Abstract
Objectives
The COVID-19 pandemic highlighted the importance of routine syndromic surveillance of respiratory infections, specifically new cases of severe acute respiratory infection (SARI). This surveillance often relies on questionnaires carried out by research nurses or transcriptions of doctor's notes, but existing, routinely collected electronic healthcare data sets are increasingly being used for such surveillance. We investigated how patient diagnosis codes, recorded within such data sets, could be used to capture SARI trends in Scotland.
Study design
We conducted a retrospective observational study using electronic healthcare data sets between 2017 and 2022.
Methods
Sensitive, specific and timely case definition (CDs) based on patient diagnosis codes contained within national registers in Scotland were proposed to identify SARI cases. Representativeness and sensitivity analyses were performed to assess how well SARI cases captured by each definition matched trends in historic influenza and SARS-CoV-2 data.
Results
All CDs accurately captured the peaks seen in laboratory-confirmed positive influenza and SARS-CoV-2 data, although the completeness of patient diagnosis records was discovered to vary widely. The timely CD provided the earliest detection of changes in SARI activity, whilst the sensitive CD provided insight into the burden and severity of SARI infections.
Conclusions
A universal SARI surveillance system has been developed and demonstrated to accurately capture seasonal SARI trends. It can be used as an indicator of emerging secondary care burden of emerging SARI outbreaks. The system further strengthens Scotland's existing strategies for respiratory surveillance, and the methods described here can be applied within any country with suitable electronic patient records.
Keywords: Surveillance, Severe acute respiratory infections (SARI), SARS-CoV-2, Influenza, ICD-10 codes, Electronic, Register-based, COVID-19
Introduction
Syndromic surveillance aims to monitor disease indicators in near real time to detect trends and outbreaks of disease earlier than would otherwise be possible via traditional public health methods.1 This is typically achieved using data from various sources – such as hospitals, emergency departments, primary care, health advice phone lines and pharmacies.2, 3, 4, 5
The COVID-19 pandemic has highlighted the ongoing importance of routine syndromic surveillance of respiratory infections, specifically new cases of severe acute respiratory infection (SARI).6 , 7 These cover a wide range of pathogens, including SARS-CoV-2, respiratory syncytial virus (RSV) and influenza. The World Health Organization defines a SARI case as an acute respiratory infection with history of fever or measured fever of ≥38 C° and cough, with onset within the last 10 days, that requires hospitalisation.8 SARI cases are thus defined by the presence of symptoms (fever, cough) rather than by laboratory confirmation of a pathogen.
The collection of syndromic respiratory data for surveillance often relies on questionnaires carried out by research nurses or transcriptions of doctor's notes.9, 10, 11 For example, in response to the 2009 influenza A (H1N1) pandemic, New Zealand established syndromic SARI surveillance within selected sentinel hospitals in 20129 as part of the Southern Hemisphere Influenza and Vaccine Effectiveness Research and Surveillance programme.10 Overnight inpatients with suspected respiratory infections were screened daily, and if the World Health Organization SARI definition was met, a respiratory sample was laboratory tested for influenza. Similar syndromic SARI surveillance systems have been implemented in nine Eastern European countries.11 These surveillance systems are often time consuming and labour intensive for clinicians collecting the data.
Existing, routinely collected electronic healthcare data sets are increasingly being used for real-time surveillance. Although symptoms data such as ‘cough’ or ‘fever’ are generally not routinely collected by these data sets, suspected or confirmed condition/disease (e.g. ‘pneumonia’), are often recorded as electronic diagnosis codes (e.g. International Classification of Diseases, Tenth Revision [ICD-10] codes).12 These codes can act as proxies for SARI presentations and hence be used to identify new SARI cases in real time.
John Hopkins University outlined a blueprint for SARI surveillance in the United States (Electronic Surveillance System for the Early Notification of Community-Based Epidemics).13 This combines numerous data sets, including a validated set of ICD-9 patient diagnosis codes (predecessor to ICD-10), to achieve daily outbreak surveillance at both community and hospital levels.14 , 15 In Australia, ICD-10 code–based surveillance for influenza was explored, using data from two sentinel hospitals, both before and during a pandemic.16 In Canada, ICD-10 code–based case-finding algorithms were demonstrated to successfully identify influenza hospitalisations from discharge records, using influenza-specific diagnosis codes (J09 and J10).17 Similarly, in Germany, an ICD-10 code–based SARI surveillance system was developed within a private network of sentinel hospitals.18
The ICD-10 codes J09-J22, including influenza, pneumonia and other acute lower respiratory infections, are the most commonly used for SARI and influenza-specific surveillance17 , 19 , 20 (Table 1 ). Two new ICD-10 codes specifically for SARS-CoV-2 have since been introduced, U07.1 and U07.2, for confirmed and suspected COVID-19 cases, respectively.21 , 22
Table 1.
