Abstract

Nicotiana benthamiana is a valuable plant chassis for heterologous production of medicinal plant natural products. This host is well suited for the processing of organelle-localized plant enzymes, and the conservation of the primary metabolism across the plant kingdom often provides required plant-specific precursor molecules that feed a given pathway. Despite this commonality in metabolism, limited precursor supply and/or competing host pathways can interfere with yields of heterologous products. Here, we use transient transcriptional reprogramming of endogenous N. benthamiana metabolism to drastically improve flux through the etoposide pathway derived from the medicinal plant Podophyllum spp. Specifically, coexpression of a single lignin-associated transcription factor, MYB85, with pathway genes results in unprecedented levels of heterologous product accumulation in N. benthamiana leaves: 1 mg/g dry weight (DW) of the etoposide aglycone, 35 mg/g DW (−)-deoxypodophyllotoxin, and 3.5 mg/g DW (−)-epipodophyllotoxin—up to two orders of magnitude above previously reported biosynthetic yields for the etoposide aglycone and eight times higher than what is observed for (−)-deoxypodophyllotoxin in the native medicinal plant. Unexpectedly, transient activation of lignin metabolism by transcription factor overexpression also reduces the production of undesired side products that likely result from competing N. benthamiana metabolism. Our work demonstrates that synthetic activation of lignin biosynthesis in leaf tissue is an effective strategy for optimizing the production of medicinal compounds derived from phenylpropanoid precursors in the plant chassis N. benthamiana. Furthermore, our results highlight the engineering value of MYB85, an early switch in lignin biosynthesis, for on-demand modulation of monolignol flux and support the role of MYB46 as a master regulator of lignin polymer deposition.
Keywords: plant metabolic engineering, transcriptional activation, lignin biosynthesis, heterologous biosynthesis, plant chassis, etoposide biosynthesis
Introduction
Plants possess an expansive repertoire of biosynthetic pathways to specialized metabolites with broad chemical diversity. Their structural complexity arises from the combination and modification of a relatively small set of primary metabolic building blocks such as amino acids, isoprenoid precursors, and acetyl-CoA, as well as plant-specific molecules including monolignols. The conservation of these core metabolic processes across the Plant Kingdom allows for the transfer of heterologous specialized plant metabolic pathways from difficult-to-cultivate medicinal plants into a plant chassis lab model such as Nicotiana benthamiana. However, because primary metabolism is a highly coordinated and controlled network that is optimized for concerted growth and development, it can limit flux through an engineered pathway.1 Yield optimization therefore may require redistribution of the native supply of primary metabolites into specialized metabolism ideally with minimal impacts on host viability. Successful approaches include overexpression of individual native enzymes that helped alleviate precursor supply bottleneck2,3 and the expression of orthogonal pathways that override native regulation.4,5
Tuning the endogenous transcription regulation of the plant host has been proposed as a potentially more efficient engineering strategy to feed a heterologous pathway, as it takes advantage of native coordination of primary metabolic genes.6,7 Transcription factors orchestrate the dynamic network of cellular responses to biotic and abiotic stimuli, as related to developmental stage, external stress, homeostasis, and natural metabolism.8−10 By directly binding to DNA or interacting with DNA-binding proteins, transcription factors control the metabolic network by up- or down-regulating gene expression levels. As more of the ∼2000 transcription factors found in a given plant species are functionally characterized, there has been interest in applying transcription regulation to metabolic engineering for various applications including reprogramming of differentiation in somatic cells11 and reactivation of endogenous metabolism.12,13
Changes in endogenous plant metabolism have been demonstrated with the use of transcription factors for a number of biosynthetic pathways such as anthocyanins (overexpression of snapdragon activators Del and Ros1 in tomato),13 isoprenoids (silencing of repressor MsYABBY5),14 isoflavones (transgenic Nicotiana tabacum coexpressing AtMYB12 and GmIFS1),15 and lignin (transgenic N. tabacum expressing Medicago WRKYs).12 Given this precedence, we considered that reprogramming host metabolism with transcription factors could be an effective way to boost yields of a transiently expressed heterologous pathway and sought to test this hypothesis in the context of etoposide aglycone biosynthesis in N. benthamiana.
Etoposide is a clinically used chemotherapeutic and is produced from lignans found in the medicinal plant Podophyllum spp. The etoposide aglycone (EA)2 can be produced in N. benthamiana by Agrobacterium-mediated transient expression of pathway genes. However, yields are limited by the availability of coniferyl alcohol (CA), a monolignol that is also a main building block for the abundant plant biopolymer lignin. In our previous work,3 we showed that exogenous addition of CA increased yields of (−)-deoxypodophyllotoxin (DPT) in N. benthamiana leaves. By boosting CA production through overexpression of the canonical primary metabolic enzymes, we achieved milligram-level production of DPT.3 Despite these yield improvements, the levels of the lignan products were still low relative to what has been observed for lignin biosynthesis engineering (e.g., ∼30 mg/g in transgenic tobacco expressing Medicago truncatula transcription factors).12 In contrast, the high flux through the monolignol pathway required for generating large quantities of lignin suggested that the innate capacity in plant tissue for diverting fixed carbon to CA could be very high. Although CA is a ubiquitous plant metabolite, we reasoned that it is likely not present in leaves at the time of Agrobacterium-mediated DNA transfer given that (1) epidermal cells have already fully expanded, (2) lignin biosynthesis has already ceased, and (3) the leaf parenchyma does not normally contain high levels of lignin (e.g., compared to the sclerenchyma).
In lignin biosynthesis, as related to development16−18 or defense response,19,20 transcriptional regulation is linked not only to the biosynthesis of monolignols—building blocks of lignin—but also other components of the secondary cell wall such as cellulose and xylan.21−23 Transcription factors involved in lignin biosynthesis are often identified with their ability to bind to the AC elements, common regulatory elements present in the promoter or 5′ untranslated regions of monolignol biosynthetic genes.24,25 Among those, certain transcription factors, such as MYB58, MYB85, and MYB103, are believed to be direct switches to monolignol biosynthesis,26,27 while others, such as SND1, MYB46, and VND6/7 (specific to xylem vessel formation), are considered to be master switches involved in controlling expression levels of those direct transcription factors and multiple processes.28−31 While characterization of transcription factors involved in monolignol biosynthesis has often been signified by ectopic deposition of lignin or reporter activation under the control of the promoter of a monolignol biosynthetic gene (e.g., 4CL),27 a primary metabolite flux increase has not been directly observed, because these products are believed to be readily utilized by the upregulated downstream processes (e.g., monolignols undergoing dehydrogenative polymerization to form lignin or dimerization to form lignans). For this study, we sought to reactivate early lignin biosynthesis in leaf cells during the heterologous pathway expression to improve the yields of engineering medicinal lignans. We hypothesized that overexpression of lignin biosynthesis transcription factors would increase coniferyl alcohol availability, which would translate into increased production levels of etoposide aglycone and its intermediates (Figure 1A).
