Abstract
Environmental enteropathy (EE) is a subclinical condition of the small intestine that is highly prevalent in low- and middle-income countries. It is thought to be a key contributing factor to childhood malnutrition, growth-stunting, and diminished oral vaccine responses. While EE has been shown to be the by-product of recurrent enteric infection, its full pathophysiology remains unclear. Here, we mapped the cellular and molecular correlates of EE by performing high-throughput single-cell RNA-sequencing on 33 small intestinal biopsies from 11 adults with EE in Lusaka, Zambia (8 HIV-negative, 3 HIV-positive), 6 adults without EE in Boston, USA, and 2 adults in Durban, South Africa, which we complemented with published data from 3 additional individuals from the same clinical site. By leveraging these data to reanalyze previously-defined bulk-transcriptomic signatures of reduced villus height and decreased plasma LPS levels in EE, we found that these signatures may be driven by an increased abundance of surface mucosal cells – a gastric-like subset previously implicated in epithelial repair in the gastrointestinal tract. In addition, we determined cell subsets whose fractional abundances associate with EE severity, small intestinal region, and HIV infection. Furthermore, by comparing duodenal EE samples with those from three control cohorts, we identified dysregulated WNT and MAPK signaling in the EE epithelium and increased pro-inflammatory cytokine gene expression in a T cell subset highly expressing a transcriptional signature of tissue-resident memory cells in the EE cohort. Altogether, our work illuminates epithelial and immune correlates of EE, and nominates cellular and molecular targets for intervention.
One Sentence Summary:
Using single-cell genomic profiling, we characterize the epithelial and immune correlates of environmental enteropathy (EE).
INTRODUCTION
Environmental enteropathy (EE) is a subclinical condition of the small intestine that is driven by environmental enteropathogen exposure (1). Also referred to as Environmental Enteric Dysfunction (EED), EE impacts millions of children and adults around the world and is associated with stunted growth, neurocognitive impairment, reduced oral vaccine efficacy, and increased risk of metabolic syndrome (2–4). Water, sanitation, and hygiene (WASH) interventions for preventing EE have proven ineffective, and ongoing work is assessing alternative therapeutic interventions (5). However, development of effective treatments has been hindered by a limited understanding of the underlying mechanisms of EE.
Pathologically, EE in the proximal small intestine is characterized by reduced villus height, greater crypt depth, and increased microbial translocation (1, 2, 6). However, in a study of Zambian children with EE and non-responsive growth stunting over time, reduced villus height was associated with decreased microbial translocation (7). A bulk transcriptomic study of Zambia children with enteropathy showed that genes differentially upregulated in biopsies with reduced villus height were downregulated in biopsies from participants increased microbial translocation (8). These studies suggest that EE is an adaptive response to potentially lethal enteropathogen exposure that comes at the cost of reduced absorptive capacity and impaired growth.
Histological analyses of EE have revealed increased abundance of lymphocytes, reduced goblet cell numbers, and altered Paneth cell morphology (6). Low plasma levels of tryptophan in children with growth stunting (9) and the amelioration of villus blunting in Zambian adults given amino acid supplementation suggest that amino acid deficiency plays a role in EE (10). Bulk transcriptomic studies of EE duodenal biopsies have revealed increased expression of NADPH oxidases, chemokines, mucins, matrix metalloproteases, interferon stimulated genes, and antimicrobial genes including LCN2, DUOX2, and DUOXA2 (11, 12). While previous work has lacked the single-cell resolution required to localize these changes to specific epithelial and immune cell subsets, application of single-cell RNA-sequencing (scRNA-seq) could help to resolve comprehensively the cellular and molecular changes that underlie EE pathophysiology (13).
Here, we applied the Seq-Well S3 platform for massively-parallel scRNA-seq (14) to characterize small intestinal biopsies from 11 adults from a community in Zambia where EE is known to be ubiquitous (15). Across these individuals, we profiled 27 biopsies spanning 3 small intestinal regions, HIV-positive and HIV-negative patients, and a range of EE severity scores. In addition, by comparing EE biopsies with those from control groups from South Africa and the USA, we found that EE was associated with upregulated WNT and downregulated MAPK signaling within the epithelium, as well as increased pro-inflammatory cytokine gene expression in a T cell subset highly expressing a transcriptional signature of tissue-resident memory cells. Altogether, our data provide insight into the epithelial and immune correlates of EE, suggesting several therapeutic targets for further investigation.
RESULTS
scRNA-seq of the proximal small intestine with and without EE
We collected 27 small intestinal biopsies from 11 Zambian volunteers with varying levels of EE severity (8 HIV-negative, 3 HIV-positive) (Fig. 1A, Table S3). For all 11, we profiled the duodenal bulb and distal duodenum; for a subset, we also collected jejunal samples (Table S1, S2). Due to the widespread prevalence of EE in Zambia and a lack of existing screening methods to identify patients without EE, we could not obtain control biopsies from participants without EE in Zambia. Thus, as controls, we profiled samples from 5 adults recruited from a gastrointestinal unit in Durban, South Africa (2 of which we profiled and 3 for which data were publicly available) (16, 17) as well as two cohorts of patients in Boston, USA where EE can safely be assumed not to occur (Table S1, S2). We note that EE is often contextualized to health by either comparing intermediate EE with severe EE (8), or by comparing EE patients to control cohorts in the United States or the United Kingdom (6). The validity of these international comparisons is supported by the environmental nature of EE and the resolution of EE in Peace Corps volunteers upon repatriation to the United States (18).
Figure 1: Single-cell RNA-sequencing of the small intestine with and without EE.
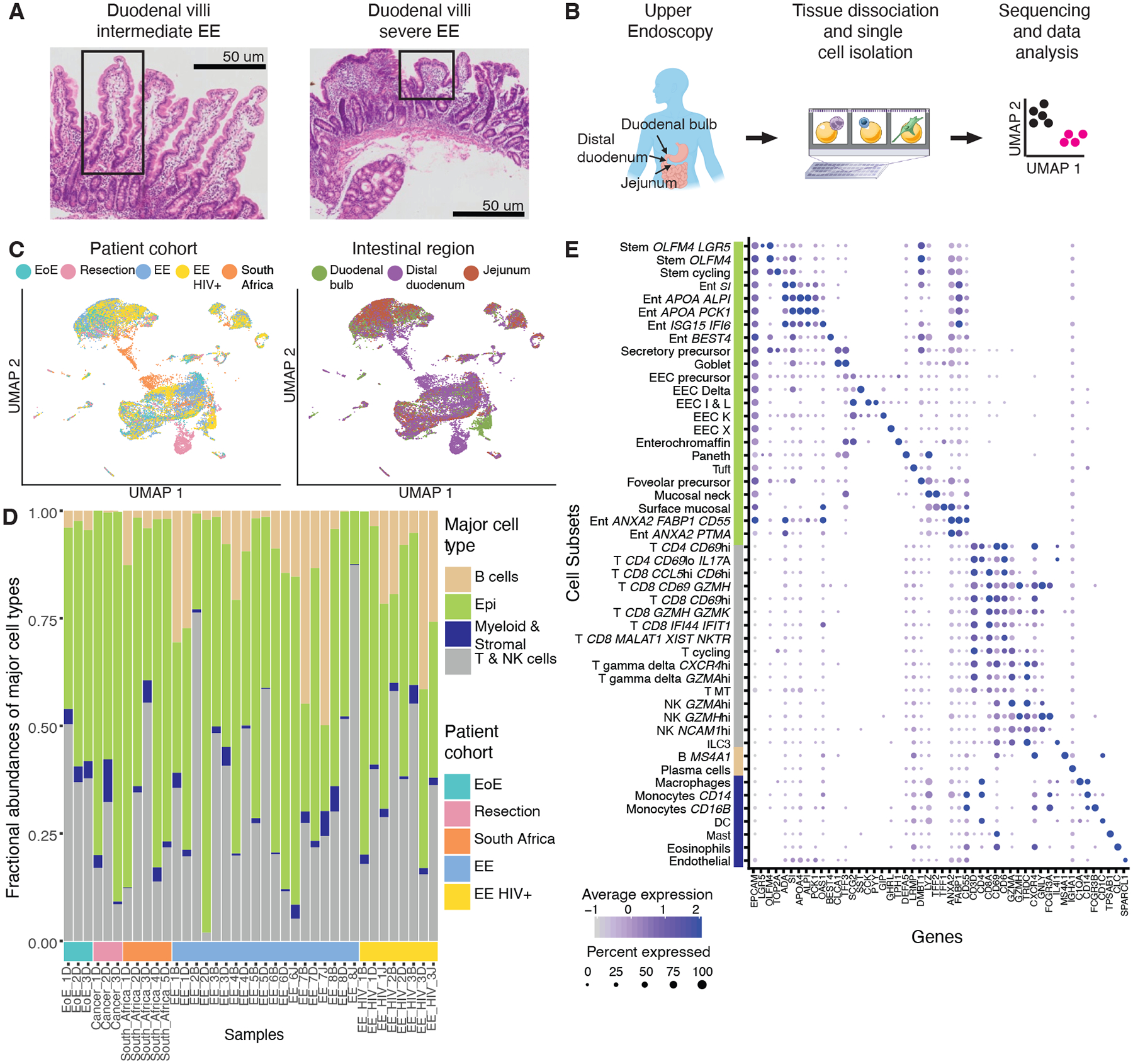
A, H&E imaging of the small intestine with intermediate and severe EE. Intestinal villi with normal morphology (left image) and with severe blunting (right image) are highlighted in boxes.
