Abstract

One of the major challenges of bottom-up synthetic biology is rebuilding a minimal cell division machinery. From a reconstitution perspective, the animal cell division apparatus is mechanically the simplest and therefore attractive to rebuild. An actin-based ring produces contractile force to constrict the membrane. By contrast, microbes and plant cells have a cell wall, so division requires concerted membrane constriction and cell wall synthesis. Furthermore, reconstitution of the actin division machinery helps in understanding the physical and molecular mechanisms of cytokinesis in animal cells and thus our own cells. In this review, we describe the state-of-the-art research on reconstitution of minimal actin-mediated cytokinetic machineries. Based on the conceptual requirements that we obtained from the physics of the shape changes involved in cell division, we propose two major routes for building a minimal actin apparatus capable of division. Importantly, we acknowledge both the passive and active roles that the confining lipid membrane can play in synthetic cytokinesis. We conclude this review by identifying the most pressing challenges for future reconstitution work, thereby laying out a roadmap for building a synthetic cell equipped with a minimal actin division machinery.
Keywords: bottom-up reconstitution, synthetic cell, cell division, actin, myosin
1. Introduction
Bottom-up synthetic biology is an emerging field at the interface of cell biology, chemistry, and physics. Several national and international initiatives have been founded recently, which are aimed at reconstituting synthetic cells that can autonomously grow and divide.1,2 As a chassis, usually giant unilamellar vesicles (GUVs) are used, which are cell-sized (5–50 μm) containers enveloped in a lipid bilayer.3−6 One of the key functions that a synthetic cell must be able to perform in order to be considered lifelike is cytokinesis,7 a process in which a cell physically splits into two daughter cells. To reconstitute cytokinesis, various strategies are being pursued, inspired by biological strategies employed by prokaryotic, archaeal, or eukaryotic cells.7,8 These biological systems have in common that cell division is accomplished by a cytoskeletal protein machinery, often ring-shaped, that assembles at the cell equator. In microbial cells (bacteria and yeast), which have a cell wall, this protein machinery has to collaborate with a complex cell wall synthesis machinery.9,10 By contrast, animal cells lack a cell wall, and cytokinesis is entirely driven by the actin cytoskeleton. Actin-based cell division could thus be an ideal basis for engineering synthetic cell division.
Bottom-up reconstitution of actin-based cell division is interesting not only from an engineering perspective but also as a means to understand how cytokinesis works at the molecular level in animal cells. Although cytokinesis is a well-studied cellular process, surprisingly many fundamental questions about its working principles remain unanswered:11 What are the relative roles of molecular motors and other components and cellular processes in force generation? How much molecular complexity is needed to ensure that the actin cortex retains its structural integrity during cytokinesis? What are the requirements for cortex–membrane interactions to promote furrow ingression? These questions are difficult to address in cell-based studies given the enormous molecular complexity of cells combined with inbuilt redundancies and substantial variation between cytokinetic mechanisms employed by different cell types and organisms.10,12,13
In this review, we propose a roadmap toward the bottom-up reconstitution of actin-driven cytokinesis in minimal cells. For brevity, we consider only the process of furrow ingression, neglecting other aspects such as membrane abscission and chromosome and cytoplasmic segregation, which are reviewed elsewhere.14−17 Based on theoretical models of cytokinesis in animal cells, we first identify four central biophysical requirements for actin-driven furrow ingression. Next we review experimental insights obtained from recent efforts to reconstitute minimal actin systems. We also emphasize the importance of controlling the surface area of the synthetic plasma membrane to enable cell division. Finally, we propose a roadmap toward building a molecular machinery that can successfully deform a minimal cell-like container.
2. Biophysical Requirements for Making a Cell Divide
Cytokinesis in animal cells is a complicated process that involves many different molecular components (lipids and proteins) whose interactions and localization are tightly regulated. At a coarse-grained level, however, it is possible to formulate general biophysical requirements for cell division based on a consideration of the mechanical forces at play. Pioneering experimental work from the 1950s onward has demonstrated that cytokinesis is accompanied by membrane furrowing,18 cortical stiffening,19,20 and the appearance of ordered filamentous structures in the cytokinetic ring.21,22 These observations have served as input for coarse-grained theoretical and computational models that describe cytokinesis as the shape evolution of a thin, viscoelastic, and active shell around a (nearly) constant volume of cytoplasm. From the models, we can infer several key requirements that a cell, living or synthetic, must fulfill in order to successfully divide (Figure 1):
Figure 1.

Four key biophysical requirements for reconstituting synthetic cell division. For cell deformation to occur, cortical activity driven by ATP hydrolysis is required (top left), which can for example be generated by myosin activity. Regulation of the cortex thickness (bottom left) is essential for control of cortical activity and is determined by the rate of actin filament turnover versus cortical flows. For cortical activity to lead to cell deformation, the symmetry of the system needs to be broken (bottom right). Finally, to accommodate the drastic change in surface-to-volume ratio during cell division, excess membrane area needs to be generated prior to or during cytokinesis (top right).
1. Cortical Activity. The actin cortex driving cytokinesis in animal cells must be active. This means that it should include elements that hydrolyze adenosine triphosphate (ATP), an energy-carrying nucleotide, to generate contractile forces that produce cellular shape changes. The viscoelastic and active nature of the cortex can be described using the framework of active gel theory as proposed by Kruse et al.(23) This formalism is typically applied in the viscous limit24−27 because cytokinesis is slow (minutes) compared to the fluidization time scale of the actin cortex (10 s).24 This effectively implies that the cortex flows on the time scale of cytokinesis, which can result from different microscopic origins such as cross-linker or filament turnover.28,29 The molecular origins of active force production are complex and depend on molecular detail, as discussed below.
2. Cortical Thickness. Active gel theory predicts that cortex activity, at least when mediated by myosin motors, is roughly proportional to cortical thickness.24−26 To maintain cortical activity, the cortex must consequently be of a controlled thickness. Cortical thickness is regulated by a balance of actin polymerization and depolymerization (or turnover) and cortical flows: cortical flows accumulate material in the cytokinetic furrow, whereas turnover redistributes actin throughout the cell. This suggests two requirements for synthetic cell division. First, components of the cortex must be laterally mobile to be effectively redistributed by cortical flows.24,26,30 Second, actin turnover rates must be low enough to allow local actin accumulation and therefore increased contractility in the furrow region. If actin is removed too rapidly, furrow constriction slows down significantly and may be halted altogether.26 On the other hand, complete lack of filament turnover in a 2D actomyosin cortex is theoretically predicted to lead to irreversible clustering of actin, inhibiting effective stress generation.31 While active gel theory has been useful to capture various aspects of the actin cortex, recent studies show that other models might be required to describe the mitotic region, where cortex thinning due to protein alignment is accompanied by increased rather than decreased cortical tension.32 Interestingly, experimental evidence suggests that in yeast cells the persistent presence of filamentous actin, rather than turnover, is key for successful contraction of the cytokinetic ring.33 This difference might be explained by the fact that in yeast the ring is an isolated one-dimensional object, for which theoretical models predict sustained contraction at both slow and rapid turnover.34
3. Cortical Symmetry Breaking. From the 1930s onward, various models have been proposed to explain the mechanical basis of cytokinesis. The early models range from active expansion of the cell poles35 through active pushing by the mitotic spindle36 to spindle-mediated relaxation of the cell poles30,37 and finally to active constriction of the cytokinetic furrow.22,26,38,39 While details vary widely between these models, they share a key characteristic: they all posit that there must be a difference in activity between the polar and equatorial regions to drive furrow ingression. After decades of research, it is now widely accepted (reviewed in ref (40)) that the main driving factor of animal cell cytokinesis is actin-based constriction at the cleavage furrow. However, in vitro reconstitution may be the ideal tool to understand actin’s role in molecular detail and to assess the extent to which other mechanisms, such as polar expansion41 or interaction with astral microtubules (reviewed in ref (42)), also contribute.
4. Regulation of Cell Surface Area and Volume. Consistent with observations in cells,43 models have generally assumed that the cytoplasm is very weakly compressible or noncompressible.26,30 The apparent cell surface-to-volume ratio, however, changes dramatically during cytokinesis.44 It follows that the cell’s (visible) surface area must change. In theoretical works this change in surface area is generally assumed to be energetically “free”, as living cells can regulate the available membrane area through a variety of processes like blebbing45 or disassembly of caveolae and membrane trafficking.46,47 This supply of membrane on demand is probably one of the most challenging aspects to recapitulate in a reconstituted system.
3. Roadmap toward Actin-Driven Synthetic Cell Division
Cytokinesis of animal cells is a highly complex and tightly regulated process. Nevertheless, as discussed earlier, fairly minimal computational models are able to recapitulate aspects of cytokinesis, suggesting that the underlying mechanisms may be recreated with simplified molecular mechanisms. Here we propose a roadmap toward reconstituting actin-driven cell division by considering lessons from recent cell and in vitro (i.e., cell-free reconstitution) studies. Basically, there are two routes for reconstitution of actin-driven cytokinesis (see Figure 2). First, cell division can be recreated via reconstitution of an actin cortex that, upon symmetry breaking, is more contractile at the cell equator than at the poles. This route is closest to cytokinesis in mammalian cells, and we therefore name it the naturalistic route. The second route is by construction of a cytokinetic ring that anchors and contracts at the cell equator, coined the engineering route. We will first discuss the design of an actin-based machinery fit for driving cytokinesis in both scenarios, and in the next section we will consider the design of the lipid membrane envelope.
Figure 2.
Routes to actin-based synthetic cell division. There are two main routes to achieve actin-driven division of a synthetic cell: by symmetry breaking of a reconstituted actin cortex, triggered by external or biochemical cues, which leads to self-enhanced furrow constriction (the “naturalistic” route, top), or by construction of a contractile ring at the cell equator (the “engineering” route, bottom). Yellow arrowheads indicate where contractile activity is concentrated. The final fission step is outside the scope of this review.
3.1. Naturalistic Route: Building a Self-Assembling Cytokinetic Ring
During interphase, mammalian cells have a continuous actin cortex that lines the plasma membrane.48 When cells enter mitosis, the cortex is remodeled and self-assembles into a contractile ring at the cell equator. Symmetry breaking and midplane localization of the cytokinetic furrow are initiated by biochemical signaling, which includes Rho-dependent myosin phosphorylation in the furrow region.26,49 The locally enhanced activation of myosin is thought to lead to cortical flows from the poles to the equator,30,50 which further accumulate and organize contractile elements in the furrow51 that drive furrow ingression.26 Such a complex self-assembling system has not been built to date, but steps have been taken along the road (Figure 3).
Figure 3.