ICD-10 codes used for severe acute respiratory infection, influenza-like illnesses and SARS-CoV-2 surveillance within the literature.
| ICD-10 code | Corresponding condition | Literature used by |
|---|---|---|
| J00 | Acute nasopharyngitis (common cold) | 17,20 |
| J01 | Acute sinusitis | 20 |
| J02 | Acute pharyngitis | 17 |
| J04∗ | Acute laryngitis and tracheitis | 17 |
| J06 | Acute upper respiratory infections of multiple and unspecified sites | 16,17,20 |
| J09∗ | Influenza due to certain identified influenza viruses | 9,17,18,20,23 |
| J10∗ | Influenza due to other identified influenza virus | 9,17,18,20,23 |
| J11∗ | Influenza due to unidentified influenza virus | 9,16, 17, 18,20,23 |
| J12∗ | Viral pneumonia, not elsewhere classified | 9,18,20,23 |
| J13∗ | Pneumonia due to Streptococcus pneumoniae | 9,18 |
| J14∗ | Pneumonia due to Haemophilus influenzae | 9,18 |
| J15∗ | Bacterial pneumonia, not elsewhere classified | 9,18 |
| J16∗ | Pneumonia due to other infectious organisms, not elsewhere classified | 9,17,18 |
| J17∗ | Pneumonia in diseases classified elsewhere | 9,18 |
| J18∗ | Pneumonia, unspecified organism | 9,16, 17, 18,20 |
| J20∗ | Acute bronchitis | 9,18,20,23 |
| J21∗ | Acute bronchiolitis | 9,18,23 |
| J22∗ | Unspecified acute lower respiratory infection | 9,16,18,20,23 |
| J40 | Bronchitis, not specified as acute or chronic | 20 |
| B34 | Viral infection of unspecified site | 16 |
| B97.4 | Respiratory syncytial virus as the cause of diseases classified elsewhere | 23 |
| R05 | Cough | 20 |
| U07.1∗ | SARS-CoV-2 confirmed | 21 |
| U07.2∗ | SARS-CoV-2 suspected | 21 |
| J80∗ | Acute respiratory distress syndrome | 24 |
ICD-10, International Classification of Diseases, Tenth Revision.
∗ICD-10 codes part of SARI surveillance in Scotland.
Set against the backdrop of the COVID-19 pandemic, Public Health Scotland developed a new surveillance system to monitor SARI presentations at hospital level within Scotland. The main goal was to use existing routinely collected ICD-10 code data, detailing Scottish hospital admissions, to monitor SARI admissions in close to real time. This would act as an indicator of the secondary care burden associated with future outbreaks of respiratory pathogens, help inform intervention policy and thereby minimise the burden on the National Health Service. In this article, we describe the development and validation of the surveillance system.
Methods
The SARI surveillance system uses ICD-10 codes listed in the routinely collected patient diagnosis fields of Scotland's two main national hospital data sets that collect data on every inpatient hospital admission in Scotland: The General Acute Inpatient and Day Case - Scottish Morbidity Record (SMR01)25 and the Rapid Preliminary Inpatient Dataset (RAPID).26 Both contain patient-level demographic details (treatment location, age, gender etc), and other variables related to a patient's stay in hospital are indexed by a Community Health Index. This unique patient identifier is used widely in other national data sets. Both SMR01 and RAPID contain six patient diagnosis fields, populated with ICD-10 codes, to document a patient's main condition and any comorbidities.
SMR01 is a validated data set with ICD-10 coding fully complete. However, records are completed after hospital discharge, and therefore, long-stay patients may not be included in the data set for a significant period, with a fully complete data set having a time lag of around 3 months.
