Abstract

Natural killer (NK) cells are a major subset of innate immune cells that are essential for host defense against pathogens and cancer. Two main classes of inhibitory NK receptors (NKR), KIR and CD94/NKG2A, play a key role in suppressing NK activity upon engagement with tumor cells or virus-infected cells, limiting their antitumor and antiviral activities. Here, we find that single-chain NKR antagonists linked to a VHH that binds the cell surface phosphatase CD45 potentiate NK and T activities to a greater extent than NKR blocking antibodies alone in vitro. We also uncovered crosstalk between NKG2A and Ly49 that collectively inhibit NK cell activation, such that CD45–NKG2A and CD45–Ly49 bispecific molecules show synergistic effects in their ability to enhance NK cell activation. The basis of the activity enhancement by CD45 ligation may reflect greater antagonism of inhibitory signaling from engagement of MHC I on target cells, combined with other mechanisms, including avidity effects, tonic signaling, antagonism of weak inhibition from engagement of MHC I on non-target cells, and possible CD45 segregation within the NK cell-target cell synapse. These results uncover a strategy for enhancing the activity of NK and T cells that may improve cancer immunotherapies.
Keywords: natural killer cell receptors, CD45, bispecific antibodies, induced proximity, protein engineering
Introduction
Natural killer (NK) cells and T cells are lymphocytes that protect against infection and cancer.1,2 The function of NK cells is tightly regulated by two classes of NK receptors, inhibitory receptors and activating NK receptors, which engage ligands, such as MHC I, and in turn tune the level of NK cell activity. Dysregulation of the balance between inhibitory and activating signaling in the context of tumor microenvironments or viral infection results in a reduced NK effector function.3−9 NKG2A and the family of Ly49 receptors are the two major inhibitory NK receptor types in mice, which have inhibitory tyrosine-based inhibition motifs (ITIMs) present in their intracellular domains (ICDs).4,10−13 One or more NKG2A and/or Ly49 family receptors are expressed on a large percentage of NK cells, as well as a small subset of CD8+ T cells.14−20 Most activated CD8 T cells upregulate NKG2A but not Ly49 receptors.20 NKG2A forms a heterodimer with CD94, whereas Ly49 receptors are displayed as homodimers on the surface of lymphocytes. Upregulation of inhibitory NK receptors and their ligands is frequently observed in different diseases.21,22 Engagement of NKG2A/CD94 or Ly49C/I receptors with MHC I molecules induces the phosphorylation of ITIM motifs in NK receptors, resulting in phosphatase binding. Recruitment of Src homology 1-containing phosphotyrosine phosphatase (SHP1) results in the transmission of inhibitory signals that repress activating NK signaling.23 In humans, NKG2A and the counterpart of Ly49 receptors, inhibitory killer cell immunoglobulin-like receptors (KIRs), play a key role in suppressing NK and T cell activities by interacting with different human leukocyte antigens (HLAs), including HLA-E, HLA-A, HLA-B, and HLA-C.24
Monoclonal antibody (mAb)-based immune checkpoint blockade (ICB) has revolutionized the treatment of cancer. Checkpoint inhibitors result in durable antitumor effects in many patients with metastatic and treatment-refractory cancers.25−28 ITIM-bearing receptors including PD-1, CTLA-4, and NKG2A have been identified as key IgSF molecules on both T cells and NK cells.14,15,29−32 Inhibition of mouse NKG2A with the 20D5 mAb restores the effector function of NK and T cells in lung cancer and lymphoma tumor models.14,15 Antibodies to inhibitory KIR are being evaluated for the treatment of cancer, and two mAbs are being tested in clinical trials in lymphoma and leukemia. IPH4102, specific for KIR3DL2, is in a clinical trial of T cell lymphoma, whereas lirilumab, specific for KIR2DL1-3, has not shown good efficacy in its phase II trial for the treatment of acute myeloid leukemia (AML).33,34 So far, anti-KIR mAbs have yet to show robust efficacy in clinical trials,33−36 suggesting that additional innovation may be required to target this interaction more effectively for cancer immunotherapy.
CD45 is a cell surface phosphatase ubiquitously expressed on immune cells that serves to modulate the extent of phosphorylation of many immunoreceptors through a variety of mechanisms including direct dephosphorylation and segregation from the immune synapse.37,38 Despite its central role in the signaling response at the membrane of immune cells, CD45 has not been actively investigated as an immunotherapeutic target. In one approach, receptor inhibition by phosphatase recruitment (RIPR), cis-ligation of CD45 with the PD-1 checkpoint receptor using bispecific molecules potentiated inhibition of the checkpoint activity.39 By cross-linking PD-1 to CD45, PD-1-RIPR suppressed both tonic and ligand-activated signaling in T cells to enhance T cell effector functions. Additional mechanisms may also play a role, such as potentially “passenger-like” sequestration of the PD1–CD45 conjugate away from the immune synapse. Here, we asked if CD45 ligation to inhibitory NKR could potentiate NKR antagonism, more effectively reversing inhibition through these receptors. We find that a series of CD45-targeted NK receptor antagonists potentiate NK and CD8+ T cell activities in vitro. We also find synergy between Ly49 and NKG2A antagonism on mouse NK cells, suggesting that maximally efficacious NKR targeting may require combination approaches.
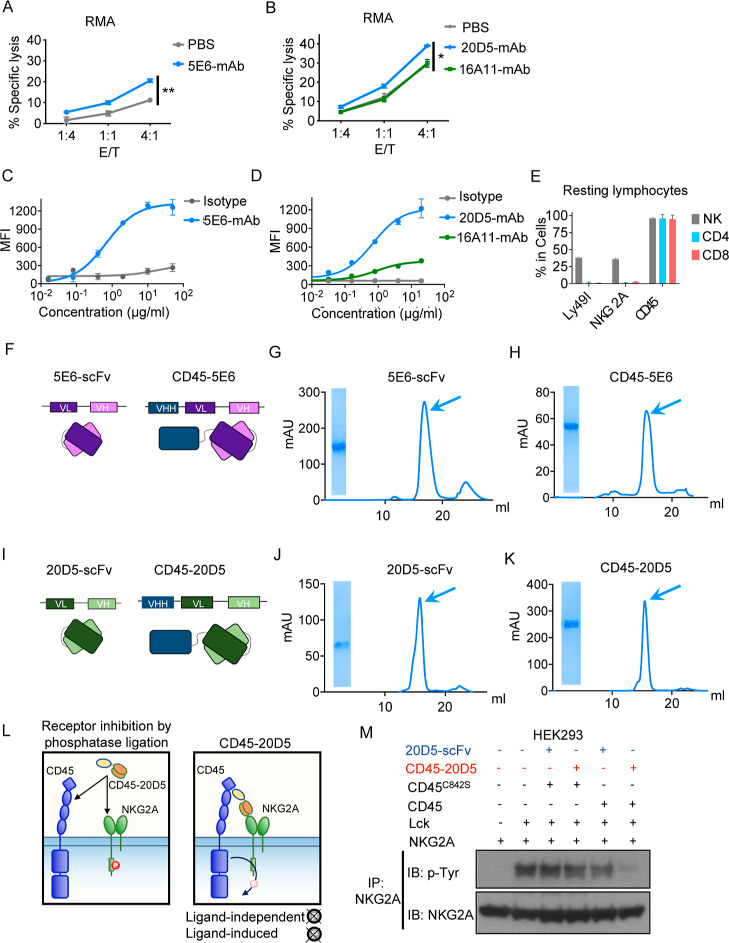
Results
Ligation of MHC I or Qa-1b to Ly49 or NKG2A receptors on NK cells, respectively, results in strong inhibition of NK activities.15,40,41 The MHC molecules present in the C57BL/6 mice examined in this study engage Ly49C, Ly49I, and NKG2A. To compare the impacts of blocking Ly49I or NKG2A on NK function, we sorted Ly49I+ or NKG2A+ NK cells and co-cultured them with RMA tumor cells, which express MHC I. The RMA cells for co-culturing with NKG2A+ NK cells were pre-cultured in IFNγ to induce Qa-1b expression. Target cell lysis by Ly49I+ NK cells was modestly augmented by the Ly49C/I-specific 5E6-mAb, whereas lysis by NKG2A+ NK cells was modestly augmented by the NKG2A-C-specific 20D5-mAb but not by another NKG2A-specific antibody, 16A11, which may not interfere with binding to Qa-1b (Figures 1A,B and S1A). Both 5E6-mAb and 20D5-mAb bound well to primary NK cells (Figure 1C,D). Given that nearly all NK cells that express NKG2A or Ly49I also express CD45 (Figure 1E), we re-engineered 20D5 and 5E6 antibodies into bispecific molecules by fusing an anti-CD45 single-domain antibody fragment (VHH) to the N terminus of 20D5 or 5E6-derived scFvs using a flexible (Gly–Ser)4 linker (Figure 1F,I). We sequenced the variable heavy (VH) and variable light (VL) chain regions of antibodies from hybridomas producing these antibodies and re-formatted VH and VL domains into scFv, as well as CD45 VHH-NKR scFv-fused molecules (Figure S1B). Following the production of these recombinant proteins from insect cells (Figure 1G,H,J,K), we checked the cell binding of 5E6-scFv and 20D5-scFv in Ly49+ and NKG2A+ NK cells and observed dose-dependent binding to NK cells, with 20D5-scFv appearing to be of higher affinity (Figure S1C). To show whether CD45 can modulate NKG2A phosphorylation, HEK293 cells expressing NKG2A, Lck, and CD45 or kinase-dead CD45 (C842S) were treated with either CD45–20D5 or 20D5-scFv. The presence of CD45–20D5 molecules promoted dephosphorylation of NKG2A as measured by tyrosine phosphorylation of NKG2A (Figure 1L,M). Dephosphorylated NKG2A was not observed with the addition of 20D5-scFV or when cells had been transfected with mutant CD45 (C842S) as opposed to WT CD45.