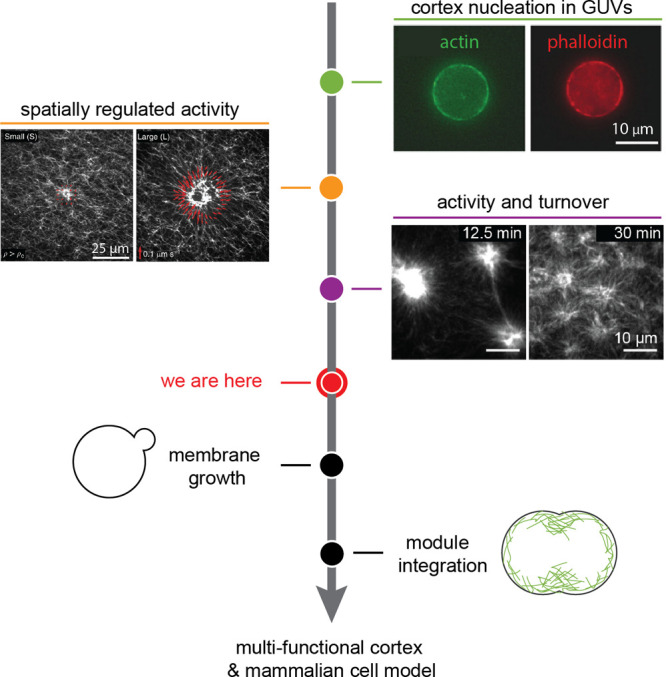
Roadmap to division with an actin cortex. Green: successful nucleation of an actin cortex inside GUVs (reproduced with permission from ref (67), copyright 2009 Biophysical Society). Filament formation of actin (green) is confirmed by colocalization of the filament-binding peptide phalloidin (red). Orange: spatiotemporal control of myosin activity by light-induced inactivation of the myosin inhibitor blebbistatin was used to generate network contraction over different length scales, from small (left) to large (right) (from ref (68), CC BY 4.0). Purple: combination of myosin activity with actin filament turnover generates sustained network contraction (from ref (63), CC BY 4.0). In the coming time, steps need to be taken to engineer membrane growth and finally to integrate the different modules inside a GUV.
3.1.1. Reconstitution of Active Actin Networks
Both cell-free experiments and theoretical models of cortex-like disordered actin networks have been used to elucidate why disordered actomyosin networks are contractile in the first place. The detailed mechanisms have been reviewed elsewhere,52−54 but they broadly comprise two scenarios. Actin filaments are semiflexible polymers with a thermal persistence length of 10–15 μm, which is on the same order as their typical contour length in in vitro studies.55 It should be noted that cortical actin filaments are much shorter in vivo, ranging from 120 to 1200 nm depending on the nucleator and cell type.56 The first contraction scenario, which is relevant for well-connected networks of long filaments, is that the anisotropic mechanical force–extension response of actin filaments causes them to buckle and break under motor-induced compressive stress.57,58 The second scenario, which is relevant for networks with short actin filaments, is that the structural polarity of actin filaments in combination with the tendency of myosin II motors to dwell at the filament plus end before detachment causes contraction via polarity sorting.25,59,60 In the actin cortex of mammalian cells there may be a combination of both mechanisms since distinct populations of short and long filaments are present there.56
Notably, the combined effect of contractile motor activity and actin turnover remains poorly explored. Theoretical models generally assume that the cytokinetic cortex does undergo actin turnover24,26,61 and have even indicated that turnover is key for sustained stress generation during furrow ingression.31 Experimentally, besides one study with a cell extract,62 only one minimal in vitro study to date has combined actin turnover and myosin activity.63 That work showed that myosin activity alone can be sufficient to induce turnover in minimal actin networks (see Figure 3, purple). Myosin-driven compaction and fragmentation of Arp2/3-nucleated actin led to the removal of actin from the network and subsequent redistribution and reincorporation of network components, creating a cortex in a dynamic steady state. Strikingly, actin turnover rates were observed to be much lower here than typical rates in cells, with actin turning over within tens of minutes rather than tens of seconds.64,65 This discrepancy is likely due to the absence of dedicated actin-severing proteins in the minimal system. More rapid turnover has been observed in vitro in volume-spanning entangled actin networks where filaments were severed by cofilin and polymerization was driven by formin.29 Combining more rapid turnover with motor activity in vitro may open a rich field of network behaviors, with complex implications for the regulation of both cortical thickness and stress propagation and relaxation.66
To build and control a system that allows actin to turn over, we can turn to the growing body of work studying the functions of various actin regulators on the single-molecule or filament level. Research into the two key nucleators of cortical actin, Arp2/3 and formins,56,69,70 has uncovered new complexities in recent years. Both the processivity and the actin filament elongation rate of different formins have been shown to be regulated by the physicochemical environment, the presence of profilin, and mechanical stress.71−73 Even more complex coregulation of formin with other barbed-end binding proteins is emerging.74 Regulation of Arp2/3 by profilin75,76 as well as by actin filament curvature77 has been known for a number of years. However, the true diversity and complexity of the various isoforms of Arp2/3, which itself is a protein complex consisting of seven protein subunits, is only just emerging.78 In addition, there are regulating factors that control cortex architecture by modulating both formin and Arp2/3 activity.70 Besides formin and Arp2/3, other actin nucleators such as the recently identified spire79 have barely been used in reconstitution experiments and may offer yet other routes toward reconstituting a minimal dynamic cortex. Actin depolymerization can equally be controlled by various factors. Disassembly of filamentous actin in vitro is usually mediated by proteins of the ADF/cofilin family.80 The activity of ADF/cofilin proteins has been shown to depend on cooperation with other proteins81,82 and on actin cross-linking.83 ADF/cofilin also facilitates debranching in actin networks nucleated by Arp2/3,80 which is furthermore sensitive to force and actin filament age.84
3.1.2. Reconstitution of Actin Cortices Inside GUVs
Controlled actin encapsulation in GUVs has proven to be a challenge. Over the years, many different methods have been explored for protein encapsulation, based on either lipid swelling85 or emulsion transfer67,86−88 (reviewed in ref (3)). Of these, methods based on emulsion transfer are currently the most successful, although the encapsulation efficiency and the ability to scale-up the number of encapsulates remain to be characterized.87 Most prior GUV studies focused on the effect of cross-link proteins and myosin motors on bulk-nucleated actin. By contrast, membrane-nucleated actin networks with turnover in GUVs remain poorly explored. Early works from the Sykes lab67,89 demonstrated that Arp2/3-nucleated cortices can be reconstituted at the inner leaflet of GUVs (Figure 3, green) and that such cortex-bearing vesicles reproduce aspects of the mechanics of living cells. More recently, Dürre et al. demonstrated that Arp2/3-nucleated cortices can induce local deformations of the GUV membrane by either polymerization forces alone or in combination with contractility induced by nonmuscle myosin-II.90 New work from the Liu lab shows that membrane-bound Arp2/3 in combination with fascin and α-actinin is sufficient to yield ringlike membrane-bound actin networks.91 Myosin-initiated contraction of these networks resulted in membrane constriction, thus getting one step closer to cell division.
More extensive work, especially with myosin-driven cortices, has been performed with stable actin filaments anchored to the membrane by streptavidin- or actin-binding membrane proteins. In such systems, cortical tension was shown to depend on the ratio of active versus passive cross-linkers,92 and excessive cortical tension was shown to cause full or partial detachment of the cortex from the membrane.92,93 Recently, Litschel et al. demonstrated the formation of actomyosin rings in GUVs.86 However, these structures were unable to deform the GUV membrane on large length scales because they slipped on the membrane. Based on our understanding of cell division, this is likely due to (at least) three missing factors: cortex turnover, symmetry breaking between the poles and equator of the synthetic cell, and a severely limited supply of extra membrane area. Symmetry breaking is likely necessary for productive and sustained membrane deformations. There are several artificial means by which symmetry breaking could be triggered in synthetic cells. Myosin activity could, for instance, be locally light-activated by targeting either the light-sensitive myosin inhibitor blebbistatin68,94,95 (see Figure 3, orange) or myosin-II directly.96 Similar approaches could be used to locally modulate the cross-link density of the actin cortex or the interaction strength of the cortex with the synthetic cell membrane. Finally, it would likely help to make GUVs shape-asymmetric, for instance by using microfluidic channels.97
Conceptually, building a dynamic actin cortex and pushing it toward self-assembly of a cytokinetic furrow is very appealing. Such a system would mimic many core attributes of the cortex of living animal cells. Furthermore, the continuous nature of such a cortex would allow it to take on a triple function: as a mechanoprotective module for the synthetic cell, as a dynamic control of cortical and membrane tension, and as a division apparatus. Its versatility sets the actin cortex apart from other cytoskeletal systems such as FtsZ.98 A lifelike actin cortex offers the opportunity to test existing theoretical models of cell division and to tease out the essential functions needed for cytokinesis in living cells. On the other hand, a dynamic actin cortex will necessarily comprise more proteins and hence a higher level of complexity than one composed of stable actin filaments. From an experimental perspective, reconstituting sustained actin turnover in combination with motor activity in particular will be challenging, as it requires fine control over both stoichiometry and activity of cytoskeletal components. In addition, timing self-assembly in connection with other cellular events, such as chromosome segregation, cell size doubling, and fission, remains a challenge to date.
3.2. Engineering Route: Building an Isolated Contractile Ring
A more engineering-type approach to synthetic cell division may also be interesting: instead of building a cortex that self-organizes into a ring, one could build an isolated ring directly (Figure 2, bottom). This would inherently fulfill the requirement for different activities in polar and equatorial contractility, as by definition the poles are not contractile in such a case. If a sufficient supply of long actin filaments throughout furrow ingression can be ensured, the need for controlled turnover may be diminished, and the complexities of such regulated filament assembly and disassembly may be avoidable. This approach will need to address three key challenges: (1) building an actin ring, (2) making it contractile, and (3) maintaining the ring’s midcenter position during contraction such that membrane invagination rather than ring slippage occurs.
3.2.1. Building an Isolated Ring
Actin filaments can be bundled and bent into ringlike structures in various ways (Figure 4, green). Most simply, ring formation can be induced by entropic effects through macromolecular crowding99 or by cross-linking with multivalent ions.100 Alternatively, proteins can be used to bend actin into rings. Septins spontaneously bend actin into ringlike structures101 and are recruited to the cytokinetic ring, where they cooperate with anillin in actin–membrane binding.102−106 Anillin itself also promotes the formation of actin rings by promoting overlap between filaments.107 Furthermore, the IQGAP fragment “curly” has recently been shown to bend actin into rings on model membranes by bending single actin filaments.108 The fact that all three of these proteins are enriched in the cytokinetic furrow109 suggests that these ring-forming capabilities may provide a cellular mechanism to promote successful cytokinesis.
Figure 4.

Roadmap toward synthetic cell division using a contractile actin ring. Green: actin rings can be formed by depletion interactions using macromolecular crowders (reproduced with permission from ref (99), copyright 2009 IOP Publishing), by proteins that combine actin binding with curvature generation such as curly (from ref (108), CC BY 4.0), or simply by confinement of actin bundles (reproduced with permission from ref (85), copyright 2015 Royal Society of Chemistry). Orange: constriction of actin rings can be executed using myosin motors (reproduced with permission from ref (110), copyright 2015 Nature Publishing Group) or actin cross-linkers like anillin (from ref (107), CC BY 4.0). Purple: the actin ring can be positioned using curvature-sensing anchors (left: septin binds preferentially to membranes of higher curvatures as shown with membrane-coated beads; from ref (111), CC BY-NC-SA 3.0) or by mechanical deformation (right: microfluidic traps deform GUVs, leading to rearrangement of FtsZ rings; from ref (112), CC BY 4.0). In the next steps toward achieving synthetic cell division, membrane growth needs to be reconstituted, and all separate modules have to be integrated.
