly enhances drug transport into the brain. J. Pharmacol. Exp. Ther. 2006;317:1246–1253. doi: 10.1124/jpet.105.097139. [DOI] [PubMed] [Google Scholar]
- Milani S., Baldelli Bombelli F., Pitek A.S., Dawson K.A., Rädler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- Moghimi S.M., Andersen A.J., Ahmadvand D., Wibroe P.P., Andresen T.L., Hunter A.C. Material properties in complement activation. Adv. Drug Deliv. Rev. 2011;63:1000–1007. doi: 10.1016/j.addr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Mohammad-Beigi H., Hayashi Y., Zeuthen C.M., Eskandari H., Scavenius C., Juul-Madsen K., Vorup-Jensen T., Enghild J.J., Sutherland D.S. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 2020;11:4535. doi: 10.1038/s41467-020-18237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monopoli M.P., Walczyk D., Campbell A., Elia G., Lynch I., Baldelli Bombelli F., Dawson K.A. Physical−chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- Monopoli M.P., Åberg C., Salvati A., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotech. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- Moyano D.F., Goldsmith M., Solfiell D.J., Landesman-Milo D., Miranda O.R., Peer D., Rotello V.M. Nanoparticle hydrophobicity dictates immune response. J. Am. Chem. Soc. 2012;134:3965–3967. doi: 10.1021/ja2108905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natte K., Friedrich J.F., Wohlrab S., Lutzki J., von Klitzing R., Österle W., Orts-Gil G. Impact of polymer shell on the formation and time evolution of nanoparticle–protein corona. Colloids Surf. B: Biointerfaces. 2013;104:213–220. doi: 10.1016/j.colsurfb.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Neagu M., Piperigkou Z., Karamanou K., Engin A.B., Docea A.O., Constantin C., Negrei C., Nikitovic D., Tsatsakis A. Protein bio-corona: critical issue in immune nanotoxicology. Arch. Toxicol. 2017;91:1031–1048. doi: 10.1007/s00204-016-1797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.H., Lee B.-J. Protein corona: a new approach for nanomedicine design. Int. J. Nanomedicine. 2017;12:3137–3151. doi: 10.2147/IJN.S129300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström A.M., Fadeel B. Safety assessment of nanomaterials: implications for nanomedicine. J. Control. Rel. Drug Deliv. Res. Europe. 2012;161:403–408. doi: 10.1016/j.jconrel.2012.01.027. [DOI] [PubMed] [Google Scholar]
- Ottonelli I., Duskey J.T., Rinaldi A., Grazioli M.V., Parmeggiani I., Vandelli M.A., Wang L.Z., Prud’homme R.K., Tosi G., Ruozi B. Microfluidic technology for the production of hybrid nanomedicines. Pharmaceutics. 2021;13:1495. doi: 10.3390/pharmaceutics13091495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D.E., Peppas N.A. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Park S.J. Protein–nanoparticle interaction: corona formation and conformational changes in proteins on nanoparticles. Int. J. Nanomedicine. 2020;15:5783–5802. doi: 10.2147/IJN.S254808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partikel K., Korte R., Mulac D., Humpf H.-U., Langer K. Serum type and concentration both affect the protein-corona composition of PLGA nanoparticles. Beilstein J. Nanotechnol. 2019;10:1002–1015. doi: 10.3762/bjnano.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partikel K., Korte R., Stein N.C., Mulac D., Herrmann F.C., Humpf H.-U., Langer K. Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles. Eur. J. Pharm. Biopharm. 2019;141:70–80. doi: 10.1016/j.ejpb.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Pederzoli F., Tosi G., Genovese F., Belletti D., Vandelli M.A., Ballestrazzi A., Forni F., Ruozi B. Qualitative and semiquantitative analysis of the protein coronas associated to different functionalized nanoparticles. Nanomedicine (London) 2018;13:407–422. doi: 10.2217/nnm-2017-0250. [DOI] [PubMed] [Google Scholar]
- Pederzoli F., Ruozi B., Duskey J., Hagmeyer S., Sauer A.K., Grabrucker S., Coelho R., Oddone N., Ottonelli I., Daini E., Zoli M., Vandelli M.A., Tosi G., Grabrucker A.M. Nanomedicine against Aβ aggregation by β-sheet breaker peptide delivery. In Vitro Evidence. Pharmaceut. 2019;11:E572. doi: 10.3390/pharmaceutics11110572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Minbo H., Fang C., Xi L., Chaocan Z. The interaction between cholesterol and human serum albumin. Protein Pept. Lett. 2008;15:360–364. doi: 10.2174/092986608784246542. [DOI] [PubMed] [Google Scholar]
