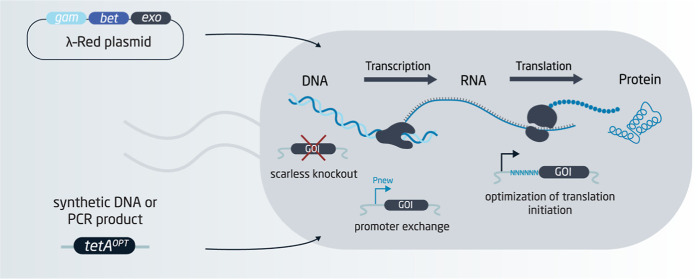
Abstract
Engineering of bacterial genomes is a fundamental craft in contemporary biotechnology. The ability to precisely edit chromosomes allows for the development of cells with specific phenotypes for metabolic engineering and for the creation of minimized genomes. Genetic tools are needed to select for cells that underwent editing, and dual-selection markers that enable both positive and negative selection are highly useful. Here, we present an optimized and easy-to-use version of the tetA dual-selection marker and demonstrate how this tetAOPT can be used efficiently to engineer at different stages of the central dogma of molecular biology. On the DNA level, tetAOPT can be used to create scarless knockouts across the Escherichia coli genome with efficiency above 90%, whereas recombinant gene integrations can be achieved with approximately 50% efficiency. On the RNA and protein level, we show that tetAOPT enables advanced genome engineering of both gene translation and transcription by introducing sequence variation in the translation initiation region or by exchanging promoters. Finally, we demonstrate the use of tetAOPT for genome engineering in the industrially relevant probiotic strain E. coli Nissle.
Keywords: genome engineering, Escherichia coli, tetA, dual-selection marker, recombineering, Escherichia coli Nissle
Introduction
Simple genome editing tools are in high demand to facilitate the Design–Build–Test–Learn cycle of bioengineering. The entire flow of information through the central dogma of molecular biology needs to be considered: for example, on the genome level, we need tools to make knockouts and edit on a large scale to create organisms with minimized genomes. In Escherichia coli, such strains have been constructed using several genetic selection markers to obtain scarless deletions.1−6 The manipulation of gene transcription is commonly achieved by exchanging or mutating promoter sequences,7−9 and translation can be modulated by introducing mutations in the 5′UTR of the mRNA, as this region is a major determinant of the rate of translation initiation.10−14
Numerous genetic tools for the model bacterium E. coli have been developed to speed up strain development.15−23 Commonly, chromosomal integration is achieved by using homologous recombination systems such as λ-Red recombineering or Rec/ET.16,20,23 These systems are based on phage proteins that enable the integration of double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA, typically provided in the form of synthetic oligonucleotides).24 Due to the low efficiency of these systems, elaborate PCR screening or genetic markers are needed to select for the recombinant strains. Antibiotic resistance markers are commonly used, but they have inherent disadvantages such as the risk of environmental spreading25 and the metabolic burden they can pose when expressed.26 For this reason, selection markers are usually removed from the final construct.
Different approaches have been developed to remove selection markers after integration. The Flp/FRT system relies on the expression of a site-specific recombinase called flippase (Flp), which promotes recombination at two flippase recognition target (FRT) sites.27,28 FRT sites, in combination with a kanamycin resistance cassette, were successfully used for generating a collection of almost 4000 E. coli single-gene knockout mutants.18 Similar to the Flp/FRT system, the Cre/loxP system uses a recombinase (Cre) that promotes recombination between loxP sites, leaving one loxP site on the genome.29 However, both Flp/FRT and Cre/loxP leave DNA sequence “scars” in the genome, which might lead to genetic instability due to homologous recombination when multiple sites of the genome are edited. Recently, advances in CRISPR/Cas9 genome editing allow for scarless integration of dsDNA fragments by utilizing CRISPR/Cas9 counterselection instead of co-integration of a selection marker.19,21,22,30
Selection markers can also be combined with counter-selectable traits that facilitate their elimination. Genes such as galK, tolC, and tetA are particularly attractive as the single genes can be selected both for and against (dual selection).31−37 However, in contrast to galK and tolC, tetA does not require a knockout of the native gene on the genome of E. coli. tetA encodes for a transmembrane protein that confers resistance to tetracycline, but its expression also leads to sensitivity toward lipophilic chelating agents, such as fusaric or quinaldic acid, as well as cadmium and nickel cations.36,37 Nevertheless, tetA is often combined with an additional counterselection marker such as sacB,38 indicating that it performs poorly as a counterselection marker.
Here, we describe multiple workflows for genome engineering in E. coli using an optimized tetA dual-selection marker. We introduced mutations in the translation initiation region (TIR) of tetA to reach different expression levels and demonstrate that an optimized tetAOPT enables efficient engineering throughout the central dogma: at the DNA level by creating multiple gene knockouts, at the transcriptional level by exchanging promoters on the genome, and on the translation level by modulating gene translation initiation.
Results
Optimization of a tetA Expression Cassette for Dual Selection in E. coli
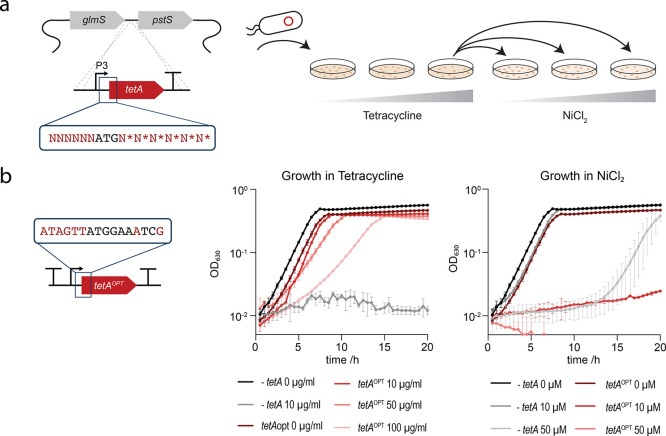
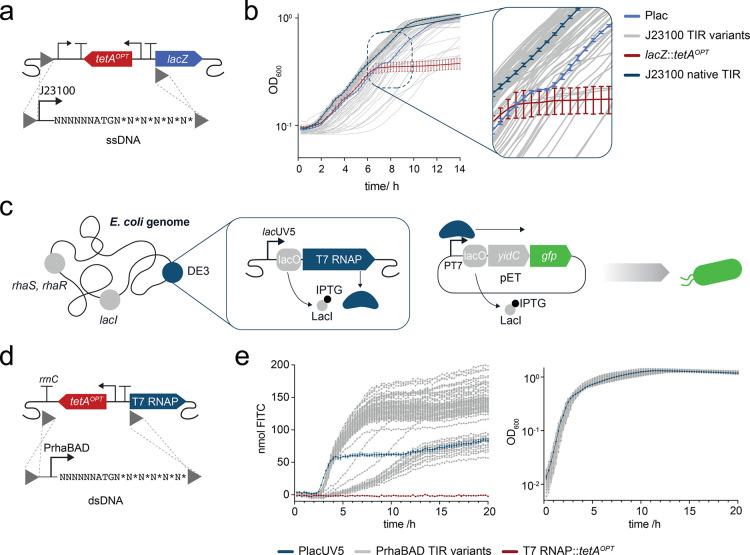
Our early observations on tetA suggested that it performed poorly because of its very narrow dynamic range of action: for example, high tetracycline concentrations were not tolerated when the resistance gene was used in a translational coupling device,13 and initial attempts to increase expression by exchanging promoters failed likely due to toxic overexpression of the tetA membrane transporter protein.39,40 We hypothesized that it would be beneficial to tune the translation of tetA so that expression levels are high enough to confer NiCl2 sensitivity but low enough to avoid a significant burden to the cell. To this end, we created a DNA sequence library of the tetA TIR.12 The TIR spans from the Shine–Dalgarno sequence to the fifth codon of the gene of interest41 and has a major impact on the overall production level of a protein.12,42−46 Randomization of the TIR has previously been successfully used to optimize the expression of different genes.12,44 For optimizing the tetA TIR, using PCR, we randomized the six nucleotides upstream of the start codon as well as the first two codons downstream of the ATG to all synonymous codons12 (Figure 1a). Additionally, we included the P3 promoter into the tetA cassette because it previously enabled genomic tetA expression.34
Figure 1.