Confinement of actin filaments inside spherical droplets or vesicles tends to promote the formation of actin rings because the confinement forces the semiflexible filaments to minimize the filament bending energy.113 Entangled or cross-linked actin networks inside emulsion droplets and inside lipid vesicles form peripheral cortex-like networks,91,114−118 while bundled actin forms one or more closed rings.85,91,110,116,117 Single rings form when the container size is smaller than the persistence length of the actin filament or bundle.91,117 Recent theoretical119 and experimental86 work has shown that ring formation can be further enhanced by introducing actin–membrane adhesion. It should be noted, however, that ring formation requires a subtle balance of filament–filament and filament–wall adhesion as well as size and stiffness of the confinement and is not trivial to precisely control experimentally.
3.2.2. Making the Isolated Ring Contractile
Contracting a once-formed actin ring can again proceed in different ways (Figure 4, orange). The classical purse-string model posits a well-organized cytokinetic ring that closes by myosin-mediated translocation of actin filaments.22,120 Although this model does not appear to hold in all cell types,121−123 recent super-resolution and electron microscopy studies showed convincing evidence that it does apply in at least some cell types.124,125 Contracting actin–myosin rings have been successfully reconstituted on supported lipid bilayers (SLBs)108 and inside water-in-oil droplets110 and GUVs.86 The efficiency of ring closure is likely determined by the orientation and arrangement of the actin filaments in the ring, which can be tuned by varying the cross-linker composition and concentration.58,117,126−128
Alternatively, ring contraction may be driven by mechanisms that do not require molecular motors. For instance, anillin was recently shown to drive actin bundle contraction even though it is a passive cross-linker.107 Contraction was attributed to an energetically driven process whereby actin filaments increase their overlap as long as energy can be gained by accumulating diffusive cross-linkers in the overlap region.107 This mechanism was enhanced when anillin was combined with actin depolymerization. Since contraction driven by passive cross-linkers does not consume energy from an external energy source such as ATP, it can only bring the system into a configuration of minimal free energy, at which point rearrangement will stop.129 Intriguingly, recent theoretical modeling130 suggests that a cross-linker that consumes ATP to unbind from actin filaments but does not actively translocate them like myosin could in principle induce contraction indefinitely. In this case, the consumption of an energy carrier breaks detailed balance in the system, and in combination with the asymmetric mechanical properties of actin, overall contractile forces can arise.
3.2.3. Keeping the Isolated Ring in Place
Although contractile actin rings have been successfully reconstituted inside GUVs, to date none of these efforts have yielded anything close to furrow-like membrane invaginations. The rings either detached or slipped along the membrane upon myosin activation,86,92,93,110 at best producing rare instances of slight membrane deformation.86 In cells, positioning of the cytokinetic ring is ensured by a complex and poorly understood interplay between the actin and microtubule cytoskeleton, local changes in lipid composition, and soluble signaling molecules.131,132 Reconstituting this interplay in GUVs seems too technically challenging to be expected in the coming years. We therefore expect that simpler, if less biological, solutions may be more promising. To the best of our knowledge, no such efforts have been reported to date. However, a few options present themselves (Figure 4, purple): curvature-sensing or -inducing scaffolding proteins such as septins111 or I-BAR-domain proteins133,134 may help in templating a furrow and inhibiting slippage of contractile actin rings. These proteins may have to be combined with more engineering-type solutions designed to deform GUVs from the outside, either by confinement in traps97,112 or by membrane-binding complexes.135−137
Building an isolated contractile actin ring in principle offers an elegant way to drive synthetic cytokinesis. The formation of such a ring requires only few components, and tuning ring contractility is certainly subtle but most likely achievable. The biggest technical challenge in this approach is to localize the ring at the equator and keep it in place during contraction in order to foster productive membrane deformation. On a more conceptual level, reconstituting isolated contractile rings likely will not bring us much insight into the mechanisms of cytokinesis in animal cells. However, it may be a valid strategy to understand mechanisms in yeast cytokinesis, in tandem with top-down work on yeast cell ghosts.138
4. Involving the Membrane
So far, we have largely ignored an important assumption in the key requirements that we set out earlier, which is that the GUV membrane and actin cortex are intrinsically coupled. However, it is far from trivial that actomyosin contraction is followed by deformation of the cellular membrane. While actomyosin networks and membranes have separately been thoroughly investigated by biophysicists, their interplay has received much less attention and presents a crucial challenge to address in the coming years.
4.1. Membrane–Cortex Anchoring
In vivo, a multitude of cytoplasmic proteins are known to be involved in actin–membrane adhesion, many of which have binding sites for both actin and plasma membrane lipids. These proteins include ERM (ezrin, radixin, moesin) proteins, myosin 1b, anillin, and septins.142−145 How these proteins cooperate in adhesion and how they are spatially organized at the membrane remains elusive. Electron microscopy and super-resolution microscopy have revealed that the distance between the filamentous actin and the plasma membrane is surprisingly large, ranging from 10 to 20 nm in the cell cortex of animal cells146 and from 60 to 160 nm in the cytokinetic ring of fission yeast.147,148 It is unclear how this large gap, which is often wider than the distance that known linker proteins span, arises. There is evidence that the actin cortex itself is stratified, with myosin filaments being restricted toward the cytoplasmic side of the cortex due to steric exclusion from the dense cortex.149 Interestingly, a recent in vitro reconstitution study showed that actin–myosin networks on SLBs spontaneously self-organize into radial actin structures (asters) with myosin at the core and layered atop to relieve steric constraints.150
Mechanical measurements on cells indicate that the cortex adheres to the membrane via a high density of weak links. With optical tweezers, one can pull membrane tubes from cells with membrane-bound beads. These tubes can easily be moved over the cell surface,151 indicating that the membrane easily zips off the cortex and quickly rebinds. Various tube-pulling experiments have shown that the force required for tube extrusion is dependent on the levels of ezrin152 and phosphatidylinositol-4,5-bisphosphate (PIP2) lipids.153 PIP2 lipids specifically interact with many actin-binding proteins, including ezrin (reviewed in ref (154)). In Schizosaccharomyces pombe cells, PIP2 depletion causes sliding of the cytokinetic ring, indicating that PIP2-dependent actin–membrane adhesion is essential for anchoring of the ring.155 Although PIP2–protein interactions are individually weak, their high density collectively causes a tight yet dynamic seam between the bilayer and the cytoskeleton.
In stark contrast to the reversible actin–membrane binding observed in vivo, in vitro reconstitution efforts have mostly relied on anchoring interactions with unphysiologically high binding affinity (Figure 5, left). Many studies used either direct coupling of biotinylated actin filaments to biotinylated lipids via streptavidin86,92,156 or indirect coupling using His-tagged actin-binding proteins coupled to Ni-NTA lipids.93,157 These bonds are virtually permanent and unbreakable.158−160 Nevertheless, at low anchor densities, actomyosin cortices anchored in this manner still detach from the membrane upon myosin activation,92,93 resulting from anchor slippage161 or pulling out of lipids.92 In two studies with high anchor density, the actomyosin cortex did remain attached to the membrane upon contraction, but it slid toward one side so that the membrane was only minimally deformed.86,92 Cortex slippage is likely due to the fluid nature of the lipid bilayer membrane. Actin and microtubule gliding assays with motor proteins anchored onto SLBs have shown that motor activity is accompanied by lipid slippage.162,163 The interplay between the dynamics of the actin cortex and the dynamics of the lipids is complicated. Adhesion to the actin cortex slows lipid diffusion,164,165 while myosin-driven actin cortex contraction can actively cluster lipids into microdomains.166−171 Altogether, it remains poorly understood what conditions are necessary for the actin cortex to remain stably anchored and cause sustained membrane deformation.
Figure 5.
Membrane engineering for synthetic cell division. A schematic overview of possibilities for membrane design is shown. Anchoring of the actin cortex (left) can be done either via filament nucleation from the membrane or via filament binding to the membrane. Binding can be done using strong permanent linkers or weaker transient links. Membrane shaping (middle) can be done by generating spontaneous curvature, for example with membrane-bound DNA nanostars139 or physiological curvature-generating proteins such as BAR proteins140 or septin.141 Otherwise, when lipids can be spatially separated, local elevation of PIP2 levels can increase cortical thickness via regulating actin nucleation and severing proteins. To provide excess area during cytokinesis (right), new membrane area can be added by fusion of small vesicles or by in situ synthesis of phospholipids. Alternatively, membrane area could be stored in reservoirs that become accessible upon furrow ingression.
Dynamic actin–membrane linkages have to date been reconstituted only on SLBs. Using ezrin recruited to the bilayer via PIP2 lipids, a dynamic actin network was created that could be remodeled by passive filament cross-linkers.172 Bead tracking microrheology showed that ezrin serves as a dynamic cross-linker for the membrane-attached actin layer, with the network stiffness being controlled by the pinning point density.173 Ezrin-anchored actin filaments could diffuse over the membrane, but longer filaments were immobilized, being pinned by a larger number of actin–membrane links.174 This indicates that collective binding with transient links can fix cytoskeletal structures in place on top of a fluid membrane. Other promising candidates for in vitro transient actin binding are septins and anillin. Septins themselves can bind to membranes and self-assemble into filamentous scaffolds.175 Membrane binding is curvature-sensitive,141,176 which renders septins interesting candidates for spatial control of actin organization in synthetic cells. In solution, septins can bind and cross-link actin filaments into curved bundles.101 This could explain the role of septins in the formation and stabilization of contractile actomyosin rings observed in vivo.101 However, the simultaneous interplay of septins with lipid membranes and actin has yet to be reconstituted in vitro. Like septins, also anillin possesses both actin-binding and membrane-binding domains. Anillin has been shown by reconstitution to be able to anchor actin filaments to lipid membranes in a RhoA-dependent manner.177 In combination with anillin’s ability to bundle and constrict actin rings via condensation forces,107 it would be interesting to explore anillin’s ability to promote synthetic cell division. Besides protein-based binding, actin filaments can also be bound to lipid membranes by electrostatic interactions that can be tuned by the choice of ions, offering an alternative route for studying and modulating transient actin–membrane binding.178
Besides actin–membrane linkers, also membrane-localized actin nucleation contributes to cortex–membrane adhesion. The main nucleators of cortical actin filaments in vivo are Arp2/3 and formin.69 In combination with membrane-bound nucleation promoting factors such as WASP, Arp2/3 is responsible for the formation of branched actin filament arrays, whereas formins nucleate linear filaments. Actin nucleation has been successfully reconstituted in vitro both with formins, often for simplicity with constitutively active mutants,179 and with Arp2/3, often activated by WASP fragments such as VCA.67,180,181 Actin turnover can be introduced by addition of severing proteins such as ADF/cofilin.182
It is unknown how filament nucleation in conjunction with actin–membrane anchoring by dynamic linker proteins such as ezrin will influence the ensemble mechanics of the actin–membrane composite. Tailoring actin-based division machineries toward synthetic cell division will require careful tuning of the cortex itself, the anchoring strategy, and also the membrane physicochemical properties.
