Abstract

Our current understanding of the chemistry of the primordial genetic material is fragmentary at best. The chemical replication of oligonucleotides long enough to perform catalytic functions is particularly problematic because of the low efficiency of nonenzymatic template copying. Here we show that this problem can be circumvented by assembling a functional ribozyme by the templated ligation of short oligonucleotides. However, this approach creates a new problem because the splint oligonucleotides used to drive ribozyme assembly strongly inhibit the resulting ribozyme. We explored three approaches to the design of splint oligonucleotides that enable efficient ligation but which allow the assembled ribozyme to remain active. DNA splints, splints with G:U wobble pairs, and splints with G to I (Inosine) substitutions all allowed for the efficient assembly of an active ribozyme ligase. Our work demonstrates the possibility of a transition from nonenzymatic ligation to enzymatic ligation and reveals the importance of avoiding ribozyme inhibition by complementary oligonucleotides.
Introduction
During the origin of life, the chemical replication of the primordial genome may have allowed for the emergence of the first ribozymes and then progressively more complex forms of RNA-based life in the RNA world.1−4 Demonstrating both nonenzymatic RNA replication and ribozyme-catalyzed RNA replication continue to remain grand challenges.5,6 However, the transition from chemical to enzymatic replication is also an important, but less studied, consideration. Because effective and prebiotically plausible pathways for the propagation of RNA templates long enough to encode an RNA replicase are yet to be discovered, we have explored the assembly of a ribozyme ligase by the nonenzymatic, template-directed ligation of oligonucleotide substrates. The substrates themselves are short enough that they may be replicable by nonenzymatic means. The fundamental hypothesis that we raise is that the genome of a very primitive protocell could consist of fairly short oligonucleotides, only 8 to 12 nucleotides in length. We suggest that these oligonucleotides could assemble by ligation into longer sequences capable of selective advantageous ribozyme activity, even though the ribozyme itself is too long to be chemically replicated.
As a model system to explore this problem we used short 5′-phosphoro-(2-methyl)imidazole activated oligoribonucleotides ending in 3′-amino-2′,3′-dideoxyribonucleotides as ligators. Both 3′-amino nucleotides and oligonucleotides have been employed extensively in origin of life studies7−10 and for the copying of RNA templates by iterated ligation11 in which the 3′-amino group of an upstream ligator attacks the 2-methylimidazole activated 5′-phosphate of the downstream ligator. Here we show that 3′-amino oligonucleotides 10–12 nucleotides long can efficiently assemble by template-directed nonenzymatic ligation into an active ribozyme ligase. Interestingly, we found that the assembled ribozyme is strongly inhibited by the very template oligonucleotides (referred to as splints) used to direct its assembly. This problem arises because each splint must recognize and bind the 3′ and 5′ ends of both oligonucleotides to be ligated. Thus, the splint binds even more tightly to the products of ligation, including the assembled ribozyme. Similar template oligonucleotide inhibition frustrated early efforts to demonstrate oligonucleotide replication by ligation.12 Here we present three distinct strategies by which such splint inhibition can be overcome, so that ribozyme activity is observed postassembly.