- Petry R., Saboia V.M., Franqui L.S., de Holanda C.A., Garcia T.R.R., de Farias M.A., de Souza Filho A.G., Ferreira O.P., Martinez D.S.T., Paula A.J. On the formation of protein corona on colloidal nanoparticles stabilized by depletant polymers. Mater. Sci. Eng. C. 2019;105 doi: 10.1016/j.msec.2019.110080. [DOI] [PubMed] [Google Scholar]
- Pozzi D., Colapicchioni V., Caracciolo G., Piovesana S., Capriotti A.L., Palchetti S., De Grossi S., Riccioli A., Amenitsch H., Laganà A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells. Nanoscale. 2014;6:2782–2792. doi: 10.1039/c3nr05559k. [DOI] [PubMed] [Google Scholar]
- Pozzi D., Caracciolo G., Digiacomo L., Colapicchioni V., Palchetti S., Capriotti A.L., Cavaliere C., Chiozzi R.Z., Puglisi A., Laganà A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale. 2015;7:13958–13966. doi: 10.1039/C5NR03701H. [DOI] [PubMed] [Google Scholar]
- Prapainop K., Witter D.P., Wentworth P. A Chemical approach for cell-specific targeting of nanomaterials: small-molecule-initiated misfolding of nanoparticle corona proteins. J. Am. Chem. Soc. 2012;134:4100–4103. doi: 10.1021/ja300537u. [DOI] [PubMed] [Google Scholar]
- Rahman M., Laurent S., Tawil N., Yahia L., Mahmoudi M. Protein-Nanoparticle Interactions, Springer Series in Biophysics. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2013. Nanoparticle and protein corona; pp. 21–44. [DOI] [Google Scholar]
- Rampado R., Crotti S., Caliceti P., Pucciarelli S., Agostini M. Recent advances in understanding the protein corona of nanoparticles and in the formulation of “stealthy” nanomaterials. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Andrikopoulos N., Velonia K., Tang H., Cai R., Ding F., Ke P.C., Chen C. Chemical and biophysical signatures of the protein corona in nanomedicine. J. Am. Chem. Soc. 2022;144:9184–9205. doi: 10.1021/jacs.2c02277. [DOI] [PubMed] [Google Scholar]
- Reschly E.J., Sorci-Thomas M.G., Davidson W.S., Meredith S.C., Reardon C.A., Getz G.S. Apolipoprotein A-I α-helices 7 and 8 modulate high density lipoprotein subclass distribution *. J. Biol. Chem. 2002;277:9645–9654. doi: 10.1074/jbc.M107883200. [DOI] [PubMed] [Google Scholar]
- Rezaei G., Daghighi S.M., Haririan I., Yousefi I., Raoufi M., Rezaee F., Dinarvand R. Protein corona variation in nanoparticles revisited: a dynamic grouping strategy. Colloids Surf. B: Biointerfaces. 2019;179:505–516. doi: 10.1016/j.colsurfb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- Rigon L., Salvalaio M., Pederzoli F., Legnini E., Duskey J.T., D’Avanzo F., De Filippis C., Ruozi B., Marin O., Vandelli M.A., Ottonelli I., Scarpa M., Tosi G., Tomanin R. Targeting brain disease in MPSII: preclinical evaluation of IDS-loaded PLGA nanoparticles. Int. J. Mol. Sci. 2019;20:2014. doi: 10.3390/ijms20082014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P., Farrar D., Perry C.C. Interpretation of protein adsorption: surface-induced conformational changes. J. Am. Chem. Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- Saha K., Rahimi M., Yazdani M., Kim S.T., Moyano D.F., Hou S., Das R., Mout R., Rezaee F., Mahmoudi M., Rotello V.M. Regulation of macrophage recognition through the interplay of nanoparticle surface functionality and protein corona. ACS Nano. 2016;10:4421–4430. doi: 10.1021/acsnano.6b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati A., Pitek A.S., Monopoli M.P., Prapainop K., Bombelli F.B., Hristov D.R., Kelly P.M., Åberg C., Mahon E., Dawson K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotech. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Segets D., Marczak R., Schäfer S., Paula C., Gnichwitz J.-F., Hirsch A., Peukert W. Experimental and theoretical studies of the colloidal stability of nanoparticles−a general interpretation based on stability maps. ACS Nano. 2011;5:4658–4669. doi: 10.1021/nn200465b. [DOI] [PubMed] [Google Scholar]
- Sempf K., Arrey T., Gelperina S., Schorge T., Meyer B., Karas M., Kreuter J. Adsorption of plasma proteins on uncoated PLGA nanoparticles. Eur. J. Pharm. Biopharm. 2013;85:53–60. doi: 10.1016/j.ejpb.2012.11.030. [DOI] [PubMed] [Google Scholar]