Optimization of tetA for dual selection. (a) Workflow for tetA TIR randomization and screening of DNA sequence library variants. (b) Growth of the selected tetAOPT TIR variant (red lines) in M9 medium supplemented with tetracycline and NiCl2. The parental strain SIJ19 (E. coli K12 MG1655)47 (-tetA, gray lines) was included as a control. Data represent the mean of biological triplicates or duplicates with standard deviations, for -tetA and tetAOPT, respectively. See Figure S1 for growth analysis of the initial TIR library variants.
For genomic integration, tetA was amplified by PCR from a SEVA plasmid (pSEVA51148) using a degenerated oligonucleotide pool that theoretically contained 32786 different nucleotide sequences and integrated with λ-Red recombineering. The bacterial cells were plated on LB agar with different concentrations of tetracycline for selection of the best performing TIR variants. The number of colonies decreased with increasing tetracycline, and the highest concentration where colonies were still observed was 50 μg/mL. From this agar plate, 96 colonies were picked and transferred to M9 agar with different concentrations of NiCl2. The colonies that showed the highest sensitivity toward NiCl2 were then selected and further characterized (Figure S1). All tetA sequence variants selected this way exhibited resistance up to 100 μg/mL tetracycline and were sensitive to 50 μM NiCl2 and above. Since the control strain without tetA was severely impaired in growth at 100 μM NiCl2, 50 μM was selected as the concentration for counterselection. One of the library variants was selected for further studies, and a terminator (L3S3P2249) was introduced upstream of tetA to avoid polar effects on tetA expression from adjacent genes when used for further engineering (Figure 1b). As upstream terminator sequences can influence expression levels,7 this final construct, tetAOPT, was re-analyzed for its resistance toward tetracycline and sensitivity toward NiCl2 (Figure 1b).
While working with tetAOPT, we made different observations that helped ensure its functionality. In M9 medium, the concentration of NiCl2 that is necessary to ensure counterselection is 50 μM, whereas the required concentrations to inhibit growth in LB were as high as 2 mM. Differences in osmolarity might be responsible for the varying sensitivity of tetAOPT harboring cells toward NiCl250. Since the LB medium composition can vary between batches, we recommend using M9 medium for more reproducible results. Further, for counterselection, it is important to wash the cells in sterile water before they are plated on NiCl2 to remove traces of LB medium. Before plating, cells are recovered overnight before they are plated on NiCl2—likely to allow for a cell population without the membrane protein TetA to establish. A shorter 4 h recovery is possible, but the efficiencies decrease. Last, plates should not be incubated for longer than 3 days since background colonies that still contain tetAOPT appear on the plates. This might be due to efflux pumps in E. coli that are active after a prolonged incubation.51 Nonetheless, even if background colonies appear, correct colonies are easily distinguishable based on their larger size.
Characterization of the tetAOPT Cassette Performance for Gene Deletions in E. coli
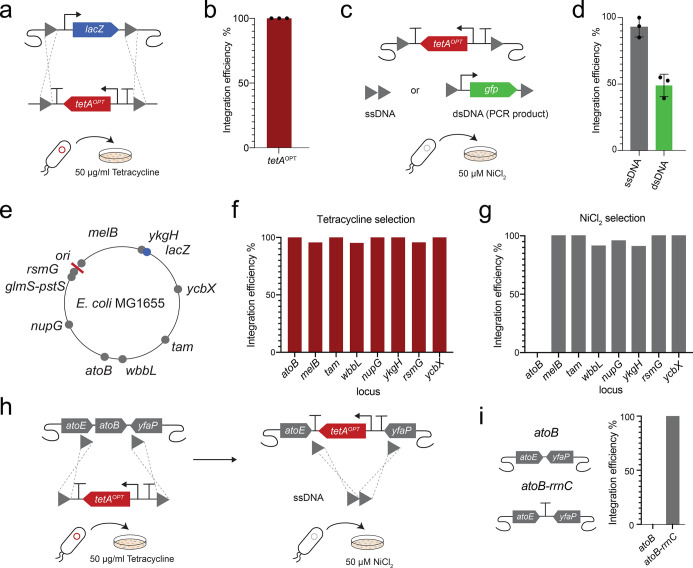
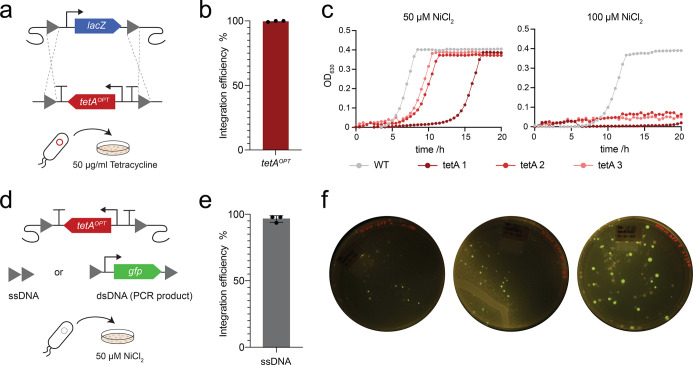
To further test the optimized tetAOPT variant, we chose to perform knockouts of different genes across the genome of E. coli K12 MG1655. We started with lacZ, enabling simple estimation of the efficiency of editing by blue-white screening with the beta-galactosidase substrate X-Gal. First, we amplified tetAOPT with 50 bp homology to two areas flanking the lacZ gene and transformed the PCR product into E. coli hosting the λ-Red recombineering system and incubated the cells on LB agar supplemented with tetracycline and X-Gal (Figure 2a). No blue colonies were observed, demonstrating a 100% efficiency of the procedure (Figures 2b andS2a). For tetAOPT removal in the second step, λ-Red proteins were re-expressed, and the cells transformed with either a single-stranded oligonucleotide (ssDNA) or a double-stranded PCR product (dsDNA), both harboring 50 bp homology to the regions up- and downstream of tetAOPT. This creates either a scarless gene deletion (with the ssDNA) or integration of a new gene (here gfp with the dsDNA) (Figure 2c), and we observed that using ssDNA was 93% efficient (Figure 2d), whereas using dsDNA was 49% efficient (Figures 2d and S2b).
