Abstract
Our understanding of the ubiquitin code has greatly evolved from conventional E1, E2 and E3 enzymes that modify Lys residues on specific substrates with a single type of ubiquitin chain to more complex processes that regulate and mediate ubiquitylation. In this Review, we discuss recently discovered endogenous mechanisms and unprecedented pathways by which pathogens rewrite the ubiquitin code to promote infection. These processes include unconventional ubiquitin modifications involving ester linkages with proteins, lipids and sugars, or ubiquitylation through a phosphoribosyl bridge involving Arg42 of ubiquitin. We also introduce the enzymatic pathways that write and reverse these modifications, such as the papain-like proteases of severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2. Furthermore, structural studies have revealed that the ultimate functions of ubiquitin are mediated not simply by straightforward recognition by ubiquitin-binding domains. Instead, elaborate multivalent interactions between ubiquitylated targets or ubiquitin chains and their readers (for example, the proteasome, the MLL1 complex or DOT1L) can elicit conformational changes that regulate protein degradation or transcription. The newly discovered mechanisms provide opportunities for innovative therapeutic interventions for diseases such as cancer and infectious diseases.
Subject terms: Ubiquitylation, Electron microscopy
Recent studies have expanded our understanding of the mechanisms and functions of ubiquitylation. Pathogens rewrite ubiquitylation to promote infection through unconventional ubiquitylation involving lipids and sugars, and structural studies have revealed that ubiquitin functions involve elaborate multivalent interactions that regulate transcription or protein degradation.
Introduction
Eukaryotic regulation of cellular processes is achieved through modular post-translational modification (PTM) cascades, whereby ‘writers’, which catalyse PTMs, and ‘readers’, which recognize them, collaboratively transduce input signals into specific cellular outputs1. Much of our understanding of the principles underlying modular signalling derives from studies of kinase or acetyltransferase catalytic domains, and the motifs that read their phosphorylation or acetylation marks, respectively. Such signalling has been likened to a ‘code’, determined by the arrangements of writer enzymatic domains, their targets, reader motifs and regulatory sequences within multidomain proteins and higher-order assemblies that execute PTM pathways. Some of the underlying modules are sufficiently well understood that PTM signalling pathways can be designed de novo, through the generation of artificial proteins built from mix-and-match regulatory, writer and reader elements. Although the modification with ubiquitin and ubiquitin-like proteins (UBLs), such as interferon-stimulated gene 15 (ISG15), small ubiquitin-like modifier (SUMO) and neural precursor cell-expressed developmentally downregulated protein 8 (NEDD8)) differs from other PTMs in that the modifiers are proteins, the ubiquitin and UBL code is often interpreted using modular signalling principles.
Ubiquitin and UBL modifications are written by combinations of E2 conjugating enzymes and E3 ligases with hallmark catalytic domains, for example the well-studied really interesting new gene (RING) domain, homologous to the E6-AP carboxy terminus (HECT) domain or RING-between-RING (RBR) domain, with unique enzymatic mechanisms (reviewed in refs.2–4). E2 and E3 enzymes catalyse modifications ranging from a single ubiquitin or UBL site-specifically linked to a particular target to polyubiquitin ‘chains’ wherein multiple ubiquitins are linked to each other. Ubiquitin and UBL modifications are ultimately read by downstream machineries that selectively bind and alter the fates of modified proteins (Fig. 1). Many polyubiquitin chain readers, including the 26S proteasome, display tandem ubiquitin-binding domains that each bind weakly to ubiquitin but that are arranged within a reader to synergistically recognize multiple ubiquitins in a linkage-specific manner. These properties are sufficiently robust to have enabled the design of linkage-selective ubiquitin chain sensors5,6.
Fig. 1. Elements of the ubiquitin code.
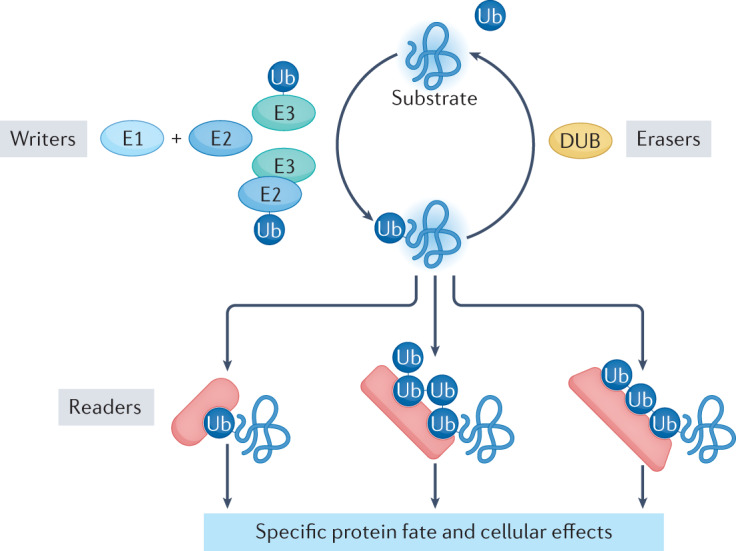
E3 enzymes (in combination with E2 and E1 enzymes) function as writers of the ubiquitin code. E3 enzymes are endowed with substrate specificity and — in a multistep mechanism that also engages E1 and E2 enzymes — attach ubiquitin to one or more residues of the substrate. Ubiquitin was primarily thought to modify Lys residues until recent studies discovered modification of many moieties on proteins and other macromolecules. This reaction can be repeated using a Lys of ubiquitin for attachment of the next ubiquitin molecule, giving rise to a ubiquitin chain. Depending on the E2–E3 pair that catalyses the last step of the ubiquitylation reaction, different Lys residues of ubiquitin are used, resulting in chains with different linkage types. Besides the seven Lys residues of ubiquitin, linkage can also occur with the amino-terminal Met. While ubiquitylation can be reversed by deubiquitylating enzymes (DUBs) that function as erasers of the ubiquitin code, it can be expanded by post-translational modifications of ubiquitin itself (Box 1). Distinct ubiquitin chains (and distinctly modified ubiquitin chains) have been shown to encode different cellular functions. They are decoded by readers equipped with ubiquitin-binding domains that are able to distinguish ubiquitin modifications and link the ubiquitylated substrate to downstream events, such as protein degradation, relocation, formation of multiprotein complexes and activation of enzymatic pathways. Ub, ubiquitin.
This Review summarizes the current knowledge of the ubiquitin code, including unprecedented ubiquitin modifications, new targets (such as sugars and lipids) and unique approaches by which bacteria and viruses reconfigure the ubiquitin code to promote infection. We also describe a fascinating set of novel structural, molecular and functional discoveries that revealed how the ubiquitin code depends on multivalent protein interactions to regulate cellular functions, as well as how its dysregulation promotes pathogenesis.
Ubiquitylation beyond Lys
Most studies of ubiquitylation have focused on linkage to amino groups, initially on Lys residues7. It seems likely that the successful identification of more than 100,000 modified Lys residues in human cells relied on the inherent stability of isopeptide bonds. More recent studies appreciated that protein amino termini are also amino groups that become linked to ubiquitin8–10. Indeed, from a chemical perspective, thioester bonds, which typically link the carboxy terminus of ubiquitin and the active site Cys in E2 and E3 enzymes, are highly reactive with properly placed amino groups11,12. Nonetheless, over the past four decades, there have been sporadic reports that the activities of viral E3 ligases13,14, endoplasmic reticulum-associated degradation15,16, peroxisomal protein translocation17–19, transcriptional regulation of cell fate20 and other processes involve ubiquitin modification of Cys, Ser and/or Thr side chains on targeted proteins (Box 1). However, such conclusions were largely based on indirect methods, for example ubiquitin modifications succumbing to reductive or hydrolytic chemical methods that destroy thioester or ester bonds, or loss of ubiquitylation upon mutation of Cys, Ser and/or Thr side chains. It was also not clear how specificity could be established for non-Lys side chains.
In the past four years, our understanding of endogenous ubiquitylation of non-Lys residues has greatly improved thanks to fortuitous discoveries facilitated by chemical biology and gene editing. Nuclear magnetic resonance spectroscopy and technical advances in proteomics allow direct detection of ester linkages between ubiquitin and a specific molecule, while deep probing of E3 ligases has established mechanisms specifying ubiquitin linkage to non-Lys moieties.
Box 1 The ubiquitin code and its modifications.
The ubiquitin code that is established by different linkage types, including linear and branched types of polyubiquitin chains and chains containing ubiquitin mixed with ubiquitin-like proteins (UBLs), is significantly expanded by the use of non-Lys residues for attachment of ubiquitin150 and post-translational modifications of ubiquitin107,151 such as phosphorylation of several Ser and Thr residues152, acetylation of six of seven Lys residues and deamidation by bacterial effectors (see the figure). Although proteomics studies show that all these ubiquitin modifications exist in cells, few have been studied functionally. Phosphorylated Ser65 (pSer65), which has been shown to change the structure of ubiquitin153,154, may have several effects on the ubiquitin code:
Phosphomimetic S65E ubiquitin shows altered affinity for ubiquitin-binding domains152.
In yeast, the phosphomimetic S65E ubiquitin is preferentially used for Lys6 chains and Lys11 chains, whereas other linkage types are disfavoured152.
Several E2–E3 pairs seem to be impaired in chain synthesis using pSer65-ubiquitin153.
Polyubiquitin chains containing pSer65-ubiquitin appear to be stabler as the activity of deubiquitylating enzymes is impeded153,155.
Acetylation of ubiquitin occurs on Lys residues and therefore interferes with ubiquitin chain formation. Hence, it might affect protein degradation mediated by Lys6-, Lys11- and/or Lys48-linked ubiquitin chains and/or shift polyubiquitin modification towards oligoubiquitylation or monoubiquitylation156 that may engage different readers. Indeed, ubiquitin acetylated at Lys6 or Lys48 not only inhibited chain elongation at Lys6 and Lys48, respectively, but also at Lys residues that are not acetylated (Lys11 and Lys63). Recently, ubiquitin acetylated at Lys11, Lys27, Lys33, Lys48 or Lys63 was also found to impair transthiolation from E1 to ubiquitin-conjugating enzyme E2 D1 (UBE2D1)157.
Whereas ubiquitin-binding domains with higher affinity for pSer65-ubiquitin have been identified, binding domains that specifically recognize acetylation of ubiquitin have not yet been reported. It is, however, likely that acetylation of ubiquitin may affect the interaction with ubiquitin-binding domains recognizing unmodified ubiquitin. Using NMR, acetylation of ubiquitin at specific Lys residues was shown to enable ubiquitin to adopt distinct conformations that modulated its binding properties, leading to acetylation-site-specific ubiquitin interactomes. Acetylation at different Lys residues of ubiquitin also hampered the capacity of several E3s to undergo autoubiquitylation158.
Writers, readers159 and erasers160,161 can be post-translationally modified to change their ubiquitin-dependent activity in many ways. For example, N acetylation of the E2 enzyme ubiquitin-conjugating enzyme 12 (UBC12) critically contributes to its interaction with the neural precursor cell-expressed developmentally downregulated protein 8 (NEDD8) E3 ligase defective in cullin neddylation 1 protein (DCN1), thereby enhancing neddylation of the DCN1 substrate cullin 1 (CUL1)162. TANK-binding kinase 1 (TBK1) and casein kinase 2 (CK2) phosphorylate the ubiquitin-binding domains of autophagy receptors (readers), thereby increasing their affinity for ubiquitin163–166. Lys99 of the deubiquitylating enzyme ubiquitin-specific protease 25 (USP25) can be either ubiquitylated or sumoylated. Both modifications cause different intramolecular interactions that either activate (ubiquitin) or inhibit (small ubiquitin-like modifier (SUMO)) the deubiquitylating activity of USP25 (refs.167,168).