B, Experimental workflow: small intestinal biopsies from the duodenal bulb, distal duodenum, and jejunum were obtained via endoscopy or tissue resection, dissociated into cells, loaded onto a Seq-Well array, processed for single-cell sequencing, and analyzed.
C, UMAP visualization by patient cohort and intestinal region for all 26,556 high quality cells from 38 samples and 22 patients.
D, Fractional abundances of major cell types amongst all single cells analyzed per sample.
E, Expression of marker genes for final cell subsets. Dot size represents the fraction of a cell subset (rows) expressing a given gene (columns). Dot hue represents the scaled average expression by gene column. For clarity, dots for genes expressed in 5% or less of cells within a given subset are not shown.
In total, we analyzed 26,556 high quality single-cell transcriptomes across 38 samples from the Zambian, U.S., and South African cohorts (Fig. 1B). After data pre-processing, UMAP visualization of the entire dataset revealed differences in cellular distribution by patient cohort, intestinal region, and HIV status (Fig. 1c). To identify cellular subsets, we applied an existing pipeline for iterative clustering of cell subsets to the Zambian and U.S. datasets (19). Next, we integrated this data with the South African data to account for potential batch effects (20). These analyses revealed that the expected major cell types were represented in almost all biopsies and that epithelial cells were the most abundant major cell type (Fig. 1D). Along with more standard QC metrics (Fig. S1–S4), this indicated consistent sample quality. In total, we identified 48 detailed cellular subsets that varied in abundance across samples (Fig. 1E, Fig. S1–S4, Table S4) (13, 21, 22).
Surface mucosal cells expressing DUOX2 in the EE epithelium
When identifying cell subsets, we noticed a similarity between the marker genes of surface mucosal cells (a cell subset most commonly found in the distal stomach) and existing gene signatures of reduced villus height and decreased plasma LPS levels in EE (Fig. S5A) (8, 23). Relative to all other cell subsets, surface mucosal cells were significantly enriched for both gene signatures (p < 1015, Wilcoxon test) (Fig. 2A, B, Fig. S5B, C). In addition, the genes in these signatures overlapped with surface mucosal cell marker genes and three antimicrobial genes (DUOX2, DUOXA2, LCN2) recently identified as histological markers of EE (Fig. 2C, D) (12, 23). Thus, our data suggest these bulk gene signatures may have been driven by an increase in surface mucosal cell abundance. In our data, the vast majority of surface mucosal cells came from duodenal bulb samples of patients with EE (Fig. S2C). In these tissues, surface mucosal cell fractional abundances were significantly correlated (Permutation test, p < 0.05) with those of the stem cycling subset and the Ent ANXA2 PTMA subset which highly expressed marker genes of dedifferentiating enterocytes (PTMA) (24) and wound associated epithelial cells (ANXA2) (Fig. 2E) (25). To further examine potential relationships between these cell subsets that co-occurred with surface mucosal cells, we inferred differentiation trajectories for the epithelial cells in our dataset with PAGA (Fig. 2F) (26) The wound healing-like epithelial subsets and surface mucosal cells all lay in between mature enterocyte and stem cell subsets in the inferred differentiation hierarchy. Running the RNA velocity package Velocyto produced similar results (Fig. S5D, E) (27). In addition, immunohistochemical staining revealed that DUOX2 localized to the villus tip in blunted villi (Fig. 2G, Fig. S6). Together, these results suggest that surface mucosal cells occur at the villus tip in EE and are associated with the presence of intermediate wound healing-like cell populations. Furthermore, we observed that surface mucosal cells uniquely expressed MUC5AC–a marker of H. pylori infection (Fig. S5A, Table S4) (28). Applying the metagenomic classification tool Kraken 2, we found that relative to the control cohort samples, 6 samples from 4 participants in the EE cohort contained significantly higher levels of H. pylori mapping reads (Fig. S5). These samples were predominantly from the duodenal bulb (Fig. S5G). Thus, the presence of surface mucosal cells in EE may be associated with H. pylori infection.
Figure 2: Surface mucosal cells uniquely express DUOX2 and are associated with a wound healing-like phenotype.
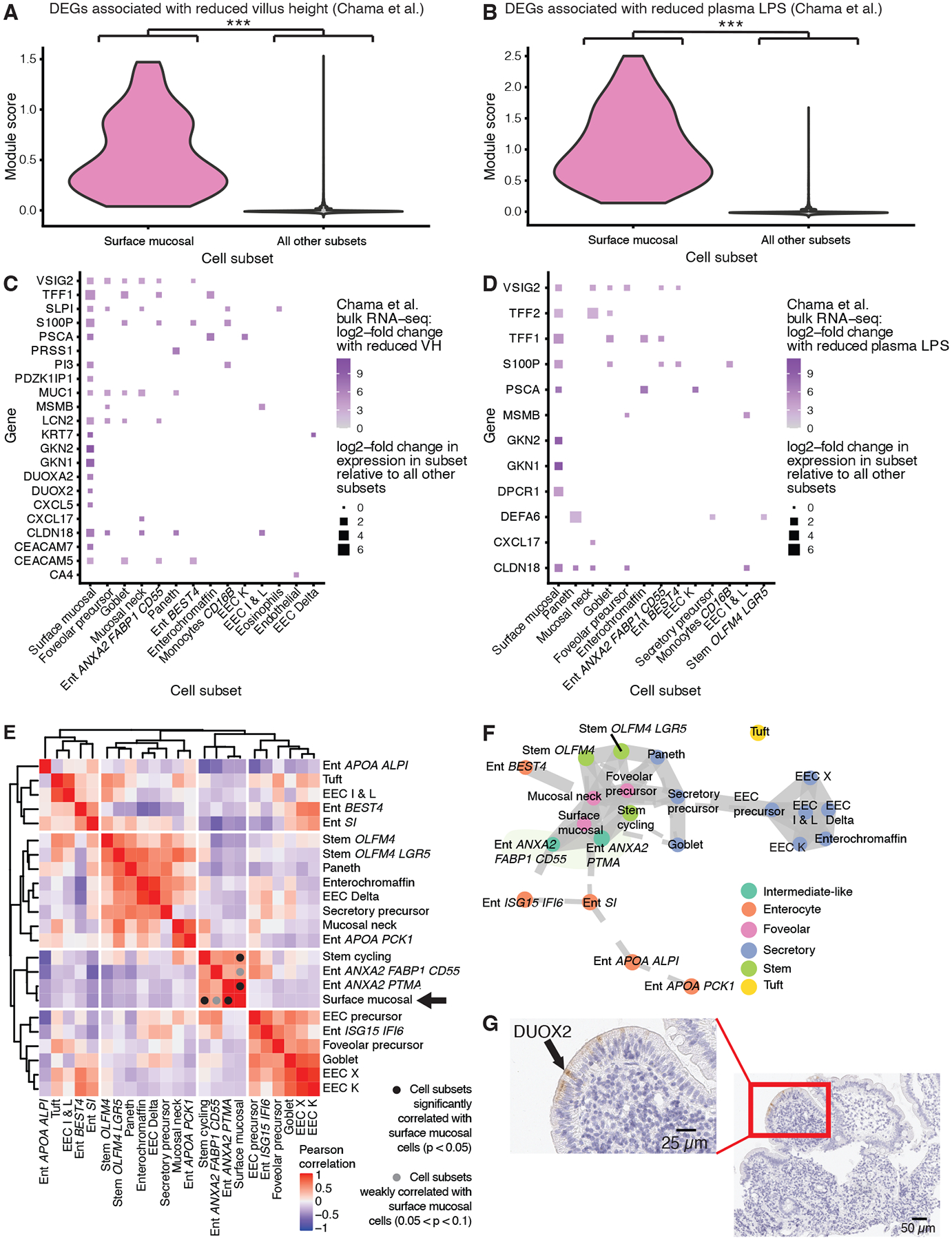
A, Violin plot of a module score generated from genes upregulated in EE samples with reduced villus height (VH) in Chama et al. (8) (***, p < 0.001; Wilcoxon test).
B, Violin plot of a module score generated from genes upregulated in EE samples wwith decreased plasma LPS concentrations in Chama et al. (8) (***, p < 0.001; Wilcoxon test).
C, Dot plot of cell subset marker genes that overlapped with the genes used to generate the module scores in panel A
D, Dot plot of cell subset marker genes that overlapped with the genes used to generate the module scores in panel B.
E, Hierarchically clustered heatmap of the Pearson correlations between the fractional abundances of epithelial cells within duodenal bulb samples from patients with EE. Cell subsets significantly correlated with surface mucosal cells are highlighted with a black circle (Permutation testing, p < 0.05). Cell subsets weakly correlated with surface mucosal cells are highlighted with a grey circle (0.05 < p < 0.1; Permutation testing).
F, PAGA trajectory visualization of epithelial subsets.
G, H&E (purple) and immunohistochemical staining for DUOX2 protein (brown) on an EE biopsy from the duodenal bulb.
Cellular correlates of intestinal region, disease severity, and HIV infection in EE
We next identified cell subsets whose fractional abundance shifted across intestinal region within HIV-negative EE patients (Fig. 3A). Duodenal bulb samples were enriched for surface mucosal cells, mucosal neck cells, and enterocytes highly expressing ANXA2, FABP1, and CD55, as well as three T cell subsets expressing markers of immune activation (IL17A, CXCR4, GZMA) (29, 30). Distal duodenal samples were enriched for enterocytes, goblet cells, and stem cells highly expressing OLFM4.