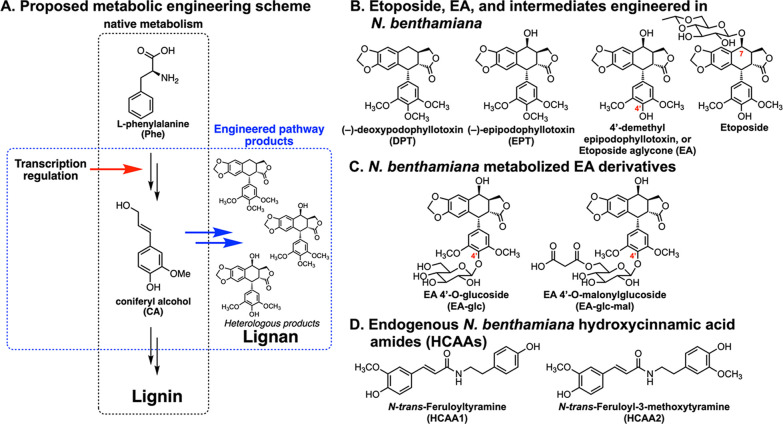
Figure 1.
Proposed scheme for transcriptional regulation engineering and etoposide-related heterologous products. (A) Proposed transcriptional regulation engineering to divert native metabolism for high-value heterologous product biosynthesis. The red arrow represents activation, black arrows represent the native metabolic pathway, and blue arrows represent the heterologous pathway. (B–D) Chemical structures of desired heterologous products and other N. benthamiana endogenous metabolites and metabolic products. O-Glycosylation on C7 is a synthetic modification of EA to produce etoposide, while undesired 4′-O-glycosylation (shown in panel C) results from native metabolism in N. benthamiana.
Results and Discussion
Repurposing of Lignin-Associated Transcription Factors for Lignan Biosynthesis
With ∼2000 transcription factors (TFs) identified in the Arabidopsis thaliana genome32−35 and a wealth of genetic and functional characterization of those TFs available,36,37 we chose to focus on heterologous expression of A. thaliana TFs in N. benthamiana using Agrobacterium-mediated transient DNA delivery for engineering the transcriptional regulation of lignan biosynthesis. Six TFs previously characterized and associated with lignin and monolignol biosynthesis were selected for initial study: AtMYB85,26AtMYB46,28AtMYB103,26AtMYB58,27AtMYB63,27 and AtVND6.30,31 Previously, we have found that individual overexpression of each CA pathway enzyme with DPT biosynthetic enzymes resulted in a yield of 4.3 mg/g dry weight (DW) for the lignan DPT,3 comparable to the biosynthetic yield of 4.5 mg/g DW found in the roots of the native plant Podophyllum hexandrum.38 We sought to test if expression of a single TF alone instead could further improve pathway flux beyond the precedent.
When each transcription factor was coexpressed with the DPT biosynthetic pathway enzymes via Agrobacterium-mediated transient expression, AtMYB46 and AtMYB85 promoted the highest yields for DPT production (Figure 2 and Figure S1). Moreover, with CYP82D61 converting DPT to EPT, milligram-scale yields were obtained with either AtMYB46 or AtMYB85, up to 60-fold improvement from the biosynthetic yield of 57.8 μg/g DW achieved with exogenous addition of the precursor (+)-pinoresinol.2 Finally, with both CYP82D61 and CYP71BE54 present to convert DPT to EA, AtMYB85 coexpression resulted in a striking improvement in EA biosynthetic yield at 0.98 mg/g DW, a 95-fold increase from the precursor-supplied biosynthetic yield of 10.3 μg/g DW.2 Interestingly, we observed significant accumulation of intermediates only when AtMYB85 was expressed (Figure S2). The accumulation of intermediate compounds in the biosynthetic pathway indicated that the precursor supply increase induced by AtMYB85 surpassed the capacity of pathway throughput by overexpression of the transgenes. That is, with AtMYB85 reactivation of monolignol biosynthesis, the precursor supply level was no longer limiting. Additionally, while DPT consumption was noted in samples expressing the full EA pathway with AtMYB46 or AtMYB85, EA-associated metabolite peaks were among the most enriched mass features in the AtMYB85-expressing samples, but not with AtMYB46 (Figure S2C,D). These data suggest that other pathways induced by AtMYB46 may compete with EA production.
Figure 2.