Most of our current knowledge of the ubiquitin code owes much to the stability of the isopeptide bond of typical ubiquitylation and stems from ‘ubiquitinomics’ studies169 comprising mass spectrometry-based proteomics studies170, biochemical and antibody-based methods, and chemical and protein probes171. However, it is very likely that a large part of the ubiquitin code remains unknown. New tools are needed to identify additional types of modification, to quantify and characterize them functionally and to understand their complexity in vivo. ADPR, ADP-ribose; PR, phosphoribosyl; Ub, ubiquitin.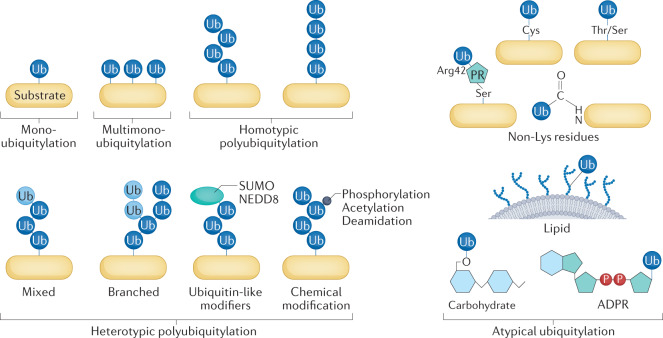
Writers forging ester bonds between ubiquitin and Ser or Thr hydroxyls
Thr was discovered as the preferred site of modification by a novel ‘RING–Cys–relay’ (RCR) catalytic mechanism used by the human E3 ligase MYCBP2 through a remarkable series of experiments initiated for unrelated purposes. MYCBP2 was found to react with a chemical probe resembling an E2–ubiquitin intermediate21. The probe was designed to stably capture E3 catalytic Cys through reaction with an electrophile positioned between the E2 conjugating enzyme and ubiquitin21–23. Proteomics studies revealed that when the probe was applied to cell lysates, it reacted with almost all E3s with a catalytic Cys known to receive ubiquitin from an E2 (that is, HECT-family and RBR-family E3s). However, MYCBP2 stood out as having a RING domain, but lacking a recognizable E3 domain with a catalytic Cys. That changed with the identification of the Cys that reacted with the probe. Elegant biochemical and structural studies showed that this Cys and another one nearby in the sequence are both catalytic in the newly discovered RCR catalytic domain, where the RING element binds an E2–ubiquitin intermediate, ubiquitin is transferred from the E2 Cys to one and then the other MYCBP2 catalytic Cys, and ubiquitin is ultimately ligated to Thr21,23 (Fig. 2a). Thr, not Lys or Ser, preferentially discharged ubiquitin from the RCR domain of MYCBP2. Crystal structures and mutational analyses revealed how the RCR domain catalyses various steps in this process. Most remarkably, an active site pocket, adjacent to the loop harbouring the second Cys, binds and positions the Thr for ubiquitylation. Knock-in mice harbouring a mutation that abolishes the RCR E3 mechanism and non-Lys ubiquitylation have a striking neurodevelopmental phenotype23. Furthermore, axons are protected in models of injury, suggesting that non-Lys ubiquitylation activity has important neuronal functions and that its inhibition might be of therapeutic value24.
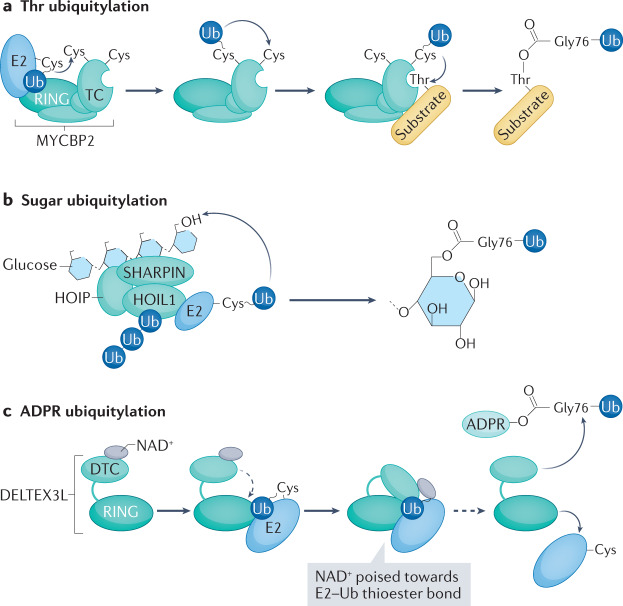
Fig. 2. Unconventional ubiquitylation.
a | Thr ubiquitylation by the E3 MYCBP2. The RING–Cys–relay (RCR) mechanism depends on the RING domain, which binds E2–ubiquitin, and on the tandem Cys (TC) domain, which contains two catalytic Cys residues. Upon E2–E3 transthiolation and E2 dissociation, ubiquitin is relayed from Cys4520 to Cys4572 within the TC domain by intramolecular transthiolation. The TC domain also positions the substrate to allow Thr esterification. b | Linear ubiquitin assembly complex (LUBAC)-mediated ubiquitylation of unbranched glucosaccharides. The LUBAC components haem-oxidized IRP2 ubiquitin ligase 1 (HOIL1)-interacting protein (HOIP) and SHANK-associated RH domain-interacting protein (SHARPIN) bind glucosaccharides, whereas HOIL1 ubiquitylates the C6 hydroxyl moiety of glucose in one catalytic step. The reaction is accelerated by non-covalent binding of unconjugated Met1-linked or Lys63-linked ubiquitin oligomers to the RING-between-RING (RBR) domain of HOIL1. c | Ubiquitylation of ADP-ribose (ADPR) by DELTEX3L. E2–ubiquitin recruitment by the RING domain of DELTEX3L promotes a conformational change that positions the NAD+ bound to the carboxy-terminal domain (DTC) in close proximity to E2–ubiquitin, which facilitates ubiquitylation of ADPR at Gly76. Ub, ubiquitin.
Regulation of ester linkage of ubiquitin to amino acid side chains gained a solid footing when haem-oxidized IRP2 ubiquitin ligase 1 (HOIL1; also known as RBCK1) was identified as another E3 catalysing such modifications25. HOIL1, along with the linear ubiquitin chain forming E3 HOIL1-interacting protein (HOIP) and regulator SHANK-associated RH domain-interacting protein (SHARPIN), form a multifunctional trimeric linear ubiquitin assembly complex (LUBAC). HOIL1 is an RBR-family E3 ligase, but its direct substrates were unclear until knock-in mice harbouring a HOIL1 active site mutation were generated to understand the physiological role of HOIL1 in immune signalling. Biochemical studies of lysates from bone marrow-derived macrophages showed a curious defect of the active site mutation: they lacked a slowly migrating form of HOIL1 that disappeared upon chemical hydrolysis of ester linkages. Proteomics revealed wild type HOIL1 is ubiquitylated on a specific Ser. HOIL1 substrates — including subunits of the Myddosome inflammatory regulatory complex — are also apparently modified by ubiquitin linkages to hydroxy group-containing amino acids, as they are sensitive to chemical treatments that hydrolyse ester bonds.
Endogenous eukaryotic ubiquitylation of non-protein molecules
HOIL1 modifies not only hydroxyls of amino acid side chains, but also primary hydroxyls of specific sugars26 (Fig. 2b). The connection to carbohydrates was indicated by a mysterious feature of HOIL1 mutant disease in humans and mouse models: accumulation of aberrant starch-like polysaccharide polyglucosan bodies, which may ultimately cause death. Elegant in vitro biochemical reconstitutions showed that HOIL1 ubiquitylates glycogen and maltoheptose, but not some other sugars. Nuclear magnetic resonance spectroscopy identified the hydroxyl functionality on the C6 carbon as the attachment site. Moreover, the binding properties of LUBAC subunits suggested how this ubiquitylation of sugars may be regulated: both HOIP and SHARPIN were found to bind to amylose resin. This result indicates that these subunits can directly localize the LUBAC E3 to ubiquitylate sugars. Also, sugar ubiquitylation was substantially increased upon HOIL1 binding to polyubiquitin chains, which may portend a feedforward mechanism whereby some polyubiquitylated moiety fuels HOIL1-catalysed ubiquitylation of nearby carbohydrate hydroxyls.
A different E3 was discovered to link the C terminus of ubiquitin to ADP-ribose (ADPR). The RING E3 ligase DELTEX3L was found to bind the ADP-ribosyltransferase PARP9, suggesting a connection between these two PTMs. Indeed, in the presence of E1, E2 and the DELTEX3L–PARP9 complex, the C terminus of ubiquitin was ADP-ribosylated27. This reaction depends on NAD+. Subsequent biochemical studies showed that DELTEX3L alone encompasses E3 ligase activity towards ADPR. Structural studies28 showed that DELTEX3L has an ADPR-binding domain, and this is positioned relative to the RING domain to promote ubiquitin transfer from a RING-bound E2 enzyme to ADPR (Fig. 2c). Notably, ADP-ribosylation blocks the C terminus of ubiquitin, and thus inhibits its conventional E1–E2–E3-dependent linkage to other proteins or macromolecules, although the modification can be reversed by deubiquitylating enzymes (DUBs)28. Given the sufficiency of DELTEX3L for these reactions, the precise role of the DELTEX3L–PARP9 complex will require further study. The interaction of DELTEX3L with PARP9 may be required for localizing DELTEX3L to substrates. Indeed, proteins that are ADP-ribosylated by PARP are recruited to the ADPR-binding site in DELTEX-family E3s for ubiquitylation28,29 both in vitro and in cells.
We anticipate that future studies will show that many E3s modify non-proteinaceous molecules. A particularly tantalizing candidate for such an E3 is the aforementioned MYCBP2, which was found to discharge ubiquitin to glycerol as well as to Thr21.
Erasing ester and thioester ubiquitin linkages
PTM ‘codes’ depend not only on writers but also on ‘eraser’ DUBs that remove modifications30. Previously, it was not readily possible to systematically assay DUB activity towards different bond types because the technology was not robust enough to generate comparable probes with the C terminus of ubiquitin linked to various amino acids31. This challenge was overcome by taking advantage of the ability of the RCR domain of MYCBP2 to forge ester bonds32. By use of a chemo-enzymatic approach to prepare a suite of reagents with ubiquitin linked to a Thr hydroxyl, Ser hydroxyl or Cys thiol, DUB activity was detected by mass spectrometry, or by a change in fluorescence polarization from the starting ubiquitin-bound 5-carboxytetramethylrhodamine substrates upon liberation of fluorescent 5-carboxytetramethylrhodamine (much like the classic DUB substrate ubiquitin–7-amido-4-methylcoumarin)33, or by SDS–PAGE. With these reagents, 53 recombinant DUBs were tested for removing ubiquitin from the various amino acid side chains. The initial screen showed that DUBs in the ubiquitin-specific protease (USP) and ubiquitin C-terminal hydrolase (UCH) families are active towards both ubiquitin–Lys and ubiquitin–Thr substrates; many in the ovarian tumour (OTU) family are specific for ubiquitin–Lys substrates, with the notable exceptions of a viral DUB and TRAF-binding domain-containing protein (TRABID). These latter two DUBs, along with those in the so-called Machado–Joseph domain-containing protease (MJD) family, were superior at removing the ubiquitin from a Thr than from a Lys32. Follow-up studies showed that a subset of DUBs with esterase activity could also remove ubiquitin from a Cys in the context of an unlabelled glutathione molecule. When tested towards peptide substrates, the MJD DUBs showed maximal activity in removing ubiquitin from the primary hydroxyl in a Ser side chain32. These intrinsic specificity differences raise the possibility that DUBs may preferentially remove ubiquitin from hydroxyls or thiols in different contexts, and possibly not only from proteins, but also from lipids or sugars. This may be particularly true for the MJD DUBs, as this entire family showed strong de-esterification activity that presumably is directed towards specific substrates in vivo.
Pathogen-induced ubiquitylation
Pathogens have developed molecular strategies to subvert and/or co-opt ubiquitin signalling for their own purposes (that is, to create a niche permissive for their intracellular replication). For this, pathogens secrete virulence factors, also known as pathogenic effectors, that function as DUBs, E3 ubiquitin ligases (such as the novel E3 ligase (NEL) family or the IpaH family) or enzymes that modify ubiquitin and UBLs (such as ISG15, SUMO and NEDD8) (reviewed in refs.34,35). These pathogen-mediated modifications either block normal ubiquitin functions in cells or create unique ubiquitin conjugations to various substrates. Both scenarios alter and expand the ubiquitin code. Bacteria and viruses apply similar principles to affect host ubiquitin and UBL function, but the spectrum of chemical entities used by specific bacteria or viruses for this purpose is surprisingly diverse and not always found in the mammalian proteome.