Figure 1.
Engineering strategy of CD45-NKR. (A) NK cells were expanded by 100 IU of mouse IL-2. Ly49I+ NK cells were sorted and allowed to recover for 2 days. Cytolysis of RMA cells expressing MHC I by sorted Ly49I+ NK cells in the presence or absence of 100 nM Ly49C/I-specific 5E6-mAb was tested by Annexin V/7-AAD staining. (B) NK cells were expanded by 100 IU mouse IL-2 for 5 days. NKG2A+ NK cells were sorted and allowed to recover for 2 days. RMA cells were stimulated with 20 ng/μL of IFNγ for 2 days for upregulating Qa-1b. NKG2A+ NK cells were tested for cytolysis of RMA cells in the presence or absence of NKG2A-specific 16A11-mAb and 20D5-mAb. (C) Representative titration of 5E6-mAb on sorted mouse NK cells. The titration data shown are median fluorescence intensity (MFI) over a range of anti-IgG-AF647 antibody concentrations. (D) Representative titration of 20D5-mAb vs 16A11 mAb on sorted mouse NK cells. The titration data shown are MFI over a range of anti-IgG-AF647 antibody concentrations. (E) Splenocytes from wild-type C57BL/6 mice were harvested and stained with a panel of antibodies against lymphocyte markers, and the cells were analyzed by flow cytometry. Quantification of percentage of Ly49I, NKG2A, and CD45 expression on splenic NK cells (CD45.2+, CD3–, and NK1.1+), CD4+ T cells (NK1.1–, CD3+, and CD4+), and CD8+ T cells (NK1.1–, CD3+, and CD8+). (F) Schematic representation of 5E6-scFv and CD45-5E6 depicting the connection between the CD45 VHH and VH and VL chains of 5E6-scFv. (G,H) Size exclusion chromatography profiles of 5E6-scFv (G) and CD45–5E6 (H). Arrowheads indicate the collected fraction analyzed by SDS-PAGE. (I) Schematic representation of 20D5-scFv and CD45–20D5, depicting the connection between the CD45 VHH and VH and VL chains of 20D5-scFv. (J,K) SEC profiles of 20D5-scFv (J) and CD45–20D5 (K). Arrowheads indicate the collected fraction analyzed by SDS-PAGE. (L) Schematic depiction of CD45-NKR-mediated receptor inhibition. (M) HEK293 cells were transiently transfected with HA-tagged mouse NKG2A, Lck, and CD45 or mutant CD45C842S. After 24 h, the cells were incubated with 100 nM CD45-20D5 or 20D5-scFv for 30 min at 37 °C. Phospho-NKG2A was detected by immunoprecipitation of HA-NKG2A, followed by immunoblotting with the phosphotyrosine antibody. All curves show mean ± SD of n = 3 per group. Groups were compared by unpaired t-test, and data are representative of two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We tested whether the Ly49-targeted CD45–5E6 could enhance NK-mediated cell killing (Figure 2A,B). Notably, when sorted Ly49I+ NK cells were co-cultured with MHC I+ RMA cells, the Ly49C/I-blocking 5E6-scFv augmented target cell killing by 7% at the highest concentration tested, whereas the Ly49C/I-targeting CD45–5E6 augmented target cell killing by more than 20% (Figures 2A,B and S2A,B). We next determined the effects of CD45–5E6 on NK activation and degranulation. Recruitment of SHP1 by the MHC I-Ly49 signaling axis results in suppression of proximal/NK activation receptor signaling, which includes CD69 upregulation, degranulation, and cytokine production. In a NK and RMA target cell co-culture assay, we performed a dose-series stimulation of CD45–5E6 and 5E6-scFv molecules. Compared to 5E6-scFv, CD45–5E6 was much more efficient at augmenting degranulation and IFNγ production (the latter determined by intracellular cytokine staining), as well as induction of CD69 expression, by Ly49I+ NK cells (Figures 2C–E and S2C). By replacing the linker connecting CD45 VHH and 20D5 scFv with a 3C cleavage site, LEVLFQGP, we tested the effect of intact CD45–5E6 versus the separate CD45 and 5E6 components after cleavage with HRV 3C protease. Consistent with our hypothesis of induced proximity, 3C treatment greatly diminished the activity of CD45–5E6 (Figure 2F,G). To summarize, we re-engineered the antibodies against NKG2A or Ly49C/I into CD45-targeted versions, which show a superior capacity to enhance NK cytolysis, degranulation, IFNγ production, and NK-activating receptor signaling compared to NKR blocking antibodies alone. From these experiments, it is not clear whether the effect is due to more efficient interference with inhibition mediated by interactions with target cell MHC I, repression of tonic signaling, enhanced avidity due to two-site binding, or possibly enhanced segregation of NKR from the NK cell synapse.
Figure 2.
Functional properties of CD45–5E6 in mouse NK cells. (A) Schematics of the NK cell killing assay of RMA cells expressing MHC I. (B) Sorted Ly49I+ NK cells were co-cultured with RMA cells in the presence or absence of 5E6-scFv or CD45–5E6 for 4 h and assayed for dose-dependent cytolysis of RMA cells by Annexin V/7-AAD staining. (C) IL-2-expanded NK cells were co-cultured with RMA cells in the presence or absence of 5E6-scFv or CD45–5E6 for 4 h and assayed for dose-dependent activation by cell surface staining of CD107a in Ly49I+ NK cells. (D) IL-2-expanded NK cells were co-cultured with RMA cells in the presence or absence of 5E6-scFv or CD45–5E6 for 4 h and assayed for dose-dependent IFNγ secretion by intracellular staining of IFNγ in Ly49I+ NK cells. (E) IL-2-expanded NK cells were co-cultured with RMA cells in the presence or absence of 5E6-scFv or CD45–5E6 for 4 h and assayed for dose-dependent degranulation by cell surface staining of CD69 in Ly49I+ NK cells. (F) HRV 3C protease was incubated with CD45–3C–5E6 at 1:100 for 24 h at 4 °C. Intact and 3C-treated CD45–3C–5E6 were analyzed by SDS-PAGE, followed by Coomassie blue staining. (G) Sorted Ly49I+ NK cells were co-cultured with RMA cells in the presence or absence of 5E6-scFv, CD45–3C–5E6, or 3C-treated CD45–3C–5E6 for 4 h and assayed for dose-dependent cytolysis of RMA cells by Annexin V/7-AAD staining. All curves show mean ± SD of n = 3 per group. Groups were compared by the unpaired t-test, and data are representative of two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
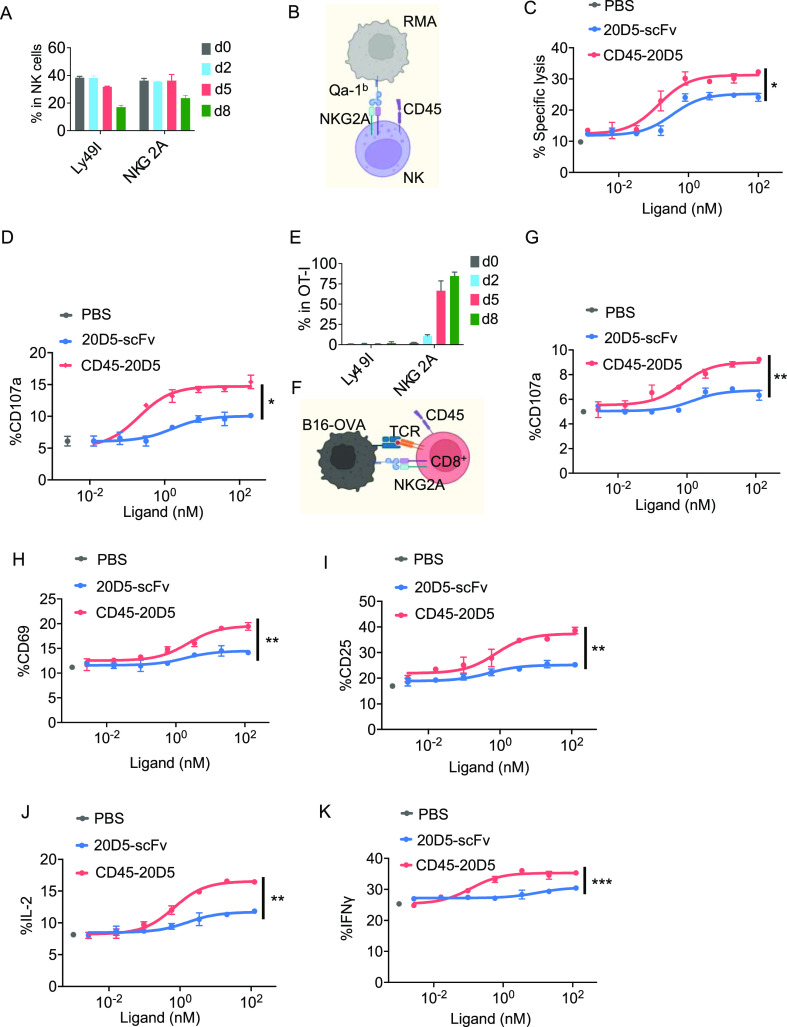
NKG2A is expressed by primary resting and IL-2 cultured NK cells to a similar extent as Ly49I (Figure 3A). We therefore employed the same strategy as that used for generating the CD45–5E6 bispecific molecule to generate and test an NKG2A-targeted CD45–20D5 bispecific molecule (Figures 1I–K and 3B). Notably, treatment with CD45–20D5 resulted in a higher maximal target cell lysis and degranulation of NK cells compared to 20D5-scFv (Figure 3C,D). NKG2A is expressed by naïve NK cells and not naïve CD8 T cells but is strongly upregulated in virus-specific CD8+ T cells and tumor antigen-specific T cells.15,20 To examine whether CD45–20D5 plays a role in CD8+ T cells, we tested its effects in a system of MHC class I-restricted, ovalbumin-specific, TCR transgenic CD8+ T cells (OT-I) and B16-OVA target cells, which had been cultured with IFNγ to induce MHC I and Qa-1b (Figure S3A–E). In this system, the mouse B16F10 melanoma cells express a model antigen, chicken ovalbumin, and therefore present the OVA peptide antigen and activate OVA-specific CD8+ OT-I cells. For generating effector OT-I cells, splenocytes from OT-I mice were stimulated with anti-CD28 and OVA257-264 octapeptide, SIINFEKL, for 8 days. Although NKG2A was expressed at low levels in resting OT-I cells, it was robustly upregulated in effector OT-I cells following activation (Figure 3E). Surface staining of CD107a, CD25, and CD69 on OT-I cells following co-culture with B16-OVA cells indicated that CD45–20D5 promotes greater OT-I degranulation and induces greater TCR signaling than 20D5-scFv (Figures 3F–I and S3F). Moreover, CD45–20D5 stimulation resulted in greater production of the effector cytokines IL-2 and IFNγ as compared to 20D5-scFv (Figure 3J–K).