4.2. Membrane Engineering
The membrane should not be considered just a passive player in cytokinesis. In contrast, membrane properties can be exploited to aid cytokinesis, for example by shaping the contractile network (Figure 5, middle). In vivo, the plasma membrane in the cleavage furrow has a distinct lipid composition that is thought to contribute to cytokinesis by biochemical signaling and perhaps also by induction of spontaneous curvature.183 Elevated PIP2 levels at the cleavage furrow probably contribute to furrow ingression by recruiting anillin, septins, and ERM proteins.184 Furthermore, PIP2-mediated signaling promotes the formation and maintenance of a stable actin cortex by promoting actin nucleation and slowing actin filament severing via actin regulatory proteins.185 Other membrane components such as gangliosides and cholesterol also accumulate in the cleavage furrow, where they regulate and bind the cortex.184 In addition, the distribution of phosphatidylethanolamine (PE) lipids over the two bilayer leaflets changes significantly during cell division: while PE lipids reside in the inner leaflet during interphase, they are exposed in the outer leaflet of the cleavage furrow during cytokinesis.186 This asymmetric distribution of PE lipids has been shown to be important for the disassembly of the contractile ring after cytokinesis.186 It is possible that the specialized lipid composition of the cleavage furrow also directly affects cytokinesis by changing the mechanical properties of the membrane, but this remains to be shown.
For engineering artificial cell division, it could be useful to exploit known mechanical effects of lipids. An important characteristic of lipid bilayers is that asymmetries between the two membrane leaflets give rise to membrane spontaneous curvature. Asymmetries can be generated in many different ways (reviewed in ref (187)), such as by different lipid compositions or different numbers of lipids in the two leaflets,188 binding of proteins to one leaflet,137 insertion of membrane-anchored DNA oligomers into one leaflet,139 or different solutes on the two sides of the membrane.189 In the context of actomyosin-based synthetic cell division, spontaneous curvature effects could be exploited for spatial control and symmetry breaking. Binding of proteins to the outer leaflet of vesicles can be used to make vesicles dumbbell-shaped and to constrict and even split the neck.137 Generation of negative membrane curvature could be used to locally recruit septins, which selectively bind to membrane areas with micrometric curvature.111,141 In addition, membrane-binding proteins that not only sense but also generate curvature could be used, such as BAR-domain proteins.190 I-BAR proteins were shown to directly bind to actin in fission yeast134 and are therefore interesting candidates for promoting actomyosin-driven membrane invagination. Interestingly, I-BAR domain proteins promote ezrin enrichment in negatively curved membrane protrusions,133 providing further prospects for boosting membrane invagination in vitro.
4.3. Addition of New Membrane Area
To create two daughter cells from a single mother cell, assuming spherical geometry, the cell surface area has to increase by 28%.26,44In vivo, this extra membrane area is delivered to the cleavage furrow by targeted endosomal transport.191 This mechanism not only leads to a local area increase but also allows fast and localized delivery of specific lipids and regulatory proteins (reviewed in ref (192)). For reconstitution of cell division, various strategies can be followed to increase the membrane area (Figure 5, right). First, GUV membranes can be grown by external addition of small unilamellar vesicles (SUVs), which can be forced to fuse with the GUV using fusogenic peptides, DNA, or charge-based interactions.193−196 Second, lipid membranes can be grown by in situ synthesis of lipids from their precursors. Examples are non-enzymatic reactions from synthetic reactive precursors197 or enzyme-catalyzed biosynthesis using either purified proteins198 or in vitro transcription–translation.199 Although there is evidence that mammalian cells do not use area reservoirs, such as microvilli, to supply extra membrane area for division,200,201 this mechanism could be exploited to engineer division in synthetic cells. Asymmetries between the two leaflets of the bilayer generated by different means (see the preceding section) can be used to store excess area in membrane tubes and buds.137,189,202,203 Low forces suffice to access these reservoirs.189,203 To achieve synthetic cell division, it will be important to match the timing of membrane areal growth with the timing of actin-driven constriction. To achieve multiple cycles of division, it will moreover be important to build in a mechanism to maintain lipid homeostasis.
5. Challenges Ahead
In the past decades, our knowledge of cell division and its molecular actors has increased tremendously. To understand the physical mechanisms governing actomyosin-driven cell division, focus is put increasingly on bottom-up reconstitution experiments. Bulk and SLB experiments have helped us to understand the mechanics of active actomyosin networks in two and three dimensions. However, translating these insights to the process of cell division is not trivial. To summarize, we list here the critical challenges that need to be overcome before we can reconstitute a minimal version of actin-driven cell division.
First, we need to understand how actin network contraction is sustained to drive division all the way. This will require myosin activity working in concert with actin turnover. While activity and turnover have been studied to great extents individually, we still have minimal understanding of how they together govern actin network mechanics and contractility. Not only is this a challenging system to understand from a physical and biological perspective, but it is also difficult to recapitulate from an experimental perspective, as it involves a large number of components whose concentration and activity need to be tightly controlled. More in vitro work in this direction, in both two and three dimensions, will be essential to explore the parameter space.
Second, it remains elusive how the actomyosin network should be anchored onto the membrane in order to achieve membrane deformations. A multitude of anchoring strategies have been developed and investigated, but only minimally in combination with a deformable membrane. Combined with our limited understanding of cortex–membrane molecular organization in vivo, this might prove to be one of the most important challenges. Future studies need to focus on understanding the influence of linker density and strength as well as membrane composition and organization. In addition, the individual contributions of actin–membrane linkers and membrane-bound filament nucleators need to be delineated. After successful deformation, size stability between the two forming daughter cells, driven by the Laplace pressure, needs to be ensured.204
Third, attention must be paid to the supply of extra membrane area during constriction. Additional area can be present in membrane reservoirs, synthesized, or added by fusion of small vesicles. However, none of these approaches have to our knowledge been co-reconstituted with actin-driven contraction and resulting membrane deformation.
Fourth, to date there has been only a minimal body of work on contractile actomyosin networks in GUVs. Confining the system in GUVs requires that all of the components be encapsulated at the right concentrations and stoichiometric ratios while preserving functionality. Although there are numerous GUV formation techniques, they have been minimally characterized for their potential to encapsulate complex mixtures of biochemically active components. More work in this direction is crucial to perform controlled reconstitution in GUVs and also to be able to extrapolate findings from bulk and SLB experiments to vesicle systems.
Fifth, spatial and temporal control of the components and their activity is crucial. In the short term, some of the involved challenges may be bypassed by taking a semiautonomous approach to synthetic cell division. For example, optogenetics, external mechanical or chemical cues, and fusion-based delivery of components with small vesicles provide handles to control the system even after encapsulation of the components inside GUVs. However, if the goal is to create a synthetic cell that divides fully autonomously, reconstitution will be more complicated, requiring for example feedback loops, signaling molecules, and internal clocks.
As a concluding remark, we note that the most pressing challenges to achieve in vitro actin-driven cell division require integration of modules. Only when actomyosin studies meet membrane biophysics, when myosin motor activity is combined with actin turnover, and when protein biochemistry becomes integrated in GUV formation can we start thinking about reconstituting cell division. In the coming years, perspectives from experimental work, theoretical studies, and simulations need to be combined to guide future work with the ultimate goal of developing a full understanding of actin-driven synthetic cell division.
Acknowledgments
We thank Ilina Bareja, Gerard Castro-Linares, and Fred MacKintosh for useful discussions about actin cross-linkers. We acknowledge financial support from The Netherlands Organization of Scientific Research (NWO/OCW) Gravitation Program Building a Synthetic Cell (BaSyC) (024.003.019).
Author Contributions
† L.B. and L.v.B. contributed equally. All of the authors together designed and wrote the manuscript.
The authors declare no competing financial interest.