Phosphoribose-linked Ser ubiquitylation
The effector arsenal of Legionella pneumophila, which causes Legionnaires’ disease, masters a unique form of ubiquitylation that ignores several principles of endogenous, canonical ubiquitylation. First, ubiquitin is conjugated not via the highly conserved C-terminal Gly76 but via Arg42, which is unprecedented; second, conjugation is via linkage to Ser residues in the substrate instead of Lys residues; third, conjugation proceeds without ATP and E1, E2 and E3 enzymes, and instead uses only one bacterial enzyme that catalyses the entire ubiquitylation reaction36. This chemically and mechanistically unique type of ubiquitylation involves a phosphoribosyl bridge between the substrate and ubiquitin and is mediated by the members of the SidE family (SdeA, SdeB, SdeC and SidE), which act as all-in-one enzymes replacing E1, E2 and E3 activities, thus being entirely independent of the host ubiquitylation system36. Instead, SidE proteins use NAD+ as a cofactor via their mono-ADP-ribosyltransferase (mART) domain and a phosphodiesterase (PDE) domain37 for an atypical two-step hydrolysis reaction38,39 (Fig. 3a). Neither phosphoribosyl-ubiquitylated proteins nor the reaction intermediate ADPR–ubiquitin can be regulated by the host’s DUBs, nor can they be used by the host’s ubiquitylation machinery. Phosphoribosylated ubiquitinome analyses show that SidE family enzymes target more than 180 structurally and functionally diverse substrates40, including Golgi apparatus proteins41, mitochondrial proteins, cytoskeletal proteins, components of the autophagy machinery and endoplasmic reticulum-associated36,40,42 and lysosomal proteins43. Thus, L. pneumophila-mediated phosphoribosyl-ubiquitylation has wide-ranging and pleiotropic effects on host cells.
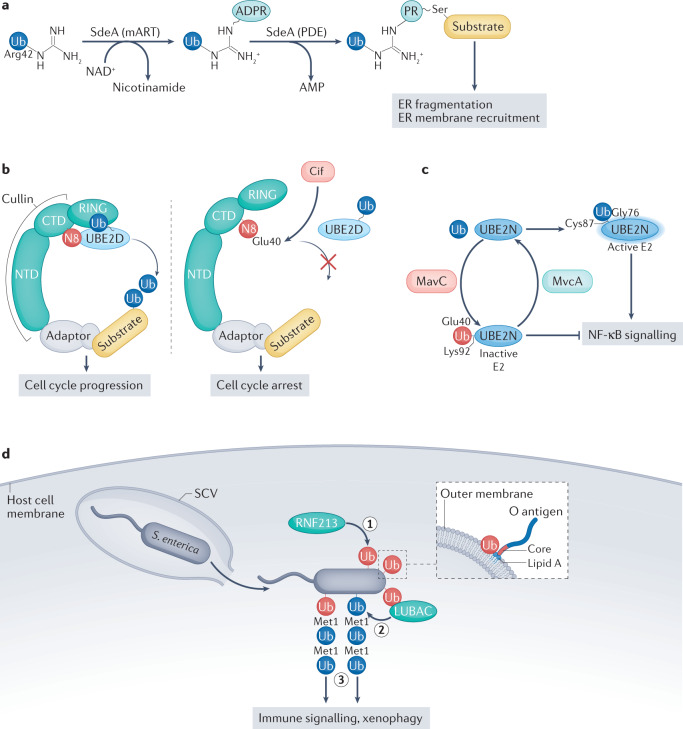
Fig. 3. Pathogen-induced ubiquitin modifications.
a | Ser ubiquitylation catalysed by the Legionella pneumophila effector SdeA. Using its mono-ADP-ribosyltransferase (mART) activity, SdeA first generates ADP-ribose (ADPR)–ubiquitin by transferring ADPR from NAD+ to Arg42 of ubiquitin. Subsequently, the phosphodiesterase (PDE) domain conjugates ADPR–ubiquitin to a Ser residue on substrates, thereby generating a ubiquitin-phosphoribosylated (PR) protein. The substrates modified in this way lead to pleiotropic changes in the host cell, including enhanced endoplasmic reticulum (ER) fragmentation and recruitment of ER membrane to L. pneumophila-containing vesicles. b | The carboxy-terminal domain (CTD) of cullin is modified by neural precursor cell-expressed developmentally downregulated protein 8 (NEDD8; N8), which leads to conformational changes that allow interactions between N8 and ubiquitin-conjugating enzyme E2 D (UBE2D) family members, the RING domain and ubiquitin. Deamidation of N8 at Gln40 (resulting in conversion to Glu40) by the enteropathogenic Escherichia coli (EPEC) virulence factor cycle-inhibiting factor (Cif) causes disruption of these mutual allosteric regulations of cullin–RING ligases (CRLs) and N8 that are required for CRL enzymatic activity. CRL substrates accumulate and cause cell cycle arrest. c | In normal conditions, ubiquitin is transferred to the catalytic Cys87 of UBE2N through a thioester bond involving Gly76 of ubiquitin. In this state, E2 is active and can participate in ubiquitylation reactions that trigger NF-κB signalling. The L. pneumophila effector MavC acts as a transglutaminase that links Ub via Gln40 to Lys92 or Lys94 of UBE2N, thereby inactivating E2 and preventing it from stimulating NF-κB signalling. The highly homologous effector MvcA reverses the reaction by hydrolysing the isopeptide bond created by MavC. d | Ubiquitylation of lipids by E3 ubiquitin ligase RING finger protein 213 (RNF213). Following escape from a damaged Salmonella enterica-containing vacuole (SCV), RNF213 is the first E3 ligase to attack cytosolic S. enterica, targeting a lipopolysaccharide composed of lipid A, core sugars and O antigen, in the outer bacterial membrane. Using an atypical zinc-finger domain, RNF213 attaches ubiquitin to a hydroxyl group of lipid A very close to the outer bacterial membrane (1). This first ubiquitin molecule then serves as the docking site for linear ubiquitin assembly complex (LUBAC), which builds linear ubiquitin chains (linked through Met1) on pre-existing ubiquitin molecules (2). The assembled ubiquitin coat is recognized by autophagy receptors such as p62, NDP52 and the optineurin effector protein NF-κB essential modulator (NEMO), which activate xenophagy (p62 and NDP52) and NF-κB-dependent immune signalling (optineurin), respectively (3). NTD, amino-terminal domain; Ub, ubiquitin. In part c, blue Ub, Ub linked via Gly76; red Ub, Ub linked via Gln40. In part d, blue Ub, ubiquitin molecule attached by LUBAC; red Ub, ubiquitin molecule attached by RNF213.
Whereas phosphoribosyl-ubiquitylation cannot be erased by host DUBs, L. pneumophila secretes factors that regulate the levels of phosphoribosyl-ubiquitylation: deubiquitylase for phosphoribosyl-ubiquitylation A (DupA) and DupB (also known as LaiE and LaiF, respectively) function as phosphoribosyl-ubiquitin-specific DUBs40,44. Surprisingly, the deubiquitylating activity of DupA and DupB is mediated by a PDE domain that structurally corresponds to the PDE domain of SidE enzymes but catalyses the opposite ligation reaction. This is possible due to the different substrates offered to DupA and DupB and their kinetic parameters. Whereas the PDE domains of SidE enzymes do not bind to phosphoribosyl-ubiquitylated substrates and have moderate binding affinity for ubiquitin, DupA and DupB show strong and selective affinity for ubiquitin and phosphoribosyl-ubiquitylated peptides. Indeed, point mutations weakening the affinities of DupA and DupB PDE domains for phosphoribosyl-ubiquitylated peptides are sufficient to convert them into SidE-type ubiquitin ligases40. This indicates that through subtle changes, pathogens can tweak their arsenal of effectors significantly.
DupA and DupB seem to spring into action at later stages of infection, possibly because L. pneumophila might actively seek to regulate the extent of phosphoribosyl-ubiquitylation to avoid the toxic effects of sustained high phosphoribosyl-ubiquitylation levels, which have been shown in both yeast and mammalian cells45. This hypothesis is also supported by the function of the L. pneumophila effector SidJ, a glutamylase that inactivates the catalytic site of the ART domains of SidE and that reduces the intracellular replication of L. pneumophila when deleted45–47. Interestingly, SidJ activity depends on its interaction with calmodulin, a eukaryote-specific protein. In this way, SidJ is activated only in the host cells, not within bacteria, and its binding to calmodulin is additionally regulated by intracellular Ca2+ levels in host cells47.
Deamidation of ubiquitin and NEDD8
Another way pathogens target the surface of ubiquitin, or the UBL and closest homologue of ubiquitin NEDD8, is by glutamine deamidation48. This virulence strategy is successfully used by Burkholderia pseudomallei, enteropathogenic Escherichia coli and L. pneumophila, which secrete effectors that possess a papain-like hydrolytic fold able to deamidate the conserved Gln40, resulting in conversion to Glu40. Cycle-inhibiting factor (Cif), the effector secreted from enteropathogenic E. coli, specifically targets NEDD8 (ref.49). As discussed later, NEDD8 modification of cullin–RING ligases (CRLs) activates their E3 activities. However, Cif-mediated deamidation inhibits this NEDD8-dependent activation of CRL ubiquitylation49–51. Mechanistically, NEDD8 deamidation does not impair the conjugation of NEDD8 onto cullin proteins, but precludes the non-covalent interactions between NEDD8 and cullins that trigger conformational changes and binding to ubiquitin-carrying enzymes52–55 (Fig. 3b). As a result, ubiquitylation and degradation of multiple CRL substrates, such as the cyclin-dependent kinase inhibitors p21 and p27, are abolished, disrupting the host’s cell cycle and arresting cell growth either at the G2–M transition or at the G1–S transition48. Similarly to Cif, Cif homologue in B. pseudomallei (CHBP) blocks cell cycle progression of infected cells by deamidation of NEDD8, but it also targets Gln40 of ubiquitin with similar efficiency. Deamidated ubiquitin cannot be transferred from E2 to the acceptor Lys49, and thus substrates such as NF-κB inhibitor-α (IκBα) escape proteasomal degradation.
An interesting example of a ubiquitin deamidation cycle is used by L. pneumophila, which secretes the effectors MavC and MvcA, which robustly deamidate Gln40 of ubiquitin but not of NEDD8 (ref.56). MavC and MvcA are structurally similar to Cif and CHBP but have an additional binding domain that targets host proteins. MavC interacts with the E2 conjugating enzyme UBE2N (also known as UBC13), which is involved in the formation of non-degradative Lys63 polyubiquitin chains. Strikingly, MavC was found not only to deamidate Gln40 but also to catalyse a transglutamination reaction that covalently links Gln40 of ubiquitin to Lys92 or Lys94 of UBE2N in the absence of E1 enzyme or ATP57–60. The resulting atypical UBE2N–ubiquitin complex does not possess E2 activity and accumulates in infected cells61 (Fig. 3c). Intriguingly, MvcA, which is 50% identical to MavC, was found to act as a DUB that specifically hydrolyses the isopeptide bond between ubiquitin and UBE2N62. In doing so at later stages of infection, MvcA restores UBE2N activity. As described earlier, highly homologous catalytic domains that mediate chemically opposite reactions appear to be a recurring concept in bacterial effectors.
Ubiquitylation of lipopolysaccharides
Work on antipathogenic strategies of mammalian cells has led to the discovery of a novel type of writer in the host proteome. It has long been assumed that so far unidentified bacterial membrane proteins serve as initial targets for ubiquitin conjugation, which then activates antibacterial responses. However, a recent study63 showed that the lipid A moiety of bacterial lipopolysaccharide (LPS), serves as the first site of ubiquitylation. LPS is the first example of a lipid modified by ubiquitin. This very unconventional modification is catalysed by the host E3 ubiquitin ligase RING finger protein 213 (RNF213) and is obligatory for the recruitment of the LUBAC E3 ligase, which adds Met1-linked ubiquitin chains onto the pre-existing ubiquitin moieties. These, in turn, serve as docking sites for autophagy receptors that function as readers of the attached ubiquitin modification and trigger the host’s autophagic response as shown in Salmonella enterica-infected cells64 (Fig. 3d).