Results
We first tested whether ribozyme assembly could be directed by short RNA template splints. We chose to assemble an RNA ligase ribozyme since successful assembly could represent a key intermediate stage in the transition from chemical RNA replication to ribozyme catalyzed replication. Recently, an RNA polymerase ribozyme was assembled by the nonenzymatic ligation of a collection of 20 to 30-nt long activated RNA oligonucleotides in 0.5% yield.13 Because of the low yield, T7 RNA polymerase transcription of an RT-PCR amplification product was needed to obtain sufficient ribozyme to measure its activity. Furthermore, this process removes the 20-mer to 30-mer splints, which would inhibit ribozyme activity.14,15
We consider a simpler and more prebiotically relevant model system that uses much shorter oligonucleotides to assemble a smaller ribozyme (Figure 1a). We chose a 52-nt long, highly efficient RNA ligase ribozyme that can ligate a 14-nt 5′-triphosphate RNA substrate to its 3′-hydroxyl end.16 We tested whether we could generate this ligase from five different oligomers, 10 to 12 nucleotides long (Figure 1b, Figure 2a), promoted by four different 10 nucleotide long splints. Each splint is complementary to 5 nt at the 3′ end of the upstream ligator and 5 nt at the 5′ of the downstream ligator. To monitor the reaction, the 5′-end of ligator 1 was labeled with a Cy3 fluorophore. We visualized the appearance of progressively longer nonenzymatic ligation products by denaturing polyacrylamide gel electrophoresis over time. We observed efficient assembly of full-length ribozyme (Figure 2b, left). However, no ligase activity could be detected (Figure 2b, right). The estimated melting temperature of the four 10-mer splints ranges from 46 to 62 °C17,18 (see Table S2 for details), while the ribozyme ligase is assayed at 48 °C. Heat denaturation followed by a renaturation step still yielded no ligase activity (Figure S1), suggesting that fully complementary 10-nt RNA splints inhibit this ribozyme.
Figure 1.
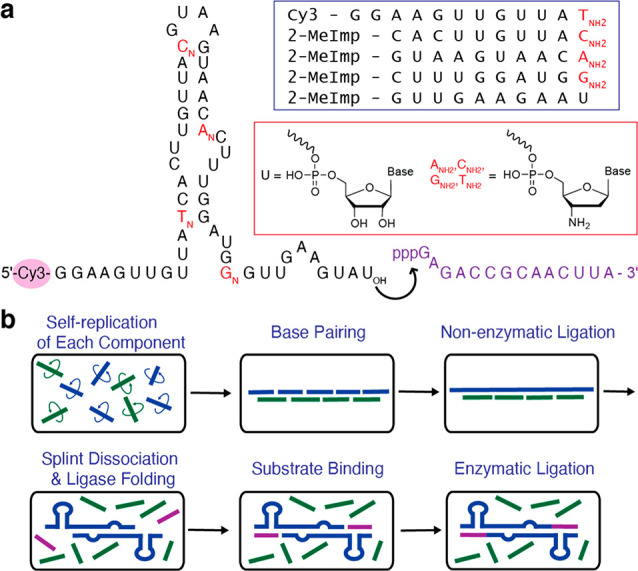
Model for the transition from nonenzymatic ligation to ribozyme mediated ligation. (a) Secondary structure of the ribozyme ligase16 used in all experiments in this paper. Black letters are ribonucleotides; red letters represent 3′-amino-2′,3′-dideoxyribonucleotides (T is used instead of U as a 3′-amino dideoxyribonucleotide in this study for synthetic convenience). The corresponding set of five ligator oligonucleotides is shown inside the box. The purple sequence is the 5′-triphosphate substrate of the ribozyme ligase. (b) Schematic representation of the experimental design. Potentially self-replicating oligonucleotides shown in blue take part in multistep ligation reactions templated by the green splint oligonucleotides to generate the full length ligase sequence. Following dilution, the splint oligonucleotides dissociate in the 48 °C ribozyme reaction mixture, allowing folding of the ligase ribozyme. The functional ligase then ligates itself to the 5′-triphosphate oligonucleotide substrate.
Figure 2.