References
- Staufer O.; et al. Building a community to engineer synthetic cells and organelles from the bottom-up. eLife 2021, 10, e73556 10.7554/eLife.73556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmon C.; Sorenson C.; Winikoff M.; Adamala K. P. Build-a-Cell: Engineering a Synthetic Cell Community. Life 2021, 11, 1176. 10.3390/life11111176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulla Y.; Aufderhorst-Roberts A.; Koenderink G. H. Shaping up synthetic cells. Phys. Biol. 2018, 15, 041001. 10.1088/1478-3975/aab923. [DOI] [PubMed] [Google Scholar]
- Spoelstra W. K.; Deshpande S.; Dekker C. Tailoring the appearance: what will synthetic cells look like?. Curr. Opin. Biotechnol. 2018, 51, 47–56. 10.1016/j.copbio.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Gaut N. J.; Adamala K. P. Reconstituting Natural Cell Elements in Synthetic Cells. Adv. Biol. 2021, 5, 2000188. 10.1002/adbi.202000188. [DOI] [PubMed] [Google Scholar]
- Litschel T.; Schwille P. Protein Reconstitution Inside Giant Unilamellar Vesicles. Annu. Rev. Biophys. 2021, 50, 525–548. 10.1146/annurev-biophys-100620-114132. [DOI] [PubMed] [Google Scholar]
- Olivi L.; Berger M.; Creyghton R. N.; De Franceschi N.; Dekker C.; Mulder B. M.; Claassens N. J.; ten Wolde P. R.; van der Oost J. Towards a synthetic cell cycle. Nat. Commun. 2021, 12, 4531. 10.1038/s41467-021-24772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S.; Ganzinger K. A.; Franquelim H. G.; Schwille P. Synthetic cell division via membrane-transforming molecular assemblies. BMC Biol. 2019, 17, 43. 10.1186/s12915-019-0665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahone C. R.; Goley E. D. Bacterial cell division at a glance. J. Cell Sci. 2020, 133, jcs237057. 10.1242/jcs.237057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Okada H.; Bi E. Comparative Analysis of the Roles of Non-muscle Myosin-IIs in Cytokinesis in Budding Yeast, Fission Yeast, and Mammalian Cells. Front. Cell Dev. Biol. 2020, 8, 593400. 10.3389/fcell.2020.593400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Nine unanswered questions about cytokinesis. J. Cell Biol. 2017, 216, 3007–3016. 10.1083/jcb.201612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes D. B.; Dawes A.; Liu J.; Nickaeen M.; Strychalski W.; Maddox A. S. Unite to divide – How models and biological experimentation have come together to reveal mechanisms of cytokinesis. J. Cell Sci. 2018, 131, jcs203570. 10.1242/jcs.203570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite J.; Osorio D. S.; Sobral A. F.; Silva A. M.; Carvalho A. X. Network Contractility During Cytokinesis-from Molecular to Global Views. Biomolecules 2019, 9, 194. 10.3390/biom9050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M. L.; Berlin I.; Neefjes J. On the move: organelle dynamics during mitosis. Trends Cell Biol. 2015, 25, 112–124. 10.1016/j.tcb.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Addi C.; Bai J.; Echard A. Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr. Opin. Cell Biol. 2018, 50, 27–34. 10.1016/j.ceb.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Horváth P.; Müller-Reichert T. A Structural View on ESCRT-Mediated Abscission. Front. Cell. Dev. Biol. 2020, 8, 586880. 10.3389/fcell.2020.586880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjur-Dietrich M. I.; Kelleher C. P.; Needleman D. J. Mechanical Mechanisms of Chromosome Segregation. Cells 2021, 10, 465. 10.3390/cells10020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts H. S. Mechanisms of Cytokinesis: A Critical Review. Q. Rev. Biol. 1961, 36, 155–177. 10.1086/403395. [DOI] [PubMed] [Google Scholar]
- Mitchison J. M.; Swann M. M. The mechanical properties of the cell surface: III. The sea-urchin egg from fertilization to cleavage. J. Exp. Biol. 1955, 32, 734–750. 10.1242/jeb.32.4.734. [DOI] [Google Scholar]
- Wolpert L. The mechanical properties of the membrane of the sea urchin egg during cleavage. Exp. Cell Res. 1966, 41, 385–396. 10.1016/S0014-4827(66)80146-5. [DOI] [PubMed] [Google Scholar]
- Arnold J. M. Cleavage furrow formation in a teoloecithal egg (Loligo Pealii) I. Filaments in Early Furrow Formation. J. Cell Biol. 1969, 41, 894–904. 10.1083/jcb.41.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. E. Preliminary notes: Cytokinesis: filaments in the cleavage furrow. Exp. Cell Res. 1968, 53, 272–316. 10.1016/0014-4827(68)90373-X. [DOI] [PubMed] [Google Scholar]
- Kruse K.; Joanny J. F.; Jülicher F.; Prost J.; Sekimoto K. Generic theory of active polar gels: A paradigm for cytoskeletal dynamics. Eur. Phys. J. E 2005, 16, 5–16. 10.1140/epje/e2005-00002-5. [DOI] [PubMed] [Google Scholar]
- Salbreux G.; Prost J.; Joanny J. F. Hydrodynamics of cellular cortical flows and the formation of contractile rings. Phys. Rev. Lett. 2009, 103, 058102. 10.1103/PhysRevLett.103.058102. [DOI] [PubMed] [Google Scholar]
- Zumdieck A.; Lagomarsino M. C.; Tanase C.; Kruse K.; Mulder B.; Dogterom M.; Julicher F. Continuum description of the cytoskeleton: Ring formation in the cell cortex. Phys. Rev. Lett. 2005, 95, 258103. 10.1103/PhysRevLett.95.258103. [DOI] [PubMed] [Google Scholar]
- Turlier H.; Audoly B.; Prost J.; Joanny J.-F. F. Furrow constriction in animal cell cytokinesis. Biophys. J. 2014, 106, 114–123. 10.1016/j.bpj.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A.-C.; Staniscia F.; Erzberger A.; Salbreux G.; Grill S. W. Cortical flow aligns actin filaments to form a furrow. eLife 2016, 5, e17807 10.7554/eLife.17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Friedrich E.; Toyoda Y.; Cattin C. J.; Müller D. J.; Hyman A. A.; Jülicher F. Rheology of the Active Cell Cortex in Mitosis. Biophys. J. 2016, 111, 589–600. 10.1016/j.bpj.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall P. M.; MacKintosh F. C.; Kovar D. R.; Gardel M. L. Cofilin drives rapid turnover and fluidization of entangled F-actin. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 12629–12637. 10.1073/pnas.1818808116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G.; Borisy G. G. On the Mechanisms of Cytokinesis in Animal Cells. J. Theor. Biol. 1983, 101, 289–316. 10.1016/0022-5193(83)90342-9. [DOI] [PubMed] [Google Scholar]
- Hiraiwa T.; Salbreux G. Role of Turnover in Active Stress Generation in a Filament Network. Phys. Rev. Lett. 2016, 116, 188101. 10.1103/PhysRevLett.116.188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P.; Clark A. G.; Smith M. B.; Cassani D. A. D.; Dierkes K.; Ragab A.; Roux P. P.; Charras G.; Salbreux G.; Paluch E. K. Actin cortex architecture regulates cell surface tension. Nat. Cell Biol. 2017, 19, 689–697. 10.1038/ncb3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew T. G.; Huang J.; Palani S.; Sommese R.; Kamnev A.; Hatano T.; Gu Y.; Oliferenko S.; Sivaramakrishnan S.; Balasubramanian M. K. Actin turnover maintains actin filament homeostasis during cytokinetic ring contraction. J. Cell Biol. 2017, 216, 2657–2667. 10.1083/jcb.201701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy B.; Thiyagarajan S. Mechanisms of contractile ring tension production and constriction. Biophys. Rev. 2018, 10, 1667–1681. 10.1007/s12551-018-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann M. M.; Mitchison J. M. The mechanism of cleavage in animal cells. Biol. Rev. 1958, 33, 103–135. 10.1111/j.1469-185X.1958.tb01409.x. [DOI] [Google Scholar]
- Dan J. C. On the Mechanism of Astral Cleavage. Phys. Zool. 1948, 21, 191–218. 10.1086/physzool.21.3.30151997. [DOI] [PubMed] [Google Scholar]
- Wolpert L. The Mechanics and Mechanism of Cleavage. Int. Rev. Cytol. 1960, 10, 163–216. [Google Scholar]
- Marsland D. The mechanisms of cell division, temperature-pressure experiments on the cleaving eggs of arbacia punctulata. J. Cell. Comp. Physiol. 1950, 36, 205–227. 10.1002/jcp.1030360207. [DOI] [PubMed] [Google Scholar]
- Yoneda M.; Dan K. Tension at the surface of the dividing sea-urchin egg. J. Exp. Biol. 1972, 57, 575–587. 10.1242/jeb.57.3.575. [DOI] [PubMed] [Google Scholar]
- Green R. A.; Paluch E.; Oegema K. Cytokinesis in Animal Cells. Annu. Rev. Cell Dev. Biol. 2012, 28, 29–58. 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Gudejko H. F. M.; Alford L. M.; Burgess D. R. Polar expansion during cytokinesis. Cytoskeleton 2012, 69, 1000–1009. 10.1002/cm.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-L. The Mechanism of Cytokinesis: Reconsideration and Reconciliation. Cell Struct. Funct. 2001, 26, 633–638. 10.1247/csf.26.633. [DOI] [PubMed] [Google Scholar]
- Hiramoto Y. A quantitative description of protoplasmic movement during cleavage in the sea-urchin egg. J. Exp. Biol. 1958, 35, 407–425. 10.1242/jeb.35.2.407. [DOI] [Google Scholar]
- Frey F.; Idema T. More than just a barrier: using physical models to couple membrane shape to cell function. Soft Matter 2021, 17, 3533–3549. 10.1039/D0SM01758B. [DOI] [PubMed] [Google Scholar]
- Sedzinski J.; Biro M.; Oswald A.; Tinevez J. Y.; Salbreux G.; Paluch E. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature 2011, 476, 462–468. 10.1038/nature10286. [DOI] [PubMed] [Google Scholar]
- Sinha B.; et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 2011, 144, 402–413. 10.1016/j.cell.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R.; Riggs B.; Sullivan W. Membrane traffic: A driving force in cytokinesis. Trends Cell Biol. 2005, 15, 92–101. 10.1016/j.tcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Chugh P.; Paluch E. K. The actin cortex at a glance. J. Cell Sci. 2018, 131, jcs186254. 10.1242/jcs.186254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano S.; Hamao K.; Hosoya H. Direct evidence for roles of phosphorylated regulatory light chain of myosin II in furrow ingression during cytokinesis in HeLa cells. Genes Cells 2009, 14, 555–568. 10.1111/j.1365-2443.2009.01288.x. [DOI] [PubMed] [Google Scholar]
- Bray D.; White J. G. Cortical flow in animal cells. Science 1988, 239, 883–888. 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- Najafabadi F. R.; Leaver M.; Grill S. W. Orchestrating Non-muscle myosin II filament assembly at the onset of cytokinesis. Mol. Biol. Cell 2022, 33, ar74. 10.1091/mbc.E21-12-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink G. H.; Paluch E. K. Architecture shapes contractility in actomyosin networks. Curr. Opin. Cell Biol. 2018, 50, 79–85. 10.1016/j.ceb.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Mendes Pinto I.; Rubinstein B. Y.; Li R. Force to divide: Structural and mechanical requirements for actomyosin ring contraction. Biophys. J. 2013, 105, 547–554. 10.1016/j.bpj.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell M.; Oakes P. W.; Lenz M.; Gardel M. L. Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 2015, 16, 486–498. 10.1038/nrm4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.; Bradley M. J.; McCullough B. R.; Pierre A.; Grintsevich E. E.; Reisler E.; De La Cruz E. M. Identification of cation-binding sites on actin that drive polymerization and modulate bending stiffness. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 16923–16927. 10.1073/pnas.1211078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M.; Erlenkämper C.; Moeendarbary E.; Charras G.; Kruse K. Actin kinetics shapes cortical network structure and mechanics. Sci. Adv. 2016, 2, e1501337 10.1126/sciadv.1501337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell M. P.; Gardel M. L. F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 20820–20825. 10.1073/pnas.1214753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz M.; Thoresen T.; Gardel M. L.; Dinner A. R. Contractile units in disordered actomyosin bundles arise from f-actin buckling. Phys. Rev. Lett. 2012, 108, 238107. 10.1103/PhysRevLett.108.238107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse K.; Jülicher F. Actively Contracting Bundles of Polar Filaments. Phys. Rev. Lett. 2000, 85, 1778–1781. 10.1103/PhysRevLett.85.1778. [DOI] [PubMed] [Google Scholar]