RNF213 is a susceptibility factor for Moyamoya disease, a severe cerebrovascular disorder65–67, and seems to be involved in lipid droplet formation, lipotoxicity, hypoxia and NF-κB signalling65. Very recently, RNF213 was found to counteract infections by various microorganisms, including Listeria monocytogenes, herpes simplex virus 1, human respiratory syncytial virus and coxsackievirus B3. This function of RNF213 is induced by type I interferons and involves its isgylation and oligomerization on lipid droplets, where it acts as a specific sensor for isgylated proteins68. Interestingly, RNF213 contains a classic RING domain sequence that typically cooperates with an E2 conjugating enzyme to transfer ubiquitin to Lys residues of substrate proteins. However, ubiquitylation of LPS diverges from this well-established mechanism and does not require the RNF213 RING domain. Instead, it depends on other domains, including a dynein-like AAA+ domain69, which could be involved in accessing hidden substrates or may promote the recruitment of RNF213 to bacteria. Also, although the precise mechanism of ubiquitin attachment remains unclear, initial studies indicate that a non-canonical zinc-finger domain (containing the strictly conserved Cys4462) might function as the E3 active site for LPS ubiquitylation70. Moreover, E3 activity seems to be allosterically regulated by ATP70. How RNF213 recognizes LPS or which functional groups of lipid A (potentially its hydroxyl groups, its phosphate groups or both) are modified is currently unknown. It is very likely that in addition to RNF213 and bacterial LPS, other examples of lipid ubiquitylation will be discovered in the future.
Viral impact on the ubiquitin code
In addition to ubiquitylation and neddylation, viruses specifically manipulate the UBL ISG15, which is encoded by one of the most highly and rapidly host-induced genes in response to viral infection71,72. ISG15 becomes attached to both host and viral proteins, with the net effect of inhibiting viral stability, transport and assembly. Moreover, isgylation stabilizes key host antiviral proteins, thereby enhancing immune responses, whereas at later stages of viral infection, isgylation appears to attenuate the cellular immune response by inhibiting or destabilizing certain host proteins73,74. Also, unconjugated ISG15, which is not attached to a target protein, can affect viral replication and host responses through non-covalent protein interactions75 and its function as a cytokine76,77, respectively (Box 2).
To escape the ubiquitin-mediated and ISG15-mediated antiviral responses, viruses have acquired different strategies, including the reduction of ISG15 transcription by sequestering signal transducer and activator of transcription 2 (STAT2) by human cytomegalovirus, the inhibition of ISG15 conjugation by influenza B virus78,79, the sequestration of isgylated host proteins that would otherwise impede viral RNA synthesis also by influenza B virus80 or the secretion of enzymes that deconjugate ISG15 from target proteins81–85. The last of these approaches is used by coronaviruses, porcine reproductive and respiratory syndrome virus and equine arteritis virus, which secrete OTU-containing proteases. The leader protease (Lb(pro)) from foot-and-mouth disease virus is even able to irreversibly inactivate ISG15 by cleaving the peptide bond before the C-terminal diGly motif, leaving the Gly–Gly dipeptide attached to the substrate85.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the cause of the ongoing COVID-19 pandemic, and its close relatives Middle East respiratory syndrome coronavirus and SARS-CoV are able to deconjugate as many as three different UBLs: ubiquitin, NEDD8 and ISG15. Intensive research during the COVID-19 pandemic revealed that the papain-like proteases (PL(pro)) of SARS-CoV and SARS-CoV-2 have remarkably divergent specificities in cleaving ubiquitin, NEDD8 and ISG15 (refs.83,86–90). Although the enzymes are almost identical, each pathogen has evolved subtle changes that — quite unpredictably — affect their specificity, thus altering the UBL code and disabling the host’s immune system in different ways. SARS-CoV preferentially cleaves Lys48-ubiquitylated substrates, whereas SARS-CoV-2 PL(pro) prefers ISG15-conjugated substrates over ubiquitylated ones. These differences may contribute to the cellular response to viral attack and might be relevant for the development of treatment strategies and anti-COVID-19 drug design. Several SARS-CoV-2 variants of concern that were circulating in 2021 carried a mutation that affected a region of the enzyme that senses ubiquitin and ISG15 and enhances Lys48–ubiquitin chain cleavage compared with the original variant91. Further, de-isgylation by PL(pro) generates free ISG15, enhancing the secretion and extracellular signalling function of ISG15. This in turn promotes pro-inflammatory cytokine production from cells of the immune system that contributes to the devasting cytokine storm reported in some patients with severe COVID-19 (refs.77,92,93). Together these findings underline how flexibly pathogens can transform the host’s ubiquitin code and the need to understand the cellular functions encoded by pathogen-rewired ubiquitin codes.
Box 2 The roles of free ubiquitin and ISG15 molecules.
Ubiquitin and interferon-stimulated gene 15 (ISG15) exert their function not only while being conjugated to substrate proteins but also in their free (unconjugated) form, both intracellularly and extracellularly76,172–174. Intracellularly, ISG15 is conjugated to substrate proteins in a mechanism analogous to ubiquitylation. The expression of ISG15 is directly induced by type I interferons or by viral or bacterial pathogen-associated molecular patterns72,78,175. Although ISG15 induction leads to the isgylation of hundreds of intracellular proteins, various cell types, including fibroblasts, neutrophils, monocytes and lymphocytes, can release free ISG15 via an atypical secretory pathway. Secreted ISG15 acts as a cytokine-like protein that influences different types of immune cell176–179 and stimulates interferon production76,174,180. Leukocyte function-associated antigen 1 (LFA1) has been identified as a receptor for extracellular ISG15 (ref.177). Its expression on natural killer cells and T cells enables ISG15 autocrine signalling to drive IFNγ production from natural killer cells and T cells. The fact that influenza B viruses prevent ISG15 secretion by sequestering it via non-structural protein 1 (NS1)78,79 underlines the importance of extracellular ISG15 in antiviral defence. Moreover, since isgylation of intracellular targets triggers potent immune response, many viruses have evolved their own deisgylases as a mechanism to evade the immune system. However, by reversing intracellular isgylation, viral de-isgylases enhance secretion of free ISG15 (ref.77), which may contribute to strong pro-inflammatory responses93 (see also “Viral impact on ubiquitin code” in the main text). The potential pro-inflammatory effect of extracellular ISG15 has been demonstrated predominantly in cell culture models. However, when mice were infected with chikungunya virus75 or vaccinia virus181, unconjugated ISG15 was shown to negatively regulate the production of pro-inflammatory cytokines and chemokines. More precisely, the absence of ISG15 led to increased cytokine production, reminiscent of a cytokine storm, suggesting that free ISG15 functions in vivo not as an antiviral factor but as an immunomodulatory protein that negatively regulates the expression of pro-inflammatory cytokines and chemokines.
Intracellularly, free ISG15 binds the ubiquitin E3 ligase neural precursor cell-expressed developmentally downregulated protein 4 (NEDD4) and blocks its interaction with ubiquitin-loaded E2 enzymes. As a result, NEDD4 is no longer able to ubiquitylate viral matrix proteins, which significantly reduces the release of Ebola virus VP40 particles from cells182,183. Free ISG15 can also interact with the cytosolic histone deacetylase 6, which is involved in aggregate formation and clearance of ubiquitylated misfolded proteins through aggrephagy (autophagy of aggresomes)184.
Free extracellular ISG15 was also shown to have antitumoural properties by facilitating intratumoural infiltration of natural killer cells, and tumour regression in nude mice185, as well as protumorigenic, immunosuppressive properties when released by nasopharyngeal carcinoma cells into the tumour microenvironment186. There is also evidence that the antitumour immune response may be determined by the ratio of free (extracellular) to conjugated intracellular ISG15. Isgylation of intracellular proteins, such as p53, has a protumorigenic effect187,188. Tumour cells are able to reduce levels of extracellular ISG15, likely by blocking its secretion through conjugation to cellular proteins.
Compared with ISG15, relatively little is known about free ubiquitin and polyubiquitin, which is considered toxic inside cells, because it could interfere with crucial ubiquitin-dependent processes, such as proteasomal degradation of polyubiquitylated proteins. Therefore, it is assumed that non-conjugated polyubiquitin is rapidly degraded by deubiquitylating enzymes189. Nevertheless, several reports show that free ubiquitin chains may play a role in immune signalling. For example, certain E2–E3 pairs, such as tripartite motif-containing protein 6 (TRIM6) and ubiquitin-conjugating enzyme E2 K (UBE2K), can generate de novo Lys48-linked ubiquitin chains, which interact with inhibitor of NF-κB kinase subunit-ε (IKKε) for a full interferon–IKKε-mediated antiviral response190. Unconjugated Lys63-linked chains have also been found to act as endogenous signalling molecules in antiviral innate immunity. They are sensed through the tandem caspase recruitment domains (CARDs) of the retinoic acid-inducible gene I protein (RIG-I) receptor, which functions as a detector of invading viral RNA, activating the NF-κB pathway191.
Advances in understanding readers
The ubiquitin code is ultimately deciphered by ‘readers’, which bind ubiquitin-modified proteins to trigger a downstream output. Outputs include degrading the ubiquitylated protein, modifying the ubiquitylated protein with another PTM and stimulating the enzyme activity of the reader. Early studies focused on how isolated ubiquitin-binding domains recognize either monoubiquitin or particular polyubiquitin chains94–97. However, it soon became clear that there is great diversity in how readers recognize modified targets98–101. Some readers primarily bind ubiquitin or a ubiquitin chain. Such readers include the degradative machinery — the 26S proteasome — and Cdc48 in yeast (also known as p97 in humans) that act on thousands of unrelated proteins whose common feature is modification by ubiquitin. Other readers recognize only a specific protein ubiquitylated on a particular Lys100,102–106. In yet other cases, recognition depends on the modified protein undergoing ubiquitin (or UBL)-dependent conformational changes107–109. All these types of interaction with a ubiquitin-modified protein have been visualized by recent structural studies.
Readers that regulate numerous different ubiquitylated proteins
Cryogenic electron microscopy (cryo-EM) structures and biophysical studies have now shown how the 26S proteasome and Cdc48 recognize, unfold and in the case of the proteasome also degrade ubiquitylated proteins110–113. The 26S proteasome consists of two subcomplexes, arranged in layers (reviewed in refs.114–116). The outermost layer is the 19S regulatory particle, which is a multiprotein complex that contains many regulatory proteins and recruits even more regulators, and it also contains an AAA-ATPase motor91,112,113,117. The 19S regulatory particle is responsible for recruiting ubiquitylated proteins via at least three distinct intrinsic ubiquitin receptor subunits, each of which displays one or more distinct ubiquitin-binding motifs114–116. These proteasome-intrinsic subunits can bind directly to ubiquitylated substrates. Alternatively, ubiquitylated substrates can be delivered to the proteasome by so-called shuttle factors (or extrinsic receptors), which contain a ubiquitin-associated domain that binds ubiquitylated substrates, and a flexibly tethered ubiquitin-like domain that binds to the proteasome-intrinsic receptors. The 19S regulatory particle also contains three DUBs with distinct activities. Meanwhile, AAA ATPases are hexameric doughnut-shaped assemblies that transduce hydrolysis of ATP in the different subunits into mechanical motion. In the case of the 26S proteasome, this involves grasping the substrate, pulling it through the AAA ATPase pore to unfold it and ultimately feeding the unfolded substrate into the third subcomplex, the barrel-shaped 20S proteolytic chamber.