In contrast, the RAPID data set is updated weekly in each of the 14 local health authorities in Scotland, with ICD-10 codes continuously added for a fixed period, after which no further updates are made. This period varies by a local health authority, from 2 to 8 weeks. The level of completeness of the ICD-10 codes therefore also varies by health authority, ranging from <5% to >90%, although overall in RAPID, the patient diagnosis fields are completed for around 30% of all hospital admissions. However, no local health authority could be identified that could act as a sentinel health authority on its own, as those with the highest levels of completeness tended to have the longest time lag. Because RAPID is an unvalidated data set, it is also possible that ICD-10 codes are updated for SMR01.
Using the ICD-10 codes likely to be representative of a SARI case (J09-J22, J80, U07.1, U07.2 and J04; Table 1), three SARI case definitions (CDs) were defined using either SMR01 or RAPID:
-
1.
Sensitive case definition (CD1): Any patient discharged from a Scottish hospital who had at least one of the specified ICD-10 codes listed in any of the six patient diagnosis fields in their SMR01 record for each individual hospital stay.
-
2.
Specific case definition (CD2): Any patient discharged from a Scottish hospital who had one of the specified ICD-10 codes listed in the main/first patient diagnosis field in their SMR01 record for each individual hospital stay.
-
3.
Timely case definition (CD3): Any patient admitted to a Scottish hospital who had at least one of the specified ICD-10 codes listed in any of the six patient diagnosis fields in their RAPID records for each individual hospital stay.
To assess how well their results matched known trends in seasonal data (representativeness), each CD was applied to historic SMR01 and RAPID data covering the period from 2017 to 2022. The number of weekly SARI cases collected by each CD was then compared with historical influenza and SARS-CoV-2 hospitalisation trends over these four seasons. These included all patients with a laboratory-confirmed test result either during their stay or within 7 days before admission for influenza and 14 days before admission for SARS-CoV-2. Patients with multiple laboratory-confirmed test results were counted separately for each infection.
In the absence of gold-standard symptoms data to definitively confirm an SARI diagnosis, we made the assumption that CD1 case counts most closely reflected the trueSARI hospital burden as all six patient diagnoses fields were used. The proportion of overall SARI cases captured by each CD (sensitivity) was estimated by calculating the weekly proportions of SARI cases (as a percentage) captured by the sensitive CD (CD1), which were also captured by the specific (CD2) and timely (CD3) CDs. The annual average of these percentages was then determined for each season (International Organization for Standardization (ISO) week 40 to week 39). A correlation analysis was conducted to examine the relation between the three CDs (CD1 vs CD2, CD1 vs CD3 and CD1 vs CD3 excluding weeks 10–23, 2020), by week.
As a further step in validation, the SARI cases were linked via Community Health Index number to the ECOSS (Electronic Communication of Surveillance in Scotland) data set that provides all laboratory test data for respiratory viruses (including SARS-CoV-2, influenza and RSV). SARS-CoV-2, influenza and RSV-confirmed SARI cases were defined as SARI cases with a laboratory-confirmed test result either during their hospital stay or 7 days before admission for influenza and RSV and 14 days before admission for SARS-CoV-2. These respiratory viruses were selected, as they are some of the most common viruses associated with SARI.27, 28, 29
R statistical software was used for all data analysis and the generation of the graphs.30 Week 53 for non-leap years presented in the graphs was dealt with by averaging data for week 52 and week 1 of the subsequent year.