Figure 3.
Functional properties of CD45–20D5 in mouse NK cells and CD8+ T cells. (A) Quantification of percentage of Ly49I+ and NKG2A+ NK cells cultured in 100 IU mouse IL-2 for days indicated. (B) Schematic of co-culture of NKG2A+ NK cells with RMA cells expressing Qa-1b. (C) RMA cells were treated with IFNγ for upregulating Qa-1b. IL-2-expanded NK cells were co-cultured with RMA cells in the presence or absence of 20D5-scFv or CD45–20D5 for 4 h and assayed for dose-dependent cytolysis of RMA cells by Annexin V/7-AAD staining. (D) RMA cells were treated with IFNγ for upregulating Qa-1b. IL-2-expanded NK cells were co-cultured with RMA cells in the presence or absence of 20D5-scFv or CD45–20D5 for 4 h and assayed for dose-dependent degranulation by cell surface staining of CD107a in NKG2A+ NK cells. (E) Quantification of percentage of Ly49I and NKG2A expression during generation of OT-I effector cells by incubating with 0.5 μg/mL anti-CD28 and 1 μg/mL SIINFEKL peptides. (F) Schematic of co-culture of NKG2A+ OT-I cells with B16-OVA cells expressing Qa-1b. (G) B16-OVA cells were treated with IFNγ for upregulating Qa-1b. Purified CD8+ OT-I cells were co-cultured with B16-OVA cells in the presence or absence of 20D5-scFv and CD45–20D5 for 4 h and assayed for dose-dependent degranulation by cell surface staining of CD107a in NKG2A+ OT-I cells. (H,I) Dose–response curves showing CD69 (H) and CD25 (I) expression in response to 20D5-scFv and CD45–20D5 stimulation. (J,K) Dose–response curves showing intracellular staining of IL-2 (J) and IFNγ (K) in response to 20D5-scFv and CD45–20D5 stimulation. All curves show mean ± SD of n = 3 per group. Groups were compared by the unpaired t-test, and data are representative of two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Previous studies have shown that many T, B, and NK cell receptors can in some cases signal even when the ligand is absent on the target cell (32, 35–37). Such effects may have different explanations, including tonic signaling, where the receptor signals were independent of the ligand, and cis-signaling, where the receptor engages the ligand on the membrane of the NK or T cell itself, or possibly when the receptor is engaged by the ligand on a third party cell in the vicinity. To investigate whether the impact of CD45-NKR is fully dependent on target cell engagement of the NK receptor ligand, we first examined the expression of Qa-1b and MHC I on NK cells by flow cytometry and found that both Qa-1b and MHC I can be detected on the surface of NK cells (Figure S4A,B). We used a peptide–MHC I-deficient target cell line, RMA-B2m KO, to address whether the impact of CD45-NKRs is dependent on MHC I expression by target cells (Figures 4A and S4C,D). We found that the CD45–5E6 significantly augmented the killing of MHC I-deficient RMA cells (Figure 4A) albeit to a smaller extent than it augmented the killing of MHC I+ RMA cells and A20 cells (Figure 4B,C). 5E6-scFv and mAb had no effect. Similarly, CD45–20D5 modestly augmented the killing of RMA target cells that lacked Qa-1b expression (RMA–Qa-1b KO) (Figure 4D) albeit to a smaller extent than it augmented the killing of RMA, A20, or B16 target cells that expressed Qa-1b (Figure 4E–G). 20D5-scFv and mAb, in contrast, did not augment the killing of Qa-1b-deficient cells but did augment the killing of WT cells. These data suggest that enhanced antagonism by CD45-NKRs reflects antagonism of at least two separable inhibitory effects of the NK receptors: inhibitory signaling resulting from engagement of MHC I on target cells and inhibitory signaling that is independent of target cell MHC I expression. The weaker inhibition that is independent of target cell MHC I expression may be due to engagement of MHC I on other cells or could reflect “tonic” inhibitory signaling that the receptor confers without engaging MHC I.
Figure 4.
CD45-NKR potentiates NK killing of RMA cells. (A,B) NK cells were expanded by mouse IL-2 for 5 days. Ly49I+ NK cells were sorted and allowed to recover for 2 days. Cytolysis of RMA B2m KO cells (A) and wild-type RMA cells (B) by sorted Ly49I+ NK cells in the presence or absence of 100 nM 5E6-scFv, CD45–5E6, or 5E6-mAb. Percent lysis was determined by Annexin V/7-AAD staining. (C) A20 cells were treated with IFNγ for MHC-I upregulation. Cytolysis of A20 cells was determined by Annexin V/7-AAD staining. (D,E) NKG2A+ NK cells were sorted and allowed to recover for 2 days. Wild-type RMA cells were treated with IFNγ for Qa-1b upregulation. Cytolysis of RMA Qa-1b KO (D) and wild-type RMA cells (E) by NKG2A+ NK cells in the presence or absence of 20D5-scFv, CD45–20D5, or 20D5-mAb at 100 nM. (F) A20 cells were treated with IFNγ for Qa-1b upregulation. Cytolysis of A20 cells by NKG2A+ NK cells in the presence or absence of 20D5-scFv or CD45–20D5 at 100 nM was assayed by Annexin V/7-AAD staining. (G) B16 cells were treated with IFNγ for Qa-1b upregulation. Cytolysis of B16 cells by NKG2A+ NK cells in the presence or absence of 20D5-scFv or CD45-20D5 at 100 nM was tested by detecting caspase-3/7 activity. All curves show mean ± SD (n = 3) and were analyzed by one-way ANOVA relative to 5E6-scFv or 20D5-scFv. Multiple comparisons were corrected using Dunnett’s test. Data are representative of two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
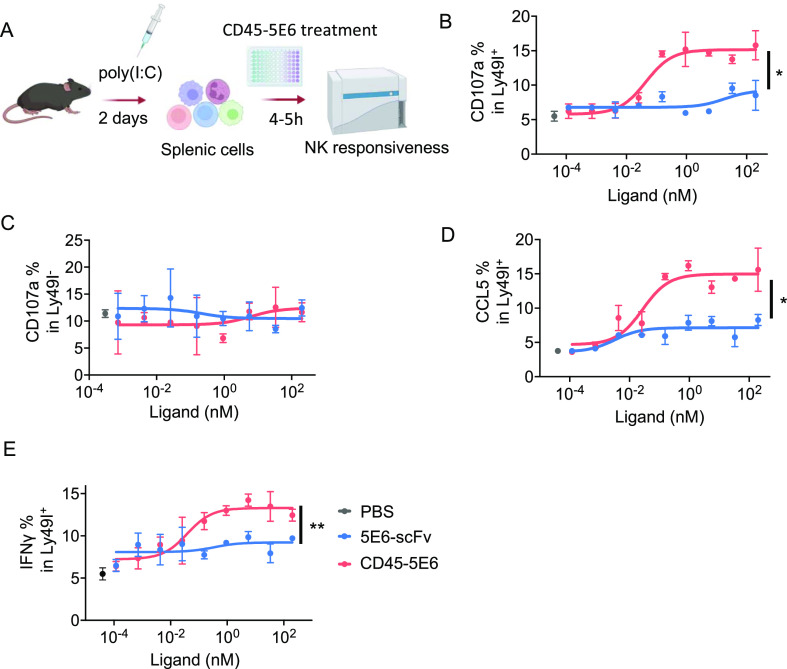
The activation state of NK cells is tightly regulated by the receptors they express, such that NK cells lacking all inhibitory receptors for self MHC I exist in a hyporesponsive state.42,43 Thus, we further investigated the impact of CD45-NKRs on responses of freshly isolated NK cells in co-cultures with target cells. Preactivated NK cells were harvested from mice pretreated with poly(I/C) and were co-cultured with target cells (Figures 5A and S5). When compared to 5E6-scFv, CD45–5E6 induced a substantially greater response in Ly49I+ NK cells (Figure 5B). Importantly, Ly49I+ NK cells, but not Ly49I– NK cells, showed significantly enhanced responsiveness in the presence of CD45–5E6 (Figure 5B,C). Moreover, CD45–5E6 stimulated many more of the Ly49I+ NK cells to produce effector cytokines and chemokines, such as IFNγ and CCL5, than did 5E6-scFv (Figure 5D,E).