A major challenge when one is trying to visualize the 26S proteasome in action is that ubiquitylated substrates are processed too rapidly to be observed by existing structural methods. Several research groups have overcome this challenge by designing optimal ubiquitylated substrates and inhibiting specific steps in the multifaceted activities of the proteasome in a variety of ways110,113. One study generated proteasome complexes with substrate polypeptide threaded into the proteolytic chamber by the motor until the substrate could not be further pulled because it was stuck by its linked ubiquitin firmly bound to the inactivated DUB Rpn11 (ref.110). Subjecting these substrate-engaged 26S proteasome complexes to cryo-EM showed the subcomplexes concentrically aligned, and the substrate traversing a direct path from its linkage to the Rpn11-bound ubiquitin, through the centre of the AAA ATPase and into the proteolytic chamber that catalyses degradation. Structures were obtained for multiple conformations, which provided a model for how the receptors in the 19S regulatory particle bind ubiquitin and how individual subunits in the heterohexameric AAA ATPase grab onto substrates and move in a coordinated manner to pull them through the centre of the motor and push them into the proteolytic chamber.
But what is the role of ubiquitin? Although the substrates used for structural studies had long polyubiquitin chains, only the proximal ubiquitin directly linked to the substrate could be visualized. Thus, it seems likely that ubiquitins within the chain dynamically interact with multiple receptors in the regulatory particle. Furthermore, the structures, together with findings from biochemical and single-molecule fluorescence resonance energy transfer studies118, revealed further intricate regulation by ubiquitin that includes impacting the rates of switching between functionally distinct 26S proteasome conformations. Ubiquitin mainly seems to affect substrate engagement and capture, in part by allosteric modulation that ultimately facilitates the ubiquitylated substrate entering and being grasped by the AAA-ATPase motor118 (Fig. 4a).
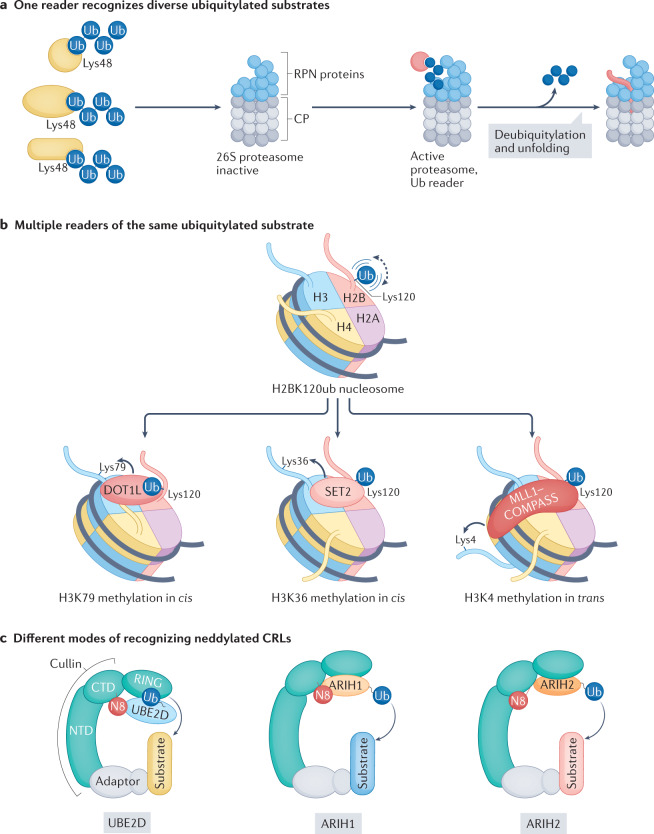
Fig. 4. Different readers of the ubiquitin code — three examples.
a | Recognition of multiple diverse ubiquitylated substrates by one reader, the 26S proteasome. Lys48-linked polyubiquitin chain-tagged proteins are recognized by ubiquitin receptors (RPN1, RPN10 and RPN13) that are part of the base subcomplex of the proteasome. Interactions between ubiquitin and the ubiquitin-binding domains of the ubiquitin receptors regulate the proteasome conformations that activate unfolding, deubiquitylation and degradation of the substrate. b | Multiple downstream machineries recognize the ubiquitylated substrate, histone H2B. Ubiquitin attached to Lys120 of H2B can flexibly adopt different positions to bind various chromatin-modifying methyltransferases. The positioning of ubiquitin in combination with additional binding sites of the methyltransferase on the nucleosome activates methylation of specific Lys residues. DOT1L and SET2 target two different Lys residues of histone H3, Lys 79 (H3K79) and H3K36, respectively, in cis, whereas MLL1-including complex of proteins associated with Set1 (COMPASS) methylates H3K4 in trans. c | Different modes through which various ubiquitin-carrying enzymes recognize neddylated cullin–RING ligases (CRLs). Neural precursor cell-expressed developmentally downregulated protein 8 (NEDD8; N8) can allosterically regulate the conformation of the cullin it modifies, and vice versa, the cullin affects the positioning of N8 within the complex so that different readers, even homologous readers (ARIH1 and ARIH2) are accommodated distinctly on different CRLs. CP, core particle; CTD, carboxy-terminal domain; H2BK120ub, Lys120-ubiquitinylated histone H2B; H4, histone H4; NTD, amino-terminal domain; Ub, ubiquitin; UBE2D, ubiquitin-conjugating enzyme E2 D.
Before degradation, many ubiquitylated proteins (for example, those embedded in membranes or complexes) must first be extracted by the unfoldase Cdc48 (p97 in humans). Cdc48 is a homohexameric AAA ATPase that associates with several interchangeable multidomain cofactors, which in turn bind substrates and regulators104,119–123. Cdc48 mediates unfolding of cofactor-recruited substrates through ratcheting through its central AAA-ATPase pore, much like the unfolding process mediated by the distinct 26S proteasomal AAA ATPases124–126.
The Npl4–Ufd1 cofactor complex is evolutionarily conserved. Recent structures of complexes of yeast proteins showed how Npl4–Ufd1 recruits substrates marked with Lys48-linked polyubiquitin chains127,128. Npl4–Ufd1 binds three ubiquitins in an arrangement attainable only by Lys48 linkages. An additional ubiquitin-binding domain is poised to bind yet another distally linked ubiquitin, although this last ubiquitin was not visible by cryo-EM. One possibility is that this fourth ubiquitin-binding site might interchangeably capture various ubiquitins at the distal end of a chain, resulting in conformational heterogeneity that would preclude observation by cryo-EM. The ubiquitin chain linked to the substrate was not observed by cryo-EM of human p97–NPL4–UFD1, suggesting this is also a conformationally dynamic complex126.
A landmark cryo-EM study visualized budding yeast Cdc48 in the act of unfolding a ubiquitylated substrate. The structure was made possible by trapping the otherwise dynamic process through use of a model substrate and inhibiting the AAA-ATPase motor by mutation or use of various nucleotide analogues128. Remarkably, a relatively proximal ubiquitin within the Lys48 chain is itself unfolded124,128. This is extraordinary considering that ubiquitin is often thought of as an exceptionally stable protein — it can be purified from a cell lysate owing to its ability to refold properly even after boiling the lysate129 — but a groove in Npl4 binds unfolded ubiquitin. The structures also showed part of the unfolded ubiquitin extended and threaded into the central pore of the Cdc48 barrel.
Together, the structures of these two molecular machines provide the first glimpses into how they distinctly recognize ubiquitin. Ubiquitin alone mediates interactions, which explains how ubiquitylation is sufficient to direct unfolding and degradation of an extraordinary array of diverse proteins. Although a substrate-linked monoubiquitin is sufficient to effectively anchor the substrate to the 26S proteasome for its translocation into the catalytic chamber for degradation130, both the 26S proteasome and Cdc48 contain multiple ubiquitin-binding sites that contribute to regulation. A recent study showed human p97–NPL4–UFD1 can associate with other p97-binding cofactors that also bind ubiquitin to reduce the threshold number of ubiquitin molecules required for substrate unfolding131. Moreover, different types of ubiquitin modification — especially branched ubiquitin chains such as those linked through both Lys11 and Lys48 — are particularly efficient at promoting degradation132. We anticipate future structures will visualize how different types of ubiquitin modification are read by the proteasome to influence the degradation process.
Ubiquitin-modified histone H2B is recognized by multiple readers
Whereas the 26S proteasome and Npl4–Ufd1-bound Cdc48 (and p97) recognize numerous ubiquitylated targets, in other cases ubiquitin directs some site-specifically modified targets to many distinct partners. One such example is the ubiquitylation of human histone H2B Lys120 (Lys123 in yeast)133. One function of ubiquitin linkage to H2B Lys120 is to trigger further histone modifications. Recent cryo-EM structures showed how the human H2B Lys120–ubiquitin-modified nucleosome binds the MLL1 complex, which methylates histone H3 Lys4 (ref.134) (and the related yeast H2B Lys123–ubiquitin-modified nucleosome bound to complex of proteins associated with Set1 (COMPASS)135,136), and Set2 (from Chaetomium thermophilum), which methylates H3 Lys36 (ref.137). Structural studies also showed how the human H2B Lys120–ubiquitin-modified nucleosome binds and activates the enzyme DOT1L, which methylates histone H3 Lys79 (refs.138–141). Interestingly, the different H2B Lys120-linked ubiquitin-bound methyltransferases target different molecules of histone H3 within the histone octamer (within each histone octamer there are two molecules of histone H2B and two molecules histone H3 — the active site of DOT1L engages Lys79 on the histone H3 located in cis relative to H2B’s ubiquitylated Lys120; the active sites of MLL1 and COMPASS face the opposite direction, to methylate Lys4 from the so-called trans molecule of histone H3) (Fig. 4b).
Ubiquitin linked to Lys120 of histone H2B is flexibly tethered to the nucleosome. As such, the globular domain of ubiquitin adopts different relative positions to bind distinct partners, which also uniquely contact nucleosomal DNA and the histone octamer. The distinct structure of each of these methyltransferases (or methyltransferase complexes) — and the collection of interactions with ubiquitin and the nucleosome — uniquely positions their active sites. It seems that multivalent interactions, including with ubiquitin, generally lower the energy barrier for attaining their active conformations. However, H2B Lys120 ubiquitylation impacts the function of each distinct partner in a different way. For MLL1, ubiquitin binding favours the active orientation and may promote formation of the complex134. Meanwhile, ubiquitin binding stabilizes the conformation of COMPASS that is competent for nucleosome binding135,136, and restricts the conformational search of DOT1L, facilitating access to H3 Lys79 (refs.138–141).
Allosteric regulation of ubiquitin-modified proteins
Ubiquitin and UBLs can modulate their target proteins by stimulating conformational changes. Early studies emphasized the inhibitory functions of such interactions. Many monoubiquitylated proteins — or proteins modified by a single UBL (for example, SUMO) — also contain ubiquitin-binding or UBL-binding domains. Intramolecular interactions between a protein’s ubiquitin-binding domain and its linked ubiquitin (or SUMO-binding domain and a linked SUMO) allosterically blocked association with other binding partners142–144.
Could intramolecular interactions between ubiquitin or a UBL and its modified protein also stimulate binding to new partners? Such allosteric interactions contribute to the functions of the UBL NEDD8. NEDD8 is approximately 60% identical to ubiquitin but has its own targets and readers. The best understood function of NEDD8 is to modify a specific Lys in the C-terminal domains of cullin proteins, which are subunits of the large family of CRL E3s94,145,146. As mentioned earlier, neddylation promotes CRL binding to ubiquitin-carrying enzymes, which are the direct mediators of ubiquitin transfer to CRL-bound substrates. Neddylated CRLs partner with different types of ubiquitin-carrying enzyme, including some E2 enzymes and members of the Ariadne RBR E3 ubiquitin protein ligase (ARIH) subfamily of RBR E3 enzymes147,148. In addition to being ubiquitin writers, such E2s and ARIH-family E3s are also readers of NEDD8 linked to a cullin-family protein.
Three core principles were revealed by structures of NEDD8-modified cullin 1 (CUL1)-containing CRLs bound to ubiquitin-conjugating enzyme E2 D (UBE2D) family members or E3 ARIH1 (refs.53,54) (Fig. 4c). First, NEDD8 engages in specific non-covalent interactions with the CUL1 domain to which it is linked. Notably, it is these interactions that are impaired by Cif-mediated deamidation of Gln40 described earlier. Second, these non-covalent interactions allosterically stabilize particular conformations of NEDD8, which, like ubiquitin, on its own normally interconverts between alternative conformations149. But when bound to CUL1, NEDD8 adopts only a so-called loop-out conformation, with its Ile44 hydrophobic patch exposed. Notably, UBE2D family members and ARIH1 specifically recognize the loop-out conformation of NEDD8. Third, in the context of the reader-bound complexes, NEDD8 also facilitates conformational changes of the CRL. The connection between the neddylated domain and the rest of CUL1 is a flexible tether, which allows distinct positioning relative to the rest of the CRL when bound to different readers. From the findings taken together, NEDD8 allosterically modulates the CRL, but cullin also allosterically modulates NEDD8 to promote its binding directly to UBE2D or ARIH1.