- Wollrab V.; Belmonte J. M.; Baldauf L.; Leptin M.; Nédeléc F.; Koenderink G. H. Polarity sorting drives remodeling of actin-myosin networks. J. Cell Sci. 2019, 132, jcs219717. 10.1242/jcs.219717. [DOI] [PubMed] [Google Scholar]
- Berthoumieux H.; Maître J. L.; Heisenberg C. P.; Paluch E. K.; Jülicher F.; Salbreux G. Active elastic thin shell theory for cellular deformations. New J. Phys. 2014, 16, 065005. 10.1088/1367-2630/16/6/065005. [DOI] [Google Scholar]
- Malik-Garbi M.; Ierushalmi N.; Jansen S.; Abu-Shah E.; Goode B. L.; Mogilner A.; Keren K. Scaling behaviour in steady-state contracting actomyosin networks. Nat. Phys. 2019, 15, 509–516. 10.1038/s41567-018-0413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonal; Ganzinger K. A.; Vogel S. K.; Mücksch J.; Blumhardt P.; Schwille P. Myosin-II activity generates a dynamic steady state with continuous actin turnover in a minimal actin cortex. J. Cell Sci. 2019, 132, jcs219899. 10.1242/jcs.219899. [DOI] [PubMed] [Google Scholar]
- Fritzsche M.; Lewalle A.; Duke T.; Kruse K.; Charras G. Analysis of turnover dynamics of the submembranous actin cortex. Mol. Biol. Cell 2013, 24, 757–767. 10.1091/mbc.e12-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin F. B.; McFadden W. M.; Yao B.; Munro E. M. Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat. Methods 2014, 11, 677–682. 10.1038/nmeth.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzash S.; McCall P. M.; Feng J.; Gardel M. L.; MacKintosh F. C. Stress relaxation in F-actin solutions by severing. Soft Matter 2019, 15, 6300–6307. 10.1039/C9SM01263J. [DOI] [PubMed] [Google Scholar]
- Pontani L. L.; Van Der Gucht J.; Salbreux G.; Heuvingh J.; Joanny J. F.; Sykes C. Reconstitution of an actin cortex inside a liposome. Biophys. J. 2009, 96, 192–198. 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmeier I.; Banerjee S.; Oakes P. W.; Jung W.; Kim T.; Murrell M. P. Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nat. Commun. 2016, 7, 12615. 10.1038/ncomms12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovellan M.; et al. Cellular control of cortical actin nucleation. Curr. Biol. 2014, 24, 1628–1635. 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; et al. SPIN90 associates with mDia1 and the Arp2/3 complex to regulate cortical actin organization. Nat. Cell Biol. 2020, 22, 803–814. 10.1038/s41556-020-0531-y. [DOI] [PubMed] [Google Scholar]
- Zimmermann D.; Homa K. E.; Hocky G. M.; Pollard L. W.; De La Cruz E. M.; Voth G. A.; Trybus K. M.; Kovar D. R. Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat. Commun. 2017, 8, 703. 10.1038/s41467-017-00445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Kerleau M.; Suzuki E. L.; Wioland H.; Jouet S.; Guichard B.; Lenz M.; Romet-Lemonne G.; Jegou A. Modulation of formin processivity by profilin and mechanical tension. eLife 2018, 7, e34176 10.7554/eLife.34176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C.; Carroll R. T.; Burke T. A.; Christensen J. R.; Bestul A. J.; Sees J. A.; James M. L.; Sirotkin V.; Kovar D. R. Profilin regulates F-Actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 2015, 32, 43–53. 10.1016/j.devcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S.; Kerleau M.; Kühn S.; Pernier J.; Romet-Lemonne G.; Jégou A.; Carlier M. F. Formin and capping protein together embrace the actin filament in a ménage à trois. Nat. Commun. 2015, 6, 8730. 10.1038/ncomms9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R. D.; Kelleher J. F.; Xu J.; Pollard T. D. Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol. Biol. Cell 1998, 9, 841–852. 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L.; Pollard T. D.; Mullins R. D. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 2000, 10, 1273–1282. 10.1016/S0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Risca V. I.; Wang E. B.; Chaudhuri O.; Chia J. J.; Geissler P. L.; Fletcher D. A. Actin filament curvature biases branching direction. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 2913–2918. 10.1073/pnas.1114292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abella J. V. G.; Galloni C.; Pernier J.; Barry D. J.; Kjær S.; Carlier M. F.; Way M. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 2016, 18, 76–86. 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- Quinlan M. E.; Heuser J. E.; Kerkhoff E.; Dyche Mullins R. Drosophila Spire is an actin nucleation factor. Nature 2005, 433, 382–388. 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Chan C.; Beltzner C. C.; Pollard T. D. Cofilin Dissociates Arp2/3 Complex and Branches from Actin Filaments. Curr. Biol. 2009, 19, 537–545. 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotila T.; Wioland H.; Enkavi G.; Kogan K.; Vattulainen I.; Jégou A.; Romet-Lemonne G.; Lappalainen P. Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 2019, 10, 5320. 10.1038/s41467-019-13213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S.; Collins A.; Golden L.; Sokolova O.; Goode B. L. Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J. Biol. Chem. 2014, 289, 30732–30742. 10.1074/jbc.M114.601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wioland H.; Jegou A.; Romet-Lemonne G. Torsional stress generated by ADF/cofilin on cross-linked actin filaments boosts their severing. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 2595–2602. 10.1073/pnas.1812053116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit N. G.; Cao W.; Bibeau J.; Johnson-Chavarria E. M.; Taylor E. W.; Pollard T. D.; De La Cruz E. M. Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 13519–13528. 10.1073/pnas.1911183117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.-C.; Koenderink G. Shape control of lipid bilayer membranes by confined actin bundles. Soft Matter 2015, 11, 8834–8847. 10.1039/C5SM01583A. [DOI] [PubMed] [Google Scholar]
- Litschel T.; Kelley C. F.; Holz D.; Adeli Koudehi M.; Vogel S. K.; Burbaum L.; Mizuno N.; Vavylonis D.; Schwille P. Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 2021, 12, 2254. 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Cauter L.; Fanalista F.; van Buren L.; De Franceschi N.; Godino E.; Bouw S.; Danelon C.; Dekker C.; Koenderink G. H.; Ganzinger K. A. Optimized cDICE for Efficient Reconstitution of Biological Systems in Giant Unilamellar Vesicles. ACS Synth. Biol. 2021, 10, 1690–1702. 10.1021/acssynbio.1c00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Wubshet N.; Litschel T.; Schwille P.; Liu A. P. Rapid Encapsulation of Reconstituted Cytoskeleton inside Giant Unilamellar Vesicles. J. Vis. Exp. 2021, 177, e63332 10.3791/63332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell M.; Pontani L. L.; Guevorkian K.; Cuvelier D.; Nassoy P.; Sykes C. Spreading dynamics of biomimetic actin cortices. Biophys. J. 2011, 100, 1400–1409. 10.1016/j.bpj.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürre K.; Keber F. C.; Bleicher P.; Brauns F.; Cyron C. J.; Faix J.; Bausch A. R. Capping protein-controlled actin polymerization shapes lipid membranes. Nat. Commun. 2018, 9, 1630. 10.1038/s41467-018-03918-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Moghimianavval H.; Liu A. P. Encapsulated actomyosin patterns drive cell-like membrane shape changes. iScience 2022, 25, 104236. 10.1016/j.isci.2022.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho K.; Tsai F.-C. C.; Lees E.; Voituriez R.; Koenderink G. H.; Sykes C. Cell-sized liposomes reveal how actomyosin cortical tension drives shape change. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 16456–16461. 10.1073/pnas.1221524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau E.; Schneider J. A. M.; Keber F. C.; Pelzl C.; Massiera G.; Salbreux G.; Bausch A. R. Shape remodeling and blebbing of active cytoskeletal vesicles. Sci. Adv. 2016, 2, e1500465 10.1126/sciadv.1500465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T.; Limouze J.; Combs C. A.; Straight A. F.; Sellers J. R. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry 2005, 44, 584–588. 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- Schuppler M.; Keber F. C.; Kröger M.; Bausch A. R. Boundaries steer the contraction of active gels. Nat. Commun. 2016, 7, 13120. 10.1038/ncomms13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K.; Miura H.; Ishida M.; Mii Y.; Kinoshita N.; Takada S.; Ueno N.; Sawai S.; Kondo Y.; Aoki K. Optogenetic relaxation of actomyosin contractility uncovers mechanistic roles of cortical tension during cytokinesis. Nat. Commun. 2021, 12, 7145. 10.1038/s41467-021-27458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanalista F.; Birnie A.; Maan R.; Burla F.; Charles K.; Pawlik G.; Deshpande S.; Koenderink G. H.; Dogterom M.; Dekker C. Shape and Size Control of Artificial Cells for Bottom-Up Biology. ACS Nano 2019, 13, 5439–5450. 10.1021/acsnano.9b00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows J. M.; Goley E. D. FtsZ dynamics in bacterial division: What, how, and why?. Curr. Opin. Cell Biol. 2021, 68, 163–172. 10.1016/j.ceb.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. W. C.; Prasad A.; Dogic Z. Condensation of isolated Semi-flexible filaments driven by depletion interactions. Europhys. Lett. 2009, 87, 48006. 10.1209/0295-5075/87/48006. [DOI] [Google Scholar]
- Tang J. X.; Käs J. A.; Shah J. V.; Janmey P. A. Counterion-induced actin ring formation. Eur. Biophys. J. 2001, 30, 477–484. 10.1007/s002490100178. [DOI] [PubMed] [Google Scholar]
- Mavrakis M.; Azou-Gros Y.; Tsai F.-C. C.; Alvarado J.; Bertin A.; Iv F.; Kress A.; Brasselet S.; Koenderink G. H.; Lecuit T. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat. Cell Biol. 2014, 16, 322–334. 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- Kinoshita M.; Field C. M.; Coughlin M. L.; Straight A. F.; Mitchison T. J. Self- and actin-templated assembly of mammalian septins. Dev. Cell 2002, 3, 791–802. 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- Field C. M.; Coughlin M.; Doberstein S.; Marty T.; Sullivan W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 2005, 132, 2849–2860. 10.1242/dev.01843. [DOI] [PubMed] [Google Scholar]