Figure 5.
CD45-5E6 potentiates responsiveness by freshly isolated Ly49I+ NK cells. (A) Schematics of the NK responsiveness assay. WT mice were treated with 200 μg of poly(I/C) for 2 days, splenocytes were harvested and co-cultured with RMA cells in the presence of 5E6-scFv or CD45–5E6 for 5 h, and NK responsiveness was tested by detecting surface expression of CD107a in Ly49I+ NK cells. (B,C) NK cells were assayed for dose-dependent CD107a in Ly49I+ (B) and Ly49I– NK (C) cells by surface staining. (D,E) Dose–response curves showing percentage of CCL5+ (D) and IFNγ+ (E) in Ly49I+ NK cells in response to 5E6-scFv or CD45–5E6 stimulation. Curves show mean ± SD (n = 3) and were analyzed by one-way ANOVA relative to 5E6-scFv. Multiple comparisons were corrected using Dunnett’s test. Bar graphs show mean ± SD (n = 4) and were analyzed by two-way ANOVA. Multiple comparisons were corrected using Tukey’s test. Data are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Primary NK cells can be divided into eight distinct subpopulations based on the surface expression and co-expression of Ly49C, Ly49I, and NKG2A (Figures 6A,B and S6A). To address how these different NK cell subpopulations respond to the CD45-NKRs, we gated the cells stained with antibodies against Ly49C, Ly49I, and NKG2A. The responses by each of the eight subpopulations of primary NK cells to CD45-NKRs and scFvs are shown in Figure S6. Figure 6 shows a simpler depiction, representing the responses by four populations of NK cells (Figure 6B): those that co-expressed NKG2A with either Ly49I and/or Ly49C; those that expressed Ly49I and/or Ly49C but not NKG2A; those that expressed NKG2A alone (Ly49C/I-negative); and those that lacked all three of these receptors. Interestingly, NK cells that co-expressed NKG2A with either Ly49I and/or Ly49C did not respond to either CD45–5E6 or CD45–20D5 separately but responded well when both CD45-NKRs were present (Figures 6C and S6B,D,E). Treatments with CD45–5E6 alone potentiated responses of NK cells that expressed Ly49C and/or Ly49I, but only if they lacked NKG2A (Figure 6D vs 6C and Figure S6C,F,G). Conversely, treatments with CD45–20D5 alone potentiated responses of NK cells that expressed NKG2A, but only if they lacked Ly49C and Ly49I (Figure 6E vs 6C and Figure S6H). In contrast to the results with CD45-NKRs, 5E6-scFv was ineffective with Ly49C/I+NKG2A– NK cells (Figures 6H and S6C,F,G) and the combination of 5E6-scFv and 20D5-scFv was ineffective with Ly49C/I+NKG2A+ NK cells (Figures 6G and S6B,D,E). However, the 20D5 scFv had similar activity to CD45–20D5 in activating Ly49C/I–NKG2A+ NK cells (Figure 6I vs 6E and Figure S6G).
Figure 6.
Combination of CD45–5E6 and CD45–20D5 potentiates NK activation. (A) FACS distributions showing NKG2A, Ly49C, or Ly49I expression in resting splenic NK cells. (B) Four different NK cell subpopulations defined by expression and co-expression of Ly49I, Ly49C, and NKG2A in WT C57BL/6 mice that were pretreated with 200 μg of poly(I/C) for 2 days before stimulating the cells with RMA target cells in the presence or absence of CD45-NKRs or NKR-scFvs. RMA cells were precultured with 20 ng/mL of IFNγ for upregulating Qa-1b and were labeled with 1 μM CTV. 1 × 106 Splenocytes of poly(I/C)-treated mice were co-cultured with 2 × 105 RMA Qa-1b+MHC I+ target cells in the presence of anti-CD107a-PE, brefeldin A, monensin, 5E6-scFv, CD45–5E6, 20D5-scFv, CD45–20D5, a combination of 5E6-scFv and 20D5-scFv, or a combination of CD45–5E6 and CD45–20D5 for 5 h. The center plot shows co-staining of gated NK cells for Ly49C and Ly49I. Gated NK cell populations that were positive for Ly49I and/or Ly49C, or negative for both, were then gated further based on NKG2A staining (right and left plots). (C–J) Each of the four populations co-cultured in the presence of CD45 bispecifics (C–F) vs scFv controls (G–J) was analyzed to determine the percentages of CD107a+ cells among Ly49C+ and/or Ly49I+ NK cells that also expressed NKG2A (Ly49C/I+NKG2A+) (C,G), Ly49C+ and/or Ly49I+ NK cells that did not express NKG2A (D,H), NKG2A+ NK cells that did not express Ly49C or Ly49I (E,I), and NK cells that did not express any of the three receptors (F,J). Bar graphs show mean ± SD (n = 4) and were analyzed by two-way ANOVA. Multiple comparisons were corrected using Tukey’s test. Data are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Not surprisingly, NK cells that lacked all three receptors failed to respond to any of these molecules, in CD45-NKR or scFv format (Figures 6F,J and S6H). Furthermore, the responses of the various subpopulations of NK cells to MHC I-deficient target cells were not much affected by the CD45-NKRs, alone or in combination (Figure S7), consistent with the analysis of NK cell killing, which showed that the effects of CD45-NKRs were much weaker when MHC I-deficient target cells were employed.
The synergistic effect of the CD45-NKR combinations on some NK cells suggests that combining CD45-NKRs that target these receptors is a potential strategy to improve their immunotherapeutic efficacy. Not only does the combination activate a subpopulation of NK cells that CD45-NKR alone cannot activate well (approximately 30% of NK cells that co-expressed NKG2A and Ly49C/I) but also each CD45-NKR activates unique NK subsets, all of which can be simultaneously activated by the CD45-NKR combination. As a result of these two features, the subpopulations that respond to the CD45-NKR combination together comprise approximately 80% of NK cells, whereas the populations that respond to each CD45-NKR separately comprise only 21–32% of NK cells.
While the NKG2A/CD94 receptor complex is conserved between humans and mice, the functional counterparts of Ly49 receptors in humans are the KIR receptors, which also bind to MHC I complexes. To investigate CD45-NKR activity on human NK cells, we designed new CD45-NKR molecules targeting human NKG2A (CD45–Z270), human KIR3DL (CD45–AZ158), and human KIR2DL (CD45-1-7F9), by fusing NK receptor antibodies with the VH and VL regions of an anti-human CD45 antibody in the form of a diabody (Figures 7A–F and S8A). To explore how these human CD45-NKR molecules impact NK cell activity, we co-cultured human NK cells from human peripheral blood mononuclear cells (PBMCs) with target cells expressing human HLA ligands (Figure S8B) and found that the CD45–Z270 significantly enhanced the killing of K562-GpHLA-E cells (Figure 7G). While Z270-scFv-stimulated NK cells showed around 3–5% higher degranulation relative to PBS-treated controls, the percentage of CD107a+ cells was markedly increased (15–18%) following treatment with CD45–Z270 (Figure 7H). Analysis of effector cytokine production further revealed that CD45–Z270 potentiated the secretion of IFNγ and TNF in NKG2A+ NK cells (Figures 7I,J and S8C,D). Similarly, while the addition of AZ158-scFv resulted in greater NK degranulation and granzyme B production than observed in the PBS control group, CD45–AZ158 induced much stronger degranulation and granzyme B production compared to AZ158-scFv (Figure 7K–M). We next investigated whether CD45–Z270 could attenuate the impact of NKG2A on activating signaling in NK cells, specifically the inhibition of ERK and S6 phosphorylation mediated by NKG2A engagement in NK cells. For this purpose, NK cells were co-cultured with K562 cells expressing HLA-E. The presence of CD45–Z270 markedly potentiated both ERK and S6 phosphorylation (Figure 7N,O). In a HEK293 dephosphorylation assay, we treated HEK293 transiently expressing human NKG2A, CD45, and Lck with CD45–Z270 and Z270-scFv molecules. Compared to Z270-scFv, CD45–Z270 induced a greater level of NKG2A dephosphorylation (Figure 7P). Thus, the CD45-NKR is also an effective strategy for enhancing human NK effector activities.
Figure 7.