Unexpected cullin-specific allosteric regulation by NEDD8 was revealed by comparison of the structures of NEDD8-modified CUL1-containing CRLs bound to ARIH1 with the structure of a NEDD8-modified CUL5-containing CRL bound to the reader ARIH2 (ref.55). Surprisingly, ARIH2 neither contacts nor approaches NEDD8 linked to CUL5. Instead, NEDD8 interacts with multiple CUL5 domains to elicit conformational changes, which generate new surfaces that bind ARIH2. These data demonstrate that homologously modified proteins can bind homologous readers in different ways. Moreover, this latter structure shows that a UBL can allosterically modulate the conformation of a target protein to indirectly promote binding to a reader.
Concluding remarks and future challenges
Upon discovery of ubiquitin-directed protein degradation in the 1980s, it was impossible to envision the breadth of ubiquitin modifications or regulation depending on ubiquitin. Forty years later, we know that beyond protein degradation, ubiquitin directly or indirectly affects virtually every cellular process with surprising specificity. This is possible because the ubiquitylation machinery can generate a versatile code from the small universal ubiquitin molecule that acquires unique abilities through differently linked and branched ubiquitin chains, multiple acceptors (also non-Lys targets) and a variety of PTMs. Reading and interpreting this expanded ubiquitin code is readily accomplished by a growing body of highly specialized ubiquitin-binding and UBL-binding receptors in cells. Decoding the biological importance of such elaborate networks could help researchers exploit the ubiquitin code for tools and therapeutic treatments (Box 3).
Many of the recently discovered types of ubiquitylation involve relatively labile linkages between the C terminus of ubiquitin — or even a ubiquitin side chain — and hydroxyl or thiol moieties on the substrate. Unlike in the case of the ubiquitin modification of Lys residues, tools are lacking for identifying such modifications in a high-throughput manner. Rather, identification of ester-linked, thioester-linked and ADPR-linked ubiquitin has largely relied on studies of particular pathways, raising the question of how many other types of ubiquitin modification remain elusive. A future challenge to be overcome will be the development of tools and methods for detecting and modulating these, and potentially other, ubiquitin modifications. Going forward, we anticipate that artificial intelligence and machine learning will gain traction in assigning E3 ligases to specific ubiquitin modifications and vice versa, and in predicting structural mechanisms of ubiquitin and UBL recognition by their reader machineries.
Importantly, the accumulated knowledge described in this Review provides a new basis for the development of innovative ubiquitin-based therapeutics, including proteolysis-targeting chimeras and molecular glues. Their efficacy and specificity will depend on tissue selectivity of action of ubiquitin-modifying enzymes and regulation of conjugation–deconjugation dynamics in cells. The expanding lexicon of ubiquitin provides vast opportunities for scientists from different fields to join and make unexpected and therapeutically important discoveries in the future.
Box 3 Modulating the ubiquitin code therapeutically.
Ubiquitylation controls the abundance, localization and activity of many disease-related proteins, and therefore has moved into the spotlight of the pharmaceutical industry for new drug development. The proteasome, a large protein complex that degrades ubiquitylated proteins, was the first target of therapeutic strategies. Proteasomal inhibitors, such as bortezomib (VELCADE) and carfilzomib (KYPROLIS)192, are used in the treatment of different cancers, in particular multiple myeloma. However, adverse side effects caused by such global interference, as well as drug resistance, are limiting their use.
Identifying more specific drugs such as inhibitors of E3 enzymes, the substrate-selective component of the ubiquitylation machinery, is challenging, as E3 ligases act in a tissue-specific and/or tumour-specific manner, and their regulatory systems are complex. Nevertheless, various small-molecule inhibitors of human E3 ligases have been developed and entered clinical trials. The only substance approved by the US Food and Drug Administration thus far is bendamustine, which targets the E3 ligase linear ubiquitin assembly complex (LUBAC)193. Bendamustine is used for the treatment of chronic lymphocytic leukaemia, multiple myeloma, non-Hodgkin lymphoma, ovarian cancer and other diseases. Similarly, deubiquitylating enzymes (DUBs), particularly ubiquitin-specific protease 7 (USP7), have been considered as therapeutic targets, albeit with limited success, as researchers face the same challenges as with the E3 inhibitors. To date, only two DUB inhibitors have made it into clinical trials: KSQ-4279 (NCT05240898), a USP1 inhibitor, and VLX1570 (NCT02372240), which targets the proteasomal DUB USP14 and therefore should actually be considered a proteasomal inhibitor194.
Owing to problems such as toxicity, unsatisfactory efficacy, lack of selectivity, drug resistance and the fact that, for some disease-related proteins, the cognate E3 and DUB are unknown, researchers are moving away from approaches that target the ‘occupancy-driven’ action of traditional enzyme inhibitors towards ones that target an ‘event-driven’ mechanism of action. This is achieved by using bivalent molecules, such as proteolysis-targeting chimeras (PROTACs) or molecular glues. Close proximity is induced between an E3 ligase and (theoretically) any protein of interest that is expressed in the same cell, thereby achieving its ubiquitylation and degradation (reviewed in refs.195,196). PROTACs are rather large modular molecules, consisting of an E3-binding module and a target-binding module that are connected by a linker. Molecular glues are low-molecular-weight molecules that create a new binding surface on a given protein by binding to it, thereby adding a desired interaction partner (for example, an E3 ligase) to its interactome. In addition to their high specificity, these ‘molecular degraders’ surpass classical inhibitors by being active at substoichiometric concentrations, as they can be recycled after the target protein has been ubiquitylated. However, drug resistance and on-target toxicities remain a problem197,198. Several PROTACs, including ARV-110 and ARV-471, which target the androgen receptor and oestrogen receptor, displayed promising results in cell lines and mouse models for prostate cancer and breast cancer, respectively, and have thus entered clinical trials199. Multiple variations of PROTACs are currently being developed to significantly expand the pool of potential substrates, such as autophagy-targeting chimeras200, autophagosome-tethering compounds201, lysosome-targeting chimeras202 and antibody-based PROTACs203, with the last two enabling the degradation of secreted and cell surface proteins. The emerging knowledge of the variations in the ubiquitin code and their consequences already has, and will continue to have, profound implications for the development of new therapies.
Acknowledgements
The authors thank D. Höller for critical reading, text editing and figure preparations, and C. Wolberger and A. Martin for helpful comments. This work was supported by the Deutsche Forschungsgemeinschaft (project number 259130777-SFB 1177), the Cluster Project ENABLE funded by the Hessian Ministry for Science and the Arts and European Research Council grant H2020 UbBAC 742720 to I.D., and by the Max Planck Gesellschaft, European Research Council grant H2020 NEDD8Activate 789016 to B.A.S. and the Leibniz Prize from the Deutsche Forschungsgemeinschaft (SCHU 3196/1-1) to B.A.S.
Glossary
- Ubiquitin-like proteins
(UBLs). A family of proteins that share key structural features with ubiquitin and can be either conjugated to substrates (type I) such ubiquitin or exist as protein domains (type II) in larger proteins.
- Really interesting new gene (RING) domain
A zinc-finger protein domain that serves as a docking site for ubiquitin-loaded E2 enzymes and allosterically activates the thioester bond between E2 and ubiquitin to facilitate ubiquitin transfer.
- Homologous to the E6-AP carboxy terminus (HECT) domain
A protein domain that contains a conserved Cys that accepts ubiquitin from E2–ubiquitin and then transfers it directly to the substrate Lys.
- RING-between-RING (RBR) domain
A protein domain present in E3s that utilize a really interesting new gene (RING) domain–homologous to the E6-AP carboxy terminus (HECT) domain hybrid mechanism for ubiquitin conjugation.
- Nuclear magnetic resonance spectroscopy
A structural biology technique that allows monitoring interactions and perturbations of NMR-active nuclei. This method allows identification through bond and through space interactions to determine structures of organic molecules, including proteins.
- Myddosome
Large intracellular signalling complexes also called ‘supramolecular organizing centres’ assembled after recognition of microorganisms that function as a signalling platform.
- ADP-ribose
(ADPR). An ester formed between the aldehydic carbon of ribose and the terminal phosphate of adenosine diphosphate that either can be attached to proteins as a post-translational modification regulating, for example, DNA repair processes or can function as a second messenger on its own, for example by activating cation channels.
- Legionnaires’ disease
A severe form of pneumonia first described after an outbreak among attendants at a convention of the American Legion in 1976. It is caused by the bacterium Legionella pneumophila inhaled from contaminated water or soil droplets and can lead to life-threatening complications.
- Isgylation
Ubiquitylation-like process in which the ubiquitin-like modifier interferon-stimulated gene 15 (ISG15) is covalently conjugated to Lys residues of substrate proteins.
- Single-molecule fluorescence resonance energy transfer
A means of measuring distances at the 1–10-nm scale in a single biomolecule allowing the quantification and characterization of binding events, intramolecular transitions (for example, during protein folding), and kinetics and dwell times for single molecules.
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Yogesh Kulathu and the other, anonymous, reviewers for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ivan Dikic, Email: dikic@biochem2.uni-frankfurt.de.