- Piekny A. J.; Glotzer M. Anillin Is a Scaffold Protein That Links RhoA, Actin, and Myosin during Cytokinesis. Curr. Biol. 2008, 18, 30–36. 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Piekny A. J.; Maddox A. S. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 2010, 21, 881–891. 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Garno C.; Irons Z. H.; Gamache C. M.; McKim Q.; Reyes G.; Wu X.; Shuster C. B.; Henson J. H. Building the cytokinetic contractile ring in an early embryo: Initiation as clusters of myosin II, anillin and septin, and visualization of a septin filament network. PLoS One 2021, 16, e0252845 10.1371/journal.pone.0252845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučera O.; Janda D.; Siahaan V.; Dijkstra S. H.; Pilátová E.; Zatecka E.; Diez S.; Braun M.; Lansky Z. Anillin propels myosin-independent constriction of actin rings. Nat. Commun. 2021, 12, 4595. 10.1038/s41467-021-24474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S.; Ghosh S.; Ivorra-Molla E.; Clarke S.; Suchenko A.; Balasubramanian M. K.; Köster D. V. Calponin-homology domain mediated bending of membrane-associated actin filaments. eLife 2021, 10, e61078 10.7554/eLife.61078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biro M.; Romeo Y.; Kroschwald S.; Bovellan M.; Boden A.; Tcherkezian J.; Roux P. P.; Charras G.; Paluch E. K. Cell cortex composition and homeostasis resolved by integrating proteomics and quantitative imaging. Cytoskeleton 2013, 70, 741–754. 10.1002/cm.21142. [DOI] [PubMed] [Google Scholar]
- Miyazaki M.; Chiba M.; Eguchi H.; Ohki T.; Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat. Cell Biol. 2015, 17, 480–489. 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- Bridges A. A.; Jentzsch M. S.; Oakes P. W.; Occhipinti P.; Gladfelter A. S. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 2016, 213, 23–32. 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzinger K. A.; Merino-Salomón A.; García-Soriano D. A.; Butterfield A. N.; Litschel T.; Siedler F.; Schwille P. FtsZ Reorganization Facilitates Deformation of Giant Vesicles in Microfluidic Traps. Angew. Chem. 2020, 132, 21556–21560. 10.1002/ange.202001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G.; Thirumalai D. Semiflexible chains in confined spaces. Phys. Rev. E 2009, 79, 011924. 10.1103/PhysRevE.79.011924. [DOI] [PubMed] [Google Scholar]
- Soares E Silva M.; Alvarado J.; Nguyen J.; Georgoulia N.; Mulder B. M.; Koenderink G. H. Self-organized patterns of actin filaments in cell-sized confinement. Soft Matter 2011, 7, 10631–10641. 10.1039/c1sm06060k. [DOI] [Google Scholar]
- Claessens M. M. A. E.; Tharmann R.; Kroy K.; Bausch A. R. Microstructure and viscoelasticity of confined semiflexible polymer networks. Nat. Phys. 2006, 2, 186–189. 10.1038/nphys241. [DOI] [Google Scholar]
- Limozin L.; Sackmann E. Polymorphism of Cross-Linked Actin Networks in Giant Vesicles. Phys. Rev. Lett. 2002, 89, 168103. 10.1103/PhysRevLett.89.168103. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y.; Redford S. A.; Lorpaiboon C.; Groaz A.; Moghimianavval H.; Litschel T.; Schwille P.; Hocky G. M.; Dinner A. R.; Liu A. P. Actin crosslinker competition and sorting drive emergent GUV size-dependent actin network architecture. Commun. Biol. 2021, 4, 1136. 10.1038/s42003-021-02653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer E.; Vache M.; Kliesch T. T.; Janshoff A. Mechanical response of adherent giant liposomes to indentation with a conical AFM-tip. Soft Matter 2015, 11, 4487–4495. 10.1039/C5SM00191A. [DOI] [PubMed] [Google Scholar]
- Adeli Koudehi M.; Rutkowski D. M.; Vavylonis D. Organization of associating or crosslinked actin filaments in confinement. Cytoskeleton 2019, 76, 532–548. 10.1002/cm.21565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. E. The contractile ring II. Determining its brief existence, volumetric changes, and vital role in cleaving arbacia eggs. J. Cell Biol. 1972, 53, 419–434. 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Kovaćs M.; Conti M. A.; Wang A.; Zhang Y.; Sellers J. R.; Adelstein R. S. Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 4509–4514. 10.1073/pnas.1116268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichl E. M.; Ren Y.; Morphew M. K.; Delannoy M.; Effler J. C.; Girard K. D.; Divi S.; Iglesias P. A.; Kuo S. C.; Robinson D. N. Interactions between Myosin and Actin Crosslinkers Control Cytokinesis Contractility Dynamics and Mechanics. Curr. Biol. 2008, 18, 471–480. 10.1016/j.cub.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishkind D. J.; Wang Y. L. Orientation and three-dimensional organization of actin filaments in dividing cultured cells. J. Cell Biol. 1993, 123, 837–848. 10.1083/jcb.123.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenix A. M.; Taneja N.; Buttler C. A.; Lewis J.; Van Engelenburg S. B.; Ohi R.; Burnette D. T. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell 2016, 27, 1465–1478. 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. H.; Ditzler C. E.; Germain A.; Irwin P. M.; Vogt E. T.; Yang S.; Wu X.; Shuster C. B. The ultrastructural organization of actin and myosin II filaments in the contractile ring: New support for an old model of cytokinesis. Mol. Biol. Cell 2017, 28, 613–623. 10.1091/mbc.e16-06-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz M. Geometrical origins of contractility in disordered actomyosin networks. Phys. Rev. X 2014, 4, 041002. 10.1103/PhysRevX.4.041002. [DOI] [Google Scholar]
- Ennomani H.; Letort G.; Guérin C.; Martiel J. L.; Cao W.; Nédélec F.; De La Cruz E. M.; Théry M.; Blanchoin L. Architecture and Connectivity Govern Actin Network Contractility. Curr. Biol. 2016, 26, 616–626. 10.1016/j.cub.2015.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwayer C.; Sikora M.; Slováková J.; Kardos R.; Heisenberg C. P. Actin Rings of Power. Dev. Cell 2016, 37, 493–506. 10.1016/j.devcel.2016.05.024. [DOI] [PubMed] [Google Scholar]
- Odde D. J. Mitosis, diffusible crosslinkers, and the ideal gas law. Cell 2015, 160, 1041–1043. 10.1016/j.cell.2015.02.048. [DOI] [PubMed] [Google Scholar]
- Chen S.; Markovich T.; MacKintosh F. C. Motor-free Contractility in Active Gels. Phys. Rev. Lett. 2020, 125 (20), 208101. 10.1103/PhysRevLett.125.208101. [DOI] [PubMed] [Google Scholar]
- Rizzelli F.; Malabarba M. G.; Sigismund S.; Mapelli M. The crosstalk between microtubules, actin and membranes shapes cell division. Open Biol. 2020, 10, 190314. 10.1098/rsob.190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster O. M.; Baum B. Shaping up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin. Cell Dev. Biol. 2014, 34, 109–115. 10.1016/j.semcdb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Tsai F. C.; Bertin A.; Bousquet H.; Manzi J.; Senju Y.; Tsai M. C.; Picas L.; Miserey-Lenkei S.; Lappalainen P.; Lemichez E.; Coudrier E.; Bassereau P. Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner. eLife 2018, 7, e37262 10.7554/eLife.37262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones G. A.; Jin J.; Oro A. E. I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J. Cell Biol. 2010, 189, 353–367. 10.1083/jcb.200910136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W.; Kocabey S.; Liedl T. DNA nanostructures in vitro, in vivo and on membranes. Nano Today 2019, 26, 98–107. 10.1016/j.nantod.2019.03.001. [DOI] [Google Scholar]
- Czogalla A.; Kauert D. J.; Franquelim H. G.; Uzunova V.; Zhang Y.; Seidel R.; Schwille P. Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles. Angew. Chem., Int. Ed. 2015, 54, 6501–6505. 10.1002/anie.201501173. [DOI] [PubMed] [Google Scholar]
- Steinkühler J.; Knorr R. L.; Zhao Z.; Bhatia T.; Bartelt S. M.; Wegner S.; Dimova R.; Lipowsky R. Controlled division of cell-sized vesicles by low densities of membrane-bound proteins. Nat. Commun. 2020, 11, 905. 10.1038/s41467-020-14696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M.; Kashiwazaki J.; Takagi T.; Srinivasan R.; Huang Y.; Balasubramanian M. K.; Mabuchi I. In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat. Cell Biol. 2013, 15, 853–859. 10.1038/ncb2781. [DOI] [PubMed] [Google Scholar]
- De Franceschi N.; Pezeshkian W.; Fragasso A.; Bruininks B. M. H.; Tsai S.; Marrink S. J.; Dekker C. A synthetic membrane shaper for controlled liposome deformation. bioRxiv 2021, 10.1101/2021.12.22.473854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T.; Boucrot E. Membrane curvature at a glance. J. cell science 2015, 128, 1065–1070. 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beber A.; Taveneau C.; Nania M.; Tsai F. C.; Di Cicco A.; Bassereau P.; Lévy D.; Cabral J. T.; Isambert H.; Mangenot S.; Bertin A. Membrane reshaping by micrometric curvature sensitive septin filaments. Nat. Commun. 2019, 10, 420. 10.1038/s41467-019-08344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger A. V.; Baum B.; Matthews H. K. The Mechanics of Mitotic Cell Rounding. Front. Cell Dev. Biol. 2020, 8, 687. 10.3389/fcell.2020.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prospéri M. T.; Lépine P.; Dingli F.; Paul-Gilloteaux P.; Martin R.; Loew D.; Knölker H. J.; Coudrier E. Myosin 1b functions as an effector of EphB signaling to control cell repulsion. J. Cell Biol. 2015, 210, 347–361. 10.1083/jcb.201501018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G.; Krauss M. Septin Remodeling During Mammalian Cytokinesis. Front. Cell Dev. Biol. 2021, 9, 768309. 10.3389/fcell.2021.768309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie K. A.; Bermeister A.; Robertson N. O.; Goodchild S. C.; Curmi P. M. G. Two Sides of the Coin: Ezrin/Radixin/Moesin and Merlin Control Membrane Structure and Contact Inhibition. Int. J. Mol. Sci. 2019, 20, 1996. 10.3390/ijms20081996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen M. P.; Colin-York H.; Schneider F.; Eggeling C.; Fritzsche M. Dissecting the actin cortex density and membrane-cortex distance in living cells by super-resolution microscopy. J. Phys. D: Appl. Phys. 2017, 50, 064002. 10.1088/1361-6463/aa52a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius M. T.; Nguyen L. T.; Ladinsky M. S.; Ortega D. R.; Aich S.; Mishra M.; Jensen G. J. Structure of the fission yeast actomyosin ring during constriction. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E1455–E1464. 10.1073/pnas.1711218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald N. A.; Lind A. L.; Smith S. E.; Li R.; Gould K. L. Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. eLife 2017, 6, e28865 10.7554/eLife.28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong Quang B. A.; Peters R.; Cassani D. A. D.; Chugh P.; Clark A. G.; Agnew M.; Charras G.; Paluch E. K. Extent of myosin penetration within the actin cortex regulates cell surface mechanics. Nat. Commun. 2021, 12, 6511. 10.1038/s41467-021-26611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.; Bhat A.; Sknepnek R.; Köster D.; Mayor S.; Rao M. Stratification relieves constraints from steric hindrance in the generation of compact actomyosin asters at the membrane cortex. Sci. Adv. 2020, 6, eaay6093. 10.1126/sciadv.aay6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M. P. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2001, 2, 392–396. 10.1038/35073095. [DOI] [PubMed] [Google Scholar]
- Paraschiv A.; Lagny T. J.; Campos C. V.; Coudrier E.; Bassereau P.; Šarić A. Influence of membrane-cortex linkers on the extrusion of membrane tubes. Biophys. J. 2021, 120, 598–606. 10.1016/j.bpj.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D.; Stauffer T.; Chen W.; Shen K.; Guo S.; York J. D.; Sheetz M. P.; Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 2000, 100, 221–228. 10.1016/S0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Janmey P. A.; Xian W.; Flanagan L. A. Controlling cytoskeleton structure by phosphoinositide-protein interactions: Phosphoinositide binding protein domains and effects of lipid packing. Chem. Phys. Lipids 1999, 101, 93–107. 10.1016/S0009-3084(99)00058-4. [DOI] [PubMed] [Google Scholar]