Human CD45-NKRs potentiate NK degranulation and activation. (A) Schematic representation of Z270-scFv and CD45–Z270, depicting the connection between the VH/VL of anti-CD45 and VH/VL of anti-NKG2A. (B,C) SEC profiles of Z270-scFv (B) and CD45–Z270 (C). Arrowheads indicate the collected fraction analyzed by SDS-PAGE. (D) Schematic representation of AZ158-scFv and CD45–AZ158 depicting the connection between the VH/VL of anti-CD45 and VH/VL of anti-KIR3DL. (E,F) SEC profiles of AZ158-scFv (E) and CD45–AZ158 (F). Arrowheads indicate the collected fraction analyzed by SDS-PAGE. (G) Human primary NK cells were expanded in vitro for 10 days. NKG2A+ NK cells were sorted and allowed to recover for 2 days. Cytolysis of K562-GpHLA-E cells by sorted NKG2A+ NK cells in the presence or absence of 100 nM Z270-scFv or CD45–Z270. Percent lysis determined by Annexin V/7-AAD staining. (H–J) Quantification of human NKG2A+ NK degranulation (H), IFNγ (I), and TNF (I) production in response to Z270-scFv and CD45–Z270. (K–M) Quantification of human KIR3DL+ NK degranulation (K,L), and granzyme B (M) production in response to Z270-scFv and CD45–Z270. (N,O) Phospho-flow detection of pErk (N) and pS6 (O) signaling in human NKG2A+ NK cells in response to Z270-scFv and CD45–Z270. (P) HEK293 cells were transfected with HA-tagged human NKG2A, Lck, CD45, or mutant CD45C853S. After 24 h, the cells were incubated with 20 and 100 nM CD45–Z270 or Z270-scFv for 30 min at 37 °C. Phospho-NKG2A was detected by immunoprecipitation of HA-NKG2A, followed by immunoblotting with the phospho-tyrosine antibody. Bar graphs and curves show mean ± SD (n = 3) and were analyzed by one-way ANOVA. Multiple comparisons were corrected using Dunnett’s test. Data are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
Induced proximity of cell surface proteins is gaining increasing interest for modulating cell surface receptor signaling. Strategies for down-tuning receptor signaling include recruitment of cell surface E3 Ub ligases, cell surface phosphatases, and mannose-6-phosphatase receptors.39,44,45 While E3 Ub ligase and MAN-6-P recruitment result in degradation of the target proteins, phosphatase recruitment exploits the intrinsically high activity of phosphatases such as CD45 to intracellularly dephosphorylate targets within close proximity. On immune cells, CD45 is the principal cell surface phosphatase and serves as a counterbalance to Src kinase actions on ITAM/ITIM/ITSM receptors to control the phosphorylation state of many important antigen receptors and checkpoint receptors.46,47 CD45 also plays other important roles in modulating TCR signaling by being segregated from the immunological synapse, thus potentiating the actions of Src kinases that act on the antigen receptor complex.37,38 Thus, ligation of inhibitory immunoreceptors to CD45 can not only reduce their phosphorylation state directly but also potentially enhance signaling through co-segregation with CD45 from the synapse, although this has not been directly shown. Additional mechanisms may also be at play.
Here, we find that induced proximity between inhibitory NKR and CD45, using single-chain bispecific agents, more effectively reverses inhibitory signaling by NKR, enhancing signaling and target cell killing by NK cells. We also show that the CD45-targeted NKG2A antagonist, CD45–20D5, potentiates CD8+ effector T cells in addition to NK cells. Upon binding to ligands such as MHC I or Qa-1b, inhibitory NK receptors negatively regulate NK activation signals. This study shows that CD45-NKR molecules enhance NK activity mainly by augmenting NK activation signals, including Erk and S6 phosphorylation and upregulating cytokine receptor CD25. These data suggest that CD45-NKR efficacy reflects better antagonism of inhibitory signaling that prevents these activation signals in NK cells. The outcome manifests as improved target cell killing, degranulation, CD69 surface expression, and secretion of effector cytokines.
The efficacy of the CD45-NKR molecules could reflect any of several non-mutually exclusive mechanisms, including more efficient termination of inhibitory signaling due to the recruitment of the phosphatase activity to the ITIM motifs of the NKR, an avidity effect of coligation to CD45, segregation of the NKR from the NK cell synapse with the target cells, or interference with tonic inhibitory signals from the NKR that do not depend on MHC I engagement.
Unlike PD-1, where evidence of tonic signaling exists,39 tonic signaling by inhibitory NKR has not been demonstrated.48 In our hands, CD45-NKR increased NK responses partially in some assays even when the target cells were MHC I-deficient. This finding is consistent with not only reduced tonic signaling but also other mechanisms, such as inhibition due to engagement of MHC I on the NK cells themselves or on neighboring NK cells or other cells.
An interesting and potentially useful finding of our work is that separate inhibition of Ly49 or NKG2A with CD45-NKR largely failed to augment NK cell activation in the case of a large (∼30%) subpopulation of NK cells that co-express NKG2A and Ly49C/I. These results are best explained by suggesting that these NK cells are fully inhibited by engagement of either NKG2A or Ly49C/I by Qa-1b or MHC I, respectively, such that interfering with each separately is insufficient to boost NK cell activity. Consistent with this interpretation, combining the two CD45-NKRs activated this NK cell subpopulation synergistically.
CD45–5E6 molecules were effective with NK cells that expressed Ly49C/I but not NKG2A. Conversely, CD45–20D5 molecules were most effective with NK cells that expressed NKG2A but not Ly49C or Ly49I. Each of these populations represents only 25–30% of NK cells. Hence, targeting each of these receptor types separately may be insufficient to stimulate the most impactful NK cell responses. Combining the two CD45-NKRs led to activation of both of these subpopulations as well as the aforementioned subpopulation that co-expressed Ly49C/I and NKG2A. In total, these subpopulations represent approximately 80% of all NK cells. In contrast, the subpopulations that respond to CD45–20D5 or CD45–5E6 separately each represent only 21–32% of NK cells. These considerations lead to the proposal that combining CD45-NKR that interfere with both types of receptors may provide much stronger efficacy than either alone in cancer immunotherapy.
Two anti-KIR blocking antibodies, lirilumab and IPH4102, have been tested in clinical trials; however, lirilumab has not shown good efficacy in the treatment of AML.33,34 Our findings suggest that the efficacy of lirilumab in cancer therapy might be enhanced by inhibition of NKG2A, and most pertinently that combining CD45-NKR that interferes with NKG2A and KIR signaling may provide the greatest efficacy in the context of cancer therapy, although this remains to be tested. Collectively, this work uncovers new strategies to design NKR antagonists and new mechanisms of inhibitory NKR signaling.
Methods
Cells
Primary NK cells, primary T cells, RMA, K562, and 721.221 cell lines were kept in a humidified incubator at 37 °C with 5% CO2 and cultured in complete RPMI 1640 medium supplemented with 2 mM GlutaMAX (Invitrogen), 10% FBS (Sigma),10 mM HEPES pH 8.0 (Thermo Fisher), 1 mM sodium pyruvate (Gibco), and 50 U/mL penicillin and streptomycin (Thermo Fisher). B16F10 and B16-OVA cells were cultured in complete DMEM medium containing 10% FBS, 50 U/mL penicillin and streptomycin, 2 mM GlutaMAX, and dissociated with TrypLE Express Enzyme (Thermo Fisher) when splitting every other day. Expi293 cells (female-derived kidney cell line) were grown in Expi293 Expression Medium (Thermo Fisher) at 37 °C, 125 rpm, 5% CO2 atmosphere with 80% humidity in flasks with ventilated caps. Hi5 cells were cultured in ESF 921 media with 10 mg/L of gentamicin sulfate at 27 °C on a shaker. SF9 cells were grown in SF900 media, which contained 10% FBS, 10 mg/L of gentamicin sulfate, and 2 mM GlutaMAX at 27 °C.
Mice
C57BL/6 wild-type mice and C57BL/6-Tg (TcraTcrb) 1100Mjb/J (OT-I) transgenic mice were purchased from the Jackson Laboratory. Mice were housed in animal facilities at Stanford University Medical School and University of California, Berkeley, and maintained according to protocols approved by the Institutional Animal Care and Use Committees on Animal Care.
Design of CD45-Targeted NK Receptor Antagonists
The hybridomas of clone 5E6 were kindly gifted by Dr. Wayne M. Yokoyama, and of clones 20D5 and 16A11 were kindly gifted by Dr. David Raulet. The VL and VH sequences of 5E6, 20D5, and 16A11 antibodies were extracted from the sequencing results provided by GenScript PROBIO. A mouse CD45-NKR molecule is composed of an anti-mouse CD45 nanobody49 fused to an anti-mouse NK receptor scFv using a GGGGTGGS linker. The sequences of human antibodies against NKG2A, KIR2DL, KIR3DL, or CD45 were extracted from patents.50−53 The bispecific diabody construct of human CD45-NKRs has the following arrangement: VH1-(G4S)-VL2-(G4S)3-VH2-(G4S)-VL1 and 1 and 2 corresponding to anti-CD45 and anti-NK receptor scFv.
Protein Expression and Purification from Insect Cells
SF9 cells were transfected with plasmids making P0 viruses. P1 viruses were made from P0 infected SF9 cells. 1 or 2 L of Hi5 cells at 2 × 106/mL were infected with 1–3 mL P1 viruses. After 2–3 days of protein production, the supernatants were harvested and purified by incubating with Ni-NTA resin for 4 h at room temperature. The resin was collected and washed with 1 × HBS containing 20 mM imidazole. Proteins were eluted with 1 × HBS containing 250 mM imidazole, filtered, and purified by size-exclusion chromatography (SEC). Protein fractions were collected and concentrated. After snap-freezing in liquid nitrogen, the proteins were stored in a −80 °C freezer. The purity and size of the proteins were further confirmed by SDS-PAGE and Coomassie blue staining.