Figure 3: Cell subsets associated with intestinal region and histologically determined EE severity.
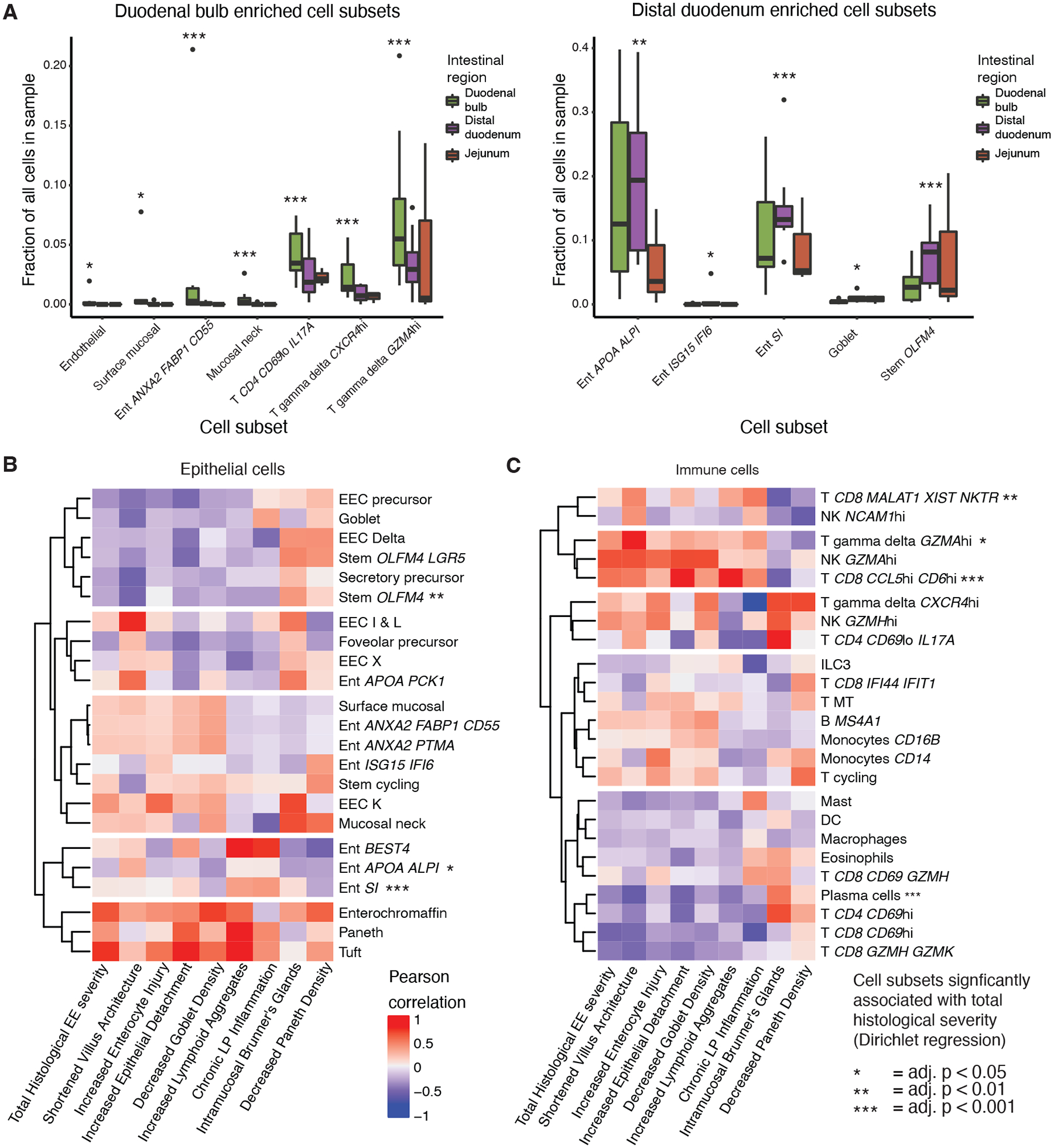
A, Cell subsets enriched in duodenal bulb and distal duodenal samples from HIV-negative EE patients (*, adj. < 0.05; **, adj. p < 0.01; ***, adj. p < 0.001, Fisher’s exact test).
B, Hierarchically clustered heatmap of HIV-negative EE epithelial cell subset relative abundance Pearson correlations with component scores of the total EE histological severity score (*, adj. p < 0.05; **, adj. p < 0.01,; ***, adj. p < 0.001; Dirichlet regression).
C, Hierarchically clustered heatmap of HIV-negative EE immune cell subset relative abundance Pearson correlations with component scores of the total EE histological severity score. (*, adj. p < 0.05; **, adj. p < 0.01; ***, adj. p value < 0.001; Dirichlet regression).
We then mapped the correlates of histologically determined EE severity in eleven biopsies (Fig. 3B,C; Supplementary Methods; Table S5) (6). In the epithelium, greater severity was associated with lower fractional abundances of the mature enterocyte and stem OLFM4 subsets, as well as higher fractional abundances of immature enterocytes. In the immune compartment, greater severity associated with a higher abundance of two T cell subsets expressing markers associated with inflammation in the intestine (GZMA and CD6) (29) and one T cell subset with high expression of MALAT1 and the lowest median number of UMIs of all T cell subsets, which together suggest that this subset may represent low quality pre-apoptotic cells (Fig. S2D) (31). In line with past findings, plasma cells abundances decreased with EE severity (32). These results suggest that severe EE is associated with an intermediate-like epithelial phenotype and inflammatory lymphocyte subsets.
Next, we sought to characterize the impact of antiretroviral-treated HIV infection on EE pathology. We found that HIV-positive samples displayed higher EE severity than HIV-negative samples (Wilcoxon test, p = 0.034) (Fig. S7A). Examining the shifts in cell subset fractional abundances with HIV infection, we found known features of HIV biology including decreased fractional abundances of CD4hi T cells and increased fractional abundances of γδ T cells highly expressing the HIV co-receptor CXCR4 (Fig. S7B, C) (16). In addition, within duodenal bulb samples, HIV pathology associated with increased fractional abundances of enterocytes highly expressing ANXA2, FABP1, and CD55, suggesting that HIV pathology may contribute to the presence of this wound healing-like subset within the duodenal bulb (Fig. S7D).
Epithelial correlates of EE
We next sought to identify features that distinguished HIV-negative EE distal duodenal samples from matched samples from participants in South Africa and the USA. While histology was not available for the South African dataset, H&E staining of a duodenal biopsy from a separate patient at the same clinical site revealed features of EE, including villus blunting, goblet cell depletion, and Paneth cell depletion, but no signs of inflammation (Fig. S8A). To investigate whether samples from this site displayed features of EE, we performed a pairwise comparison of the fractional abundances of all cell subsets between the three geographical locations in this study (Fig. S8B–D). Relative to the U.S. cohorts, both the Zambian and South African cohorts displayed two characteristic features of EE: reduced goblet cell and increased plasma cell fractional abundances (6). However, plasma cells and T cell subsets expressing markers of inflammation (IL17A, GZMA) were increased in fractional abundance in the Zambia EE cohort relative to the South African cohort (29), suggesting that not all features of EE were present in the South African samples. Thus, we took two approaches to identify the distinguishing attributes of confirmed EE in the Zambian cohort. First, we compared the Zambian cohort to all control cohorts. Then, we compared the Zambian cohort with confirmed EE to only the U.S. cohorts in case our previous analysis was confounded by potential features of EE in the South African cohort.
Comparing the fractional abundances of epithelial cells from patients with confirmed EE with those from all other cohorts, we found an enrichment of stem OLFM4 cells, foveolar precursor cells, and enterocytes co-expressing APOA4 and ALPI in EE, as well as reduced fractional abundance of EEC K cells (Fig. 4A). Differential expression analysis between epithelial cells in EE and control cohorts revealed compartment-wide upregulation of genes (PIGR, CCL25) involved in antibody transport and lymphocyte recruitment, among others (Fig. 4B,C Table S6) (33, 34). EE epithelial cells also highly expressed CTNNB1, a key component of WNT/ß-catenin signaling. In agreement, PROGENy analysis suggested increased WNT signaling in all three stem cell subsets and decreased MAPK signaling in cycling stem cells and stem OLFM4 cells in EE (Fig. 4D) (35). To help corroborate and extend these findings, we immunohistochemically stained Zambian EE and U.S. control samples for ß-catenin. We found that ß-catenin stained at higher intensity in the EE epithelium (Fig. S9). In addition, tuft cells in EE highly expressed ALOX5AP (which is involved in inflammation via leukotriene biosynthesis (36)) and SERPINA1 (which encodes ɑ−1-antitrypsin (AAT), a biomarker of epithelial damage in EE (5)) (Fig. 4C). Finally, comparison of our data to past intestinal scRNA-seq datasets revealed no evidence for “colonification” of the small intestine in EE and showed limited overlap between the genes upregulated in EE and ulcerative colitis (UC). (Fig. S10).
Figure 4: The epithelium of EE is characterized by increased WNT signaling, and decreased MAPK signaling.