Results
Representativeness and sensitivity analyses
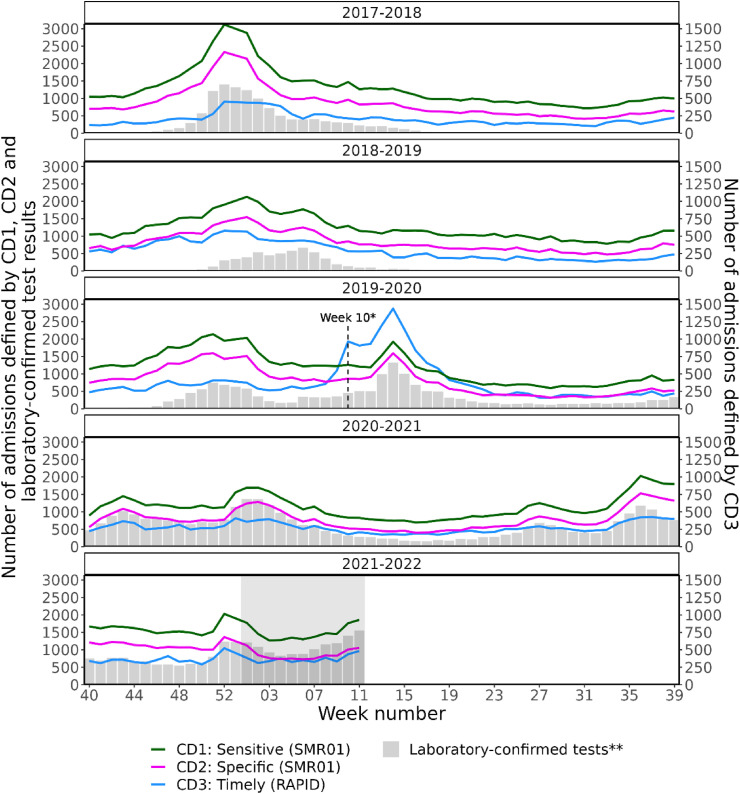
The representativeness analysis shows that all CDs identified similar seasonal peaks and trends compared with the laboratory-confirmed influenza and SARS-CoV-2 test results between week 40, 2017, and week 10, 2022 (Fig. 1 ). The number of identified SARI cases drops substantially in the most recent period, reflecting the 8- to 12-week time delay for the completion of SMR01. The decrease is less pronounced for RAPID data, which only ever captures a proportion of hospital admissions. The sensitivity analysis indicates that on average (from week 40, 2017, to week 10, 2022), CD2 captured 67.5% (42,388/62,752) of the SARI cases identified by CD1 and showed a strong correlation (r = 0.98). CD3 only returned 23.8% (14,815/62,752) of CD1's total case count (Table 2 ) and correlated moderately with CD1 (r = 0.48). However, CD3 still captured the overall trends of the other CDs, and this percentage increased to 33.3% (20,086/60,333) during the 2019–2020 season (Fig. 1). A strong correlation was found between CD1 and CD3 when weeks 10–23, 2020, were excluded, as the completion of ICD-10 codes in RAPID was significantly higher than any other period investigated (r = 0.75). The proportion of cases captured by CD2 did not vary significantly over time, but there was no additional advantage in using CD2 for the surveillance system.
Fig. 1.
Weekly number of SARI cases for each case definition and weekly number of laboratory-confirmed influenza and SARS-CoV-2 test results amongst hospital patients either during their stay or within 7 and 14 days before admission, respectively, by season, from week 40, 2017, to week 10, 2022. ∗Week 10, 2020, corresponds to the first case of community transmission of SARS-CoV-2. ∗∗Laboratory-confirmed test results of influenza and SARS-CoV-2; these correspond to positive results obtained during the patient's hospital stay or the 7 days before hospital admission for influenza or the 14 days before hospital admission for SARS-CoV-2. ∗∗∗The CD3 SARI cases are scaled by two (with an appropriate y-axis provided on the right) to emphasise the patterns the case definition captures. ∗∗∗∗The grey window highlights the maximum estimated time delay within the RAPID data set. Figures within this window (for CD3) are therefore liable to increase further.
Table 2.
Number and percentage of SARI cases captured by the sensitive case definition (CD1; n = 62,752), which were also captured by the specific (CD2) and timely (CD3) case definitions, by season, from week 40, 2017, to week 10, 2022.
| Case definition | Number and percentage of CD1 cases captured |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2017–2018 |
2018–2019 |
2019–2020 |
2020–2021 |
Average |
||||||
| n | % | n | % | n | % | n | % | n | % | |
| CD1: sensitive (SMR01) | 67,221 | 100 | 63,400 | 100 | 60,333 | 100 | 60,055 | 100 | 62,752 | 100 |
| CD2: specific (SMR01) | 44,530 | 66.2 | 42,329 | 66.8 | 41,387 | 68.6 | 41,306 | 68.8 | 42,388 | 67.5 |
| CD3: timely (RAPID) | 10,499 | 15.6 | 14,909 | 23.5 | 20,086 | 33.3 | 13,765 | 22.9 | 14,815 | 23.8 |
All CDs identified the influenza outbreak that occurred in 2017–2018 and trends during this time were similar to confirmed influenza test results of hospitalised patients. The number of new SARI cases identified through CD1 was highest during this outbreak and higher even than during the peak of the COVID-19 pandemic. During the 2018–2019 winter season, CD1 and CD2 showed an increase in SARI cases before an increase in laboratory-confirmed influenza cases was observed. Similarly, at the start of the 2019–2020 season, SARI cases identified through CD1 and CD2 increased before an increase in laboratory-confirmed influenza cases was seen. Later that season, and at the start of the pandemic, an increase in SARI cases was observed with all three CDs, with CD3 picking up the rise in SARI cases slightly before the laboratory test result data, demonstrating its potential to identify changes in SARI hospital admissions that may occur before other signals. At the start of the 2021–2022 winter season, the number of admissions identified through CD2 was almost identical to the number of SARS-CoV-2 laboratory-confirmed tests of hospitalised patients, suggesting that during this time, all hospitalised patients with SARS-CoV-2 had a U07 (SARS-CoV-2) code in their main diagnosis field in SMR01. These analyses confirm that the selected CDs capture representative trends in SARI cases.