Generation of Mouse Effector NK Cells
Spleens were dissected and minced from C57BL/6 mice. Red blood cells (RBCs) were lysed with 1 × ACK lysis buffer (BioLegend). The NK cells were purified with NK cell isolation kit (Miltenyi Biotec) according to manufacturer’s instructions, and cultured in complete RPMI1640 medium with 100 IU/mL murine IL-2 at 0.2–0.5 × 106/mL. Fresh medium was added every other day. The cells were harvested on day 5 or 6, stained with flow antibodies, YLI-90 for anti-Ly49I, 16A11 for anti-NKG2A, and sorted for live CD3–CD49b+Ly49+ or NKG2A+ NK cells using a SH800S cell sorter (Sony). The sorted cells were allowed to recover for 2 days before using as effector cells in the co-culture assays.
Antibodies for Flow Cytometry
Mouse experiments: antibodies to CD16/32 (93), CD107a (1D4B, APC-Cy7), CD69 (H1.2F3, BV785), GM-CSF (MP1-22E9, PE-Cy7), CD49b (DX5, PE), and CD94 (18d3, PerCP/Cy5.5) were purchased from BioLegend. Anti-Ly49I (YLI-90, FITC), anti-CD3 (145-2C11, PerCP/Cy5.5), anti-CD8 (53–6.7, PE-Cy7), anti-MHC I (AF6-88.5.5.3, APC) and goat anti-mouse IgG cross-adsorbed secondary antibody, and Alexa Fluor 647 were purchased from Thermo Fisher. Antibodies to Ly49C and Ly49I (5E6, FITC), Qa-1b (6A8.6F10.1A6, PE), and IFNγ (XMG1.2, Alexa Fluor 647) were purchased from BD Bioscience. Anti-HA (6E2, Pacific Blue) was purchased from Cell Signaling Technology. Anti-NKG2A (705829, Alexa Fluor 647) was purchased from R&D Systems. Human experiments: Antibodies to CD16/32/64 (TrueStain-FcX), CD3 (UCHT1, APC/Cy7), CD56 (NCAM, BV605), CD69 (FN50, PE), CD107a (H4A3, FITC), MIP-1β (D21-1351, PE), IL-2 (MQ1-17H12, FITC), HLA-E (3D12, APC), and β2M (2M2, FITC) were purchased from BioLegend. Anti-NKG2A (REA110, APC) and anti-KIR3DL1 (DX9, PE) were purchased from Miltenyi Biotec. Anti-HLA-C (DT-9, BV421) was purchased from BD Bioscience. Anti-Granzyme B (GB12, PE) was purchased from Thermo Fisher. Anti-KIR2DL1 (143211, Alexa Fluor 488) was purchased from R&D Systems. Anti-NKG2A (Z199, APC) was purchased from Beckman Coulter. Anti-phospho-Erk1/2 (Thr202/Tyr204) (197G2, Alexa Fluor 647) and anti-phospho-S6 ribosomal protein (Ser235/236) (2F9, Alexa Fluor 488) were purchased from Cell Signaling Technology. The 4LO3311 hybridoma (anti-Ly49C) was a generous gift from S. Lemieux (Quebec, Canada), and the mAb was purified from CELLine classic 1000 (Integra Biosciences, Chur, Switzerland) supernatant and then conjugated to biotin according to standard methods. Biotin-conjugated mAb were detected with streptavidin PE-Cy7 (BioLegend).
Antibodies for Cell Activation
Anti-mouse NKG2A (16A11, purified) was purchased from BioLegend. Anti-mouse NKG2A (20D5, purified) and anti-mouse CD28 (37.51, purified) were purchased from BioXcell. Anti-mouse Ly-49C and Ly-49I (5E6, purified) were purchased from BD Bioscience.
Flow Cytometry
Cells were washed in FACS buffer (2% FBS in PBS) and incubated with the Fc receptor blocking antibody for 10 min, followed by staining with anti-CD3, NK1.1, Ly49I, NKG2A for 20 min at 4 °C. Intracellular staining of IFNγ, CCL5, or IL-2 was performed according to the protocol of eBioscience intracellular fixation & permeabilization buffer set (Cat.88-8824-00, Thermo Fisher). Briefly, cells were fixed in fixation buffer for 20 min at room temperature. After washing with FACS buffer, cells were resuspended, washed in 1X permeabilization buffer, and stained with anti-IFNγ, CCL5, or IL-2 antibodies for 20 min at 4 °C. Cells were resuspended in FACS buffer after washing. Flow cytometry was performed using a CytoFlex (Beckman Coulter). Data were analyzed using FlowJo software.
NK Cytotoxicity Assay
For CD45–20D5, RMA cells were preincubated with 20 ng/mL of IFNγ for 2 days. For CD45–5E6, RMA cells did not require IFNγ stimulation. Target cells were labeled with 1 μM cell trace violet (CTV) (Thermo Fisher) for 20 min at 37 °C. Effector NK cells were co-cultured with 10,000 target cells at different effector/target ratios in the presence of 100 nM CD45-NKR molecules. After 4 h, the cells were spun down, washed with PBS, and assayed with Annexin V/7-AAD staining kit (BioLegend). The cells were stained with Annexin V, diluted in 1x Annexin V buffer for 15 min, and incubated with 2x 7-AAD for 5 min. Dead cells defined as AnnexinV+ and/or 7AAD+ cells were detected in a CytoFlex flow cytometer (Beckman Coulter). The cells were stained with Annexin V, diluted in 1x Annexin V buffer for 15 min, and incubated with 2x 7-AAD for 5 min. Dead cells defined as AnnexinV+ and/or 7AAD+ cells were detected in a CytoFlex flow cytometer (Beckman Coulter).Dead cells were defined as Annexin V+. The cells were stained in a Cytoflex cytometer with Annexin V, diluted in 1x Annexin V buffer for 15 min, and incubated with 2x 7-AAD for 5 min.
NK Degranulation and Activation Assays
Effector NK cells at 1 × 106/mL were co-cultured with target cells at 1:2 ratio for 4 h, anti-CD107a (1D4B, APC-Cy7), brefeldin A (BD), monensin (BD), and serial dilutions (6×) of CD45-NKR and antibody controls were added to the co-culture wells at the beginning of the assay. The cells were stained with a panel of antibodies for gating NK subsets and testing NK activation. Production of effector cytokines was tested by intracellular staining of IFNγ. The cells were analyzed in a Cytoflex flow cytometer. FACS results were plotted by FlowJo (TreeStar) and Prism 9.3.1 (GraphPad).
Generation of Mouse Effector OT-I Cells
Splenocytes were harvested from OT-I mice. CD8+ OT-I cells were purified by the CD8+ T cell isolation kit (Mitenyi Biotec) and cultured for 7 days in complete RPMI1640 supplemented with 0.5 μg/mL anti-CD28 (37.51, purified), 1 μg/mL OVA257-264 peptide (GenScript), and 100 IU/mL mIL-2. The cells were split when the concentration reached 1.5 × 106/mL. NKG2A expression level was confirmed by FACS staining.
OT-I Degranulation and Activation Assays
B16-OVA cells were preincubated with 20 ng/mL IFNγ for upregulation of MHC I and Qa-1b. Target cells were labeled with 1 μM CTV prior to co-culture. In the presence of anti-mouse CD107a, brefeldin A (BD), and monensin (BD), effector OT-I cells at 1 × 106/mL were co-cultured with target cells at a ratio of 1:2 by adding serial dilutions (6×) of CD45–20D5 and 20D5-scFv to the co-culture wells. 4 h later, the cells were stained with anti-CD3, CD8, NK1.1, NKG2A, CD69, and CD25. Anti-NKG2A (705829, Alexa Fluor 647) was used for gating NKG2A+ OT-I cells. Effector cytokine production was tested by intracellular staining for IFNγ and IL-2.
NK Responsiveness Assay
At day 0, C57BL/6 mice were treated with 200 μg poly(I/C) (Invivogen) for 2 days. RMA cells were treated with 20 ng/mL of IFNγ for upregulating Qa-1b. At day 2, target cells were labeled with 1 μM CTV (Thermo Fisher) for 20 min at 37 °C. The splenocytes were harvested. Splenic cells at 1 × 106 were co-cultured with target cells at a 1:2 effector to target ratio in the presence of anti-CD107a (Biolegend), brefeldin A, and monensin, and serial dilutions (6×) of CD45-NKR molecules for 5 h were added at the beginning of the assay. NK subpopulations were stained and gated by using a panel of antibodies prior to detection. Anti-Ly49I (YLI-90, FITC) and anti-NKG2A (705829, Alexa Fluor 647) were used for gating Ly49I+ and NKG2A+ cells. Cells were analyzed in a Cytoflex flow cytometer. NK responsiveness was assayed by cell surface staining of CD107a and intracellular staining of IFNγ. Results were plotted by FlowJo and Prism 9.3.1.
Combination Treatment Assay
RMA cells were treated with 20 ng/mL of IFNγ to induce upregulation of Qa-1b. 1 × 106 Splenocytes from poly(I/C)-treated mice were harvested and co-cultured with 2 × 105 RMA or RMA-B2m–/– cells in the absence or presence of 10 or 100 nM 5E6-scFv, CD45–5E6, 20D5-scFv, CD45–20D5, 5E6-scFv + 20D5-scFv, and CD45–5E6 + CD45–20D5 for 5 h. Anti-CD107a, brefeldin A, and monensin were added to the co-culture wells before the experiment. The cells were stained with FACS antibodies and analyzed in Cytoflex. Anti-Ly49I (YLI-90, FITC), anti-Ly49C (4LO3311, biotin-conjugated antibody followed by secondary staining with streptavidin–PE–Cy7), and anti-NKG2A (705829, Alexa Fluor 647) were used for gating Ly49I+, Ly49C+, and NKG2A+ cells. NK responsiveness in Ly49C+Ly49I+NKG2A+, Ly49C+Ly49I+NKG2A–, Ly49C+Ly49I–NKG2A+, Ly49C–Ly49I+NKG2A+, Ly49C+Ly49I–NKG2A–, Ly49C–Ly49I+NKG2A–, Ly49C–Ly49I–NKG2A+, and Ly49C–Ly49I–NKG2A– cells was tested by cell surface staining of CD107a. Results were plotted by FlowJo and Prism 9.3.1.