- Snider C. E.; Willet A. H.; Chen J. S.; Arpag G.; Zanic M.; Gould K. L. Phosphoinositide-mediated ring anchoring resists perpendicular forces to promote medial cytokinesis. J. Cell Biol. 2017, 216, 3041–3050. 10.1083/jcb.201705070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C.; Caorsi V.; Campillo C.; Sykes C. Interplay between membrane tension and the actin cytoskeleton determines shape changes. Phys. Biol. 2018, 15, 065004. 10.1088/1478-3975/aad1ab. [DOI] [PubMed] [Google Scholar]
- Shrivastava R.; Köster D.; Kalme S.; Mayor S.; Neerathilingam M. Tailor-made ezrin actin binding domain to probe its interaction with actin in-vitro. PLoS One 2015, 10, e0123428 10.1371/journal.pone.0123428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico F.; Russek A.; González L.; Grubmüller H.; Scheuring S. Heterogeneous and rate-dependent streptavidin–biotin unbinding revealed by high-speed force spectroscopy and atomistic simulations. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 6594–6601. 10.1073/pnas.1816909116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A.; Groves J. T. Kinetic control of histidine-tagged protein surface density on supported lipid bilayers. Langmuir 2008, 24, 4145–4149. 10.1021/la703788h. [DOI] [PubMed] [Google Scholar]
- Raghunath G.; Dyer R. B. Kinetics of Histidine-Tagged Protein Association to Nickel-Decorated Liposome Surfaces. Langmuir 2019, 35, 12550–12561. 10.1021/acs.langmuir.9b01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmazhan E.; Dunn A. R. The membrane-actin linker ezrin acts as a sliding anchor. Sci. Adv. 2022, 8, eabo2779. 10.1126/sciadv.abo2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R.; Fischer J.; Schwarz F. W.; Walter W. J.; Schwille P.; Diez S. Transport efficiency of membrane-anchored kinesin-1 motors depends on motor density and diffusivity. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E7185–E7193. 10.1073/pnas.1611398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernier J.; Kusters R.; Bousquet H.; Lagny T.; Morchain A.; Joanny J. F.; Bassereau P.; Coudrier E. Myosin 1b is an actin depolymerase. Nat. Commun. 2019, 10, 5200. 10.1038/s41467-019-13160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F.; Waithe D.; Clausen M. P.; Galiani S.; Koller T.; Ozhan G.; Eggeling C.; Sezgin E. Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS. Mol. Biol. Cell 2017, 28, 1507–1518. 10.1091/mbc.e16-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann F.; Vogel S. K.; Schwille P. Lateral Membrane Diffusion Modulated by a Minimal Actin Cortex. Biophys. J. 2013, 104, 1465–1475. 10.1016/j.bpj.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S. K.; Greiss F.; Khmelinskaia A.; Schwille P. Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife 2017, 6, e24350 10.7554/eLife.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster D. V.; Husain K.; Iljazi E.; Bhat A.; Bieling P.; Mullins R. D.; Rao M.; Mayor S. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E1645–E1654. 10.1073/pnas.1514030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigmann A.; Sadeghi S.; Keller J.; Hell S. W.; Eggeling C.; Vink R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife 2014, 3, e01671 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.; Mayor S. Active organization of membrane constituents in living cells. Curr. Opin. Cell Biol. 2014, 29, 126–132. 10.1016/j.ceb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Gowrishankar K.; Ghosh S.; Saha S.; C. R.; Mayor S.; Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 2012, 149, 1353–1367. 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Liu A. P.; Fletcher D. A. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys. J. 2006, 91, 4064–4070. 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön M.; Mey I.; Steinem C. Influence of cross-linkers on ezrin-bound minimal actin cortices. Prog. Biophys. Mol. Biol. 2019, 144, 91–101. 10.1016/j.pbiomolbio.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Nöding H.; Schön M.; Reinermann C.; Dörrer N.; Kürschner A.; Geil B.; Mey I.; Heussinger C.; Janshoff A.; Steinem C. Rheology of Membrane-Attached Minimal Actin Cortices. J. Phys. Chem. B 2018, 122, 4537–4545. 10.1021/acs.jpcb.7b11491. [DOI] [PubMed] [Google Scholar]
- Mosby L. S.; Hundt N.; Young G.; Fineberg A.; Polin M.; Mayor S.; Kukura P.; Köster D. V. Myosin II Filament Dynamics in Actin Networks Revealed with Interferometric Scattering Microscopy. Biophys. J. 2020, 118, 1946–1957. 10.1016/j.bpj.2020.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuba A.; Bano F.; Castro Linares G.; Iv F.; Mavrakis M.; Richter R. P.; Bertin A.; Koenderink G. H. Membrane binding controls ordered self-assembly of animal septins. eLife 2021, 10, e63349 10.7554/eLife.63349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon K. S.; Woods B. L.; Crutchley J. M.; Gladfelter A. S. An amphipathic helix enables septins to sense micrometer-scale membrane curvature. J. Cell Biol. 2019, 218, 1128–1137. 10.1083/jcb.201807211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Guan R.; Lee I. J.; Liu Y.; Chen M.; Wang J.; Wu J. Q.; Chen Z. Mechanistic Insights into the Anchorage of the Contractile Ring by Anillin and Mid1. Dev. Cell 2015, 33, 413–426. 10.1016/j.devcel.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer C. F.; Baldauf L.; van Buren L.; Wassenaar T. A.; Melo M. N.; Koenderink G. H.; Marrink S. J. Charge-dependent interactions of monomeric and filamentous actin with lipid bilayers. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 5861–5872. 10.1073/pnas.1914884117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S. H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004, 16, 99–105. 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Romero S.; Le Clainche C.; Didry D.; Egile C.; Pantaloni D.; Carlier M. F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 2004, 119, 419–429. 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Guevorkian K.; Manzi J.; Pontani L. L.; Brochard-Wyart F.; Sykes C. Mechanics of Biomimetic Liposomes Encapsulating an Actin Shell. Biophys. J. 2015, 109, 2471–2479. 10.1016/j.bpj.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicher P.; Sciortino A.; Bausch A. R. The dynamics of actin network turnover is self-organized by a growth-depletion feedback. Sci. Rep. 2020, 10, 6215. 10.1038/s41598-020-62942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M. M.; Chang F.; Burgess D. R. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell 2005, 9, 781–790. 10.1016/j.devcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Cauvin C.; Echard A. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2015, 1851, 832–843. 10.1016/j.bbalip.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Logan M. R.; Mandato C. A. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol. Cell 2006, 98, 377–388. 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- Emoto K.; Umeda M. Membrane Lipid Control of Cytokinesis. Cell Struct. Funct. 2001, 26, 659–665. 10.1247/csf.26.659. [DOI] [PubMed] [Google Scholar]
- Bassereau P.; et al. The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D: Appl. Phys. 2018, 51, 343001. 10.1088/1361-6463/aacb98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R.; Miettinen M. S.; Fricke N.; Lipowsky R.; Dimova R. The glycolipid GM1 reshapes asymmetric biomembranes and giant vesicles by curvature generation. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 5756–5761. 10.1073/pnas.1722320115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M.; Steinkühler J.; Roy D.; Dasgupta R.; Lipowsky R.; Dimova R. Asymmetric Ionic Conditions Generate Large Membrane Curvatures. Nano Lett. 2018, 18, 7816–7821. 10.1021/acs.nanolett.8b03584. [DOI] [PubMed] [Google Scholar]
- Simunovic M.; Voth G. A.; Callan-Jones A.; Bassereau P. When Physics Takes Over: BAR Proteins and Membrane Curvature. Trends Cell Biol. 2015, 25, 780–792. 10.1016/j.tcb.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster C. B.; Burgess D. R. Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 3633–3638. 10.1073/pnas.052342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel J. A.; Prekeris R. Membrane dynamics during cytokinesis. Curr. Opin. Cell Biol. 2013, 25, 92–98. 10.1016/j.ceb.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson Marsden H.; Elbers N. A.; Bomans P. H. H.; Sommerdijk N. A. J. M.; Kros A. A Reduced SNARE Model for Membrane Fusion. Angew. Chem. 2009, 121, 2366–2369. 10.1002/ange.200804493. [DOI] [PubMed] [Google Scholar]
- Lira R. B.; Robinson T.; Dimova R.; Riske K. A. Highly Efficient Protein-free Membrane Fusion: A Giant Vesicle Study. Biophys. J. 2019, 116, 79–91. 10.1016/j.bpj.2018.11.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S.; Wunnava S.; Hueting D.; Dekker C. Membrane Tension–Mediated Growth of Liposomes. Small 2019, 15, 1902898. 10.1002/smll.201902898. [DOI] [PubMed] [Google Scholar]
- Dreher Y.; Jahnke K.; Bobkova E.; Spatz J. P.; Göpfrich K. Division and Regrowth of Phase-Separated Giant Unilamellar Vesicles. Angew. Chem. 2021, 133, 10756–10764. 10.1002/ange.202014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara K.; Tamura M.; Shohda K. I.; Toyota T.; Suzuki K.; Sugawara T. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 2011, 3, 775–781. 10.1038/nchem.1127. [DOI] [PubMed] [Google Scholar]
- Exterkate M.; Caforio A.; Stuart M. C. A.; Driessen A. J. M. Growing Membranes in Vitro by Continuous Phospholipid Biosynthesis from Free Fatty Acids. ACS Synth. Biol. 2018, 7, 153–165. 10.1021/acssynbio.7b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanken D.; Foschepoth D.; Serrão A. C.; Danelon C. Genetically controlled membrane synthesis in liposomes. Nat. Commun. 2020, 11, 4317. 10.1038/s41467-020-17863-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. E. The origin of cleavage forces in dividing eggs: A mechanism in two steps. Exp. Cell Res. 1981, 134, 231–240. 10.1016/0014-4827(81)90480-8. [DOI] [PubMed] [Google Scholar]
- Wong G. K.; Allen P. G.; Begg D. A. Dynamics of filamentous actin organization in the sea urchin egg cortex during early cleavage divisions: Implications for the mechanism of cytokinesis. Cytoskeleton 1997, 36, 30–42. . [DOI] [PubMed] [Google Scholar]
- Bhatia T.; Agudo-Canalejo J.; Dimova R.; Lipowsky R. Membrane Nanotubes Increase the Robustness of Giant Vesicles. ACS Nano 2018, 12, 4478–4485. 10.1021/acsnano.8b00640. [DOI] [PubMed] [Google Scholar]
- Bhatia T.; Christ S.; Steinkühler J.; Dimova R.; Lipowsky R. Simple sugars shape giant vesicles into multispheres with many membrane necks. Soft Matter 2020, 16, 1246–1258. 10.1039/C9SM01890E. [DOI] [PubMed] [Google Scholar]
- Wang X.; Li L.; Shao Y.; Wei J.; Song R.; Zheng S.; Li Y.; Song F. Effects of the Laplace pressure on the cells during cytokinesis. iScience 2021, 24, 102945. 10.1016/j.isci.2021.102945. [DOI] [PMC free article] [PubMed] [Google Scholar]