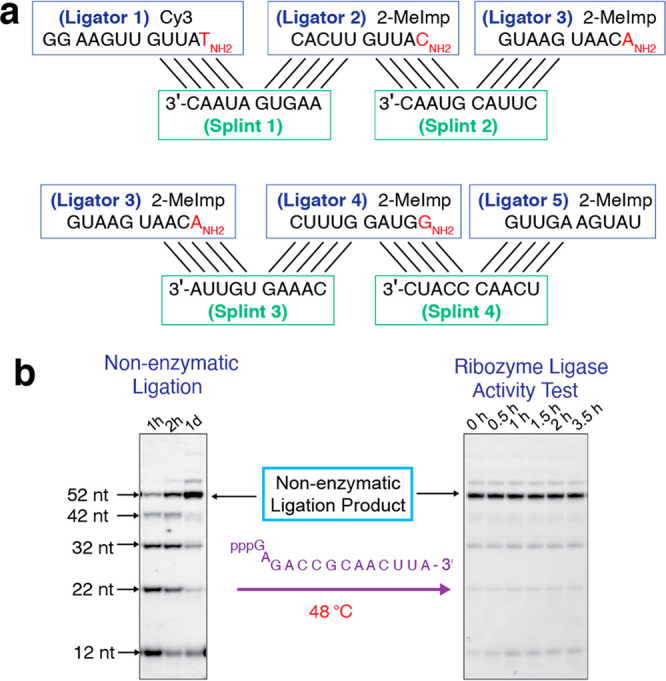
Efficient assembly but strong inhibition of a ribozyme ligase by 10-nt RNA splints. (a) Diagram of nonenzymatic ligation of five ligator oligonucleotides directed by four 10-nt RNA splint oligonucleotides (5′-AAGUGAUAAC-3′, 5′-CUUACGUAAC-3′, 5′-CAAAGUGUUA-3′, and 5′-UCAACCCAUC-3′). (b) (Left) PAGE analysis of the assembly of the ribozyme ligase by nonenzymatic ligation. Reactions contained 20 μM of ligator 1, 25 μM of ligators 2 to 5, 25 μM of splints 1 to 4, 100 mM HEPES, pH 8.0, 100 mM NaCl, 50 mM MgCl2, and 100 mM HEI (1-(2-Hydroxyethyl)imidazole). The ligation reaction was carried out on ice. (Right) PAGE analysis showing the lack of activity of the product ligase. The nonenzymatic ligation product was diluted 10-fold into the ribozyme reaction buffer: 50 mM bis-tris propane, pH 8.5, 25 mM MgCl2, and 3 μM triphosphate substrate. The ligase reaction was performed at 48 °C. No ligase activity was detected. Gels depicted are representative of triplicate experiments.
For the assembly of a functional RNA ligase in a manner that is prebiotically plausible, it is essential to prevent ribozyme inhibition by the splints. The splints must bind tightly and specifically enough for efficient assembly but have a low enough melting temperature for dissociation from the ligase. Shorter 8-nt RNA splints resulted in a severe loss of ribozyme yield (Figure S2). However, we discovered three solutions to this problem.
We first tried 10-mer DNA splints, since DNA:RNA duplexes are less stable than RNA:RNA duplexes of the same sequence19 (Figure 3a and Table S2). Potentially prebiotic pathways to DNA precursors have been reported20,21 and template-directed ligation preferentially leads to the synthesis of “all DNA” and “all RNA” oligonucleotide products.9 These studies suggest that DNA may have coexisted with RNA in the prebiotic world. This raises the possibility that distinct RNA and DNA oligonucleotides could have replicated together within protocells. With four 10-mer DNA splints, the 52nt ligase assembled with a yield of ∼60% after 1 day (Figure 3b left, Figure S3). Importantly, the assembled ribozyme efficiently catalyzed the ligation reaction (Figure 3b, right). Thus, DNA splints provide the affinity and specificity needed for efficient ribozyme assembly, while the low thermal stability of DNA:RNA duplexes allows for splint dissociation followed by ribozyme folding into the catalytically active form.
Figure 3.