Laboratory-confirmed results
During the first two-and-a-half-year observation period, seasonal patterns were visible for SARI cases identified through CD3 that had a laboratory-confirmed influenza test result, whereas seasonal patterns for those with a laboratory-confirmed RSV test result were observed in winter 2018–2019 and 2019–2020 (Fig. 2 ). Minimal influenza and RSV were observed in 2020–2021 and 2021–2022, whereas there was a clear peak in SARI cases with a laboratory-confirmed SARS-CoV-2 test result around week 14, 2020.
Fig. 2.
Weekly number of SARI cases identified by CD3, by laboratory-confirmed pathogen and season, from week 40, 2017, to week 10, 2022. ∗The grey window highlights the maximum estimated time delay within the RAPID data set. Figures within this window (for CD3) are therefore liable to increase further.
Discussion
In Scotland, the CD3 is now being routinely used in parallel with the CD1 to identify SARI cases for this new SARI surveillance system. SARI surveillance (as opposed to pathogen-based surveillance) has the important advantage of identifying increased SARI activity arising from new variants or pathogens that are not yet part of routine testing procedures. Although the time lag associated with the availability of complete SMR01 data limits its use for achieving true real-time surveillance using either CD1 or CD2, CD1 and CD3 complement each other to provide both a complete and timely SARI surveillance system for Scotland. CD3 is being used to carry out weekly identification of SARI cases within RAPID at national level. Although ICD-10 codes have not reached their full level of completeness in the most recent 8-week period, and the actual numbers of SARI cases are uncertain, any increase in SARI numbers within these most recent weeks will always represent an increase in real-life cases and can thus act as an indicator of increasing burden in secondary care. Trends in overall SARI numbers can also be cross-checked with trends in the individual local health authorities that have the shortest time lag for full completion for early signals of localised increases in SARI admissions. CD1 using SMR01 data then provides a more complete, but retrospective, picture of overall number of admissions. This enables validation of the peaks and trends captured by CD3 retrospectively and also provides supporting evidence for any recent patterns that emerged using CD3. Further data linkage provides an in-depth analysis of the SARI cases and outputs on age groups, gender, laboratory pathogens detected and ICU/HDU admission. Weekly reporting from the system is used to help inform policy decisions at both local health authority level and within national government.
The validation of the three CDs demonstrated that the number of SARI cases identified by CD3 was highest in 2019–2020 during the peak of the COVID-19 pandemic. CD3 picked up seasonal peaks after the introduction of SARS-CoV-2, but pre-SARS-CoV-2, these seasonal trends are less evident. This is likely due to the improvement of the data quality and completeness levels of ICD-10 codes in the RAPID data set as a response to the pandemic.
CD1 and CD2 demonstrate clear seasonal trends across the four seasons in line with the laboratory-confirmed influenza and SARS-CoV-2 test results. However, the number of SARI cases identified through both CDs reached their highest levels during the influenza epidemic in 2017–2018 and not during the pandemic, despite widespread reporting that health systems were experiencing unprecedented strain during this time. It may be that the threshold for an SARI hospital admission was higher during the pandemic. The Scottish population was directed to self-isolate and only seek care when absolutely essential, so only the most severe cases of COVID-19 patients were admitted to hospital, and these patients required high-level and resource-intensive care.31, 32, 33
Before the COVID-19 pandemic, it was observed that the data on laboratory-confirmed, positive influenza cases consistently fell significantly below the weekly case count numbers identified by CD1 and CD2 used by the SARI surveillance. It may be that fewer patients were tested for non-SARS-CoV-2 respiratory pathogens at this time. This changed in 2020–2021 when the increase in SARS-CoV-2 testing meant that the number of laboratory-confirmed cases was much closer to the CD1 and CD2 figures and is particularly obvious at the start of the pandemic in weeks 10–20, 2020, when the first case of community transmission was identified in Scotland,34 and in the rest of the 2020–2021 season (Fig. 1).