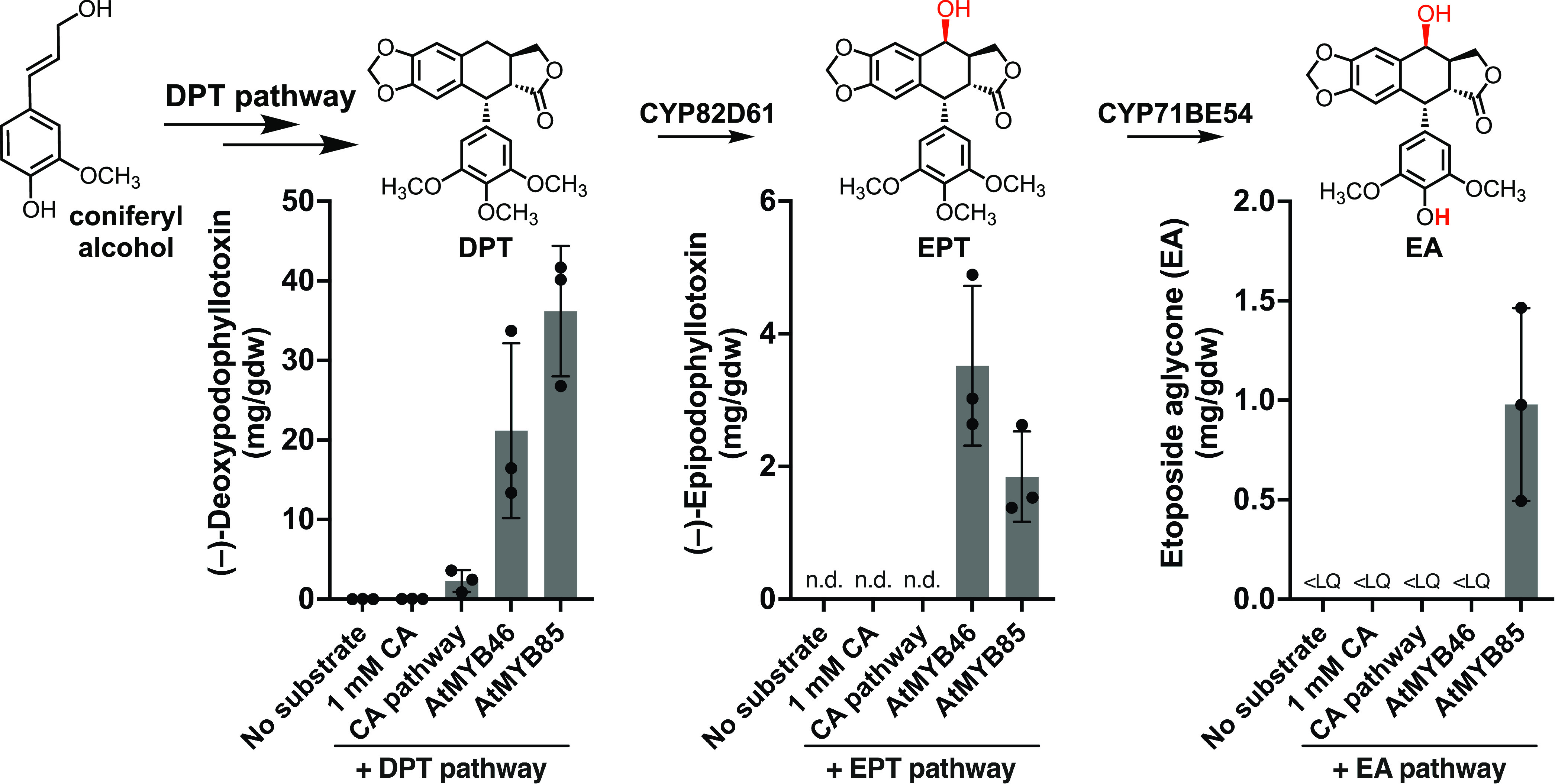
Biosynthetic yields of etoposide intermediates 7 days post infiltration promoted by Agrobacterium-mediated transient expression of CA pathway genes or single lignin-associated TFs in N. benthamiana leaves. DPT pathway: dirigent protein (PhDIR), pinoresinol-lariciresinol reductase (PhPLR), secoisolariciresinol dehydrogenase (PhSDH), PhCYP719A23, PhOMT3, PhCYP71CU1, PhOMT1, and Ph2ODD; EPT pathway: DPT pathway and PhCYP82D61; EA pathway: EPT pathway and PhCYP71BE54; No substrate: no exogenous addition of the substrate CA; 1 mM CA: exogenous addition of CA; CA pathway: eight canonical biosynthetic enzymes converting phenylalanine to coniferyl alcohol;39,40 n.d.: no data; <LQ: below limit of quantification (<0.04 mg/g DW). Biosynthetic yields were quantified based on EIC peak integrations (m/z of [M + H]+) as detected by LC–MS in comparison to standard curves. Bar heights show the means of biological triplicates, and error bars show standard deviations.
Given the marked yield improvement of late-stage etoposide intermediates by either AtMYB46 or AtMYB85, we next coexpressed the EA pathway with both AtMYB46 and AtMYB85 simultaneously to determine if there is a synergistic effect. Unexpectedly, the EA yield with both transcription factors present was significantly lower than that with AtMYB85 (Figure S3). These data suggest that the effect of AtMYB46 overexpression overrides shifts in metabolism caused by overexpression of AtMYB85 alone. Given that AtMYB46 is known to regulate downstream expression of AtMYB85 in Arabidopsis,28 it is possible that AtMYB46 regulates a larger portion of metabolism than AtMYB85, e.g., whereas AtMYB85 seems to regulate the production of lignin precursors, AtMYB46 could also control pathways that result in further metabolism of these precursors and the biosynthetic intermediates, channeling phenylpropanoid monomers toward lignin and limiting their availability for the etoposide aglycone pathway. Furthermore, we questioned whether endogenous copies of the transcription factors would activate the desired endogenous metabolism more effectively compared to their homologs from Arabidopsis. Interestingly, overexpression of N. benthamiana MYB46 and MYB85 did not boost EA production to the same extent that the Arabidopsis versions could despite their high degrees of sequence similarity in the DNA-binding domain (Figure S4 and Figure S5). Accumulation of EA and its metabolites when the EA pathway was simultaneously overexpressed with NbMYB85a or NbMYB85b did not surpass the levels obtained with AtMYB85 coexpression.
Several other parameters were tested in an attempt to optimize product yields. First, we examined the effect of introducing less AtMYB85-harboring Agrobacterium strain with respect to the Agrobacterium strains harboring the EA pathway genes. Overall, we found little difference in EA yields in the range of 0.03–0.1 OD for inoculation, but we did notice a continued decrease in the EA glycoside production with increasing inoculum levels of the AtMYB85-harboring Agrobacterium strain (Figure S6A,B,D). In a separate experiment, we tested the length of time prior to collection of leaf tissue and found that yields from leaves extracted 5–9 days post infiltration (dpi) were typically higher than those from leaves extracted 13–18 dpi (Figure S6C,E).
Suppression of Undesired Glycosylation in AtMYB85-Expressing N. benthamiana Leaves
In the course of our work, we noted that expression of AtMYB85 and AtMYB46 resulted in similar product yields for the DPT and EPT pathways, but that of AtMYB85 was uniquely effective in boosting the yield of EA when coexpressed with the EA pathway genes. To probe this result further, we analyzed the metabolites made after expression of the EA pathway in planta (Figure 3A). Previously, we found that EA can be further metabolized in N. benthamiana leaves by endogenous enzymes, reducing the desired product (EA) yield due to formation of the 4′-O-glycosylation metabolites: EA 4′-O-glucoside and EA 4′-O-malonylglucoside (see Figure 1C).2 To our surprise, not only did the EA yield improve significantly in the AtMYB85-expressing samples, but the ratios of EA and the 4′-O-glycosides had been altered relative to coexpression with MYB46 or CA pathway genes (Figure 3B). Even after adding an acid-hydrolysis step to remove glycosyl groups from EA, the net quantity of EA 4′-O-aglycone produced in leaves still remained the highest in the AtMYB85-expressing samples. Thus, the striking improvement in EA biosynthetic yields promoted by AtMYB85 overexpression may be attributed to the combination of two factors: (1) increased precursor (CA) availability in the N. benthamiana leaves funneled into EA biosynthesis and (2) the apparent suppression of undesired endogenous metabolism of EA by AtMYB85 relative to AtMYB46. In a separate effort, we also attempted to identify an endogenous glycosyltransferase responsible for the undesired metabolism but were unable to find a unique N. benthamiana enzyme (see Supplementary Discussion).