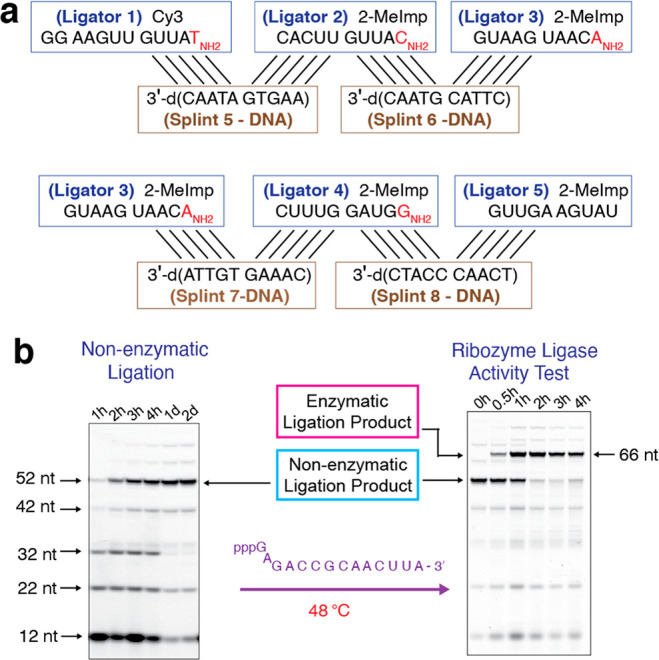
Efficient assembly of active ribozyme ligase by 10-nt DNA splints. (a) Diagram of nonenzymatic ligation of five ligator oligonucleotides directed by four 10-nt DNA splint oligonucleotides (5′-d(AAGTGATAAC)-3′, 5′-d(CTTACGTAAC)-3′, 5′-d(CAAAGTGTTA)-3′, and 5′-d(TCAACCCATC-3′)). (b) (Left) PAGE analysis of the assembly of the ribozyme ligase by nonenzymatic ligation. (Right) PAGE analysis showing the activity of the ribozyme ligase. The conditions for both reactions, including oligonucleotide concentrations, are as in Figure 2b, except for the use of DNA splints 5 to 8. Gels depicted are representative of triplicate experiments.
As an alternative to DNA splints, we redesigned the RNA splints to include several C to U changes and shortened the terminal splint to a 9-mer (Figure 4a). Changing G:C base pairs to G:U wobble pairs lowers the thermal stability of the ribozyme-splint duplexes. To maintain efficient assembly via nonenzymatic ligation, we only introduced one or two C to U substitutions in each splint. Using these splints led to ∼50% and ∼66% yields of ribozyme within 6 and 48 h, respectively (Figure 4b left, Figure S4). Importantly, the resultant ribozyme was catalytically active (Figure 4b, right).
Figure 4.
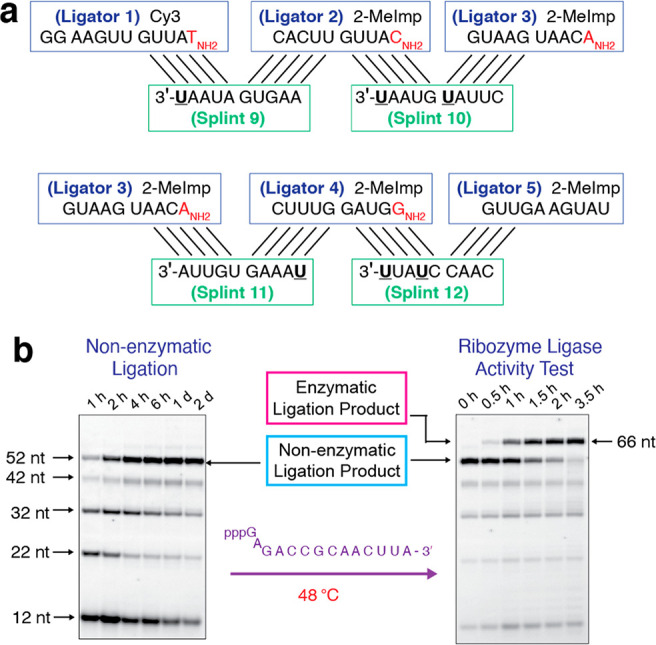
Efficient assembly of active ribozyme ligase by 10-nt and 9-nt RNA splints with G:U wobble pairing. (a) Diagram of nonenzymatic ligation of five ligator oligonucleotides directed by three 10-mer and one 9-nt RNA splint oligonucleotides with six C to U changes (5′-AAGUGAUAAU-3′, 5′-CUUAUGUAAU-3′, 5′-UAAAGUGUUA-3′ and 5′-CAACCUAUU-3′). (b) (Left) PAGE analysis of the assembly of active ribozyme ligase by nonenzymatic ligation. (Right) PAGE analysis showing the activity of the ribozyme ligase. The conditions for both reactions, including oligonucleotide concentrations, are as in Figure 2b, except for the use of splints 9 to 12 as shown. Gels depicted are representative of triplicate experiments.