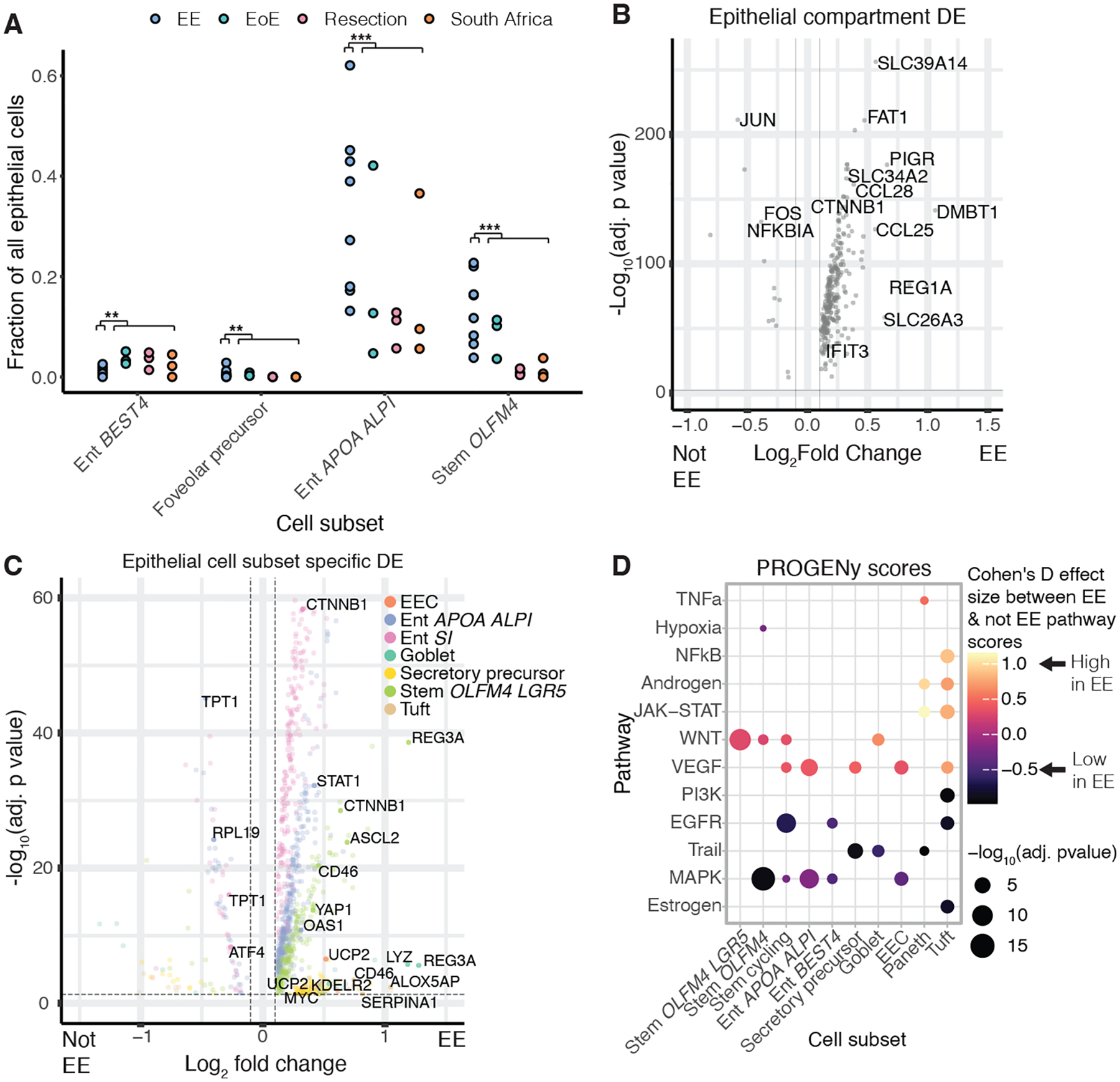
A, Cell subsets with significant shifts in relative abundances between EE and all control cohorts. (*, adj. p < 0.05; **, adj. p < 0.01; ***, adj. p < 0.001; Fisher’s exact test).
B, Genes differentially expressed in the epithelial compartment in EE relative to all control cohorts. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
C, Genes differentially expressed in EE relative to all control cohorts within specific cellular subsets. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
D, PROGENy pathway prediction scores for epithelial cells in EE relative to controls
Immune correlates of EE
Next, we compared cell proportions of immune cells between the Zambian cohort with confirmed EE and all control cohorts. This revealed that EE samples were enriched for CD8hi T cells highly expressing MALAT1 and γδ T cells highly expressing GZMA (Fig. 5A). Consistent with previous findings, plasma cells were increased in EE relative to non-EE cohorts (Fig. S10C) (32). Conducting differential expression analyses, we found that the majority of significant gene expression changes in EE immune cells occurred within the T cell compartment (Fig. 5B, Fig. S11A–C). Out of all T cell subsets, T CD8 CD69hi cells displayed the most differentially expressed genes between EE and controls, including upregulation of effector-like genes (IFNG, CCL5, IL32) in EE and downregulation of genes (IL7R, CXCR4) promoting memory T cell formation after infection (37, 38) (Fig. 5C, Table S6). As CD69 is a potential marker for T cell tissue residency, we scored all T cells subsets on a gene signature of tissue resident memory T cells and found that the T CD8 CD69hi subset scored highest (adjusted p = 4.98*10−78, one sided Wilcoxon test) (Fig. 5D) (39). Scoring all T CD8 CD69hi cells on T cell activation signatures, we found that relative to controls, cells in this subset from EE scored higher for signatures of cytotoxicity and cytokine production (Fig. 5E) (40). In agreement, immunohistochemical staining revealed more cells positive for Granzyme B in EE samples relative to controls (Fig. S12). Nominating putative ligand-receptor interactions between cellular subsets using NicheNet, we found potentially increased IFNγ signaling in the EE epithelium stemming from IFNγ production by T cells, especially the T CD8 CD69hi cells. (Fig. S13) (41). Altogether, our data reveal immune correlates of EE that may contribute to pathogenesis and reduced oral vaccine efficacy.
Figure 5: EE is associated with a shift towards activated T cell phenotypes.

A, Cell subsets with significant shifts in relative abundances between EE and all control cohorts (*, adj. p< 0.05; **, adj. p < 0.01; ***, adj. p < 0.001; Fischer’s exact test).
B, Genes differentially expressed in the T & NK cell compartment in EE relative to all control cohorts. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
C, Genes differentially expressed in in EE relative to all control cohorts within specific cellular subsets. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
D, Module scores for a tissue resident memory T cell signature in EE relative to all control cohorts.
E, T cell activation signatures enriched in T CD8 CD69hi cells from EE patients relative to all control cohorts. Proliferation: adj. p = 3.2*10−04, Cohen’s D effect size = 0.32. CD8 Cytotoxic: adj. p = 6.2*10−25, Cohen’s D effect size = 0.95. CD8 Cytokine: adj. p = 2.3*10−23, Cohen’s D effect size = 0.90
Evidence of reduced epithelial proliferation in EE relative to the U.S. cohorts
Finally, we compared only the Zambian EE and U.S. cohorts. Differential expression analysis revealed that EE samples displayed compartment-wide downregulation of genes (KLF4, ATF3, FOS, and JUN) involved in epithelial proliferation, IL-22 signaling, and goblet cell development (Fig. 6A) (42–44). Consistently, the EE samples displayed lower fractional abundances of goblet cells and ILC3s (producers of IL-22), as well as higher fractional abundances of γδ T cells (negative regulators of IL-22 production in mice fed a low protein diet) (Fig. S8B) (45, 46). Furthermore, gene set enrichment analysis (GSEA) revealed enrichment of the Reactome signature for response of EIF2AAK4 and GCN2 to amino acid deficiency in the epithelial cells (Fig. 6B, Table S8). In addition, goblet cells from patients with confirmed EE upregulated markers for lower crypt goblet cells suggesting that EE goblet cells have a more immature phenotype (Fig. 6C) (47). Furthermore, EE stem cells scored significantly lower on a gene signature of cycling human cells, and displayed lower PROGENY scores for the pro-proliferative EGFR, MAPK, and PI3K pathways (Fig. 6D–E, Table S8) (48). Upstream transcription factor activity inference with DoRothEA revealed reduced activation of ATF2 and ATF4 broadly across the epithelium, consistent with reduced IL-22 signaling (Fig. S14A, Table S9) (49). In sum, our results suggest that relative to the U.S. controls, the EE epithelium is characterized by reduced proliferation, IL-22 signaling, and goblet cell development. However, within the EE cohort, EE severity scores correlated with cycling scores in stem cells (Fig. S1B). Intriguingly, this suggested that although EE patients as a whole display lower levels of epithelial proliferation relative to U.S. controls, more severe EE leads to relatively higher epithelial proliferation than less severe EE.
Figure 6: Evidence of reduced proliferation in the EE epithelium relative to U.S. cohorts.