- Shannahan J.H., Podila R., Aldossari A.A., Emerson H., Powell B.A., Ke P.C., Rao A.M., Brown J.M. Formation of a protein corona on silver nanoparticles mediates cellular toxicity via scavenger receptors. Toxicol. Sci. 2015;143:136–146. doi: 10.1093/toxsci/kfu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanwar S., Liang L., Nechaev A.V., Bausheva D.K., Balalaeva I.V., Vodeneev V.A., Roy I., Zvyagin A.V., Guryev E.L. Controlled formation of a protein corona composed of denatured bsa on upconversion nanoparticles improves their colloidal stability. Materials. 2021;14:1657. doi: 10.3390/ma14071657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X.-R., Wei X.-Q., Song X., Hao L.-Y., Cai X.-X., Zhang Z.-R., Peng Q., Lin Y.-F. Independent effect of polymeric nanoparticle zeta potential/surface charge, on their cytotoxicity and affinity to cells. Cell Prolif. 2015;48:465–474. doi: 10.1111/cpr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg S.J., Postupalenko V., Duskey J.T., Palivan C.G., Meier W. Stimuli-responsive codelivery of oligonucleotides and drugs by self-assembled peptide nanoparticles. Biomacromolecules. 2016;17:935–945. doi: 10.1021/acs.biomac.5b01614. [DOI] [PubMed] [Google Scholar]
- Singh N., Marets C., Boudon J., Millot N., Saviot L., Maurizi L. In vivo protein corona on nanoparticles: does the control of all material parameters orient the biological behavior? Nanoscale Adv. 2021;3:1209–1229. doi: 10.1039/D0NA00863J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.R., Cicerone M.T., Meuse C.W. Tertiary structure changes in albumin upon surface adsorption observed via fourier transform infrared spectroscopy. Langmuir. 2009;25:4571–4578. doi: 10.1021/la802955w. [DOI] [PubMed] [Google Scholar]
- Sobczynski D.J., Eniola-Adefeso O. IgA and IgM protein primarily drive plasma corona-induced adhesion reduction of PLGA nanoparticles in human blood flow. Bioeng. Transl. Med. 2017;2:180–190. doi: 10.1002/btm2.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Jiang H., Xu B., Yu Y., Chen X. Effects of protein corona on active and passive targeting of cyclic RGD peptide-functionalized pegylation nanoparticles. Mol. Pharm. 2018;15:5019–5030. doi: 10.1021/acs.molpharmaceut.8b00612. [DOI] [PubMed] [Google Scholar]
- Teir M.M.A., Ghithan J., Darwish S., Abu-Hadid M.M. 2012. Multi-Spectroscopic Investigation of the Interactions between Cholesterol and Human Serum Albumin; p. 11. [Google Scholar]
- Tekie F.S.M., Hajiramezanali M., Geramifar P., Raoufi M., Dinarvand R., Soleimani M., Atyabi F. Controlling evolution of protein corona: a prosperous approach to improve chitosan-based nanoparticle biodistribution and half-life. Sci. Rep. 2020;10:9664. doi: 10.1038/s41598-020-66572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S., Docter D., Rosfa S., Wlodarski A., Kuharev J., Rekik A., Knauer S.K., Bantz C., Nawroth T., Bier C., Sirirattanapan J., Mann W., Treuel L., Zellner R., Maskos M., Schild H., Stauber R.H. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- Tenzer S., Docter D., Kuharev J., Musyanovych A., Fetz V., Hecht R., Schlenk F., Fischer D., Kiouptsi K., Reinhardt C., Landfester K., Schild H., Maskos M., Knauer S.K., Stauber R.H. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotech. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Tewabe A., Abate A., Tamrie M., Seyfu A., Abdela Siraj E. Targeted drug delivery — from magic bullet to nanomedicine: principles, challenges, and future perspectives. J. Multidiscip. Healthc. 2021;14:1711–1724. doi: 10.2147/JMDH.S313968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi G. Frontiers in Nanomedicine. BENTHAM SCIENCE PUBLISHERS. 2017. How does “protein corona” affect the in vivo efficiency of polymeric nanoparticles? State of art; pp. 199–238. [DOI] [Google Scholar]
- Tosi G., Costantino L., Rivasi F., Ruozi B., Leo E., Vergoni A.V., Tacchi R., Bertolini A., Vandelli M.A., Forni F. Targeting the central nervous system: in vivo experiments with peptide-derivatized nanoparticles loaded with Loperamide and Rhodamine-123. J. Control. Release. 2007;122:1–9. doi: 10.1016/j.jconrel.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Tsai D.-H., DelRio F.W., Keene A.M., Tyner K.M., MacCuspie R.I., Cho T.J., Zachariah M.R., Hackley V.A. Adsorption and conformation of serum albumin protein on gold nanoparticles investigated using dimensional measurements and in situ spectroscopic methods. Langmuir. 2011;27:2464–2477. doi: 10.1021/la104124d. [DOI] [PubMed] [Google Scholar]