Brenda A. Schulman, Email: schulman@biochem.mpg.de
References
- 1.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 2.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 3.Harper JW, Schulman BA. Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-Box hypothesis. Annu. Rev. Biochem. 2021;90:403–429. doi: 10.1146/annurev-biochem-090120-013613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Argiles-Castillo D, Kane EI, Zhou A, Spratt DE. HECT E3 ubiquitin ligases-emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020;133:jcs258087. doi: 10.1242/jcs.258087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Wijk SJL, Fiškin E, Dikic I. Selective monitoring of ubiquitin signals with genetically encoded ubiquitin chain-specific sensors. Nat. Protoc. 2013;8:1449–1458. doi: 10.1038/nprot.2013.089. [DOI] [PubMed] [Google Scholar]
- 6.Sims JJ, et al. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods. 2012;9:303–309. doi: 10.1038/nmeth.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Tatham MH, Plechanovová A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 2013;453:137–145. doi: 10.1042/BJ20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmar G, Winklhofer KF. Linear ubiquitin chains: cellular functions and strategies for detection and quantification. Front. Chem. 2019;7:915. doi: 10.3389/fchem.2019.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatsuka T, Sasaki T, Kaiser ET. Peptide segment synthesis catalyzed by the semisynthetic enzyme thiolsubtilisin. J. Am. Chem. Soc. 1987;109:3808–3810. doi: 10.1021/ja00246a064. [DOI] [Google Scholar]
- 12.Chu S-H, Mautner HG. Analogs of neuroeffectors. V. Neighboring-group effects in the reactions of esters, thiolesters, and selenolesters. The hydrolysis and aminolysis of benzoylcholine, benzoylthiolcholine, benzoylselenolcholine, and of their dimethylamino analogs. J. Org. Chem. 1966;31:308–312. doi: 10.1021/jo01339a069. [DOI] [Google Scholar]
- 13.Wang X, et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadwell K, Coscoy L. Biochemistry: ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell. 2010;40:917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J. Biol. Chem. 2010;285:23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho AF, et al. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 2007;282:31267–31272. doi: 10.1074/jbc.M706325200. [DOI] [PubMed] [Google Scholar]
- 18.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 19.Léon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J. Biol. Chem. 2007;282:7424–7430. doi: 10.1074/jbc.M611627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vosper JMD, et al. Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 2009;284:15458–15468. doi: 10.1074/jbc.M809366200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pao KC, et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556:381–385. doi: 10.1038/s41586-018-0026-1. [DOI] [PubMed] [Google Scholar]
- 22.Pao KC, et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat. Chem. Biol. 2016;12:324–331. doi: 10.1038/nchembio.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mabbitt PD, et al. Structural basis for RING-Cys-relay E3 ligase activity and its role in axon integrity. Nat. Chem. Biol. 2020;16:1227–1236. doi: 10.1038/s41589-020-0598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virdee S. An atypical ubiquitin ligase at the heart of neural development and programmed axon degeneration. Neural Regen. Res. 2022;17:2347–2350. doi: 10.4103/1673-5374.338992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelsall IR, Zhang J, Knebel A, Arthur SJC, Cohen P. The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc. Natl Acad. Sci. USA. 2019;116:13293–13298. doi: 10.1073/pnas.1905873116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelsall IR, et al. HOIL-1 ubiquitin ligase activity targets unbranched glucosaccharides and is required to prevent polyglucosan accumulation. EMBO J. 2022;41:e109700. doi: 10.15252/embj.2021109700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CS, et al. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell. 2017;66:503–516. doi: 10.1016/j.molcel.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatrin C, et al. Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases. Sci. Adv. 2020;6:eabc0418. doi: 10.1126/sciadv.abc0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed SF, et al. DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination. Sci. Adv. 2020;6:eabc0629. doi: 10.1126/sciadv.abc0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Reverter D. Molecular mechanisms of dubs regulation in signaling and disease. Int. J. Mol. Sci. 2021;22:986. doi: 10.3390/ijms22030986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho J, Park J, Kim EE, Song EJ. Assay systems for profiling deubiquitinating activity. Int. J. Mol. Sci. 2020;21:5638. doi: 10.3390/ijms21165638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Cesare V, et al. Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases. Proc. Natl Acad. Sci. USA. 2021;118:e2006947118. doi: 10.1073/pnas.2006947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 34.Maculins T, Fiskin E, Bhogaraju S, Dikic I. Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res. 2016;26:499–510. doi: 10.1038/cr.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange SM, Armstrong LA, Kulathu Y. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol. Cell. 2022;82:15–29. doi: 10.1016/j.molcel.2021.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Qiu J, et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhogaraju S, et al. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell. 2016;167:1636–1649. doi: 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Akturk A, et al. Mechanism of phosphoribosyl-ubiquitination mediated by a single legionella effector. Nature. 2018;557:729–745. doi: 10.1038/s41586-018-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalayil S, et al. Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature. 2018;557:734–738. doi: 10.1038/s41586-018-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin D, et al. Regulation of phosphoribosyl-linked serine ubiquitination by deubiquitinases DupA and DupB. Mol. Cell. 2020;77:164–179. doi: 10.1016/j.molcel.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, et al. Serine-ubiquitination regulates Golgi morphology and the secretory pathway upon Legionella infection. Cell Death Differ. 2021;10:2957–2969. doi: 10.1038/s41418-021-00830-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotewicz KM, et al. A single legionella effector catalyzes a multistep ubiquitination pathway to rearrange tubular endoplasmic reticulum for replication. Cell Host Microbe. 2017;21:169–181. doi: 10.1016/j.chom.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Leon JA, et al. Positive and negative regulation of the master metabolic regulator mTORC1 by two families of legionella pneumophila effectors. Cell Rep. 2017;21:2031–2038. doi: 10.1016/j.celrep.2017.10.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan M, et al. Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. Proc. Natl Acad. Sci. USA. 2019;116:23518–23526. doi: 10.1073/pnas.1916287116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhogaraju S, et al. Inhibition of bacterial ubiquitin ligases by SidJ–calmodulin catalysed glutamylation. Nature. 2019;572:382–386. doi: 10.1038/s41586-019-1440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Black MH, et al. Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science. 2019;364:787–792. doi: 10.1126/science.aaw7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan N, et al. Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature. 2019;572:387–391. doi: 10.1038/s41586-019-1439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taieb F, Nougayrède J, Philippe, Oswald E. Cycle inhibiting factors (Cifs): cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins. 2011;3:356–368. doi: 10.3390/toxins3040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui J, et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jubelin G, et al. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010;6:e1001128. doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morikawa H, et al. The bacterial effector Cif interferes with SCF ubiquitin ligase function by inhibiting deneddylation of cullin1. Biochem. Biophys. Res. Commun. 2010;401:268–274. doi: 10.1016/j.bbrc.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 52.Yu C, et al. Gln40 deamidation blocks structural reconfiguration and activation of SCF ubiquitin ligase complex by Nedd8. Nat. Commun. 2015;6:10053. doi: 10.1038/ncomms10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baek K, et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horn-Ghetko D, et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature. 2021;590:671–676. doi: 10.1038/s41586-021-03197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostrhon S, et al. CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 2021;17:1075–1083. doi: 10.1038/s41589-021-00858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valleau D, et al. Discovery of ubiquitin deamidases in the pathogenic arsenal of Legionella pneumophila. Cell Rep. 2018;23:568–583. doi: 10.1016/j.celrep.2018.03.060. [DOI] [PubMed] [Google Scholar]
- 57.Gan N, Nakayasu ES, Hollenbeck PJ, Luo ZQ. Legionella pneumophila inhibits immune signalling via MavC-mediated transglutaminase-induced ubiquitination of UBE2N. Nat. Microbiol. 2019;4:134–143. doi: 10.1038/s41564-018-0282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu Y, et al. Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC. Nat. Commun. 2020;11:1774. doi: 10.1038/s41467-020-15645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, et al. Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors. Nat. Commun. 2020;11:2751. doi: 10.1038/s41467-020-16587-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan H, et al. Molecular basis of ubiquitination catalyzed by the bacterial transglutaminase MavC. Adv. Sci. 2020;7:2000871. doi: 10.1002/advs.202000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng L, et al. Activation of the Iκb kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 62.Gan N, et al. Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination. EMBO J. 2020;39:e102806. doi: 10.15252/embj.2019102806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otten EG, et al. Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature. 2021;594:111–116. doi: 10.1038/s41586-021-03566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noad J, et al. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2017;2:17063. doi: 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda M, et al. Moyamoya disease patient mutations in the RING domain of RNF213 reduce its ubiquitin ligase activity and enhance NFκB activation and apoptosis in an AAA+ domain-dependent manner. Biochem. Biophys. Res. Commun. 2020;525:668–674. doi: 10.1016/j.bbrc.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 66.Wang C, et al. RNF213 gene silencing upregulates transforming growth factor β1 expression in bone marrow-derived mesenchymal stem cells and is involved in the onset of Moyamoya disease. Exp. Ther. Med. 2021;22:1024. doi: 10.3892/etm.2021.10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu W, et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thery F, et al. Ring finger protein 213 assembles into a sensor for ISGylated proteins with antimicrobial activity. Nat. Commun. 2021;12:5772. doi: 10.1038/s41467-021-26061-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahel J, et al. Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism. Elife. 2020;9:e56185. doi: 10.7554/eLife.56185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahel J, et al. E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP. Preprint at. bioRxiv. 2021 doi: 10.1101/2021.05.10.443411. [DOI] [Google Scholar]
- 71.Der SD, Zhou A, Williams BRG, Silverman RH. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 1992;267:7806–7813. doi: 10.1016/S0021-9258(18)42585-9. [DOI] [PubMed] [Google Scholar]
- 73.Minakawa M, Sone T, Takeuchi T, Yokosawa H. Regulation of the nuclear factor (NF)-κB pathway by ISGylation. Biol. Pharm. Bull. 2008;31:2223–2227. doi: 10.1248/bpb.31.2223. [DOI] [PubMed] [Google Scholar]
- 74.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Werneke SW, et al. ISG15 is critical in the control of chikungunya virus infection independent of UbE1l mediated conjugation. PLoS Pathog. 2011;7:e1002322. doi: 10.1371/journal.ppat.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D’Cunha J, Knight E, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl Acad. Sci. USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swaim CD, et al. Modulation of extracellular ISG15 signaling by pathogens and viral effector proteins. Cell Rep. 2020;31:107772. doi: 10.1016/j.celrep.2020.107772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan W, Aramini JM, Montelione GT, Krug RM. Structural basis for ubiquitin-like ISG 15 protein binding to the NS1 protein of influenza B virus: a protein-protein interaction function that is not shared by the corresponding N-terminal domain of the NS1 protein of influenza A virus. Virology. 2002;304:291–301. doi: 10.1006/viro.2002.1663. [DOI] [PubMed] [Google Scholar]
- 80.Zhao C, et al. Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat. Commun. 2016;7:12754. doi: 10.1038/ncomms12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Medina GN, et al. Impairment of the DeISGylation activity of foot-and-mouth disease virus Lpro causes attenuation in vitro and in vivo. J. Virol. 2020;94:e00341-20. doi: 10.1128/JVI.00341-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shin D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frias-Staheli N, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swatek KN, et al. Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proc. Natl Acad. Sci. USA. 2018;115:2371–2376. doi: 10.1073/pnas.1710617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klemm T, et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Békés M, et al. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol. Cell. 2016;62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freitas BT, et al. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 89.Báez-Santos YM, st. John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rut W, et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patchett S, et al. A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease. Cell Rep. 2021;36:109754. doi: 10.1016/j.celrep.2021.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Munnur D, et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021;22:1416–1427. doi: 10.1038/s41590-021-01035-8. [DOI] [PubMed] [Google Scholar]
- 94.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 95.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 97.Haakonsen DL, Rape M. Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol. 2019;29:704–716. doi: 10.1016/j.tcb.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 98.Oh E, Akopian D, Rape M. Principles of ubiquitin- dependent signaling. Annu. Rev. Cell Dev. Biol. 2018;34:137–162. doi: 10.1146/annurev-cellbio-100617-062802. [DOI] [PubMed] [Google Scholar]