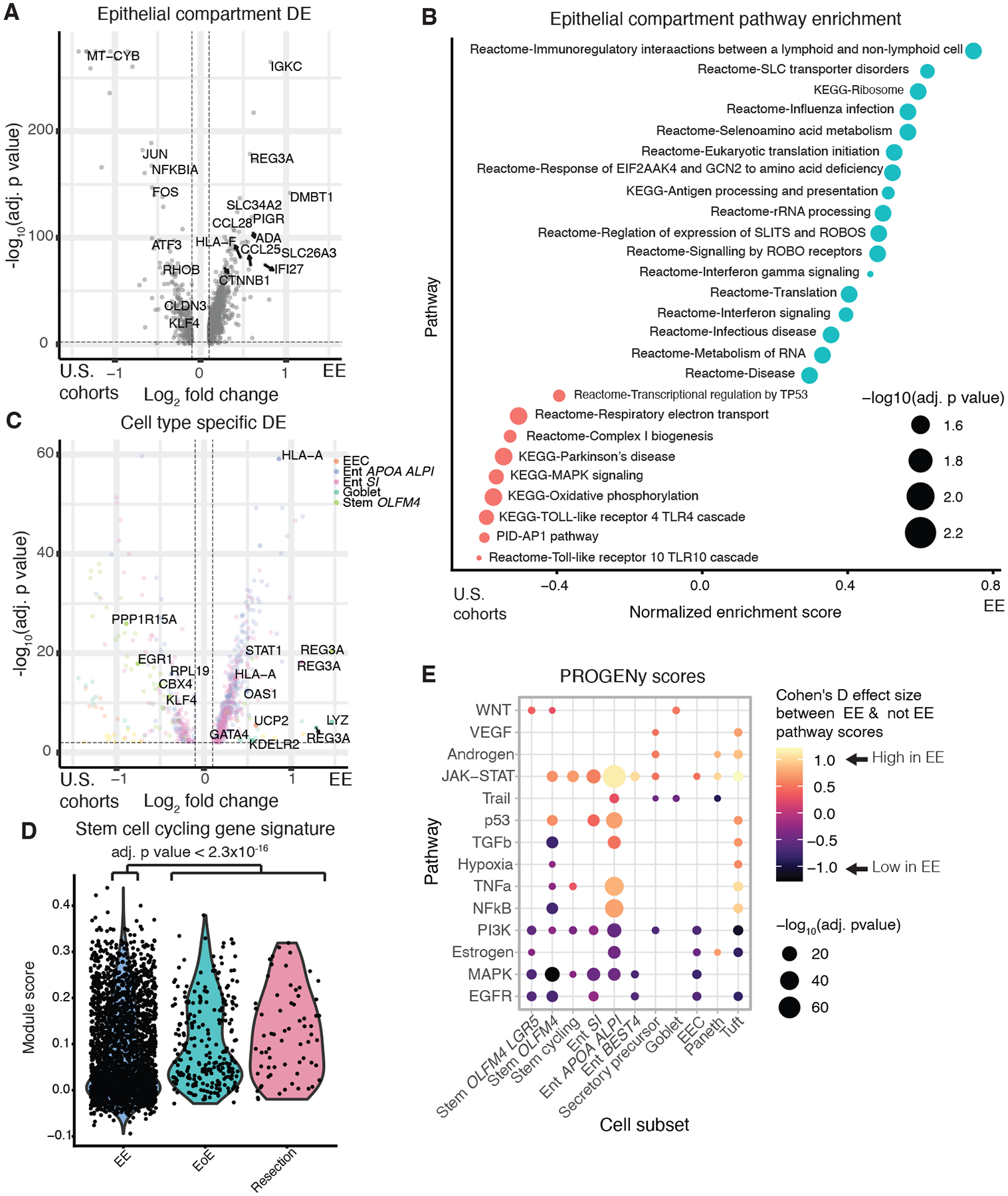
A, Genes differentially expressed in the epithelial compartment in EE relative to the U.S. cohorts. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
B, Gene set enrichment analysis of genes upregulated in epithelial compartment cells in EE relative to the U.S. cohorts.
C, Genes differentially expressed in EE relative to the U.S. cohorts within specific cellular subsets. Horizontal and vertical dashed lines respectively refer to an adjusted p value threshold of 0.01 and a log fold change threshold of 0.1.
D, Module scores for cell cycle genes in EE and U.S. cohorts in all cells from stem cell subsets.
E, PROGENy pathway prediction scores for epithelial cells in EE relative to U.S. cohorts.
DISCUSSION
Here, we profiled EE with the Seq-Well S3 platform for scRNA-seq. We thereby identified a cell subset – surface mucosal cells – which highly expressed DUOX2 and whose gene expression pattern matched existing bulk gene signatures of reduced villus height and reduced plasma LPS concentrations in EE. In addition, our dataset revealed variations in cell subset fractional abundance by small intestinal region, HIV infection, and EE severity, as well as epithelial and immune subsets differing between EE and control samples. Altogether, our work re-contextualizes past bulk-transcriptomic studies of EE and maps the cellular correlates of EE pathology.
The presence of surface mucosal cells highly expressing DUOX2 in EE may reflect remodeling of the epithelium into an intermediate wound healing-like state. Dedifferentiation of mature cells could facilitate repair of the epithelial barrier and reduce microbial translocation at the expense of reducing surface area and absorptive capacity, explaining why reduced nutrient absorption in EE is associated with decreased microbial translocation (7). This process may be due, in part, to H. pylori: as H. pylori gastritis has been shown often to be associated with duodenal colonization in children (50), H. pylori infection may explain why previous studies have found high levels of DUOX2 transcripts in the distal duodenum of some children with EE, whereas our study found DUOX2 expressing surface mucosal cells in the duodenal bulb (which is closest to the stomach where most H. pylori infection occurs) but not the distal duodenum of adults with EE.
Comparison to all control cohorts illuminated the epithelial and immune cell correlates of EE. Increased abundances of immature epithelial cell subsets, increased WNT/ß-catenin signaling, and decreased MAPK signaling suggested that the EE epithelium is biased towards an immature phenotype. Furthermore, Tuft cells upregulated genes involved in promoting and responding to inflammation (51). In addition, we found lower relative abundances of T cells expressing a transcriptional signature of tissue-resident memory T cells, but those cells had elevated expression of inflammatory cytokines (including IFNγ) in EE, suggesting that while these cells are present in a lower abundance in EE, they may be chronically activated. This, in turn, may lead to immune exhaustion and impaired responses to new immune stimuli, which may contribute a hindered response to oral vaccines (52). While IFNγ is often viewed as a pro-inflammatory cytokine in acute inflammation, numerous studies have demonstrated that in chronic inflammation, IFNγ can produce tolerogenic effects, which would be consistent with reduced oral vaccine efficacy in EE (53).
Additionally, we compared EE to only the U.S. cohorts to account for potential confounding features of moderate EE in the South African cohort. Relative to the U.S. cohorts, the EE epithelium was characterized by decreased epithelial proliferation and changes in cell subset fractional abundances, consistent with decreased IL-22 signaling (45, 46, 54). This is in line with work that found decreased abundances of transcripts from pro-proliferative pathways in the feces of Malawian children with EE (55), as well as work showing that during Cryptosporidium infection protein malnutrition leads to reduced turnover of intestinal epithelial cells (56). Consistently, reduced epithelial proliferation in EE relative to the U.S. cohorts was associated with GSEA enrichment of a response to amino acid deficiency and reduced goblet cell abundances–whose differentiation can be induced by tryptophan (57). This agrees with work demonstrating decreased tryptophan metabolism in Pakistani children with EE and work showing that amino acid supplementation ameliorates villus blunting in adults with EE (10, 12). Thus, amino acid deficiency may lead to hypoproliferative signaling in the EE epithelium characterized by reduced stem cell proliferation and goblet cell abundance. One intriguing aspect of this hypoproliferative signaling is that it stands in stark contrast to the hyperproliferative signaling observed in Crohn’s disease, which is of particular interest as limited evidence suggests lower rates of IBD in countries where EE is endemic (16, 49).
However, when comparing within the EE cohort, more severe EE was positively associated with stem cell proliferation. This may be due to the interplay of malnutrition and infection. The reduced dietary quality in the population of Zambian adults we studied may impose a proliferative constraint on enteropathy, leading to reduced stem cell turnover relative to intestinal homeostasis. However, individuals with more severe EE may have more proliferation than others in response to infective and inflammatory drivers. This discrepancy between our within and across country analyses is congruent with past work which found similar discrepancies, highlighting the necessity of comparing to an outgroup to fully understand the pathophysiology of EE (32, 58). Follow-up mechanistic studies are needed to clarify the role that malnutrition and infection play in epithelial proliferation in EE.
It is important to recognize that our study has several inherent limitations. We were not able to include a non-EE control group of age-matched adults in Lusaka, Zambia. Thus, we cannot rule out the possibility that the observed differences between EE patients and the U.S. and South African cohorts are due to unobserved variables that differed between these patient populations, especially the high burden of enteropathogens in tropical settings. In addition, our findings are primarily correlative due to the associative nature of measuring mRNA expression and the difficulties associated with mechanistic follow-up validation in humans. This is further limited by the lack of available tissue for histological analysis of samples from the South African cohort. As we saw few stromal cells in our scRNA-seq dataset, our tissue dissociation was likely biased against this subset and future work will be needed to characterize these cells in EE. In addition, the inflamed small intestinal epithelium and lamina propria are highly heterogenous environments containing several relatively rare cell types such as Paneth cells, and a variety of immune subsets which we did not have sufficient power to analyze in great detail. Furthermore, the HIV status of the participants from the U.S. cohorts was not determined. Additionally, we did not screen EE patients for Celiac disease and we cannot completely rule out the possibility that some patients may have had Celiac disease; however, we note that in Zambia the staple diet is maize (a gluten free food) and that past studies of EE in Zambian adults have found no evidence of Celiac disease (59). Also, pediatric EE may differ from EE in adults, which calls for future studies in pediatric cohorts. Finally, as EE is an endemic condition in low- and middle-income countries across the globe, it will be necessary to validate our results in cohorts with EE from geographic locations other than Zambia.