Human NK Activation
Leukoreduction system (LRS) chambers from different donors were sourced from Stanford Blood Center. Peripheral blood mononucleated cells were isolated by density gradient. Human PBMC cells at 0.8 × 106/mL were cultured in NK MACS medium supplemented with 5% human AB serum (Millipore Sigma), NK MACS supplement, and 500 IU/mL human IL-2 (Miltenyi Biotec). Human NK cells were cultivated and expanded for 10–14 days. The CD3–CD56+ NK cells were confirmed by FACS staining.
Statistical Analysis
Statistics were performed using Prism v9.3.1(350) (GraphPad Software). Data are expressed as mean ± standard deviation (SD) unless otherwise indicated. We used Student’s t-test to analyze experiments with two groups, one-way ANOVA, followed by Dunnett’s multiple comparison tests and two-way ANOVA, and followed by Tukey’s multiple comparison test to analyze experiments with more than two groups. Significance is indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
Acknowledgments
We thank Thorbald van Hall for WT RMA and RMA-Qa-1b KO cells. We acknowledge Dario Campana for sharing the K562-GpHLA-E cells. This work was supported by 2 R01 CA177684 06A1 and Emerson Collective to K.C.G. and by the NIH grant R01 AI113041 to DHR. Y.J. is the recipient of a “CRI/Amgen Postdoctoral Fellowship” (CRI award 3984). Portions of the schematics in the figures were generated by BioRender.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00337.
Engineering strategy of inhibitory CD45-NKR, validating surface expression of Ly49I and MHC I, gating scheme for target cell lysis, validating surface expression of Qa-1b and MHC I in target cells, gating scheme for NK responsiveness by detecting CD107a, additional data of CD45–5E6 and CD45–20D5 combinational treatment, additional data of human CD45-NKRs, unprocessed gels, plasmids used for protein expression, protein sequences of mouse CD45-NKRs, protein sequences of mouse NKRs, protein sequences of human CD45-NKRs, HEK293 dephosphorylation assay, and 3C linker experiment (PDF)
Author Contributions
K.C.G. conceived the project. J.R., D.H.R., and K.C.G. wrote the original manuscript. J.R., Y.J., L.L.S., and L.K.P. designed and performed the experiments. J.R. and Y.J. analyzed the data. K.C.G. and D.H.R. supervised the experiments and provided resources.
The authors declare the following competing financial interest(s): K.C.G. and J.R. are co-inventors on a patent WO/2022/081975 based upon the technology described in this manuscript. K.C.G. is the founder of Indupro Therapeutics. D.H.R. is the cofounder of Dragonfly Therapeutics.
Supplementary Material
References
- Björkström N. K.; Strunz B.; Ljunggren H.-G. Natural killer cells in antiviral immunity. Nat. Rev. Immunol. 2022, 22, 112–123. 10.1038/s41577-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.; Huang J. CD8+ T cells and NK cells: parallel and complementary soldiers of immunotherapy. Curr. Opin. Chem. Eng. 2018, 19, 9–20. 10.1016/j.coche.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N. K.; Kissiov D. U.; Raulet D. H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 2022, 1–16. 10.1038/s41577-022-00732-1. [DOI] [PubMed] [Google Scholar]
- Höglund P.; Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat. Rev. Immunol. 2010, 10, 724–734. 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- Pfefferle A.; Jacobs B.; Haroun-Izquierdo A.; Kveberg L.; Sohlberg E.; Malmberg K.-J. Deciphering Natural Killer Cell Homeostasis. Front. Immunol. 2020, 11, 812. 10.3389/fimmu.2020.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 2018, 154, 383–393. 10.1111/imm.12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiani M. D.; Aguado E.; Tarazona R.; Romero P.; Molina I.; Santamaria M.; Solana R.; Peña J. Expression of killer inhibitory receptors on cytotoxic cells from HIV-1-infected individuals. Clin. Exp. Immunol. 1999, 115, 472–476. 10.1046/j.1365-2249.1999.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biassoni R. Human Natural Killer Receptors, Co-Receptors, and Their Ligands. Curr. Protoc. Immunol. 2009, 84, 14. 10.1002/0471142735.im1410s84. [DOI] [PubMed] [Google Scholar]
- Moretta L.; Bottino C.; Cantoni C.; Mingari M. C.; Moretta A. Human natural killer cell function and receptors. Curr. Opin. Pharmacol. 2001, 1, 387–391. 10.1016/s1471-4892(01)00067-4. [DOI] [PubMed] [Google Scholar]
- Vance R. E.; Kraft J. R.; Altman J. D.; Jensen P. E.; Raulet D. H. Mouse CD94/NKG2A Is a Natural Killer Cell Receptor for the Nonclassical Major Histocompatibility Complex (MHC) Class I Molecule Qa-1b. J. Exp. Med. 1998, 188, 1841–1848. 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A.; Kubota S.; Lohwasser S.; Mager D. L.; Takei F. Diversity of NK cell receptor repertoire in adult and neonatal mice. J. Immunol. 1999, 163, 212–216. [PubMed] [Google Scholar]
- Lanier L. L. Natural Killer Cells: From No Receptors to Too Many. Immunity 1997, 6, 371–378. 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- Brooks A. G.; Posch P. E.; Scorzelli C. J.; Borrego F.; Coligan J. E. NKG2A Complexed with CD94 Defines a Novel Inhibitory Natural Killer Cell Receptor. J. Exp. Med. 1997, 185, 795–800. 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P.; Denis C.; Soulas C.; Bourbon-Caillet C.; Lopez J.; Arnoux T.; Bléry M.; Bonnafous C.; Gauthier L.; Morel A.; Rossi B.; Remark R.; Breso V.; Bonnet E.; Habif G.; Guia S.; Lalanne A. I.; Hoffmann C.; Lantz O.; Fayette J.; Boyer-Chammard A.; Zerbib R.; Dodion P.; Ghadially H.; Jure-Kunkel M.; Morel Y.; Herbst R.; Narni-Mancinelli E.; Cohen R. B.; Vivier E. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743. 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfoort N.; Borst L.; Korrer M. J.; Sluijter M.; Marijt K. A.; Santegoets S. J.; van Ham V. J.; Ehsan I.; Charoentong P.; André P.; Wagtmann N.; Welters M. J. P.; Kim Y. J.; Piersma S. J.; van der Burg S. H.; van Hall T. NKG2A Blockade Potentiates CD8 T Cell Immunity Induced by Cancer Vaccines. Cell 2018, 175, 1744–1755. 10.1016/j.cell.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J.; Wang X.; Radfar S.; Sproule T. J.; Roopenian D. C.; Cantor H. CD8+ T regulatory cells express the Ly49 Class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc. Natl. Acad. Sci. 2011, 108, 2010–2015. 10.1073/pnas.1018974108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi N.; Robbins S. H.; Ugolini S.; Georgel P.; Hoebe K.; Bouneaud C.; Ronet C.; Kaser A.; DiCioccio C. B.; Tomasello E.; Blumberg R. S.; Beutler B.; Reiner S. L.; Alexopoulou L.; Lantz O.; Raulet D. H.; Brossay L.; Vivier E. Expansion and Function of CD8+ T Cells Expressing Ly49 Inhibitory Receptors Specific for MHC Class I Molecules. J. Immunol. 2004, 173, 3773–3782. 10.4049/jimmunol.173.6.3773. [DOI] [PubMed] [Google Scholar]
- McMahon C. W.; Raulet D. H. Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol. 2001, 13, 465–470. 10.1016/s0952-7915(00)00242-9. [DOI] [PubMed] [Google Scholar]
- Ugolini S.; Vivier E. Regulation of T cell function by NK cell receptors for classical MHC class I molecules. Curr. Opin. Immunol. 2000, 12, 295–300. 10.1016/s0952-7915(00)00090-x. [DOI] [PubMed] [Google Scholar]
- McMahon C. W.; Zajac A. J.; Jamieson A. M.; Corral L.; Hammer G. E.; Ahmed R.; Raulet D. H. Viral and Bacterial Infections Induce Expression of Multiple NK Cell Receptors in Responding CD8+ T Cells. J. Immunol. 2002, 169, 1444–1452. 10.4049/jimmunol.169.3.1444. [DOI] [PubMed] [Google Scholar]
- Sun C.; Xu J.; Huang Q.; Huang M.; Wen H.; Zhang C.; Wang J.; Song J.; Zheng M.; Sun H.; Wei H.; Xiao W.; Sun R.; Tian Z. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology 2017, 6, 1264562. 10.1080/2162402X.2016.1264562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T.; Seow S. V.; Wong D.; Robinson M.; Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest. 2019, 129, 2094–2106. 10.1172/jci123955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burshtyn D. N.; Yang W.; Yi T.; Long E. O. A Novel Phosphotyrosine Motif with a Critical Amino Acid at Position −2 for the SH2 Domain-mediated Activation of the Tyrosine Phosphatase SHP-1. J. Biol. Chem. 1997, 272, 13066–13072. 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- Sternberg-Simon M.; Brodin P.; Pickman Y.; Önfelt B.; Kärre K.; Malmberg K.-J.; Höglund P.; Mehr R. Natural Killer Cell Inhibitory Receptor Expression in Humans and Mice: A Closer Look. Front. Immunol. 2013, 4, 65. 10.3389/fimmu.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanen J. B. A. G.; Robert C. Immune Checkpoint Inhibitors. Prog. Tumor. Res. 2015, 42, 55–66. 10.1159/000437178. [DOI] [PubMed] [Google Scholar]