Figure 3.

Impact on the EA 4′-O-glycosylation by AtMYB85 coexpression. (A) Extracted ion chromatograms (EIC) for EA (m/z [M + H]+: 401.1231) as detected by LC–MS in N. benthamiana leaves expressing the EA pathway along with the CA pathway, AtMYB46 or AtMYB85. Earlier eluting peaks are in-source fragmentation products of EA 4′-O-glycosides. The AtMYB85 chromatogram is to scale, and the AtMYB46 chromatogram is scaled by 20× and the CA pathway by 5×. (B) EA and its metabolic products (4′-O-glycosides) in plant extracts from N. benthamiana expressing the heterologous EA pathway. The x-axis shows the source of CA biosynthesis boosted by the CA pathway, AtMYB46, or AtMYB85 coexpression. Acid-hydrolysis treatment removes glycosyl groups from EA 4′-O-glycoside products. Data points show EIC peak integrations for corresponding ion species (m/z: 585.1579 [M + Na]+ for EA-glc, 671.1583 [M + Na]+ for EA-glc-mal, and 401.1231 [M + H]+ for EA) as detected by LC–MS. Bar heights indicate the means of biological triplicates, and error bars indicate standard deviations.
Optimal CA Biosynthesis As Regulated by AtMYB85 in N. benthamiana Leaves
In our previous work,3 we observed the highest level of DPT accumulation only when all eight primary metabolic genes from P. hexandrum that are part of the CA biosynthetic pathway (PAL, C4H, 4CL, HCT, C3H, CCoA-OMT, CCR, and CAD) were overexpressed in N. benthamiana, and omission of even a single gene resulted in at least a half-fold reduction in yield. Since overexpression of AtMYB85 resulted in DPT accumulation at higher levels than the CA pathway overexpression, we reasoned that AtMYB85-coordinated control of the CA pathway was beneficial for increasing the precursor availability. Although MYB85 is known to regulate early lignin biosynthesis, the direct induction of CA pathway genes has not been measured. Thus, we measured transcript levels of these enzymes in N. benthamiana leaves expressing GFP (control) or AtMYB85 with EA pathway enzymes by qRT-PCR. We confirmed that all eight enzymes involved in the CA pathway were upregulated by at least ∼4-fold compared to the GFP control (Figure S7). Given that transcription factor overexpression typically results in higher yields than the CA pathway overexpression, we speculate that the transcription factor control may better reflect the endogenous expression ratios for each gene of the pathway, ensuring optimal levels for proper CA biosynthesis—as opposed to individual gene overexpression at the saturating levels. In one possibility, unregulated gene expression could result in the feedback-inhibition mechanism of pathway intermediates, affecting enzyme activity upstream or downstream—a prominent phenomenon found in the monolignol biosynthesis.41−46
Reactivation of Lignin Biosynthesis in N. benthamiana Leaves by AtMYB46
While both AtMYB46 and AtMYB85 are involved in lignin biosynthesis activation, AtMYB46 is considered to be a master switch controlling other transcription regulation mechanisms,28,47 and AtMYB85 is thought to directly activate monolignol biosynthesis.26 Given the different yields obtained for DPT and EA production using these TFs, we sought to probe the differences in the mechanism of lignin biosynthesis reactivation induced by AtMYB46 or AtMYB85. Corroborating previous reports, overexpression of AtMYB46 in the N. benthamiana leaves resulted in higher accumulation of lignin compared to the empty-vector control (Figure 4). In particular, AtMYB46 resulted in a striking level of lignin accumulation with the leaves turning brittle 5–7 days post infiltration (Video S1 and Figure 4A). At the cellular level, overexpression of AtMYB46, but not AtMYB85, recapitulated lignin deposition, typically found in tracheary elements. It is noteworthy that AtMYB85-overexpressing leaves did not react to the phloroglucinol stain, unlike the AtMYB46-overexpressing samples, suggesting that lignin deposition proceeded to completion in the latter to a higher degree as previously reported in the literature.48 The differences in phenotypical outputs, both in cell wall deposition and physical properties, prompted us to investigate other metabolic signatures that could differentiate the roles of AtMYB46 and AtMYB85. We conducted untargeted metabolomic analysis and found that the metabolic profiles of N. benthamiana leaves expressing GFP, AtMYB85 + EA pathway or AtMYB46 + EA pathway were considerably different based on the principal component analysis (PCA) clustering (Figure S8). Of note, many of the mass features (metabolite signals defined by the observed m/z and retention time on reverse-phase chromatography) with significant PCA loadings for the AtMYB46-expressing leaf tissue samples appeared to be of higher molecular weights compared to mass features in AtMYB85-expressing leaf samples, which may be indicative of lignin polymerization in AtMYB46-expressing leaves. In contrast, AtMYB85 activated the biosynthesis of other small molecules (namely, hydroxycinnamic acid amides, or HCAAs, see Figure 1D) and may be involved in activation of competing metabolism other than lignin polymerization (Tables S4–S7 and Figure S9).
Figure 4.
Reactivation of lignin biosynthesis in N. benthamiana. (A) Bright field images of uninfiltrated and infiltrated leaves show that AtMYB46-expressing leaves break upon folding. Phloroglucinol stain (magenta) reveals extensive lignin deposition in AtMYB46-expressing leaves, in parallel with xylem-like lignin banding in cell walls of mesophyll cells as observed by DIC microscopy. Magnifications: bright field and phloroglucinol stain, 2×; DIC, 400×. (B) Lignin content as quantified by the acetyl bromide method. Data points show the means of triplicates from four different experiments (n = 4), and error bars indicate standard deviations. ANOVA on log-transformed data showed a statistical difference between means (***p < 0.0001). Multiple comparisons showed that lignin content in AtMYB46-expressing leaves was the only value statistically different from the rest.