Figure 2.
Gene knockout and insertion strategies using tetAOPT. The functionality of tetAOPT was tested by performing knockouts in different genomic locations in E. coli K12 MG1655. (a) Illustration of integrating tetAOPT while deleting lacZ. (b) Efficiency of tetAOPT integration was estimated by blue-white screening on LB agar supplemented with X-Gal. White colonies were counted as positive and blue as negative. (c) Illustration of tetA counterselection: tetAOPT can be removed using either a single-stranded oligonucleotide (ssDNA), which results in a clean deletion, or a double-stranded DNA (dsDNA) to integrate, for example, another gene of interest (here gfp). (d) The efficiency of tetAOPT removal was estimated using colony PCR on 50 colonies from selection plates in triplicates. (e)tetAOPT was used to knock out different genes across the genome of E. coli. Efficiencies of tetracycline (f) and NiCl2 selection (g) were estimated for each locus using colony PCR. (h + i) Since no positive colony could be found for the removal of tetAOPT in the atoB locus, a new approach removed tetAOPT while leaving the tetAOPTrrnC terminator on the genome. In panels b and d, the bars represent the mean of three biological replicates and the data points are shown. In f, g, and i, bars represent efficiencies of genome editing measured by PCR of 23 different colonies. In a + c, gray triangles depict recombination sites.
Since gene expression levels vary across the genome of E. coli and we have learned that tetA expression levels are critical for selection, we next tested the knockout of eight different genes in different locations previously shown to affect expression.52−55tetAOPT integration efficiencies for all loci were above 95%, while nickel counterselection efficiencies in most cases were above 90%. A notable exception to the latter was the atoB locus where removal of tetAOPT completely failed. We speculated that this could be due to genetic interactions at the new sequence boundary generated after tetAOPT removal. To circumvent this effect, we created a new ssDNA with homology to the downstream terminator (rrnC) of the tetAOPT construct, thereby leaving the terminator behind, which solved the problem and increased the efficiency to 100%. In addition to deleting these eight genes, using tetAOPT, we were also able to delete the 40.7 kb fli locus in K12 MG1655 in a single step, showing that large deletions are possible with tetAOPT (Figure S3).
Tuning of Native Gene Expression Facilitated by Dual Selection with tetAOPT
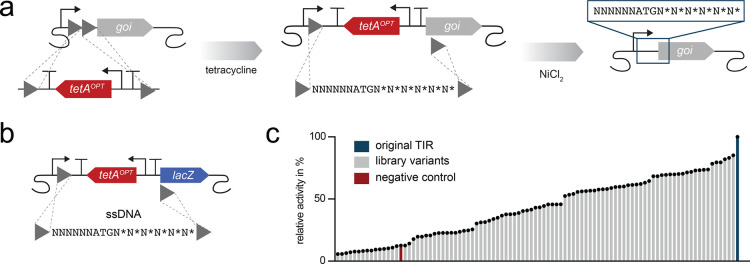
Introducing subtle DNA sequence changes, for example, in the TIR of heterologous genes is a simple and efficient way to tune protein production levels.12,13,52 Such sequence libraries do not only enable the selection of highly expressing variants but also the selection of expression levels that minimize the expression burden in the cell - our optimization of tetA being a good example of this. We wondered if a generalized tetAOPT workflow could be an efficient way to change the expression or regulation of native E. coli genes. In such a workflow, we first integrate tetAOPT directly upstream of the gene of interest and then introduce new regulatory DNA sequences, such as a TIR library, by removing tetAOPT with NiCl2 counterselection in combination with transforming a degenerated oligonucleotide or a PCR product (Figure 3a). This general approach allows first to screen for the consequence of interrupting gene expression and second to screen DNA libraries for changing the expression or regulation of the gene (Figure 3a).
Figure 3.
Engineering of translation using tetAOPT. (a) Illustration of the workflow for modulating translation initiation rates by using tetAOPT. In the first step, tetAOPT is integrated upstream of the start codon of the gene of interest (goi) by selection on tetracycline. In the next step, tetAOPT is removed using a degenerated oligonucleotide harboring homology (gray triangles; recombination sites) to the flanking regions of tetAOPT and selection on NiCl2. The degenerated oligonucleotide introduces a random sequence in the TIR. (b) Illustration of the TIR randomization of the native E. coli gene lacZ. (c) Relative beta-galactosidase activity of the TIR library variants (gray) normalized to the native TIR (dark blue). A negative control was included (tetAOPT inserted upstream of lacZ; red). In c, bar graphs of the libraries shown are based on single data points.
As a first simple demonstration, we integrated the tetAOPT cassette in front of lacZ, followed by introducing a TIR library with a short degenerate oligonucleotide. After recombineering, the cells were plated on agar plates with X-Gal and nickel and incubated overnight. The resulting colonies showed highly variable blue coloring (Figure S4), and sequencing of eight clones confirmed that a sequence library was created. Ninety four random colonies were picked and assayed for beta-galactosidase activity in liquid culture, and they similarly showed highly different activity levels (Figure 3c). Interestingly, with this approach, the native TIR seemed to provide the highest expression of beta-galactosidase. This demonstrates a highly simplistic and efficient workflow for manipulating gene expression levels directly on the E. coli genome.
Next, we tested a workflow for integration of new promoters. Since, in our experience, a larger genetic perturbation such as introduction of new genes or gene regulatory elements often compromises the TIR, we decided to introduce new promoters together with randomizing the TIR region. In this workflow, short constitutive promoters, such as the Anderson promoters, can still be integrated with single-stranded degenerate oligonucleotides (Figure 4a). We chose to exchange the native lac promoter for an Anderson collection constitutive promoter (J23100) while also introducing a TIR library. Sequencing of eight clones confirmed that a sequence library was created (Figure S5). This promoter exchange alleviated carbon catabolite repression of lacZ as observed by the absence of a diauxic shift during growth in medium supplemented with both glucose and lactose (Figure 4b). Further, the strain with the constitutive promoter and the native TIR entered the exponential growth phase earlier. Thus, by introducing a new promoter with a TIR library, strains with different growth behavior were created.
Figure 4.