Splints with G:U wobble pairs to the ligator oligonucleotides need not arise due to low fidelity replication, because if splints are independently replicating genomic fragments, their sequences would be expected to evolve in favor of optimal ribozyme assembly and activity. Here we tested this hypothesis using G:U wobble pairs as a proof of concept, but other sequence changes could have similar effects.
Since inosine has been proposed to have preceded guanosine as an RNA nucleotide in prebiotic conditions,22 we replaced guanosine with inosine to weaken the thermal stability of the splint-ribozyme complex (Table S2). We substituted G with I in splints 1, 2, and 3, to afford splints 13, 14, and 15, respectively. Due to the lack of G in splint 4, we shortened splint 4 to 8 nt to generate splint 16. Indeed, we found that the resulting splints enabled efficient ribozyme assembly and also allowed for good ribozyme activity (Figure 5). Thus, inosine-based RNA would seem to have yet another advantage over guanosine-based RNA, by minimizing the inhibition of ribozyme activity by complementary oligonucleotides. A similar scenario could be envisaged to also facilitate the replication of genomic fragments.
Figure 5.
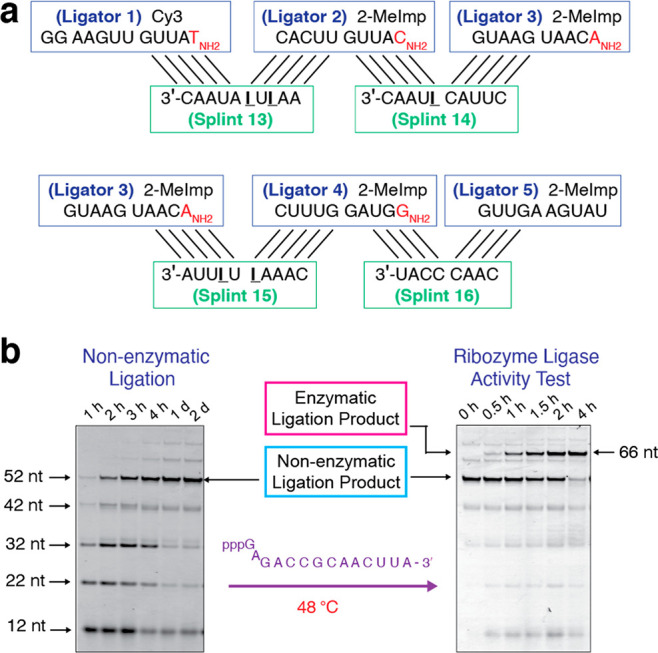
Efficient assembly of active ribozyme ligase by 10-nt and 8-nt RNA splints with I:C base pairing. (a) Diagram of nonenzymatic ligation of five ligator oligonucleotides directed by three 10-mer and one 8-nt RNA splint oligonucleotides with five G to I changes (5′-AAIUIAUAAC-3′, 5′-CUUAC IUAAC-3′, 5′-CAAAIUIUUA-3′, and 5′-CAACCCAU-3′). (b) (Left) PAGE analysis of the assembly of the ribozyme ligase by nonenzymatic ligation. (Right) PAGE analysis showing the activity of the ribozyme ligase. The conditions for both reactions, including oligonucleotide concentrations, are as in Figure 2b, except for the use of the splints 13 to 16 as illustrated above. Gels depicted are representative of triplicate experiments.