Similarly, in 2020–2021, the high number of SARS-CoV-2 tests being recorded provides a useful platform for validation of the SARI CDs. Almost all respiratory hospital admissions would have had a SARS-CoV-2 test, and so the testing figures could be assumed to capture a high proportion of SARS-CoV-2 hospital admissions. However, this is only applicable to SARI cases admitted for SARS-CoV-2 due to the low transmission of influenza and other respiratory pathogens during this same period, probably as a result of the nationwide lockdowns and social-distancing measures associated with the SARS-CoV-2 response.
The validation highlighted several periods of poor completion of ICD-10 codes within the RAPID data set. Nationally, between 2017 and 2019 (inclusive), the completion levels of these ICD-10 codes were around 10% but have increased to around 30% completion more recently. At the start of the COVID-19 pandemic, the ICD-10 completion levels in RAPID increased considerably but soon decreased when pressure in health care settings increased. This RAPID completeness issue is complex; partly caused by the collection of the ICD-10 codes not being mandatory; non-standardised policies, differing standards and multiple different reporting formats across the various regional local health authorities; alongside staffing constraints experienced within the data reporting teams. Completion of ICD-10 codes within the RAPID data set also varies between hospital and week by week, so timeliness and completeness of RAPID need to be continually monitored at local health authority level, and checks made for potentially biased completeness levels, for example, by ICD-10 code or over time. It is likely that similar variations and inconsistencies may occur in the data collection arena across countries worldwide. Thus, when developing this kind of electronic register-based surveillance, care should be taken to identify and understand any sources of variation, along with any other nuances of a chosen data set.
In summary, SARI surveillance using routinely collected, electronic hospital data sets has been shown to be viable and a valuable source of information for monitoring SARI trends across Scotland, given current levels of completion and timeliness. The quality of the real-time results could be further strengthened if levels of completeness were improved. Outputs provide detailed information without the need for additional data collection resources at hospital level and can easily be expanded upon, or linked to additional data sets, to provide further insight. This is a faster alternative for real-time SARI surveillance than traditional syndromic methods. Scotland's SARI surveillance system can act as an indicator of secondary care burden; be easily adaptable to include both existing and future emerging respiratory pathogens; reflect the broader picture of disease burden due to SARI; and provide data for further analyses, such as vaccine effectiveness for COVID-19 and influenza. SARI outputs presented here are updated routinely and presented in weekly Public Health Scotland reports. Further validation work is ongoing to assess and further enhance the performance of this surveillance system. From an international perspective, this type of surveillance could be applied within any country with similar electronic hospital data sets and could even evolve into a real-time worldwide SARI surveillance system, which could help strengthen SARI surveillance across the globe.
Author statements
Acknowledgements
The authors thank Diogo Marques from Epiconcept for his technical support and advice with the development of the surveillance system. The authors also acknowledge colleagues in Public Health Scotland who provided ongoing advice and support relating to the interpretation of the data sets.
Ethical approval
No ethical approval was required for this study.
Funding
The data of the study were originally collected as part of the project “Establishing Severe Acute Respiratory Infections (SARI) surveillance and performing hospital-based COVID-19 transmission studies,” funded by the European Centre for Disease Prevention and Control through a service contract with Epiconcept (ECD.11236 and Amendment N° 1 ECD.11810).35 The work was supported by the European SARI Network (E-SARI-NET), funded by the European Centre for Disease Prevention and Control (ECDC) and coordinated by Epiconcept, which aims to improve SARI surveillance within partner countries across Europe.
Competing interests
None declared.
Author contributions
J.W., H.M. and G.L. developed the study protocol. J.W., C.H., H.M., D.M. and N.Y. analysed the data, with support from J.J.Y. and J.E. J.W., C.H., J.J.Y. and J.E. drafted the article. L.A.W. and J.M. critically reviewed the article. All authors revised the article critically and approved the final version.