Engineering of transcription using tetAOPT. (a) Illustration of how a short constitutive promoter (J23100) and a TIR sequence library were introduced in front of the native E. coli gene lacZ. (b) Growth in M9 medium supplemented with 0.05% glucose and 0.5% lactose. (c) Illustration of YidC-GFP expression using the T7 system. The T7 RNA polymerase (T7 RNAP) is part of the DE3 element on the E. coli chromosome. T7 RNAP is expressed from the lacUV5 promoter, regulated by LacI and therefore inducible by IPTG. T7 RNAP drives expression of YidC-GFP from a T7 promoter on a pET vector. Cells that successfully express YidC will be green fluorescent. (d) Illustration of the exchange of the lacUV5 promoter with the tunable promoter PrhaBAD with a randomized TIR. (e) Absolute production of YidC-GFP (nmol FITC) and growth of associated strains (OD600) were measured for 20 h. In b + e, growth of the control strains with the native promoter (blue), the new promoter and the native TIR (dark blue), and the negative control (red) was measured in triplicates. Measurements of the TIR library variants (gray) are based on single measurements. The fluorescein standard curve used for fluorescence normalization can be found in Figure S6. In a + d, gray triangles depict recombination sites.
To demonstrate a workflow using more complex promoters that need to be amplified via PCR or ordered as a synthetic DNA fragment (dsDNA) prior to integration by tetAOPT NiCl2 counterselection, we decided to address a well-known issue with leaky expression of the T7 RNA polymerase (RNAP) from its lacUV5 promoter. Leaky expression causes problems when handling toxic proteins,52,56 and we addressed this by exchanging PlacUV5 with the rhamnose-tunable rhaBAD promoter. First, PlacUV5 was removed through tetAOPT integration. Next, a pET28 vector expressing the toxic YidC-GFP57 was introduced followed by the re-expression of the λ-Red proteins and electroporation with a PCR product encoding PrhaBAD and a TIR library (Figure 4d). Sequencing of eight clones confirmed that a sequence library was created (Figure S5). The resulting clones were screened for YidC expression based on GFP fluorescence. Expression levels of YidC-GFP in cells with the rhaBAD promoter varied greatly, and some exceeded the levels of expression from the original T7 strain with the lacUV5 promoter (Figure 4e). With increasing concentrations of rhamnose, fluorescence levels increased (Figure S6). Similarly, we constructed a PrhaBAD-T7 RNAP TIR library for cells expressing only GFP (Figure S7). In this case, we were able to reach comparable expression levels of the new PrhaBAD constructs to the original T7 strain, and GFP expression was titratable by varying rhamnose concentrations (Figure S7). This demonstrates that the tetAOPT workflow facilitates advanced genome engineering, for example, to prevent gene expression toxicity.
Performance of tetAOPT in a Probiotic E. coli Strain
While E. coli K12 MG1655 is a classical model bacterium,21,47,52E. coli Nissle is primarily used as a model probiotic strain. When it comes to engineering probiotics, removal of antibiotic resistance markers is particularly crucial to avoid spreading resistance genes to the human gut microbiota.58 For this reason, we decided to investigate if tetAOPT can be used for engineering of E. coli Nissle, as it offers the possibility to scarlessly engineer genomes.
Again, we chose to replace the native lacZ locus of E. coli Nissle with tetAOPT, using the approach described above for E. coli K12 MG1655, to calculate the efficiencies by simple blue-white screening. Counterselection efficiencies were estimated by removing tetAOPT and simultaneously creating a knockout with ssDNA or by integrating a gene, in this case, gfp, with dsDNA. The integration efficiencies were 99.7 and 96.5% for positive selection with tetracycline (50 μg/mL) and ssDNA counterselection with 50 μM NiCl2, respectively (Figures 5b,e andS8), whereas for dsDNA counterselection, a high background growth made it difficult to count colonies and therefore to calculate efficiencies. The sensitivity of E. coli Nissle, harboring tetAOPT, toward NiCl2 was further assessed in liquid medium (Figure 5c) using three colonies (tetA 1–3) from the tetracycline selection plates that were randomly selected. Indeed, the strains seemed to be insensitive toward the previously used 50 μM NiCl2, whereas increasing the concentration to 100 μM prevented the growth of tetAOPT harboring strains but not the strain without tetAOPT. Increasing the NiCl2 concentration to 100 μM on solid medium did not lead to any growth; however, with 80 μM NiCl2, we were able to reduce the background and select positive colonies, but efficiency could still not be calculated (Figure 5f). Overall, the three randomly selected strains resulting from the tetracycline selection showed a very different sensitivity profile, which was reflected in the background growth.
Figure 5.
Gene knockout strategy inE. coliNissle. (a) Illustration of integrating tetAOPT and replacing lacZ in E. coli Nissle. (b) Efficiency of tetAOPT integration was estimated by blue-white screening on LB agar supplemented with X-Gal. White colonies were counted as positive and blue as negative. (c) Growth of E. coli Nissle, harboring tetAOPT in the lacZ locus in M9 medium supplemented with NiCl2. Three colonies of the lacZ knockout were selected (tetA 1–3; red), and the E. coli Nissle wild-type strain (WT, gray) was included as a control. Measurements represent single replicates. (d) Illustration of tetA counterselection. tetAOPT can be removed by either a single-stranded oligonucleotide (ssDNA), which results in a clean knockout or a double-stranded DNA (dsDNA) to integrate another gene of interest (in this case, gfp). Selection is performed on M9 agar supplemented with 50 μM NiCl2. (e) The efficiency of tetAOPT removal was estimated using colony PCRs of 50 colonies from the selection plates in triplicates. (f) dsDNA-mediated counterselection on M9 agar supplemented with 80 μM NiCl2. gfp was integrated into the lacZ:tetAOPT strain (tetA 1–3), and green fluorescence was visualized under blue light. The three plates represent integrations into strains tetA 1–3. In a + d, gray triangles depict recombination sites.
Discussion
Previously, we developed the tetA dual-selection marker for its use in the Standardized Genome Architecture (SEGA).52 In this setup, tetAOPT is part of a standardized landing pad and allows for the integration of a gene of interest following a simple protocol. Because the optimized marker proved to work very efficiently for this purpose, we here explored more broadly using it to engineer different stages of the central dogma in molecular biology.
On the DNA level, tetAOPT facilitates efficient gene deletions and insertions, and different loci generally did not significantly affect the performance. One exception was in the atoB locus, where tetAOPT could not be removed scarlessly by counterselection. Instead, we observed that NiCl2 treatment caused mutations in tetAOPT. These mutations likely occurred because the sequence that would have been created through the removal of tetAOPT, two converging genes, posed a burden to the cell. We could show that leaving behind one of the terminators from tetAOPT resulted in 100% efficient removal of the rest of tetAOPT. This is an interesting learning lesson for, for example, the design of minimal genomes created by top-down approaches: it is important to consider not only the essentiality of the genes that are removed but also the new sequence context created by the deletions. With this lesson in mind, and with the successful deletion of the 40.7 kb fli locus, tetAOPT shows great promise as a tool for large-scale genomic rearrangements.