Discussion
Our work suggests a scenario for the origin of life in which the replicating genomic oligonucleotides in the first protocells could have been quite small. Since reasonably active ribozyme nucleases are generally longer than 30–50 nucleotides, it had generally been assumed that replicating genomic fragments had to be at least 30 nucleotides long if not longer. Even when ribozymes are assembled from multiple oligonucleotides, the oligonucleotides are typically longer than 20 nucleotides.13,23,24 Copying sequences of this length by primer extension with activated ribonucleotides remains an unsolved problem. Even if solved, complete cycles of replication would require a way for long RNA–RNA duplexes to dissociate so that the copied strands can be copied. In contrast, completely replicating 8–12-mer oligonucleotides is much more plausible, both in terms of template copying and in terms of their ability to dissociate. Such 8–12 mers are unlikely to exhibit catalytic activity individually, but multiple nonenzymatic splinted ligations could assemble longer functional oligonucleotides, as we have demonstrated.
Replicating genomic RNA and concurrently assembling longer functional ribozymes faces several obstacles. First, efficiently copying of RNA templates 8–12 nucleotides long must be achieved either by monomer polymerization, by the ligation of smaller oligomers, or some combination thereof. Second, efficient splinted ligation must be achieved with native RNA oligonucleotides (as opposed to 3′-amino-terminated oligonucleotides) so that longer functional, but nonreplicating, sequences can be assembled. Third, the duplex products of oligonucleotide copying must dissociate in order to allow for iterative cycles of replication. Finally, the affinity of splint-templates for their ligator oligonucleotides must be high enough to allow for efficient ligation but low enough to allow for splint dissociation, so that the assembled product strand can fold into an active ribozyme.
The first two problems would benefit from an effective way of increasing the nucleophilicity of the RNA 3′-hydroxyl, perhaps by a means of delivering an appropriate catalytic metal ion to the reaction center. The third problem might be overcome if either strand displacement25 or strand separation could be triggered by transient environmental changes, for instance, temperature fluctuations, pH fluctuations,26 or salt concentration fluctuations following wet–dry cycles.27
RNA splints that had only 3–4 nucleotides of complementarity to both ligator oligonucleotides led to slow and/or inaccurate ligation and thus inefficient assembly. On the other hand, splints with longer regions of complementarity failed to dissociate, leading to ribozyme inhibition. We found that the introduction of wobble pairs or inosine allowed RNA splints to accurately direct ribozyme assembly through iterated ligation events without inhibiting the assembled ribozyme. Additionally, DNA splints exhibited excellent specificity with low enough affinity to avoid ribozyme inhibition. Prebiotically, it is possible that the admixture of low levels of deoxy-, arabino-, or threo-nucleotides into RNA oligonucleotides could have had the same effect, as well as potentially decreasing duplex stability enough to facilitate multiple cycles of replication.9,28 Important roles for weakly binding oligonucleotides are also found in a previously described mechanism for the homeostatic modulation of ribozyme activity. Here too, short oligonucleotides inhibit ribozyme folding at high concentrations but dissociate effectively and thereby lead to ribozyme activation during protocell growth.14
Summary
We have demonstrated a potential path from nonenzymatic ligation to enzymatic ligation by showing that a ribozyme ligase can be assembled from a set of short oligonucleotides via splint-templated nonenzymatic ligation. Although the ribozyme was strongly inhibited by complementary RNA splint oligonucleotides, we found that inhibition could be avoided by using either DNA splints, splints with G:U wobble pairs, or splints with I:C base pairs. Our study provides prebiotically plausible solutions to the long-standing problem of ribozyme inhibition by complementary oligonucleotides. Solving the related problems of oligonucleotide replication and splint-directed ribozyme assembly may lead to further insights into the origins of life in an RNA world.
Acknowledgments
We thank Constantin Giurgiu, Dr. Thomas Wright, Dr. Seohyun Chris Kim, Dr. Li Li, and Dr. Victor Lelyveld for helpful discussions and technical assistance and Prof. Yamuna Krishnan for helpful comments on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c06722.