Conclusions
Two hypotheses were considered to explain the unexpected suppression of EA product glycosylation observed in experiments with AtMYB85 overexpression: (1) AtMYB85 induces the biosynthesis of endogenous hydroxycinnamic metabolites (e.g., HCAAs), which could serve as glycosyltransferase substrates, and might act as competitive inhibitors to etoposide aglycone intermediates for glycosylation, and (2) AtMYB85 represses the expression of certain UDP-glycosyltransferases (UGTs) that exhibit off-target activity on the heterologous products. In line with the first hypothesis, in our optimization efforts, we observed biosynthetic yields of EA glycosides inversely correlated with the production of the HCAA side products (Figure S6A,C). For the second hypothesis, we identified NbUGT73A24 that can glycosylate EA and its intermediates, but the expression level of the gene did not decrease with AtMYB85 overexpression (see Supplementary Discussion). However, it is possible that AtMYB85 regulates the expression of other, not yet identified, NbUGTs that contribute to EA glycosylation. Alternatively, there might be more complex metabolic regulation in place, for example, as is observed in flavonol biosynthesis and glycosylation.49 Examples of TFs that differentially regulate two competing branches of metabolism50 include A. thaliana MYB TFs that activate lignin biosynthesis while inhibiting flavonoid biosynthesis, including MYB85.51 In conclusion, our data show that overexpression of AtMYB85 increases the yield of EA, and two synergistic mechanisms are possible: direct increase in endogenous precursor supply and suppression of the competing metabolism (Figure 5).
Figure 5.
Model of transcription regulation engineering described in this study. Red dashed arrows indicate transcriptional regulation previously described in the literature, blue dotted arrows indicate proposed transcriptional regulation based on metabolite profiling, and black and green arrows indicate metabolite conversion.
Here, we demonstrate the value of transient transcription factor expression to improve plant specialized metabolite biosynthesis in a plant heterologous host. As more plant pathways become targets for metabolic engineering, this approach is likely to be a valuable and convenient tool for altering the base metabolism in the plant chassis and could be applied to enhance production of other common pathway precursors in addition to monolignols (e.g., aliphatic amines for alkaloid biosynthesis or isoprenes for complex terpene production). AtMYB85 overexpression with the EA pathway enzymes led to the highest biosynthetic yields of etoposide aglycone observed at ∼1 mg/g DW. Previously, precursor supply was observed to be more limiting than enzyme activity in EA biosynthesis in N. benthamiana, as evidenced by no intermediate accumulation in the secondary metabolism. Quantification of EA pathway intermediates in N. benthamiana leaves indicates that precursor availability is no longer limiting. Our findings reported here show promise for application of synthetic transcription regulation for metabolic engineering of biosynthetic pathways in plants.
Methods
Safety Statement
Handling of acetyl bromide was entirely conducted inside a chemical fume hood. No other unexpected or unusually high safety hazards were encountered.
Candidate Gene Selection and Cloning
General Procedure
Q5 High-Fidelity 2X Master Mix was used for all PCR amplification steps, except for colony PCR, for which Hot Start Taq 2X Master Mix was used instead. All enzymes used for cloning were purchased from New England BioLabs, unless otherwise noted. Oligonucleotide primers were purchased from Integrated DNA Technologies. Plasmid constructs were assembled in an isothermal DNA assembly reaction as described by Gibson et al.52 NEB 5-alpha Competent Escherichia coli cells were used for plasmid storage and isolation. Plasmid DNAs were isolated from the liquid cultures of E. coli using the ZR Plasmid Miniprep kit (Zymo Research). Isolated plasmids and PCR amplicons were confirmed for correct sequences by Sanger DNA sequencing performed by Elim Biopharm.
pEAQ-HT Constructs for N. benthamiana Expression
E. coli strains harboring AtMYB85 (TAIR accession: AT4G22680; clone name: PYAT4G22680), AtMYB46 (TAIR accession: AT5G12870; clone name: U16973), AtMYB63 (TAIR accession: AT1G79180; clone name: PYAT1G79180), AtMYB103 (TAIR accession: AT1G63910; clone name: PYAT1G63910), AtMYB58 (TAIR accession: AT1G16490; clone name: DKLAT1G16490), and AtVND6 (TAIR accession: AT5G62380; clone name: DQ056734) were purchased from Arabidopsis Biological Resource Center (ARBC). The pET28a construct for SrUGT71E1 and the N. benthamiana cDNA templates for N. benthamiana genes (NbMYB46a [NCBI GenBank accession: OP121090], NbMYB46b [OP121091], NbMYB85a [OP121092], and NbMYB85b [OP121093]) served as templates. The full-length coding sequences (CDS) were PCR-amplified from the plasmids with corresponding primers (see Table S1) using Q5 High-Fidelity 2X Master Mix (NEB), Gibson-assembled into pEAQ-HT53 predigested with AgeI and XhoI, and then transformed into E. coli 5-alpha chemically competent cells. Sequence-validated pEAQ-HT constructs were transformed into Agrobacterium tumefaciens (GV3101:pMP90) using the freeze–thaw method.
Agrobacterium-Mediated Transient Expression in N. benthamiana
Agrobacterium strains prepared as described above were grown on LB plates supplemented with 10 μg/mL gentamicin and 50 μg/mL kanamycin for 1–2 days. The cells were resuspended in LB media and centrifuged at 8000g for 5 min, after which the supernatant was discarded. The cells were induced in 500 μL of induction media (10 mM MES buffer at pH 5.6 with 10 mM MgCl2 and 150 μM acetosyringone) for 1–2 h at room temperature. The cell suspension was further diluted with the induction media to the desired inoculum level (OD600 = 0.2, unless otherwise noted) based on the measured optical density at 600 nm and infiltrated on the underside of the N. benthamiana leaves with a needleless 1 mL syringe. Plants were 4–5 weeks old at the time of infiltration, grown under a 16 h light cycle. Biological replicates consisted of leaves of different ages (based on the stemming location on the plant, counting from the bottom) from different N. benthamiana plants from the same batch, unless otherwise noted. For metabolite extraction, leaves were harvested on 5–7 dpi.
Methanol Extraction and LCMS Analysis of Plant Leaf Extracts
N. benthamiana leaves were flash-frozen in liquid nitrogen and lyophilized to dryness. The dry samples were homogenized in a ball-mill homogenizer with 5 mm-diameter stainless steel beads at 25 Hz for 2 min. Twenty microliters of 80% (v/v) methanol/water was added per mg of dry sample, and the samples were refluxed at 65 °C for 10 min. The resulting plant extracts were filtered with 0.45 μm PTFE filters prior to LCMS injection.