The observed efficiencies for dsDNA-mediated selection against tetAOPT are lower than for ssDNA. This has previously been shown and may be related to the observation that ssDNA-mediated recombineering only requires the beta protein.34,52,59tetAOPT can also be used to tune the expression of native genes on the chromosome on the transcriptional and translational level. Our experimental setup of changing both translation and transcription allows us to screen the phenotype of a gene, as its expression is first interrupted by tetAOPT integration. This can give initial valuable information that a gene influences a specific phenotype, and the approach can easily be expanded to screen many genes for those that show the biggest effect. However, this approach also comes with the drawback that essential genes or operons with downstream essential genes cannot be targeted.
By introducing a TIR library in front of lacZ, we could create strains with different beta-galactosidase levels in the cell. Interestingly, the native TIR still seemed to provide the highest expression level. This opens the question whether the TIR of native genes always evolved toward the highest possible translation level based on the given level of transcription from its promoter. We did a simple in silico analysis of predicted translation levels from similar Shine-Dalgarno sequence libraries49 that suggest that this might be the case at least for genes involved in carbohydrate utilization.
On the transcriptional level, we used tetAOPT to exchange the promoters driving the expression of lacZ and the gene encoding T7 RNA polymerase. For lacZ, the exchange alleviated the diauxic shift, thereby demonstrating a potential of tetAOPT for highly specific decoupling of carbon catabolite repression, enabling better utilization of mixed carbon sources. When exchanging the lacUV5 promoter of the T7 RNAP to the rhaBAD promoter, we achieved higher production levels of the toxic YidC protein for the PrhaBAD constructs.
Most tetAOPT applications were demonstrated in E. coli K12 MG1655-a model strain for chromosomal engineering.52E. coli Nissle is commonly used as a probiotic strain, and its engineering is interesting for the development of microbiome therapeutics.60−63 In Nissle, tetAOPT-mediated selection and counterselection were as efficient as for K12 MG1655 with oligonucleotides. However, counterselection using dsDNA suffers from a high background growth in the presence of nickel. This demonstrates a need for further optimization of tetA for use in this strain background and, more generally, that ideally such genetic tools should be optimized for every individual organism.
Overall, the simple protocols as well as the high efficiencies have made tetAOPT our current preferred method for E. coli chromosomal engineering. tetAOPT is simple to make, either by PCR amplifying it from common plasmid backbones with the promoter and terminators being part of oligonucleotide overhangs or by ordering a SEGA strain and amplifying it from the chromosome. SEGA strains can be ordered from the Belgian Co-ordinated Collections of Microorganisms (bccm.belspo.be/genecorner-hosts/SEGA), and the DNA sequence of the optimized tetA can be found in the Supporting Information, on sega-genomes.com or in sbol format as part of the SEGA collection on synbiohub.org.
Methods
Strains, Cultivation, and Media Composition
Strains used in this study are listed in Table S1. E. coli K12 MG1655 or E. coli Nissle were used for all genome modifications. For initial TIR optimization of tetA and integration into the genome, the strain SIJ1947 was used. All strains were cultivated in lysogeny broth (LB) at 37 °C or 30 °C in case cells harbored pSIM19. If required, media were supplemented with antibiotics. Unless otherwise stated, antibiotics were used in the following concentrations: spectinomycin (50 μg/mL) and tetracycline (50 μg/mL). For recombineering purposes, all strains were made electrocompetent by washing the cells twice in ice-cold sterile water and centrifugation at 11000 rpm for 30 s. M9 agar plates were used for NiCl2 counterselection and contained 2 g/L glucose, 1x M9 salts, 2 mM MgSO4, 100 μM CaCl2, 1X trace elements, and 0.5 μg/mL thiamine. 10X M9 salts consist of 68 g/L Na2PO4, 30 g/L KH2PO4, 5 g/L NaCl, and 10 g/L NH4Cl. 1000X trace elements consist of 10 g/L FeCl3x6H2O, 2 g/L ZnSO4x7H2O, 0.4 g/L CuCl2x2H2O, 1 g/L MnSO4xH2O, 0.6 g/L CoCl2x6H2O, 3.2 mL/L 0.5 M Na2EDTA, and pH8. For screening beta-galactosidase expression, X-gal (0.02 mg/mL) was added to the M9 plates. IPTG (1 mM) or l-rhamnose (5 mM) was added for induction, if necessary.
Genetic Manipulation of E. coli
Genetic manipulations were performed as previously described.50 Detailed protocols about tetA dual selection in E. coli K12 MG1655 and Nissle can be found at protocols.io (dx.doi.org/10.17504/protocols.io.5jyl892y7v2w/v1). PCRs were performed using Phusion Hot Start II Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) or Neq2X7 polymerase.64 All oligonucleotides were ordered from Integrated DNA Technologies (IDT, Coralville, IA, USA) and are listed in Table S3. PCR products were visualized on 1% agarose gels using the iBright Imaging System (Thermo Fisher Scientific, Waltman, MA, USA), running the iBright gel imager software (v1.6.0). PCR purifications and plasmid isolations were performed using the NucleoSpin Gel and PCR Clean-up Kit and the NucleoSpin Plasmid Purification Kit (Macherey-Nagel, Düren, Germany), respectively. For the creation of gene knockouts, tetAOPT was amplified via PCR from strain K12 MG1655-TetAopt with oligonucleotides that each contains 50 bp homology to the integration site (#106-#163). Cells were plated on LB agar supplemented with 50 μg/mL tetracycline, 0.02 mg/mL X-Gal, and 1 mM IPTG. Blue-white screening was performed to identify correct colonies. The ssDNA targets the lagging strand and was designed using MODEST65 (modest.biosustain.dtu.dk). Removal of tetAOPT was achieved by electroporation of an oligonucleotide (ssDNA) or a PCR-amplified J23100-BCD-gfp construct (dsDNA) with 50 bp homology to the up- and downstream regions (#108-#161). Cells were plated on M9 agar supplemented with 50 μM NiCl2 for selection. For ssDNA-mediated counterselection, colony PCR of 23 colonies and a negative control for each locus was performed to estimate the efficiencies (oligonucleotides #106-#163). Colony PCRs were visualized using the LabChip GX Touch Nucleic Acid Analyzer (PerkinElmer, Waltham, MA, USA) and a DNA 5 KK Assay LabChip (PerkinElmer, Waltham, MA, USA). For dsDNA-mediated counterselection, efficiencies were estimated by calculating the ratio of fluorescent to non-fluorescent cells.