Experimental procedures, strong ligase inhibition by 10-nt RNA splints, poor efficiency of ribozyme assembly with 8-nt RNA splints, yield of full-length RNA ligase assembly by nonenzymatic ligation using DNA splints, yield of full-length RNA ligase assembly by nonenzymatic ligation using G:U wobble pairing RNA splints, yield of full-length RNA ligase assembly by nonenzymatic ligation using I:C wobble pairing RNA splints, summary of oligonucleotide sequences, and estimated melting temperatures of the splints and the nonenzymatic ligation product (PDF)
Author Present Address
∥ Alnylam Pharmaceuticals, 300 Third Street, Cambridge, Massachusetts 02142, USA.
Author Contributions
L.Z. and D.K.O. contributed equally. The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.
J.W.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by a grant (290363) from the Simons Foundation and by a grant from the NSF (CHE-1607034) to J.W.S. D.K.O. is a recipient of a Postdoctoral Research Scholarship (B3) from the Fonds de Recherche du Quebec–Nature et Technologies (FRQNT), Quebec, Canada, and a Postdoctoral Fellowship from Canadian Institutes of Health Research (CIHR) from Canada.
The authors declare no competing financial interest.
Supplementary Material
References
- Woese C. R.The Genetic Code the Molecular Basis for Genetic Expression; Harper & Row: New York, 1967. [Google Scholar]
- Crick C. The Origin of the Genetic Code. J. Mol. Biol. 1968, 38, 367–379. 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Orgel L. E. Evolution of the Genetic Apparatus. J. Mol. Biol. 1968, 38 (3), 381–393. 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Origin of Life: The RNA World. Nature 1986, 319 (6055), 618–618. 10.1038/319618a0. [DOI] [Google Scholar]
- Szostak J. W. The Narrow Road to the Deep Past : In Search of the Chemistry of the Origin of Life Minireviews. Angew. Chem., Int. Ed. 2017, 56 (37), 11037–11043. 10.1002/anie.201704048. [DOI] [PubMed] [Google Scholar]
- Wachowius F.; Attwater J.; Holliger P. Nucleic Acids: Function and Potential for Abiogenesis. Q. Rev. Biophys. 2017, 50, e4. 10.1017/S0033583517000038. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Zhang N.; Blain J. C.; Szostak J. W. Synthesis of N3′-P5′-Linked Phosphoramidate DNA by Nonenzymatic Template-Directed Primer Extension. J. Am. Chem. Soc. 2013, 135 (2), 924–932. 10.1021/ja311164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Blain J. C.; Zielinska D.; Gryaznov S. M.; Szostak J. W. Fast and Accurate Nonenzymatic Copying of an RNA-like Synthetic Genetic Polymer. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (44), 17732–17737. 10.1073/pnas.1312329110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik S.; Krishnamurthy R. The Role of Sugar-Backbone Heterogeneity and Chimeras in the Simultaneous Emergence of RNA and DNA. Nat. Chem. 2019, 11 (11), 1009–1018. 10.1038/s41557-019-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty D. K.; Zhou L.; Szostak J. W. Nonenzymatic Template-Directed Synthesis of Mixed-Sequence 3′-NP-DNA up to 25 Nucleotides Long Inside Model Protocells. J. Am. Chem. Soc. 2019, 141 (26), 10481–10488. 10.1021/jacs.9b04858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.; O’Flaherty D. K.; Szostak J. W. Template-Directed Copying of RNA by Non-Enzymatic Ligation. Angew. Chem., Int. Ed. 2020, 59 (36), 15682–15687. 10.1002/anie.202004934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann T. N.; Strohbach A.; Seitz O. Achieving Turnover in DNA-Templated Reactions. ChemBioChem 2008, 9 (14), 2185–2192. 10.1002/cbic.200800290. [DOI] [PubMed] [Google Scholar]