Examining our work as a whole, a potential picture of EE pathogenesis emerges. Relative amino acid deficiency due to a low-quality diet may lead to reduced epithelial proliferation and differentiation towards goblet cells, which would diminish anti-microbial mucosal defense, leading to increased pathogen-mediated damage of the enterocyte. This may then lead to epithelial remodeling towards an intermediate wound healing-like phenotype associated with the presence of surface mucosal cells. In addition, enteropathogen mediated damage would further exaggerate the pathogen-induced IFNγ response and may explain the observed pro-inflammatory polarization of CD8 CD69hi T cells in our data. Together, these findings nominate several therapeutic axes for inducing healthy epithelial proliferation and immune efficacy in EE.
MATERIALS AND METHODS
Study design
Adult volunteers were recruited from a disadvantaged community in Lusaka, Zambia, in which we have carried out previous studies of environmental enteropathy (7, 11). All volunteers gave written, fully informed consent. The study was approved by the University of Zambia Biomedical Research Ethics Committee (reference 006-11-15, dated 12th January 2016). From July 2018 to August 2018, volunteers underwent endoscopy with a Pentax 2990i gastroscope or a Pentax VSB2990i enteroscope, in the endoscopy unit of the University Teaching Hospital, Lusaka, under sedation with diazepam and pethidine. Duodenal tissue was collected from eosinophilic esophagitis (EoE) patients undergoing surveillance gastroscopy at Massachusetts General Hospital (MGH), Boston. Informed consent was obtained from EoE patients under a protocol approved by MGH. Resection samples were obtained from patients undergoing duodenal resection for pancreatic cancer (but in whom no local spread was apparent) in accordance with MGH IRB guidance under Mass General Brigham Protocol 2010P000632. Informed consent was obtained from participants recruited into this study at the Inkosi Albert Luthuli Central Hospital in Durban, South African. No randomization or blinding was done in this study. No power analyses were conducted due to the observational nature of this study and the lack of pre-existing scRNA-seq datasets of EE. The number of samples used is presented in the figures, supplementary figures, and supplementary tables.
Biopsy handling, immunohistochemical staining, and tissue digestion
Biopsies from the patients with environmental enteropathy were collected into normal saline, then orientated under a dissecting microscope, fixed in buffered formal saline, and processed to 3μm sections for haematoxylin/eosin staining. These sections were scanned using an Olympus VS120 scanning microscope, measured for average villus height (VH) and crypt depth (CD), and scored for EE severity using a recently published methodology (6). Duodenal bulb samples from EE patients were stained for DUOX2 protein and distal duodenal samples from EE patients and from normal tissue obtained from Mass General Brigham were stained for ß-catenin and GZMB protein; for more details see supplementary methods. Biopsies from EE, EoE, resection, and South African patients were dissociated into single-cell suspensions using a modified version of a previously published protocol (21). For further detail see the supplementary methods.
Single-cell RNA-sequencing with Seq-Well S3:
Please refer to the supplementary methods for further detail. Briefly, the epithelial and lamina propria layers of the biopsies were dissociated into single-cell suspensions. Then, single-cells were loaded onto a functionalized polydimethylsiloxane (PDMS) array preloaded with uniquely-barcoded mRNA capture beads (Chemgenes; MACOSKO-2011–10), and sequencing libraries were obtained and sequenced on an Illumina Next-Seq. Sequencing read alignment and demultiplexing was performed on the cumulus platform using snapshot 6 of the Drop-seq pipeline (48), resulting in a cell barcode by UMI digital gene expression (DGE) matrix. QC was performed to remove low quality cell barcodes and doublet cells. Bam files from sequencing were classified with Kraken2 to find metagenomic mapping reads. To identify cell subsets, we adopted an existing pipeline for automated iterative clustering of single-cell data that has been shown to identify batch effects without collapsing distinct rare cell types (19). We applied this pipeline to the data from the EE and U.S. cohorts. To correct for batch effects between data collected in different laboratories, we integrated our data with the South African dataset (20). All subsets were scored on gene signatures of reduced villus height and decreased and plasma LPS signatures from Chama et al. (8) using the AddModuleScore function in Seurat and a Wilcoxon test was used to assess significance. Epithelial trajectories were inferred with PAGA (26). RNA velocity trajectories were inferred with velocyto (27).
Analyses examining variation within samples from the EE cohort
To assess the epithelial subsets associated with surface mucosal cells in duodenal bulb samples from EE patients, we calculated the Pearson correlation between the fractional abundances of all epithelial subsets within these subsets. We then hierarchically clustered the resulting correlations with the ComplexHeatMap R package. Changes in the relative abundances of cell subsets by differing HIV infection status and small intestinal region were detected by a leave-one-out approach in order to avoid identifying patient specific effects using a Fisher’s exact test. Cell subsets significantly associated with histological EE severity were identified by running Dirichlet Regression. Further details are provided in the supplementary methods.
Comparison of distal duodenal samples from HIV-negative EE and control cohorts
Distal duodenal samples from HIV-negative patients iwth EE were compared to matched samples from control cohorts. We sought to identify biological features driven by variation in EE biology relative to the control cohorts (as opposed to identifying biology that distinguished only one cohort from EE). Thus, in all subsequent analyses we required that results pass the following two criteria: 1) Result significant when comparing EE vs all control cohorts 2) Result displayed the same direction of change between EE and each control cohort. Further details are provided in the supplementary methods.
Statistical analysis
The statistical test used for each comparison is denoted in the corresponding figure legend. Tests were conducted in R, and a Benjamini-Hochberg adjusted p value of 0.05 was used for significance.
Supplementary Material
Acknowledgements
We thank the participants in this study; B. Mead, S. Nguyen, A. Rubin, and R. Xavier for insightful feedback and copy editing; Chola Mulenga for histological processing and morphometry; Rose Banda for recruitment of the Zambian volunteers and sample collection; and Joyce Sibwani and Rose Soko for expert endoscopy nursing. Figure 1b was generated with Biorender.com.
Funding
This work was supported, in part, by grants to PK from Barts & The London Charity (G-000907) and from the Bill & Melinda Gates Foundation (OPP1066118). A.K.S. was supported was supported, in part, by the Searle Scholars Program, the Beckman Young Investigator Program, Sloan Fellowship in Chemistry, the NIH (5U24AI118672), the Bill and Melinda Gates Foundation, and the Ragon Institute. T.W. was supported by the NIH (DK007762). J.O.M was supported by the Richard and Susan Smith Family Foundation, the HHMI Damon Runyon Cancer Research Foundation Fellowship (DRG-2274-16), the AGA Research Foundation’s AGA-Takeda Pharmaceuticals Research Scholar Award in IBD – AGA2020-13-01, the HDDC Pilot and Feasibility P30 DK034854, the Food Allergy Science Initiative, the Leona M. and Harry B. Helmsley Charitable Trust, The Pew Charitable Trusts Biomedical Scholars and The New York Stem Cell Foundation. T.K.H. was supported by the NIH (F30-AI143160-01A1). J.E.S.S. was supported by the NIH (F32DK128872).
Competing interests
J.O.M. reports compensation for consulting services with Cellarity and Hovione. A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Repertoire Immune Medicines, Hovione, Third Rock Ventures, Ochre Bio, FL82, Empress Therapeutics, Relation Therapeutics, Senda Biosciences, IntrECate biotherapeutics, and Dahlia Biosciences unrelated to this work. A.K.S. has received research support from Merck, Novartis, Leo Pharma, Janssen, the Bill and Melinda Gates Foundation, the Moore Foundation, the Pew-Stewart Trust, Foundation MIT, the Chan Zuckerberg Initiative, Novo Nordisk and the FDA unrelated to this work. T.K.H. has served as a consultant and holds equity in nference, inc. The remaining authors disclose no conflicts.
Abbreviations
- CD
Crypt depth
- EE
Environmental enteropathy
- EoE
Eosinophilic esophagitis
- SAM
Severe acute malnutrition
- scRNA-seq
single-cell RNA-sequencing
- VH
Villus height
Footnotes
Supplementary Materials
Materials and Methods
Fig. S1. Characterization of epithelial subsets.
Fig. S2. Characterization of T and NK cell subsets.
Fig. S3. Characterization of B cell, myeloid, and stromal subsets.
Fig. S4. Number of genes and UMIs per cell across samples.
Fig. S5. Characterization of surface mucosal and dedifferentiation-like subsets.
Fig. S6. Immunohistochemical staining for DUOX2 protein.
Fig. S7. Variation in EE biology associated with HIV infection.
Fig. S8. Samples from South African participants display features of EE.
Fig. S9. Immunohistochemical staining for ß-catenin.
Fig. S10. Contextualizing EE epithelial cells with existing intestinal scRNA-seq signatures.
Fig. S11. Analysis of B cells and myeloid cells between EE and controls.
Fig. S12. Immunohistochemical staining for GZMB protein.
Fig. S13. NicheNet analysis of cell-cell signaling.
Fig. S14. Epithelial signaling changes in the EE cohort relative to the U.S. cohorts.
Table S1. Clinical characteristics of donors of intestinal biopsy samples.
Table S2. Patient cohort, intestinal region, and HIV infection status of participants.
Table S3. Villus morphometry for EE distal duodenal samples.
Table S4. Marker genes for all subsets.
Table S5. Histological severity scores for EE biopsies with matched histology.
Table S6. Genes differentially expressed in EE relative to control cohorts.