Library Design and Construction
All libraries of the TIR were constructed using degenerated oligonucleotides. The six nucleotides upstream of the start codon were changed to all possible nucleotides, and the six nucleotides downstream of the start were changed to all possible synonymous codon combinations.12 For tetA TIR libraries, tetA was amplified from the plasmid backbone of pSEVA55148 via PCR with oligonucleotides #101 and #102. The oligonucleotide binding upstream (#101) was degenerated and contained 50 bp homology to the integration site as well as the P3 promoter and the 5′ UTR (see Figure 1). The downstream oligonucleotide (#102) contained 50 bp homology to the integration site. For selection of the correct tetA variant, tetA was integrated into strain SIJ1947 downstream of gfp in the intergenic region of glmS and pstS. Cells were plated on LB agar with different concentrations of tetracycline (10, 25, 50, 75, and 100 μg/mL (Figure S1). Cells that grew on 50 μg/mL were then transferred to M9 agar supplemented with different NiCl2 concentrations (10, 25, 50, 75, and 100 μM) (Figure S1). Colonies that exhibited the highest sensitivity toward NiCl2 were selected for growth analysis in liquid M9 medium supplemented with either different tetracycline or NiCl2 concentrations. Sequencing of the TIR region of these colonies was conducted with oligonucleotide #172 to confirm a successful library generation (Figure S1). To insulate the tetA cassette from surrounding genes, the synthetic terminator L3S3P2249 was integrated upstream of tetA with oligonucleotide #103, resulting in tetAOPT. TIR libraries of genes in E. coli were performed by first integrating tetAOPT upstream of the start codon in the opposite orientation of the coding sequence (oligonucleotides #164-#171). tetAOPT was removed with a degenerated oligonucleotide, corresponding to the lagging strand, harboring 50 bp homology to up- and downstream of the TIR randomization region (oligonucleotide #166) (see Figure 3a). When the native promoter was exchanged, short constitutive promoters were integrated as part of this degenerated oligonucleotide with oligonucleotide #167. For the integration of larger inducible promoters, a PCR of the promoter construct was performed with oligonucleotides #170 and #171 each harboring 50 bp homology to the flanking regions of tetAOPT.
Beta-galactosidase Assay
To measure the beta-galactosidase activity, the assay developed by Schaefer et al. (2017)66 was adapted. A culture was set up in LB medium in a 96-deepwell plate and incubated overnight at 37 °C. The preculture was used for inoculation of a new culture in a 1:100 dilution and grown for 2 h at 37 °C. Then, the cultures were induced with 1 mM IPTG and grown until an OD600 of approximately 1.3 was reached. For the assay, 80 μL of the culture was mixed with 120 μL of a custom beta-galactosidase mix (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 36 mM b-mercaptoethanol, a pinch of Lysozyme, 1x cellytic B, and 1.1 mg/mL ONPG). OD420 was measured every 60 s in a Synergy H1 Plate reader (BioTej, Winooski VT, USA) using the BioTek Gen5 software (v3.08). The slope of the substrate conversion was calculated using GraphPad Prism9 (version 9.0.0). The slope of the control E. coli sample (original TIR) was set to 100%, and the library samples were normalized to it.
Growth and Fluorescence Analysis
Growth in liquid medium was analyzed in 96-well plates using the plate reader ELx808 (BioTek, Winooski, VT, USA) for OD630 measurements only or Synergy H1 (BioTek, Winooski, VT, USA) for combined OD630 and fluorescent measurements using the BioTek Gen5 software (v3.08). Overnight cultures were diluted 1:100 in LB medium and incubated at 37 °C with continuous shaking. Growth was measured every 15 m for at least 20 h. Cultures were induced with 1 mM IPTG or 5 mM l-rhamnose after 2 h, if necessary. For diauxic growth experiments, cells were grown in LB medium supplemented with 0.05% glucose and 0.5% lactose. For YidC-GFP production, the cells were grown in LB medium and induced after 2 h with 1 mM IPTG and 5 mM l-rhamnose, unless otherwise stated. A Breathe-Easy film (Sigma-Aldrich, St. Louis, MO, USA) was used to minimize evaporation during continuous growth analysis. Library variants were grown as single clones, while control strains were grown in triplicates, if not stated otherwise. GFP fluorescence was measured from the bottom with excitation set to 485 nm and emission set to 528 nm. Growth and fluorescence curves were analyzed using GraphPad Prism version 9.1.0 (GraphPad Software, Inc., San Diego, CA, USA). GFP fluorescence values were normalized using a standard curve generated with sodium fluorescein dilutions ranging from 10 nM to 500 nM (see Figure S6).
Acknowledgments
The authors thank Morten Sommer lab (Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Kgs. Lyngby, Denmark) for providing the strain E. coli Nissle 1917. Furthermore, we want to thank Jonathan Dyrsting Sandvang for his support on constructing the lacZ TIR library. The authors acknowledge funding by the Novo Nordisk Foundation (NNF20CC0035580) and by the “Bioroboost” project under EU Horizon 2020 research and innovation program under grant agreement N820699. A.G.V.S. was supported by grant no. NNF18CC0033664 as a fellow of the Copenhagen Bioscience PhD Programme.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00345.
Additional experimental details including growth curves of tetA TIR variants; blue-white screening of agar plates to calculate lacZ knockout efficiencies; knockout of the 40 kb fli region in E. coli; selection plates of lacZ TIR libraries and TIR sequences of selected variants; sequences of TIR library variants of strains with different lacZ promoters; fluorescein standard curve used for GFP fluorescence normalization and rhamnose titration of PrhaBAD-T7 RNA polymerase strains expressing YidC-GFP; expression of GFP in strains expressing T7 polymerase from rhaBAD promoter including TIR randomization; blue-white screening of agar plates to calculate lacZ knockout efficiencies in E. coli Nissle; and strain table, plasmid table, and nucleotide sequences (PDF)
Author Contributions
The manuscript was written by C.N.B. and M.H.H.N. The design of the optimized marker was made by C.N.B., M.R., and M.N. All experiments were conducted and analyzed by C.N.B., except for experiments regarding the exchange of native promoters, which were conducted by A.G.V.S. M.H.H.N. supervised this work. All authors read and approved the content of the manuscript. Correspondence and material requests should be addressed to M.H.H.N.
The authors declare the following competing financial interest(s): Selective optimisation of a ribosome binding site for protein production is patented (EP3234146, US 10,696,963) and owned by CloneOpt AB. M.H.H.N. is shareholder in CloneOpt.