- Weiner G. J. Building better monoclonal antibody-based therapeutics. Nat. Rev. Cancer 2015, 15, 361–370. 10.1038/nrc3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanishi J.; Mandai M.; Ikeda T.; Minami M.; Kawaguchi A.; Matsumura N.; Abiko K.; Baba T.; Yamaguchi K.; Ueda A.; Kanai M.; Mori Y.; Matsumoto S.; Murayama T.; Chikuma S.; Morita S.; Yokode M.; Shimizu A.; Honjo T.; Konishi I. Efficacy and safety of anti-PD-1 antibody (Nivolumab: BMS-936558, ONO-4538) in patients with platinum-resistant ovarian cancer. J. Clin. Oncol. 2014, 32, 5511. 10.1200/jco.2014.32.15_suppl.5511. [DOI] [Google Scholar]
- Leach D. R.; Krummel M. F.; Allison J. P. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Science 1996, 271, 1734–1736. 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- Freeman G. J.; Long A. J.; Iwai Y.; Bourque K.; Chernova T.; Nishimura H.; Fitz L. J.; Malenkovich N.; Okazaki T.; Byrne M. C.; Horton H. F.; Fouser L.; Carter L.; Ling V.; Bowman M. R.; Carreno B. M.; Collins M.; Wood C. R.; Honjo T. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J. Exp. Med. 2000, 192, 1027–1034. 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic A.; Fiegler N.; Brunner-Weinzierl M.; Cerwenka A. CTLA-4 Is Expressed by Activated Mouse NK Cells and Inhibits NK Cell IFN-γ Production in Response to Mature Dendritic Cells. J. Immunol. 2014, 192, 4184–4191. 10.4049/jimmunol.1302091. [DOI] [PubMed] [Google Scholar]
- Concha-Benavente F.; Kansy B. A.; Moskovitz J.; Moy J. D.; Chandran U. R.; Ferris R. L. PD-L1 mediates dysfunction in activated PD-1+ NK cells in head and neck cancer patients. Cancer Immunol. Res. 2018, 6, 1548–1560. 10.1158/2326-6066.cir-18-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey N.; Dumas P.-Y.; Recher C.; Gastaud L.; Lioure B.; Bulabois C.-E.; Pautas C.; Marolleau J.-P.; Leprêtre S.; Raffoux E.; Thomas X.; Hicheri Y.; Bonmati C.; Quesnel B.; Rousselot P.; Castaigne S.; Jourdan E.; Malfuson J. V.; Guillerm G.; Bouhris J. H.; Ojeda M.; Hunault M.; Ifrah N.; Gardin C.; Delannoy A.; Beautier L.; Paturel C.; Andre P.; Zerbib R.; Preudhomme C.; Toubert A.; Dulphy N.; Olive D.; Pigneux A.; Dombret H. Randomized Phase 2 Trial of Lirilumab (anti-KIR monoclonal antibody, mAb) As Maintenance Treatment in Elderly Patients (pts) with Acute Myeloid Leukemia (AML): Results of the Effikir Trial. Blood 2017, 130, 889. 10.1182/blood.v130.suppl_1.889.889. [DOI] [Google Scholar]
- Daver N. G.; Garcia-Manero G.; Cortes J. E.; Basu S.; Ravandi F.; Kadia T. M.; Borthakur G.; Jabbour E.; Dinardo C. D.; Pemmaraju N.; Brandt M.; Pierce S.; Hussin N.; Kornblau S. M.; Andreeff M.; Konopleva M.; Ning J.; Allison J. P.; Sharma P.; Kantarjian H. M. Phase IB/II study of lirilumab with azacytidine (AZA) in relapsed AML. J. Clin. Oncol. 2017, 35, e18505 10.1200/jco.2017.35.15_suppl.e18505. [DOI] [Google Scholar]
- Vey N.; Bourhis J.-H.; Boissel N.; Bordessoule D.; Prebet T.; Charbonnier A.; Etienne A.; Andre P.; Romagne F.; Benson D.; Dombret H.; Olive D. A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood 2012, 120, 4317–4323. 10.1182/blood-2012-06-437558. [DOI] [PubMed] [Google Scholar]
- Vey N.; Bourhis J.-H.; Recher C.; Etienne A.; Charbonnier A.; Andre P.; Rey J.; Calmels F.; Zerbib R.; Buffet R.; Prebet T. Repeated Dosing Of Anti-KIR (IPH2101) As Maintenance Therapy In Ederly Patients With Acute Myeloid Leukemia. Blood 2013, 122, 2696. 10.1182/blood.v122.21.2696.2696. [DOI] [Google Scholar]
- Davis S. J.; van der Merwe P. A. van der. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 2006, 7, 803–809. 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- Chang V. T.; Fernandes R. A.; Ganzinger K. A.; Lee S. F.; Siebold C.; McColl J.; Jönsson P.; Palayret M.; Harlos K.; Coles C. H.; Jones E. Y.; Lui Y.; Huang E.; Gilbert R. J. C.; Klenerman D.; Aricescu A. R.; Davis S. J. Initiation of T cell signaling by CD45 segregation at “close-contacts”. Nat. Immunol. 2016, 17, 574–582. 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R. A.; Su L.; Nishiga Y.; Ren J.; Bhuiyan A. M.; Cheng N.; Kuo C. J.; Picton L. K.; Ohtsuki S.; Majzner R. G.; Rietberg S. P.; Mackall C. L.; Yin Q.; Ali L. R.; Yang X.; Savvides C. S.; Sage J.; Dougan M.; Garcia K. C. Immune receptor inhibition through enforced phosphatase recruitment. Nature 2020, 586, 779–784. 10.1038/s41586-020-2851-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. Y. L.; George T.; Dorfman J. R.; Roland J.; Kumar V.; Bennett M. The Role of Ly49A and 5E6(Ly49C) Molecules in Hybrid Resistance Mediated by Murine Natural Killer Cells Against Normal T Cell Blasts. Immunity 1996, 4, 67–76. 10.1016/s1074-7613(00)80299-x. [DOI] [PubMed] [Google Scholar]
- Vance R. E.; Jamieson A. M.; Cado D.; Raulet D. H. Implications of CD94 deficiency and monoallelic NKG2A expression for natural killer cell development and repertoire formation. Proc. Natl. Acad. Sci. 2002, 99, 868–873. 10.1073/pnas.022500599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez N. C.; Treiner E.; Vance R. E.; Jamieson A. M.; Lemieux S.; Raulet D. H. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 2005, 105, 4416–4423. 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet D. H.; Vance R. E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006, 6, 520–531. 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- Cotton A. D.; Nguyen D. P.; Gramespacher J. A.; Seiple I. B.; Wells J. A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn G.; Banik S. M.; Miller C. L.; Riley N. M.; Cochran J. R.; Bertozzi C. R. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol. 2021, 17, 937–946. 10.1038/s41589-021-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston M. L.; Xu Z.; Weiss A. CD45: A Critical Regulator of Signaling Thresholds in Immune Cells. Annu. Rev. Immunol. 2003, 21, 107–137. 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- McNeill L.; Salmond R. J.; Cooper J. C.; Carret C. K.; Cassady-Cain R. L.; Roche-Molina M.; Tandon P.; Holmes N.; Alexander D. R. The Differential Regulation of Lck Kinase Phosphorylation Sites by CD45 Is Critical for T Cell Receptor Signaling Responses. Immunity 2007, 27, 425–437. 10.1016/j.immuni.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Doucey M.-A.; Scarpellino L.; Zimmer J.; Guillaume P.; Luescher I. F.; Bron C.; Held W. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat. Immunol. 2004, 5, 328–336. 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- Rossotti M.; Tabares S.; Alfaya L.; Leizagoyen C.; Moron G.; González-Sapienza G. Streamlined method for parallel identification of single domain antibodies to membrane receptors on whole cells. Biochim. Biophys. Acta, Gen. Subj. 2015, 1850, 1397–1404. 10.1016/j.bbagen.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi N.; Gauthier L.; Morel Y.; Moretta A.; Parolini S.; Rossi B.. Anti-kir3d antibodies. U.S. Patent 20,120,064,081 A1, 2012.
- Spee P. J. L.; Padkaer S. B.. Anti-NKG2A antibodies and uses thereof. U.S. Patent 9,683,041 B2, 2014.
- Kolbinger F.; Herrera J. M. C.; Aszódi A.; Saldanha J. W.; Hall B. M.; Gregori S.; Roncarolo M. G.; Loux V.; Aversa G.; Jeschke M.. Therapeutic binding molecules. WO 2005026210 A2, 2005.
- Moretta A.; Chiesa M. D.; Andre P.; Gauthier L.; Romagne F.; Wagtmann P. A. N. R.; Svendsen I.; Zahn S.; Svensson A.; Thorolfsson M.; Padkaer S. B.; Kjaergaard K.; Spee P.; Wilken M.. Pan-KIR2DL NK-receptor antibodies and their use in diagnostik and therapy. WO 2005003172 A2, 2005.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.