Table S7. KEGG, REACTOME, and PID pathway analysis.
Table S8. PROGENY pathway activation scores.
Table S9. Transcription factors with upstream activity predictions from DoRothEA.
Data availability
All data associated with this study are in the paper or supplementary materials. All scRNA-seq data is available at the Alexandria Project, a Bill and Melinda Gates Foundation-funded portal (part of the Single-Cell Portal hosted by The Broad Institute of MIT and Harvard): https://singlecell.broadinstitute.org/single_cell/study/SCP1307. The raw data for the EE and EoE cohorts is available at Gene Expression Omnibus (GEO) accession number GSE168883. The raw data for the resection samples is available by contacting the authors and establishing a Data Use Agreement. Code is available at https://doi.org/10.5281/zenodo.6703873 (63)
References and Notes
- 1.Chen RY, Kung VL, Das S, Hossain MS, Hibberd MC, Guruge J, Mahfuz M, Begum SMKN, Rahman MM, Fahim SM, Gazi MA, Haque R, Sarker SA, Mazumder RN, Di Luccia B, Ahsan K, Kennedy E, Santiago-Borges J, Rodionov DA, Leyn SA, Osterman AL, Barratt MJ, Ahmed T, Gordon JI, Duodenal Microbiota in Stunted Undernourished Children with Enteropathy, N. Engl. J. Med 383, 321–333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrant RL, Deboer MD, Moore SR, Scharf RJ, Lima AAM, The impoverished gut - A triple burden of diarrhoea, stunting and chronic disease, Nat. Rev. Gastroenterol. Hepatol 10, 220–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Church JA, Parker EPK, Kosek MN, Kang G, Grassly NC, Kelly P, Prendergast AJ, Exploring the relationship between environmental enteric dysfunction and oral vaccine responses, Future Microbiol. 13, 1055–1070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semba RD, Shardell M, Trehan I, Moaddel R, Maleta KM, Ordiz MI, Kraemer K, Khadeer M, Ferrucci L, Manary MJ, Metabolic alterations in children with environmental enteric dysfunction, Sci. Rep 6, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tickell KD, Atlas HE, Walson JL, Environmental enteric dysfunction: A review of potential mechanisms, consequences and management strategies BMC Med. 17, 181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu TC, Vanbuskirk K, Ali SA, Kelly MP, Holtz LR, Yilmaz OH, Sadiq K, Iqbal N, Amadi B, Syed S, Ahmed T, Moore S, Ndao IM, Isaacs MH, Pfeifer JD, Atlas H, Tarr PI, Denno DM, Moskaluk CA, A novel histological index for evaluation of environmental enteric dysfunction identifies geographic-specific features of enteropathy among children with suboptimal growth, PLoS Negl. Trop. Dis 14, 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amadi B, Zyambo K, Chandwe K, Besa E, Mulenga C, Mwakamui S, Siyumbwa S, Croft S, Banda R, Chipunza M, Chifunda K, Kazhila L, Vanbuskirk K, Kelly P, Adaptation of the small intestine to microbial enteropathogens in Zambian children with stunting, Nat. Microbiol, doi: 10.1038/s41564-020-00849-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chama M, Amadi BC, Chandwe K, Zyambo K, Besa E, Shaikh N, Ndao IM, Tarr PI, Storer C, Head R, Kelly P, Transcriptomic analysis of enteropathy in Zambian children with severe acute malnutrition, EBioMedicine 45, 456–463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosek MN, Mduma E, Kosek PS, Lee GO, Svensen E, Pan WKY, Olortegui MP, Bream JH, Patil C, Asayag CR, Sanchez GM, Caulfield LE, Gratz J, Yori PP, Plasma tryptophan and the kynurenine-tryptophan ratio are associated with the acquisition of statural growth deficits and oral vaccine underperformance in populations with environmental enteropathy, Am. J. Trop. Med. Hyg 95, 928–937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis-Auguste J, Besa E, Zyambo K, Munkombwe D, Banda R, Banda T, Watson A, Mayneris-Perxachs J, Swann J, Kelly P, Tryptophan, glutamine, leucine, and micronutrient supplementation improves environmental enteropathy in Zambian adults: A randomized controlled trial, Am. J. Clin. Nutr 110, 1240–1252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly P, Besa E, Zyambo K, Louis-Auguste J, Lees J, Banda T, Soko R, Banda R, Amadi B, Watson A, Endomicroscopic and Transcriptomic Analysis of Impaired Barrier Function and Malabsorption in Environmental Enteropathy, PLoS Negl. Trop. Dis 10, 1–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberman Y, Iqbal NT, Ghandikota S, Mallawaarachchi I, Braun T, Dexheimer PJ, Rahman N, Hadar R, Sadiq K, Ahmad Z, Idress R, Iqbal J, Ahmed S, Hotwani A, Umrani F, Ehsan L, Medlock G, Syed S, Moskaluk C, Ma JZ, Jegga AG, Moore SR, Ali SA, Denson LA, Mucosal Genomics Implicate Lymphocyte Activation and Lipid Metabolism in Refractory Environmental Enteric Dysfunction, Gastroenterology (2021), doi: 10.1053/j.gastro.2021.01.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A, A single-cell survey of the small intestinal epithelium., Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Christopher Love J, Shalek AK, Seq-Well: Portable, low-cost rna sequencing of single cells at high throughput, Nat. Methods 14, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly P, Menzies I, Crane R, Zulu I, Nickols C, Feakins R, Mwansa J, Mudenda V, Katubulushi M, Greenwald S, Farthing M, Responses of small intestinal architecture and function over time to environmental factors in a tropical population, Am. J. Trop. Med. Hyg 70, 412–419 (2004). [PubMed] [Google Scholar]
- 16.Fardoos R, Asowata OE, Herbert N, Nyquist SK, Zungu Y, Singh A, Ngoepe A, Mbano IM, Mthabela N, Ramjit D, Karim F, Kuhn W, Madela FG, Manzini VT, Anderson F, Berger B, Pers TH, Shalek AK, Leslie A, Kløverpris H, HIV infection drives interferon signalling within intestinal SARS-CoV-2 target cells, JCI Insight (2021), doi: 10.1172/jci.insight.148920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asowata OE, Singh A, Ngoepe A, Herbert N, Fardoos R, Reddy K, Zungu Y, Nene F, Mthabela N, Ramjit D, Karim F, Govender K, Ndung’u T, Porterfield JZ, Adamson JH, Madela FG, Manzini VT, Anderson F, Leslie A, Kløverpris HN, Irreversible depletion of intestinal CD4+ T-cells is associated with T-cell activation during chronic HIV infection, JCI Insight (2021), doi: 10.1172/jci.insight.146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keusch GT, Rosenberg IH, Denno DM, Duggan C, Guerrant RL, Lavery JV, Tarr PI, Ward HD, Black RE, Nataro JP, Ryan ET, Bhutta ZA, Coovadia H, Lima A, Ramakrishna B, Zaidi AKM, Hay Burgess DC, Brewer T, Implications of acquired environmental enteric dysfunction for growth and stunting in infants and children living in low-and middle-income countries, Food Nutr. Bull 34, 357–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng HB, Doran BA, Kimler K, Yu A, Tkachev V, Niederlova V, Cribbin K, Fleming R, Bratrude B, Betz K, Cagnin L, McGuckin C, Keskula P, Albanese A, Sacta M, de Sousa Casal J, Taliaferro F, Ford M, Ambartsumyan L, Suskind DL, Lee D, Deutsch G, Deng X, V Collen L, Mitsialis V, Snapper SB, Wahbeh G, Shalek AK, Ordovas-Montanes J, Kean LS, A Treatment-Na{\”\i}ve Cellular Atlas of Pediatric Crohn{\textquoteright}s Disease Predicts Disease Severity and Therapeutic Response, medRxiv (2021), doi: 10.1101/2021.09.17.21263540. [DOI] [Google Scholar]
- 20.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data, Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani A-C, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis., Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, Chen H, Wang J, Tang H, Ge W, Zhou Y, Ye F, Jiang M, Wu J, Xiao Y, Jia X, Zhang T, Ma X, Zhang Q, Bai X, Lai S, Yu C, Zhu L, Lin R, Gao Y, Wang M, Wu Y, Zhang J, Zhan R, Zhu S, Hu H, Wang C, Chen M, Huang H, Liang T, Chen J, Wang W, Zhang D, Guo G, Construction of a human cell landscape at single-cell level, Nature 581, 303–309 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Goldenring JR, Pyloric metaplasia, pseudopyloric metaplasia, ulcer-associated cell lineage and spasmolytic polypeptide-expressing metaplasia: reparative lineages in the gastrointestinal mucosa, J. Pathol 245, 132–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, Van Den Born M, Korving J, De Sauvage F, Van Es JH, Van Oudenaarden A, Clevers H, Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters, Cell Stem Cell 18, 203–213 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Rankin CR, Hilgarth RS, Leoni G, Kwon M, Den Beste KA, Parkos CA, Nusrat A, Annexin A2 regulates β1 integrin internalization and intestinal epithelial cell migration, J. Biol. Chem 288, 15229–15239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, Theis FJ, PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells, Genome Biol. 20, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV, RNA velocity of single cells, Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Den Brink GR, Tytgat KMAJ, Van Der Hulst RWM, Van Der Loos CM, Einerhand AWC, Büller HA, Dekker J, H pylori colocalises with MUC5AC in the human stomach, Gut 46, 601–607 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velaga S, Ukena SN, Dringenberg U, Alter C, Pardo J, Kershaw O, Franzke A, Granzyme a is required for regulatory T-cell mediated prevention of gastrointestinal graft-versus-host disease, PLoS One 10, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman NP, Vongsa RA, Faherty SL, Salzman NH, Dwinell MB, Targeted intestinal epithelial deletion of the chemokine receptor CXCR4 reveals important roles for extracellular-regulated kinase-1/2 in restitution, Lab. Investig 91, 1040–1055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madissoon E, Wilbrey-Clark A, Miragaia RJ, Saeb-Parsy K, Mahbubani KT, Georgakopoulos N, Harding P, Polanski K, Huang N, Nowicki-Osuch K, Fitzgerald RC, Loudon KW, Ferdinand JR, Clatworthy MR, Tsingene A, Van Dongen S, Dabrowska M, Patel M, Stubbington MJT, Teichmann SA, Stegle O, Meyer KB, ScRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation, Genome Biol. 21, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell DI, Murch SH, Elia M, Sullivan PB, Sanyang MS, Jobarteh B, Lunn PG, Chronic T cell-mediated enteropathy in rural West African children: Relationship with nutritional status and small bowel function, Pediatr. Res 54, 306–311 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Campbell DJ, Butcher EC, Intestinal attraction: CCL25 functions in effector lymphocyte recruitment to the small intestine, J. Clin. Invest 110, 1079–1081 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turula H, Wobus CE, The role of the polymeric immunoglobulin receptor and secretory immunoglobulins during mucosal infection and immunity, Viruses 10, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schubert M, Klinger B, Klünemann M, Sieber A, Uhlitz F, Sauer S, Garnett MJ, Blüthgen N, Saez-Rodriguez J, Perturbation-response genes reveal signaling footprints in cancer gene expression, Nat. Commun 9 (2018), doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ualiyeva S, Lemire E, Aviles EC, Wong C, Boyd AA, Lai J, Liu T, Matsumoto I, Barrett NA, Boyce JA, Haber AL, Bankova LG, Tuft cell-produced cysteinyl leukotrienes and IL-25 synergistically initiate lung type 2 inflammation., Sci. Immunol 6, eabj0474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaix J, Nish SA, Lin W-HW, Rothman NJ, Ding L, Wherry EJ, Reiner SL, Cutting Edge: CXCR4 Is Critical for CD8 + Memory T Cell Homeostatic Self-Renewal but Not Rechallenge Self-Renewal, J. Immunol 193, 1013–1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandele A, Joshi NS, Zhu J, Paul WE, Leonard WJ, Kaech SM, Formation of IL-7Rα high and IL-7Rα low CD8 T Cells during Infection Is Regulated by the Opposing Functions of GABPα and Gfi-1, J. Immunol 180, 5309–5319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, Friedman AL, Shen Y, Farber DL, Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites, Cell Rep. 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, Farber DL, Sims PA, Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease, Nat. Commun 10, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browaeys R, Saelens W, Saeys Y, NicheNet: modeling intercellular communication by linking ligands to target genes, Nat. Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Edgar BA, Boutros M, ATF3 acts as a rheostat to control JNK signalling during intestinal regeneration, Nat. Commun 8 (2017), doi: 10.1038/ncomms14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glal D, Sudhakar JN, Lu HH, Liu MC, Chiang HY, Liu YC, Cheng CF, Shui JW, ATF3 sustains IL-22-induced STAT3 phosphorylation to maintain mucosal immunity through inhibiting phosphatases, Front. Immunol 9 (2018), doi: 10.3389/fimmu.2018.02522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH, The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon, Development 129, 2619–2628 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Victor AR, Nalin AP, Dong W, McClory S, Wei M, Mao C, Kladney RD, Youssef Y, Chan WK, Briercheck EL, Hughes T, Scoville SD, Pitarresi JR, Chen C, Manz S, Wu L-C, Zhang J, Ostrowski MC, Freud AG, Leone GW, Caligiuri MA, Yu J, IL-18 Drives ILC3 Proliferation and Promotes IL-22 Production via NF-κB, J. Immunol 199, 2333–2342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan ZA, Khoury-Hanold W, Lim J, Smillie C, Biton M, Reis BS, Zwick RK, Pope SD, Israni-Winger K, Parsa R, Philip NH, Rashed S, Palm N, Wang A, Mucida D, Regev A, Medzhitov R, γδ T cells regulate the intestinal response to nutrient sensing., Science 371 (2021), doi: 10.1126/science.aba8310. [DOI] [PubMed] [Google Scholar]
- 47.Manco R, Averbukh I, Porat Z, Halpern KB, Amit I, Itzkovitz S, Clump sequencing exposes the spatial expression programs of intestinal secretory cells, bioRxiv, 2020.08.05.237917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets, Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell N, Pantazi E, Pavlidis P, Tsakmaki A, Li K, Yang F, Parker A, Pin C, Cozzetto D, Minns D, Stolarczyk E, Saveljeva S, Mohamed R, Lavender P, Afzali B, Digby-Bell J, Tjir-Li T, Kaser A, Friedman J, Macdonald TT, Bewick GA, Lord GM, Interleukin-22 orchestrates a pathological endoplasmic reticulum stress response transcriptional programme in colonic epithelial cells, Gut 69, 578–590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonamico M, Mariani P, Magliocca FM, Petrozza V, Montuori M, Pezzella C, Luzzi I, Carpino F, Helicobacter pylori duodenal colonization in children., Acta Paediatr. 86, 356–360 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Dixon RAF, Diehl RE, Opast E, Rands E, Millerll DK, Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis, Nature 343, 282–284 (1990). [DOI] [PubMed] [Google Scholar]
- 52.Wherry EJ, Kurachi M, Molecular and cellular insights into T cell exhaustion, Nat. Rev. Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rožman P, Švajger U, The tolerogenic role of IFN-γ Cytokine Growth Factor Rev. 41, 40–53 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Keir ME, Yi T, Lu TT, Ghilardi N, The role of IL-22 in intestinal health and disease, J. Exp. Med 217, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, Ordiz MI, Stauber J, Shaikh N, Trehan I, Barnell E, Head RD, Maleta K, Tarr PI, Manary MJ, Environmental Enteric Dysfunction Includes a Broad Spectrum of Inflammatory Responses and Epithelial Repair Processes, CMGH 2, 158–174.e1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Bolick DT, Kolling GL, Fu Z, Guerrant RL, Protein malnutrition impairs intestinal epithelial cell turnover, a potential mechanism of increased cryptosporidiosis in a murine model, Infect. Immun 84, 3542–3549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarado DM, Chen B, Iticovici M, Thaker AI, Dai N, VanDussen KL, Shaikh N, Lim CK, Guillemin GJ, Tarr PI, Ciorba MA, Epithelial Indoleamine 2,3-Dioxygenase 1 Modulates Aryl Hydrocarbon Receptor and Notch Signaling to Increase Differentiation of Secretory Cells and Alter Mucus-Associated Microbiota, Gastroenterology 157, 1093–1108.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly P, Bajaj-Elliott M, Katubulushi M, Zulu I, Poulsom R, Feldman RA, Bevins CL, Dhaliwal W, Reduced gene expression of intestinal α-defensins predicts diarrhea in a cohort of African adults, J. Infect. Dis 193, 1464–1470 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amadi B, Besa E, Zyambo K, Kaonga P, Louis-Auguste J, Chandwe K, Tarr PI, Denno DM, Nataro JP, Faubion W, Sailer A, Yeruva S, Brantner T, Murray J, Prendergast AJ, Turner JR, Kelly P, Impaired Barrier Function and Autoantibody Generation in Malnutrition Enteropathy in Zambia, EBioMedicine 22, 191–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolock SL, Lopez R, Klein AM, Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data, Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, Itzkovitz S, Spatial Reconstruction of Single Enterocytes Uncovers Broad Zonation along the Intestinal Villus Axis, Cell 175, 1156–1167.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Elmentaite R, Kumasaka N, Roberts K, Fleming A, Dann E, King HW, Kleshchevnikov V, Dabrowska M, Pritchard S, Bolt L, Vieira SF, Mamanova L, Huang N, Perrone F, Goh Kai’En I, Lisgo SN, Katan M, Leonard S, Oliver TRW, Hook CE, Nayak K, Campos LS, Domínguez Conde C, Stephenson E, Engelbert J, Botting RA, Polanski K, van Dongen S, Patel M, Morgan MD, Marioni JC, Bayraktar OA, Meyer KB, He X, Barker RA, Uhlig HH, Mahbubani KT, Saeb-Parsy K, Zilbauer M, Clatworthy MR, Haniffa M, James KR, Teichmann SA, Cells of the human intestinal tract mapped across space and time. Nature. 597, 250–255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kummerlowe C. (2022). Code for single-cell profiling of environmental enteropathy reveals signatures of epithelial remodeling and immune activation. Zenodo. 10.5281/zenodo.6703873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. All scRNA-seq data is available at the Alexandria Project, a Bill and Melinda Gates Foundation-funded portal (part of the Single-Cell Portal hosted by The Broad Institute of MIT and Harvard): https://singlecell.broadinstitute.org/single_cell/study/SCP1307. The raw data for the EE and EoE cohorts is available at Gene Expression Omnibus (GEO) accession number GSE168883. The raw data for the resection samples is available by contacting the authors and establishing a Data Use Agreement. Code is available at https://doi.org/10.5281/zenodo.6703873 (63)