Supplementary Material
References
- Kolisnychenko V.; Plunkett G.; Herring C. D.; Fehér T.; Pósfai J.; Blattner F. R.; Pósfai G. Engineering a Reduced Escherichia coli Genome. Genome Res. 2002, 12, 640–647. 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B. J.; Sung B. H.; Koob M. D.; Lee C. H.; Lee J. H.; Lee W. S.; Kim M. S.; Kim S. C. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 2002, 20, 1018–1023. 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- Pósfai G.; Plunkett G.; Fehér T.; Frisch D.; Keil G. M.; Umenhoffer K.; Kolisnychenko V.; Stahl B.; Sharma S. S.; de Arruda M.; Burland V.; Harcum S. W.; Blattner F. R. Emergent properties of reduced-genome Escherichia coli. Science 2006, 312, 1044–1046. [DOI] [PubMed] [Google Scholar]
- Park M. K.; Lee S. H.; Yang K. S.; Jung S.-C.; Lee J. H.; Kim S. C. Enhancing recombinant protein production with an Escherichia coli host strain lacking insertion sequences. Appl. Microbiol. Biotechnol. 2014, 98, 6701–6713. 10.1007/s00253-014-5739-y. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Ichimura T.; Mizoguchi H.; Tanaka K.; Fujimitsu K.; Keyamura K.; Ote T.; Yamakawa T.; Yamazaki Y.; Mori H.; Katayama T.; Kato J. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005, 55, 137–49. 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi H.; Sawano Y.; Kato J.; Mori H. Superpositioning of deletions promotes growth of Escherichia coli with a reduced genome. DNA Res. 2008, 15, 277–284. 10.1093/dnares/dsn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H.; Rubin A. J.; Sauer R. T. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011, 39, 1131–1141. 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H.; Fischer C.; Nevoigt E.; Stephanopoulos G. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 12678–12683. 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armetta J.; Schantz-Klausen M.; Shepelin D.; Vazquez-Uribe R.; Bahl M. I.; Laursen M. F.; Licht T. R.; Sommer M. O. A. Escherichia coli Promoters with Consistent Expression throughout the Murine Gut. ACS Synth. Biol. 2021, 10, 3359–3368. 10.1021/acssynbio.1c00325. [DOI] [PubMed] [Google Scholar]
- Oesterle S.; Gerngross D.; Schmitt S.; Roberts T. M.; Panke S. Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli. Sci. Rep. 2017, 7, 12327. 10.1038/s41598-017-12395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N.; Yuan Z.; Zhang X.; Chen J.; Zhou S.; Deng Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Res. 2020, 48, 10602–10613. 10.1093/nar/gkaa786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh K.; Martínez V.; Toddo S.; Guntur S.; Herrgård M. J.; Elofsson A.; Nørholm M. H. H.; Daley D. O. Enhanced Protein Production in Escherichia coli by Optimization of Cloning Scars at the Vector–Coding Sequence Junction. ACS Synth. Biol. 2015, 4, 959–965. 10.1021/acssynbio.5b00033. [DOI] [PubMed] [Google Scholar]
- Rennig M.; Martinez V.; Mirzadeh K.; Dunas F.; Röjsäter B.; Daley D. O.; Nørholm M. H. H. TARSyn: Tunable Antibiotic Resistance Devices Enabling Bacterial Synthetic Evolution and Protein Production. ACS Synth Biol 2018, 7, 432–442. 10.1021/acssynbio.7b00200. [DOI] [PubMed] [Google Scholar]
- Salis H. M.; Mirsky E. A.; Voigt C. A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapa S.; Taira S.; Heikkinen E.; Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999, 27, 2777. 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaeva N. I.; Gak E. R.; Zimenkov D. V.; Skorokhodova A. Y.; Biryukova I. V.; Mashko S. V. Dual-In/Out strategy for genes integration into bacterial chromosome: a novel approach to step-by-step construction of plasmid-less marker-less recombinant E. coli strains with predesigned genome structure. BMC Biotechnol 2008, 8, 63. 10.1186/1472-6750-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F.; Cui L.; Priest D. G.; Endy D.; Dodd I. B.; Shearwin K. E. One-Step Cloning and Chromosomal Integration of DNA. ACS Synth. Biol. 2013, 2, 537–541. 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- Baba T.; Ara T.; Hasegawa M.; Takai Y.; Okumura Y.; Baba M.; Datsenko K. A.; Tomita M.; Wanner B. L.; Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006, 2, 2006. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne M. E.; Moo-Young M.; Chung D. A.; Chou C. P. Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 5103–5114. 10.1128/aem.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. H.; Isaacs F. J.; Carr P. A.; Sun Z. Z.; Xu G.; Forest C. R.; Church G. M. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 460, 894–898. 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C.; Pedersen L. E.; Sommer M. O. A.; Nielsen A. T. CRMAGE: CRISPR Optimized MAGE Recombineering. Sci. Rep. 2016, 6, 19452. 10.1038/srep19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo P. L. H.; Ronda C.; Klompe S. E.; Chen E. E.; Acree C.; Wang H. H.; Sternberg S. H. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 2021, 39, 480–489. 10.1038/s41587-020-00745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K.; Thomason L. C.; Kuznetsov S. G.; Court D. L. Recombineering: A Homologous Recombination-Based Method of Genetic Engineering. Nat. Protoc. 2009, 4, 206–223. 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol 2013, 303, 298–304. 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Cunningham D. S.; Koepsel R. R.; Ataai M. M.; Domach M. M. Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb Cell Fact 2009, 8, 27. 10.1186/1475-2859-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. C.; Wood E. A.; Cox M. M. Convenient and reversible site-specific targeting of exogenous DNA into a bacterial chromosome by use of the FLP recombinase: the FLIRT system. J. Bacteriol. 1997, 179, 6076–6083. 10.1128/jb.179.19.6076-6083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer H. P. Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. J. Mol. Microbiol. Biotechnol. 2003, 5, 67–77. 10.1159/000069976. [DOI] [PubMed] [Google Scholar]
- Abremski K.; Wierzbicki A.; Frommer B.; Hoess R. H. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J. Biol. Chem. 1986, 261, 391–396. 10.1016/s0021-9258(17)42485-9. [DOI] [PubMed] [Google Scholar]
- Zhao D.; Yuan S.; Xiong B.; Sun H.; Ye L.; Li J.; Zhang X.; Bi C. Development of a fast and easy method for Escherichia coli genome editing with CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 205. 10.1186/s12934-016-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S.; Costantino N.; Court D. L.; Jenkins N. A.; Copeland N. G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005, 33, e36 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito J. A. Recombineering with tolC as a Selectable/Counter-selectable Marker: remodeling the rRNA Operons of Escherichia coli. Nucleic Acids Res. 2008, 36, e4 10.1093/nar/gkm1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C. J.; Lajoie M. J.; Napolitano M. G.; Mosberg J. A.; Goodman D. B.; Aach J.; Isaacs F. J.; Church G. M. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Res. 2014, 42, 4779–4790. 10.1093/nar/gkt1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y. S.; Chandran S.-P.; Kim K.; Lee S. K. Oligo- and dsDNA-mediated genome editing using a tetA dual selection system in Escherichia coli. PLOS ONE 2017, 12, e0181501 10.1371/journal.pone.0181501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. K.; Buckingham J. M.; Hanners J. L.; Hildebrand C. E.; Walters R. A. Plasmid-conferred tetracycline resistance confers collateral cadmium sensitivity to E. coli cells. Plasmid 1982, 8, 86–88. 10.1016/0147-619x(82)90044-0. [DOI] [PubMed] [Google Scholar]