- Varnamkhasti B.S., Hosseinzadeh H., Azhdarzadeh M., Vafaei S.Y., Esfandyari-Manesh M., Mirzaie Z.H., Amini M., Ostad S.N., Atyabi F., Dinarvand R. Protein corona hampers targeting potential of MUC1 aptamer functionalized SN-38 core–shell nanoparticles. Int. J. Pharm. 2015;494:430–444. doi: 10.1016/j.ijpharm.2015.08.060. [DOI] [PubMed] [Google Scholar]
- Wagner S., Zensi A., Wien S.L., Tschickardt S.E., Maier W., Vogel T., Worek F., Pietrzik C.U., Kreuter J., von Briesen H. Uptake mechanism of apoe-modified nanoparticles on brain capillary endothelial cells as a blood-brain barrier model. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey C.D., Chan W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- Wang G., Chen F., Banda N.K., Holers V.M., Wu L., Moghimi S.M., Simberg D. Activation of human complement system by dextran-coated iron oxide nanoparticles is not affected by dextran/Fe ratio, hydroxyl modifications, and crosslinking. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lin Y., Nienhaus K., Nienhaus G.U. The protein corona on nanoparticles as viewed from a nanoparticle-sizing perspective. WIREs Nanomed. Nanobiotechnol. 2018;10 doi: 10.1002/wnan.1500. [DOI] [PubMed] [Google Scholar]
- Warsi M.H. Development and optimization of vitamin E TPGS based PLGA nanoparticles for improved and safe ocular delivery of ketorolac. J. Drug Deliv. Sci. Technol. 2021;61 doi: 10.1016/j.jddst.2020.102121. [DOI] [Google Scholar]
- Xiao W., Gao H. The impact of protein corona on the behavior and targeting capability of nanoparticle-based delivery system. Int. J. Pharm. 2018;552:328–339. doi: 10.1016/j.ijpharm.2018.10.011. [DOI] [PubMed] [Google Scholar]
- Yan Y., Gause K.T., Kamphuis M.M.J., Ang C.-S., O’Brien-Simpson N.M., Lenzo J.C., Reynolds E.C., Nice E.C., Caruso F. Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano. 2013;7:10960–10970. doi: 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- Yoo J., Park C., Yi G., Lee D., Koo H. Active targeting strategies using biological ligands for nanoparticle drug delivery systems. Cancers. 2019;11:640. doi: 10.3390/cancers11050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhao L., Guo C., Yan B., Su G. Regulating protein corona formation and dynamic protein exchange by controlling nanoparticle hydrophobicity. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Burnum K.E., Luna M.L., Petritis B.O., Kim J.-S., Qian W.-J., Moore R.J., Heredia-Langner A., Webb-Robertson B.-J.M., Thrall B.D., Camp D.G., II, Smith R.D., Pounds J.G., Liu T. Quantitative proteomics analysis of adsorbed plasma proteins classifies nanoparticles with different surface properties and size. PROTEOMICS. 2011;11:4569–4577. doi: 10.1002/pmic.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu T., Yu W., Ruan S., He Q., Gao H. Ligand size and conformation affect the behavior of nanoparticles coated with in vitro and in vivo protein corona. ACS Appl. Mater. Interfaces. 2018;10:9094–9103. doi: 10.1021/acsami.7b16096. [DOI] [PubMed] [Google Scholar]
- Zhang W., Cho W.C., Bloukh S.H., Edis Z., Du W., He Y., Hu H.Y., Ten Hagen T.L.M., Falahati M. An overview on the exploring the interaction of inorganic nanoparticles with microtubules for the advancement of cancer therapeutics. Int. J. Biol. Macromol. 2022;212:358–369. doi: 10.1016/j.ijbiomac.2022.05.150. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin Z., Chen Y., Gao D., Wang P., Lin Y., Wang Y., Wang F., Han Y., Yuan H. Co-delivery of Docetaxel and Resveratrol by liposomes synergistically boosts antitumor efficiency against prostate cancer. Eur. J. Pharm. Sci. 2022;174 doi: 10.1016/j.ejps.2022.106199. [DOI] [PubMed] [Google Scholar]
- Zu M., Ma Y., Cannup B., Xie D., Jung Y., Zhang J., Yang C., Gao F., Merlin D., Xiao B. Oral delivery of natural active small molecules by polymeric nanoparticles for the treatment of inflammatory bowel diseases. Adv. Drug Deliv. Rev. 2021;176 doi: 10.1016/j.addr.2021.113887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.