- Wachowius F.; Holliger P. Non-Enzymatic Assembly of a Minimized RNA Polymerase Ribozyme. ChemSystemsChem. 2019, 1, e1900004 10.1002/syst.201900004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart A. E.; Adamala K. P.; Szostak J. W. A Simple Physical Mechanism Enables Homeostasis in Primitive Cells. Nat. Chem. 2016, 8 (5), 448–453. 10.1038/nchem.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari S. H.; Famulok M. Competitive Regulation of Modular Allosteric Aptazymes by a Small Molecule and Oligonucleotide Effector. RNA 2005, 11 (10), 1514–1520. 10.1261/rna.2840805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M. P.; Joyce G. F. Highly Efficient Self-Replicating RNA Enzymes. Chem. Biol. 2014, 21 (2), 238–245. 10.1016/j.chembiol.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M.; Kierzek R.; Jaeger J. A.; Sugimoto N.; Caruthers M. H.; Neilson T.; Turner D. H. Improved Free-Energy Parameters for Predictions of RNA Duplex Stability. Proc. Natl. Acad. Sci. U. S. A. 1986, 83 (24), 9373–9377. 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznov S. M. Oligonucleotide N3′-->P5′ Phosphoramidates as Potential Therapeutic Agents. Biochim. Biophys. Acta, Gene Struct. Expression 1999, 1489 (1), 131–140. 10.1016/S0167-4781(99)00151-7. [DOI] [PubMed] [Google Scholar]
- Kankia B. I.; Marky L. A. DNA, RNA, and DNA/RNA Oligomer Duplexes: A Comparative Study of Their Stability, Heat, Hydration, and Mg2+ Binding Properties. J. Phys. Chem. B 1999, 103 (41), 8759–8767. 10.1021/jp991614x. [DOI] [Google Scholar]
- Teichert J. S.; Kruse F. M.; Trapp O. Direct Prebiotic Pathway to DNA Nucleosides. Angew. Chem., Int. Ed. 2019, 58 (29), 9944–9947. 10.1002/anie.201903400. [DOI] [PubMed] [Google Scholar]
- Xu J.; Green N. J.; Gibard C.; Krishnamurthy R.; Sutherland J. D. Prebiotic Phosphorylation of 2-Thiouridine Provides Either Nucleotides or DNA Building Blocks via Photoreduction. Nat. Chem. 2019, 11 (5), 457–462. 10.1038/s41557-019-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C.; O’Flaherty D. K.; Zhou L.; Lelyveld V. S.; Szostak J. W. Inosine, but None of the 8-Oxo-Purines, Is a Plausible Component of a Primordial Version of RNA. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (52), 13318–13323. 10.1073/pnas.1814367115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A.; Couture S.; Szostak J. W. A Multisubunit Ribozyme That Is a Catalyst of and Template for Complementary Strand RNA Synthesis. Science 1991, 251 (5001), 1605–1608. 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- Ohmori R.; Saito H.; Ikawa Y.; Fujita Y.; Inoue T. Self-Replication Reactions Dependent on Tertiary Interaction Motifs in an RNA Ligase Ribozyme. J. Mol. Evol. 2011, 73 (3), 221–229. 10.1007/s00239-011-9471-2. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Kim S. C.; Ho K. H.; O’Flaherty D. K.; Giurgiu C.; Wright T. H.; Szostak J. W. Non-Enzymatic Primer Extension with Strand Displacement. eLife 2019, 8, e51888. 10.7554/eLife.51888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani A.; Bonfio C.; Johnson C. M.; Sutherland J. D. PH-Driven RNA Strand Separation under Prebiotically Plausible Conditions. Biochemistry 2018, 57 (45), 6382–6386. 10.1021/acs.biochem.8b01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianeselli A.; Mast C. B.; Braun D. Periodic Melting of Oligonucleotides by Oscillating Salt Concentrations Triggered by Microscale Water Cycles Inside Heated Rock Pores. Angew. Chem., Int. Ed. 2019, 58 (37), 13155–13160. 10.1002/anie.201907909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C.; Zhou L.; Zhang W.; O’Flaherty D. K.; Rondo-Brovetto V.; Szostak J. W. A Model for the Emergence of RNA from a Prebiotically Plausible Mixture of Ribonucleotides, Arabinonucleotides, and 2′-Deoxynucleotides. J. Am. Chem. Soc. 2020, 142 (5), 2317–2326. 10.1021/jacs.9b11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