- Maloy S. R.; Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 1981, 145, 1110–1111. 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolsky T.; Fong S.-T.; Lee B. T. O. Direct Selection of Tetracycline-Sensitive Escherichia coli Cells Using Nickel Salts. Plasmid 1996, 36, 112–115. 10.1006/plas.1996.0038. [DOI] [PubMed] [Google Scholar]
- Li X.-T.; Thomason L. C.; Sawitzke J. A.; Costantino N.; Court D. L. Positive and negative selection using the tetA-sacB cassette: recombineering and P1 transduction in Escherichia coli. Nucleic Acids Res. 2013, 41, e204 10.1093/nar/gkt1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay G. G.; Rothstein D. M. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob. Agents Chemother. 1993, 37, 191–198. 10.1128/aac.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela M. S.; Siddiqui K. A. I.; Sarkar B. L. High Expression of Plasmid-Encoded Tetracycline Resistance Gene inE. coliCauses a Decrease in Membrane-Bound ATPase Activity. Plasmid 1996, 36, 19–25. 10.1006/plas.1996.0027. [DOI] [PubMed] [Google Scholar]
- Reeve B.; Hargest T.; Gilbert C.; Ellis T.. Predicting Translation Initiation Rates for Designing Synthetic Biology. Front. Bioeng. Biotechnol. 2014, 0. 10.3389/fbioe.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C. O.; Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry 1990, 29, 5881–5889. 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Vimberg V.; Tats A.; Remm M.; Tenson T. Translation initiation region sequence preferences in Escherichia coli. BMC Mol Biol 2007, 8, 100. 10.1186/1471-2199-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm M. H. H.; Toddo S.; Virkki M. T. I.; Light S.; von Heijne G.; Daley D. O. Improved production of membrane proteins in Escherichia coli by selective codon substitutions. FEBS Lett. 2013, 587, 2352–8. 10.1016/j.febslet.2013.05.063. [DOI] [PubMed] [Google Scholar]
- Looman A. C.; Bodlaender J.; Comstock L. J.; Eaton D.; Jhurani P.; de Boer H. A.; van Knippenberg P. H. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987, 6, 2489–2492. 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona L.; Zou Z.; Stutzman N.; Sun P. D. Influence of the Second Amino Acid on Recombinant Protein Expression. Protein Expr Purif 2010, 74, 248–256. 10.1016/j.pep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M. T.; Pedersen M.; Klausen M. S.; Jensen S. I.; Wulff T.; Harrison S.; Nielsen A. T.; Herrgård M. J.; Sommer M. O. A. Predictable tuning of protein expression in bacteria. Nat. Methods 2016, 13, 233–236. 10.1038/nmeth.3727. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R.; Martínez-García E.; Calles B.; Chavarría M.; Arce-Rodríguez A. A.; de las Heras A. D.; Kim G.; Nikel J.; Platero P. I.; de Lorenzo R.; de Lorenzo de las HerasV. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J.; Liu P.; Nielsen A. A. K.; Brophy J. A. N.; Clancy K.; Peterson T.; Voigt C. A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 2013, 10, 659–664. 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- Stavropoulos T. A.; Strathdee C. A. Expression of the tetA(C) tetracycline efflux pump in Escherichia coli confers osmotic sensitivity. FEMS Microbiol. Lett. 2000, 190, 147–150. 10.1111/j.1574-6968.2000.tb09277.x. [DOI] [PubMed] [Google Scholar]
- Rodrigue A.; Effantin G.; Mandrand-Berthelot M.-A. Identification of rcnA (yohM), a Nickel and Cobalt Resistance Gene in Escherichia coli. J. Bacteriol. 2005, 187, 2912–2916. 10.1128/jb.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer C. N.; Rennig M.; Ehrmann A. K.; Nørholm M. H. H. A standardized genome architecture for bacterial synthetic biology (SEGA). Nat. Commun. 2021, 12, 5876. 10.1038/s41467-021-26155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englaender J. A.; Jones J. A.; Cress B. F.; Kuhlman T. E.; Linhardt R. J.; Koffas M. A. G. Effect of Genomic Integration Location on Heterologous Protein Expression and Metabolic Engineering in E. coli. ACS Synth. Biol. 2017, 6, 710–720. 10.1021/acssynbio.6b00350. [DOI] [PubMed] [Google Scholar]
- Bryant J. A.; Sellars L. E.; Busby S. J. W.; Lee D. J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014, 42, 11383–11392. 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz S. A.; Diao R.; Wolfe M. B.; Fivenson E. M.; Lin X. N.; Freddolino P. L. High-Resolution Mapping of the Escherichia coli Chromosome Reveals Positions of High and Low Transcription. Cell Syst 2019, 8, 212e9–225. 10.1016/j.cels.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyde S. A. H.; Nørholm M. H. H. Tailoring the evolution of BL21(DE3) uncovers a key role for RNA stability in gene expression toxicity. Commun Biol 2021, 4, 1–9. 10.1038/s42003-021-02493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley D. O.; Rapp M.; Granseth E.; Melén K.; Drew D.; von Heijne G. von. Global Topology Analysis of the Escherichia coli Inner Membrane Proteome. Science 2005, 308, 1321–1323. 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- Wright A. von. Regulating the Safety of Probiotics - The European Approach. Curr. Pharm. Des. 2005, 11, 17–23. 10.2174/1381612053382322. [DOI] [PubMed] [Google Scholar]
- Datta S.; Costantino N.; Zhou X.; Court D. L. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A 2008, 105, 1626–1631. 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B.; Yang Y.; Tham W. L.; Chen L.; Guo J.; Zhu G. Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application. Appl. Microbiol. Biotechnol. 2016, 100, 8693–8699. 10.1007/s00253-016-7829-5. [DOI] [PubMed] [Google Scholar]
- Praveschotinunt P.; Duraj-Thatte A. M.; Gelfat I.; Bahl F.; Chou D. B.; Joshi N. S. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 2019, 10, 5580. 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.; Lin C.; Yu J.; Qi Q.; Wang Q. Bioengineered Escherichia coli Nissle 1917 for tumour-targeting therapy. Microb. Biotechnol. 2020, 13, 629–636. 10.1111/1751-7915.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Chen X.; Sheng H.; Shen X.; Sun X.; Yan Y.; Wang J.; Yuan Q. Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact. 2020, 19, 56. 10.1186/s12934-020-01318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Rollán C.; Ehrmann A. K.; Nørholm M. H. H. Neq2X7: a multi-purpose and open-source fusion DNA polymerase for advanced DNA engineering and diagnostics PCR. bioRxiv 2022, 10.1101/2022.03.14.484273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M. T.; Klausen M. S.; Anderson M. V.; Wallin A. I. N.; Wang H. H.; Sommer M. O. A. MODEST: a web-based design tool for oligonucleotide-mediated genome engineering and recombineering. Nucleic Acids Res. 2014, 42, W408–W415. 10.1093/nar/gku428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J.; Jovanovic G.; Kotta-Loizou I.; Buck M. Single-step method for β-galactosidase assays in Escherichia coli using a 96-well microplate reader. Anal. Biochem. 2016, 503, 56–57. 10.1016/j.ab.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.