- 99.Grumati P, Dikic I. Ubiquitin signaling and autophagy. J. Biol. Chem. 2018;293:5404–5413. doi: 10.1074/jbc.TM117.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 101.García-Rodríguez N, Wong RP, Ulrich HD. Functions of ubiquitin and SUMO in DNA replication and replication stress. Front. Genet. 2016;7:87. doi: 10.3389/fgene.2016.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X, Htet ZM, López-Alfonzo E, Martin A, Walters KJ. Proteasome interaction with ubiquitinated substrates: from mechanisms to therapies. FEBS J. 2021;288:5231–5251. doi: 10.1111/febs.15638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davis C, Spaller BL, Matouschek A. Mechanisms of substrate recognition by the 26S proteasome. Curr. Opin. Struct. Biol. 2021;67:161–169. doi: 10.1016/j.sbi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu X, Rapoport TA. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018;53:22–28. doi: 10.1016/j.ceb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Worden EJ, Wolberger C. Activation and regulation of H2B-Ubiquitin-dependent histone methyltransferases. Curr. Opin. Struct. Biol. 2019;59:98–106. doi: 10.1016/j.sbi.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–65. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rennie ML, Chaugule VK, Walden H. Modes of allosteric regulation of the ubiquitination machinery. Curr. Opin. Struct. Biol. 2020;62:189–196. doi: 10.1016/j.sbi.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 109.Khago D, Fucci IJ, Byrd RA. The role of conformational dynamics in the recognition and regulation of ubiquitination. Molecules. 2020;25:5933. doi: 10.3390/molecules25245933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science. 2018;362:eaav0725. doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ding Z, et al. Structural snapshots of 26S proteasome reveal tetraubiquitin-induced conformations. Mol. Cell. 2019;73:1150–1161. doi: 10.1016/j.molcel.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 112.Chen X, et al. Cryo-EM reveals unanchored M1-ubiquitin chain binding at hRpn11 of the 26S proteasome. Structure. 2020;28:1206–1217. doi: 10.1016/j.str.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dong Y, et al. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2018;565:49–55. doi: 10.1038/s41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mao Y. Structure, dynamics and function of the 26S proteasome. Subcell. Biochem. 2021;96:1–151. doi: 10.1007/978-3-030-58971-4_1. [DOI] [PubMed] [Google Scholar]
- 115.Bard JAM, et al. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018;87:697–724. doi: 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sakata E, Eisele MR, Baumeister W. Molecular and cellular dynamics of the 26S proteasome. Biochim. Biophys. Acta Proteins Proteom. 2021;1869:140583. doi: 10.1016/j.bbapap.2020.140583. [DOI] [PubMed] [Google Scholar]
- 117.Martinez-Fonts K, et al. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020;11:477. doi: 10.1038/s41467-019-13906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jonsson E, Htet ZM, Bard JAM, Dong KC, Martin A. Ubiquitin modulates 26S proteasome conformational dynamics and promotes substrate degradation. Preprint at. bioRxiv. 2021 doi: 10.1101/2021.08.18.456915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berner N, Reutter KR, Wolf DH. Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path. Annu. Rev. Biochem. 2018;87:751–782. doi: 10.1146/annurev-biochem-062917-012749. [DOI] [PubMed] [Google Scholar]
- 120.Stach L, Freemont PS. The AAA+ ATPase p97, a cellular multitool. Biochem. J. 2017;474:2953–2976. doi: 10.1042/BCJ20160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bays NW, Hampton RY. Cdc48-Ufd1-Npl4: stuck in the middle with Ub. Curr. Biol. 2002;12:R366–R371. doi: 10.1016/S0960-9822(02)00862-X. [DOI] [PubMed] [Google Scholar]
- 122.Olszewski MM, Williams C, Dong KC, Martin A. The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun. Biol. 2019;2:29. doi: 10.1038/s42003-019-0283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Buchberger A, Schindelin H, Hänzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 124.Ji Z, et al. Translocation of polyubiquitinated protein substrates by the hexameric Cdc48 ATPase. Mol. Cell. 2021;82:570–584. doi: 10.1016/j.molcel.2021.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cooney I, et al. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pan M, et al. Mechanistic insight into substrate processing and allosteric inhibition of human p97. Nat. Struct. Mol. Biol. 2021;28:614–p625. doi: 10.1038/s41594-021-00617-2. [DOI] [PubMed] [Google Scholar]
- 127.Sato Y, et al. Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4. Nat. Commun. 2019;10:5708. doi: 10.1038/s41467-019-13697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Twomey EC, et al. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365:eaax1033. doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978;81:1100–1105. doi: 10.1016/0006-291X(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 130.Sun H, et al. Diverse fate of ubiquitin chain moieties: the proximal is degraded with the target, and the distal protects the proximal from removal and recycles. Proc. Natl Acad. Sci. USA. 2019;116:7805–7812. doi: 10.1073/pnas.1822148116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fujisawa R, Labib KPM. Multiple UBX proteins reduce the ubiquitin threshold of the mammalian p97-UFD1-NPL4 unfoldase. Preprint at. bioRxiv. 2022 doi: 10.1101/2022.01.13.476277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 134.Xue H, et al. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature. 2019;573:445–449. doi: 10.1038/s41586-019-1528-1. [DOI] [PubMed] [Google Scholar]
- 135.Worden EJ, Zhang X, Wolberger C. Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. Elife. 2020;9:e53199. doi: 10.7554/eLife.53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsu PL, et al. Structural basis of H2B ubiquitination-dependent H3K4 methylation by COMPASS. Mol. Cell. 2019;76:712–723.e4. doi: 10.1016/j.molcel.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bilokapic S, Halic M. Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat. Commun. 2019;10:3795. doi: 10.1038/s41467-019-11726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell. 2019;176:1490–1501. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anderson CJ, et al. Structural basis for recognition of ubiquitylated nucleosome by Dot1L methyltransferase. Cell Rep. 2019;26:1681–1690. doi: 10.1016/j.celrep.2019.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Valencia-Sánchez MI, et al. Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. Mol. Cell. 2019;74:1010–1019. doi: 10.1016/j.molcel.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yao T, et al. Structural basis of the crosstalk between histone H2B monoubiquitination and H3 lysine 79 methylation on nucleosome. Cell Res. 2019;29:330–333. doi: 10.1038/s41422-019-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hoeller D, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 143.Flick K, et al. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 2004;6:634–641. doi: 10.1038/ncb1143. [DOI] [PubMed] [Google Scholar]
- 144.Baba D, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 145.Rusnac DV, Zheng N. Structural Biology of CRL Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020;1217:9–31. doi: 10.1007/978-981-15-1025-0_2. [DOI] [PubMed] [Google Scholar]
- 146.Wang K, Deshaies RJ, Liu X. Assembly and regulation of CRL ubiquitin ligases. Adv. Exp. Med. Biol. 2020;1217:33–46. doi: 10.1007/978-981-15-1025-0_3. [DOI] [PubMed] [Google Scholar]
- 147.Sakata E, et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat. Struct. Mol. Biol. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- 148.Kelsall IR, et al. TRIAD1 and HHARI bind to and are activated by distinct neddylated cullin-RING ligase complexes. EMBO J. 2013;32:2848–2860. doi: 10.1038/emboj.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hospenthal MK, Freund SMV, Komander D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat. Struct. Mol. Biol. 2013;20:555–565. doi: 10.1038/nsmb.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McClellan AJ, Laugesen SH, Ellgaard L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open. Biol. 2019;9:190147. doi: 10.1098/rsob.190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 152.Swaney DL, Rodríguez-Mias RA, Villén J. Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO Rep. 2015;16:1131–1144. doi: 10.15252/embr.201540298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wauer T, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gladkova C, et al. An invisible ubiquitin conformation is required for efficient phosphorylation by PINK 1. EMBO J. 2017;36:3555–3572. doi: 10.15252/embj.201797876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gersch M, et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 2017;24:920–930. doi: 10.1038/nsmb.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ohtake F, et al. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 2015;16:192–201. doi: 10.15252/embr.201439152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lacoursiere RE, Shaw GS. Acetylated ubiquitin modulates the catalytic activity of the E1 enzyme Uba1. Biochemistry. 2021;60:1276–1285. doi: 10.1021/acs.biochem.1c00145. [DOI] [PubMed] [Google Scholar]
- 158.Kienle SM, et al. Electrostatic and steric effects underlie acetylation-induced changes in ubiquitin structure and function. Nat. Commun. 2022;13:5435. doi: 10.1038/s41467-022-33087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang Y, Wang F. Post-translational modifications of deubiquitinating enzymes: expanding the ubiquitin code. Front. Pharmacol. 2021;12:685011. doi: 10.3389/fphar.2021.685011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 162.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pilli M, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Richter B, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl Acad. Sci. USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 167.Denuc A, Bosch-Comas A, Gonzàlez-Duarte R, Marfany G. The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS ONE. 2009;4:e5571. doi: 10.1371/journal.pone.0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 169.Vere G, Kealy R, Kessler BM, Pinto-Fernandez A. Ubiquitomics: an overview and future. Biomolecules. 2020;10:1453. doi: 10.3390/biom10101453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang YS, Wu KP, Jiang HK, Kurkute P, Chen RH. Branched ubiquitination: detection methods, biological functions and chemical synthesis. Molecules. 2020;25:5200. doi: 10.3390/molecules25215200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Henneberg LT, Schulman BA. Decoding the messaging of the ubiquitin system using chemical and protein probes. Cell Chem. Biol. 2021;28:889–902. doi: 10.1016/j.chembiol.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang GP, Khatoon S, Iqbal K, Grundke-Iqbal I. Brain ubiquitin is markedly elevated in Alzheimer disease. Brain Res. 1991;566:146–151. doi: 10.1016/0006-8993(91)91692-T. [DOI] [PubMed] [Google Scholar]
- 173.Takada K, et al. Serum concentrations of free ubiquitin and multiubiquitin chains. Clin. Chem. 1997;43:1188–1195. doi: 10.1093/clinchem/43.7.1188. [DOI] [PubMed] [Google Scholar]
- 174.Knight E, Cordova B. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J. Immunol. 1991;146:2280–2284. doi: 10.4049/jimmunol.146.7.2280. [DOI] [PubMed] [Google Scholar]
- 175.Radoshevich L, et al. ISG15 counteracts Listeria monocytogenes infection. Elife. 2015;4:e06848. doi: 10.7554/eLife.06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Napolitano A, et al. Cysteine-reactive free ISG15 generates IL-1β–producing CD8α+ dendritic cells at the site of infection. J. Immunol. 2018;201:604–614. doi: 10.4049/jimmunol.1701322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Swaim CD, Scott AF, Canadeo LA, Huibregtse JM. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol. Cell. 2017;68:581–590. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Recht M, Borden EC, Knight E. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 1991;147:2617–2623. doi: 10.4049/jimmunol.147.8.2617. [DOI] [PubMed] [Google Scholar]
- 179.Villarreal DO, et al. Ubiquitin-like molecule ISG15 acts as an immune adjuvant to enhance antigen-specific CD8 T-cell tumor immunity. Mol. Ther. 2015;23:1653–1662. doi: 10.1038/mt.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bogunovic D, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Eduardo-Correia B, Martinez-Romero C, Garcia-Sastre A, Guerra S. ISG15 is counteracted by vaccinia virus E3 protein and controls the proinflammatory response against viral infection. J. Virol. 2014;88:2312–2318. doi: 10.1128/JVI.03293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl Acad. Sci. USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nakashima H, Nguyen T, Goins WF, Chiocca EA. Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J. Biol. Chem. 2015;290:1485–1495. doi: 10.1074/jbc.M114.593871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Burks J, Reed RE, Desai SD. Free ISG15 triggers an antitumor immune response against breast cancer: a new perspective. Oncotarget. 2015;6:7221–7231. doi: 10.18632/oncotarget.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen RH, et al. Tumor cell-secreted ISG15 promotes tumor cell migration and immune suppression by inducing the macrophage M2-like phenotype. Front. Immunol. 2020;11:594775. doi: 10.3389/fimmu.2020.594775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Huang YF, Wee S, Gunaratne J, Lane DP, Bulavin DV. Isg15 controls p53 stability and functions. Cell Cycle. 2014;13:2199–2209. doi: 10.4161/cc.29209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li C, et al. Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget. 2014;5:8429–8441. doi: 10.18632/oncotarget.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 190.Rajsbaum R, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kumar SK, et al. Carfilzomib or bortezomib in combination with lenalidomide and dexamethasone for patients with newly diagnosed multiple myeloma without intention for immediate autologous stem-cell transplantation (ENDURANCE): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2020;21:1317–1330. doi: 10.1016/S1470-2045(20)30452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.de Cesare V, et al. The MALDI-TOF E2/E3 ligase assay as universal tool for drug discovery in the ubiquitin pathway. Cell Chem. Biol. 2018;25:1117–1127. doi: 10.1016/j.chembiol.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Gutierrez-Diaz BT, Gu W, Ntziachristos P. Deubiquitinases: pro-oncogenic activity and therapeutic targeting in blood malignancies. Trends Immunol. 2020;41:327–340. doi: 10.1016/j.it.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kannt A, Đikić I. Expanding the arsenal of E3 ubiquitin ligases for proximity-induced protein degradation. Cell Chem. Biol. 2021;28:1014–1031. doi: 10.1016/j.chembiol.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 196.Jevtić P, Haakonsen DL, Rapé M. An E3 ligase guide to the galaxy of small-molecule-induced protein degradation. Cell Chem. Biol. 2021;28:1000–1013. doi: 10.1016/j.chembiol.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 197.Hanzl A, et al. Charting functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders. Preprint at. bioRxiv. 2022 doi: 10.1101/2022.04.14.488316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Khan S, et al. Proteolysis targeting chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene. 2020;39:4909. doi: 10.1038/s41388-020-1336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li W, Elhassan RM, Hou X, Fang H. Recent advances in small molecule PROTACs for the treatment of cancer. Curr. Med. Chem. 2021;28:4893–4909. doi: 10.2174/0929867327666201117141611. [DOI] [PubMed] [Google Scholar]
- 200.Takahashi D, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell. 2019;76:797–810. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 201.Li Z, Zhu C, Ding Y, Fei Y, Lu B. ATTEC: a potential new approach to target proteinopathies. Autophagy. 2020;16:185–187. doi: 10.1080/15548627.2019.1688556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ahn G, et al. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol. 2021;17:937–946. doi: 10.1038/s41589-021-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cotton AD, Nguyen DP, Gramespacher JA, Seiple IB, Wells JA. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J. Am. Chem. Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]