For N. benthamiana-metabolized EA glycosides, acid hydrolysis was conducted as previously described with slight modifications.3 The incubation time at 95 °C was kept at 10 min instead of 2 h due to concerns about the aglycone product stability.
Methanolic extracts were analyzed by reversed-phase column chromatography on a coupled Agilent 6520 Accurate-Mass q-ToF ESI mass spectrometer using a 5 μm, 2 × 100 mm Gemini NX-C18 column (Phenomenex) with mobile phases of water with 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B). The chromatography was run at a flow rate of 0.4 mL/min with the following gradient: 0–1 min, 3% B; 1–21 min, 3–50% B; 21–22 min, 50–97% B; 22–27 min, 97% B; 27–28 min, 97–3% B; and 28–32 min, 3% B. MS parameters were as follows: mass range, 50–1700 m/z; drying gas, 300 °C and 11–12 L/min; nebulizer, 35 psig; capillary, 3500 V; fragmentor, 150 V; skimmer, 65 V; octopole 1RF Vpp, 750 V; 699.3 ms per spectrum. The eluent in the first minute of each run was discarded to avoid salt contamination in the mass spectrometer.
Histochemical Analysis of Lignification in the N. benthamiana Leaves
Lignin deposition was visualized with phloroglucinol staining. Round discs were cut from N. benthamiana leaves, bleached in 12.5% v/v acetic acid in ethanol (2–3 h with decanting and replenished solution once), and stored in 70% ethanol in water until further processing. The discs were incubated in an ethanolic solution of 2% phloroglucinol for 10 min and then transferred to 6 N HCl solution. After ∼10 min of incubation, samples were recorded with a digital camera coupled to a stereo microscope when staining of leaves and veins was observed. For higher magnification microscopy, leaf discs were cleared with Hoyer’s solution54 and analyzed by differential interference contrast (DIC) microscopy on a Leica DM2500 microscope.
Lignin Content Quantification
Lignin content in N. benthamiana leaves (WT [no agro-infiltration], or expressing EV, AtMYB46, or AtMYB85) was quantified using the acetyl bromide method as described by Lee et al.,20 with slight modifications. The leaves were flash-frozen in liquid nitrogen and lyophilized to dryness. The dried samples were homogenized in a ball-mill homogenizer (Retsch MM 400) at 25 Hz for 2 min with 5 mm-diameter stainless steel beads. The ground samples were washed serially with 70% ethanol, chloroform/methanol (1:1 v/v), and acetone. The washed cell wall materials were completely dried at 45 °C under positive air flow. Two hundred microliters of 25% acetyl bromide solution in acetic acid was added per mg of sample, and the samples were incubated at 70 °C for an hour with occasional inversion every 10 min. The samples were cooled on ice and centrifuged for 5 min at 16,000g. One hundred microliters of the supernatant (or 25% acetyl bromide solution in acetic acid as the blank) was transferred to a glass vial and combined serially with 400 μL of 2 M NaOH, 70 μL of 0.5 M hydroxylamine hydrochloride, and 430 μL of acetic acid. Two hundred microliters of the resulting solutions was transferred to each well in a UV-specific 96-well microplate, and absorbance was measured at 280 nm. Measured absorbance values were corrected by subtracting the absorbance of the blank (similar to absorbance measured on an empty well), and the percentage of acetyl bromide soluble lignin (%ABSL) was calculated by the Beer–Lambert law assuming a path length of 0.539 cm55 and an extinction coefficient for N. benthamiana of 23.077 g–1 cm–1.56
Acknowledgments
We would like to thank Bailey Schultz for helpful discussions in the early development stage of the project, Jeremy Hunt for providing the Jupyter notebook template for XCMS analysis, Alex Engel and Ryan Nett for valuable feedback on the manuscript, Prof. Michael Court (UGT Nomenclature Committee) for assigning standard names for the NbUGT genes, and Prof. Dominique Bergmann for providing access to the DIC microscope. This work was funded by the National Science Foundation Graduate Research Fellowship under grant no. DGE-1656518 (to S.S.K.) and a DARPA Young Faculty Award no. D18AP00046. E.S.S. is a Howard Hughes Medical Institute Investigator.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00289.
Author Contributions
S.S.K., D.L.W, C.J.R., and E.S.S. designed the experiments and analyzed the data. S.S.K., D.L.W., and C.J.R. performed the experiments. S.S.K. wrote the original draft of the manuscript. E.S.S., D.L.W, and C.J.R. helped write and revise, reviewed, and approved the final manuscript.
The authors declare the following competing financial interest(s): Elizabeth Sattely is on the SAB of Calyxt.
Supplementary Material
References
- Panda S.; Kazachkova Y.; Aharoni A. Catch-22 in Specialized Metabolism: Balancing Defense and Growth. J. Exp. Bot. 2021, 72, 6027–6041. 10.1093/jxb/erab348. [DOI] [PubMed] [Google Scholar]
- Lau W.; Sattely E. S. Six Enzymes from Mayapple That Complete the Biosynthetic Pathway to the Etoposide Aglycone. Science 2015, 349, 1224–1228. 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz B. J.; Kim S.-Y.; Lau W.; Sattely E. S. Total Biosynthesis for Milligram-Scale Production of Etoposide Intermediates in a Plant Chassis. J. Am. Chem. Soc. 2019, 141, 19231–19235. 10.1021/jacs.9b10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.; Stephenson M. J.; Miettinen K.; Brouwer B.; Leveau A.; Brett P.; Goss R. J. M.; Goossens A.; O’Connell M. A.; Osbourn A. A Translational Synthetic Biology Platform for Rapid Access to Gram-Scale Quantities of Novel Drug-like Molecules. Metab. Eng. 2017, 42, 185–193. 10.1016/j.ymben.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Peña R.; Sattely E. S. Rerouting Plant Terpene Biosynthesis Enables Momilactone Pathway Elucidation. Nat. Chem. Biol. 2021, 17, 205–212. 10.1038/s41589-020-00669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpoorte R.; van der Heijden R.; Memelink J. Engineering the Plant Cell Factory for Secondary Metabolite Production. Transgenic Res. 2000, 9, 323–343. 10.1023/A:1008966404981. [DOI] [PubMed] [Google Scholar]
- Broun P. Transcription Factors as Tools for Metabolic Engineering in Plants. Curr. Opin. Plant Biol. 2004, 7, 202–209. 10.1016/j.pbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Chen W.; Provart N. J.; Glazebrook J.; Katagiri F.; Chang H.-S.; Eulgem T.; Mauch F.; Luan S.; Zou G.; Whitham S. A.; Budworth P. R.; Tao Y.; Xie Z.; Chen X.; Lam S.; Kreps J. A.; Harper J. F.; Si-Ammour A.; Mauch-Mani B.; Heinlein M.; Kobayashi K.; Hohn T.; Dangl J. L.; Wang X.; Zhu T. Expression Profile Matrix of Arabidopsis Transcription Factor Genes Suggests Their Putative Functions in Response to Environmental Stresses[W]. Plant Cell 2002, 14, 559–574. 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L.Transcription Factors ofArabidopsis and Rice: A Genomic Perspective. In Annual Plant Reviews Volume 29: Regulation of Transcription in Plants; John Wiley & Sons, Ltd, 2007; pp. 28–53. [Google Scholar]
- van Verk M. C.; Gatz C.; Linthorst H. J. M.. Chapter 10 Transcriptional Regulation of Plant Defense Responses. In Advances in Botanical Research; Advances in Botanical Research; Academic Press, 2009; Vol. 51, pp. 397–438. [Google Scholar]
- Maher M. F.; Nasti R. A.; Vollbrecht M.; Starker C. G.; Clark M. D.; Voytas D. F. Plant Gene Editing through de Novo Induction of Meristems. Nat. Biotechnol. 2020, 38, 84–89. 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M. A.; He X.; Dixon R. A. Elicitor-Induced Transcription Factors for Metabolic Reprogramming of Secondary Metabolism in Medicago Truncatula. BMC Plant Biol. 2008, 8, 132. 10.1186/1471-2229-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E.; Titta L.; Giorgio M.; Mock H.-P.; Matros A.; Peterek S.; Schijlen E. G. W. M.; Hall R. D.; Bovy A. G.; Luo J.; Martin C. Enrichment of Tomato Fruit with Health-Promoting Anthocyanins by Expression of Select Transcription Factors. Nat. Biotechnol. 2008, 26, 1301–1308. 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Reddy V. A.; Panicker D.; Mao H.-Z.; Kumar N.; Rajan C.; Venkatesh P. N.; Chua N.-H.; Sarojam R. Metabolic Engineering of Terpene Biosynthesis in Plants Using a Trichome-Specific Transcription Factor MsYABBY5 from Spearmint (Mentha Spicata). Plant Biotechnol. J. 2016, 14, 1619–1632. 10.1111/pbi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A.; Misra P.; Khan M. P.; Swarnkar G.; Tewari M. C.; Bhambhani S.; Trivedi R.; Chattopadhyay N.; Trivedi P. K. Co-Expression of Arabidopsis Transcription Factor, AtMYB12, and Soybean Isoflavone Synthase, GmIFS1, Genes in Tobacco Leads to Enhanced Biosynthesis of Isoflavones and Flavonols Resulting in Osteoprotective Activity. Plant Biotechnol. J. 2014, 12, 69–80. 10.1111/pbi.12118. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K.; Scheel D. Physiology and Molecular Biology of Phenylpropanoid Metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 347–369. 10.1146/annurev.pp.40.060189.002023. [DOI] [Google Scholar]
- Davin L. B.; Lewis N. G.. Phenylpropanoid Metabolism: Biosynthesis of Monolignols, Lignans and Neolignans, Lignins and Suberins. In Phenolic Metabolism in Plants; Stafford H. A., Ibrahim R. K., Eds.; Recent Advances in Phytochemistry; Springer US: Boston, MA, 1992; pp. 325–375. [Google Scholar]
- Whetten R.; Sederoff R. Lignin Biosynthesis. Plant Cell 1995, 7, 1001–1013. 10.1105/tpc.7.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A.; Paiva N. L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H.; Jeon H. S.; Kim S. H.; Chung J. H.; Roppolo D.; Lee H.-J.; Cho H. J.; Tobimatsu Y.; Ralph J.; Park O. K. Lignin-Based Barrier Restricts Pathogens to the Infection Site and Confers Resistance in Plants. EMBO J. 2019, 38, e101948. 10.15252/embj.2019101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.; Ye Z.-H. Transcriptional Regulation of Lignin Biosynthesis. Plant Signaling Behav. 2009, 4, 1028–1034. 10.4161/psb.4.11.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem W. R.; Clay N. K. Regulation of Plant Secondary Metabolism and Associated Specialized Cell Development by MYBs and BHLHs. Phytochemistry 2016, 131, 26–43. 10.1016/j.phytochem.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M.; Zhang J.; Tschaplinski T. J.; Tuskan G. A.; Chen J.-G.; Muchero W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. 10.3389/fpls.2018.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J.; Rohde A.; Christensen J. H.; Van de Peer Y.; Boerjan W. Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers L. A.; Campbell M. M. The Genetic Control of Lignin Deposition during Plant Growth and Development. New Phytol. 2004, 164, 17–30. 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- Zhong R.; Lee C.; Zhou J.; McCarthy R. L.; Ye Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Lee C.; Zhong R.; Ye Z.-H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.; Richardson E. A.; Ye Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.; Ye Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2012, 53, 368–380. 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- Kubo M.; Udagawa M.; Nishikubo N.; Horiguchi G.; Yamaguchi M.; Ito J.; Mimura T.; Fukuda H.; Demura T. Transcription Switches for Protoxylem and Metaxylem Vessel Formation. Genes Dev. 2005, 19, 1855–1860. 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T.; Thitamadee S.; Machida Y.; Chua N.-H. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 Regulate Tracheary Element Differentiation in Arabidopsis. Plant Cell 2008, 20, 3359–3373. 10.1105/tpc.108.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.; He K.; Liu D.; Bai S.; Gu X.; Wei L.; Luo J. DATF: A Database of Arabidopsis Transcription Factors. Bioinformatics 2005, 21, 2568–2569. 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- Iida K.; Seki M.; Sakurai T.; Satou M.; Akiyama K.; Toyoda T.; Konagaya A.; Shinozaki K. RARTF: Database and Tools for Complete Sets of Arabidopsis Transcription Factors. DNA Res. 2005, 12, 247–256. 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón D. M.; Ruzicic S.; Dreyer I.; Mueller-Roeber B. PlnTFDB: An Integrative Plant Transcription Factor Database. BMC Bioinf. 2007, 8, 42. 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N.; Ohme-Takagi M. Functional Analysis of Transcription Factors in Arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L.; Heard J.; Martin G.; Reuber L.; Jiang C.-Z.; Keddie J.; Adam L.; Pineda O.; Ratcliffe O. J.; Samaha R. R.; Creelman R.; Pilgrim M.; Broun P.; Zhang J. Z.; Ghandehari D.; Sherman B. K.; Yu G.-L. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J. L.; Breton G.; Nagel D. H.; Kang S. E.; Bonaldi K.; Doherty C. J.; Ravelo S.; Galli M.; Ecker J. R.; Kay S. A. A Genome-Scale Resource for the Functional Characterization of Arabidopsis Transcription Factors. Cell Rep. 2014, 8, 622–632. 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. E.; Dewick P. M. Aryltetralin Lignans from Podophyllum Hexandrum and Podophyllum Peltatum. Phytochemistry 1984, 23, 1147–1152. 10.1016/S0031-9422(00)82628-X. [DOI] [Google Scholar]
- Weng J.-K.; Chapple C. The Origin and Evolution of Lignin Biosynthesis. New Phytol. 2010, 187, 273–285. 10.1111/j.1469-8137.2010.03327.x. [DOI] [PubMed] [Google Scholar]
- Barros J.; Serk H.; Granlund I.; Pesquet E. The Cell Biology of Lignification in Higher Plants. Ann. Bot. 2015, 115, 1053–1074. 10.1093/aob/mcv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrin J.; Dixon R. A. Stress Responses in Alfalfa (Medicago Sativa L.): II. Purification, Characterization, and Induction of Phenylalanine Ammonia-Lyase Isoforms from Elicitor-Treated Cell Suspension Cultures. Plant Physiol. 1990, 92, 447–455. 10.1104/pp.92.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K.; Tsao C. C.; Li L.; Popko J. L.; Umezawa T.; Carraway D. T.; Smeltzer R. H.; Joshi C. P.; Chiang V. L. Coniferyl Aldehyde 5-Hydroxylation and Methylation Direct Syringyl Lignin Biosynthesis in Angiosperms. PNAS 1999, 96, 8955–8960. 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S. A.; Leshkevich J.; Chiang V. L.; Tsai C.-J. Differential Substrate Inhibition Couples Kinetically Distinct 4-Coumarate:Coenzyme A Ligases with Spatially Distinct Metabolic Roles in Quaking Aspen. Plant Physiol. 2002, 128, 428–438. 10.1104/pp.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Cheng X.; Lu S.; Nakatsubo T.; Umezawa T.; Chiang V. L. Clarification of Cinnamoyl Co-Enzyme A Reductase Catalysis in Monolignol Biosynthesis of Aspen. Plant Cell Physiol. 2005, 46, 1073–1082. 10.1093/pcp/pci120. [DOI] [PubMed] [Google Scholar]
- Chen H.-C.; Song J.; Williams C. M.; Shuford C. M.; Liu J.; Wang J. P.; Li Q.; Shi R.; Gokce E.; Ducoste J.; Muddiman D. C.; Sederoff R. R.; Chiang V. L. Monolignol Pathway 4-Coumaric Acid:Coenzyme A Ligases in Populus. Trichocarpa: Novel Specificity, Metabolic Regulation, and Simulation of Coenzyme A Ligation Fluxes. Plant Physiol. 2013, 161, 1501–1516. 10.1104/pp.112.210971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P.; Naik P. P.; Chen H.-C.; Shi R.; Lin C.-Y.; Liu J.; Shuford C. M.; Li Q.; Sun Y.-H.; Tunlaya-Anukit S.; Williams C. M.; Muddiman D. C.; Ducoste J. J.; Sederoff R. R.; Chiang V. L. Complete Proteomic-Based Enzyme Reaction and Inhibition Kinetics Reveal How Monolignol Biosynthetic Enzyme Families Affect Metabolic Flux and Lignin in Populus Trichocarpa. Plant Cell 2014, 26, 894–914. 10.1105/tpc.113.120881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y.; Yamaguchi M.; Endo H.; Rejab N. A.; Ohtani M. NAC-MYB-Based Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Land Plants. Front. Plant Sci. 2015, 6, 288. 10.3389/fpls.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S.; Mitsuda N. Reconstitution of a Secondary Cell Wall in a Secondary Cell Wall-Deficient Arabidopsis Mutant. Plant Cell Physiol. 2015, 56, 299–310. 10.1093/pcp/pcu208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R.; Messner B.; Faus-Kessler T.; Hoffmann T.; Schwab W.; Hajirezaei M.-R.; von Saint Paul V.; Heller W.; Schäffner A. R. Feedback Inhibition of the General Phenylpropanoid and Flavonol Biosynthetic Pathways upon a Compromised Flavonol-3-O-Glycosylation. J. Exp. Bot. 2012, 63, 2465–2478. 10.1093/jxb/err416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Yang J.; Li H.; Chiang V. L.; Fu Y. Cooperative Regulation of Flavonoid and Lignin Biosynthesis in Plants. Crit. Rev. Plant Sci. 2021, 40, 109–126. 10.1080/07352689.2021.1898083. [DOI] [Google Scholar]
- Geng P.; Zhang S.; Liu J.; Zhao C.; Wu J.; Cao Y.; Fu C.; Han X.; He H.; Zhao Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation1 [OPEN]. Plant Physiol. 2020, 182, 1272–1283. 10.1104/pp.19.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A. III; Smith H. O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Sainsbury F.; Thuenemann E. C.; Lomonossoff G. P. PEAQ: Versatile Expression Vectors for Easy and Quick Transient Expression of Heterologous Proteins in Plants. Plant Biotechnol. J. 2009, 7, 682–693. 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- Hoyer’s Medium. Cold Spring Harb. Protoc. 2011, 2011 (), pdb.rec12429, 10.1101/pdb.rec12429. [DOI] [Google Scholar]
- Foster C. E.; Martin T. M.; Pauly M. Comprehensive Compositional Analysis of Plant Cell Walls (Lignocellulosic Biomass) Part I: Lignin. J. Visualized Exp. 2010, 37, e1745. 10.3791/1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q.; Si J.; Cui X.; Peng H.; Chen X.; Xing H.; Dou D. The Soybean Cinnamate 4-Hydroxylase Gene GmC4H1 Contributes Positively to Plant Defense via Increasing Lignin Content. Plant Growth Regul. 2019, 88, 139–149. 10.1007/s10725-019-00494-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.