References
- 1.Henning K.J. What is syndromic surveillance? Morb Mortal Wkly Rep. 2004;53(7) http://www.jstor.org/stable/23315680 [Internet]. [cited 2022 Mar 18]. Available from: [Google Scholar]
- 2.Abat C., Chaudet H., Rolain J.-M., Colson P., Raoult D. Traditional and syndromic surveillance of infectious diseases and pathogens. Int J Infect Dis. 2016 Jul;48:22–28. doi: 10.1016/j.ijid.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandl K.D., Overhage J.M., Wagner M.M., Lober W.B., Sebastiani P., Mostashari F., et al. Implementing syndromic surveillance: a practical guide informed by the early experience. J Am Med Inform Assoc. 2004 Apr;11(2):141–150. doi: 10.1197/jamia.M1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson B.J., Durrheim D.N. The remarkable adaptability of syndromic surveillance to meet public health needs. J Epidemiol Glob Health. 2013 Mar;3(1):41–47. doi: 10.1016/j.jegh.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing J., Burkom H., Tokars J. Method selection and adaptation for distributed monitoring of infectious diseases for syndromic surveillance. J Biomed Inform. 2011 Dec;44(6):1093–1101. doi: 10.1016/j.jbi.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO) 2022. End-to-end integration of SARS-CoV-2 and influenza sentinel surveillance: revised interim guidance.https://www.who.int/publications/i/item/WHO-2019-nCoV-Integrated_sentinel_surveillance-2022.1 [Internet] [cited 2022 Mar 18]. Available from: [Google Scholar]
- 7.European Centre for Disease Prevention and Control (ECDC) 2021. COVID-19 surveillance guidance - Transition from COVID-19 emergency surveillance to routine surveillance of respiratory pathogens.https://www.ecdc.europa.eu/en/publications-data/covid-19-surveillance-guidance [Internet] [cited 2022 Mar 18]. Available from: [Google Scholar]
- 8.World Health Organization (WHO) 2014. Global Epidemiological Surveillance Standards for Influenza.https://www.who.int/influenza/resources/documents/WHO_Epidemiological_Influenza_Surveillance_Standards_2014.pdf [Internet] [cited 2022 Mar 18]. Available from: [Google Scholar]
- 9.Huang Q.S., Baker M., McArthur C., Roberts S., Williamson D., Grant C., et al. Implementing hospital-based surveillance for severe acute respiratory infections caused by influenza and other respiratory pathogens in New Zealand. Western Pac Surveill Response J. 2014 Jun;5(2):23–30. doi: 10.5365/WPSAR.2014.5.1.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q.S., Turner N., Baker M.G., Williamson D.A., Wong C., Webby R., et al. Southern hemisphere influenza and vaccine effectiveness research and surveillance. Influenza Other Respir Viruses. 2015 Jul;9(4):179–190. doi: 10.1111/irv.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meerhoff T.J., Simaku A., Ulqinaku D., Torosyan L., Gribkova N., Shimanovich V., et al. Surveillance for severe acute respiratory infections (SARI) in hospitals in the WHO European region - an exploratory analysis of risk factors for a severe outcome in influenza-positive SARI cases. BMC Infect Dis. 2015 Jan 8;15:1. doi: 10.1186/s12879-014-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Classification of Diseases (ICD) [Internet] https://www.who.int/standards/classifications/classification-of-diseases [cited 2022 Feb 28]. Available from:
- 13.Lombardo J., Burkom H., Elbert E., Magruder S., Lewis S.H., Loschen W., et al. A systems overview of the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE II) J Urban Health. 2003 Jun;80(2 Suppl 1):i32–i42. doi: 10.1007/PL00022313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention (CDC) 2017. Biosense Platform Quick Start Guide to Using ESSENCE.https://www.cdc.gov/nssp/biosense/docs/biosense-platform-quick-start-guide-for-essence.pdf [Internet] [cited 2022 Mar 18]. Available from: [Google Scholar]
- 15.Burkom H.S., Loschen W.A., Wojcik R.A., Holtry R.S., Punjabi M.A., Siwek M.M., et al. 2020 Aug 17. ESSENCE, the Electronic Surveillance System for the Early Notification of Community-Based Epidemics. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore K., Black J., Rowe S., Franklin L. Syndromic surveillance for influenza in two hospital emergency departments. Relationships between ICD-10 codes and notified cases, before and during a pandemic. BMC Public Health. 2011 May 18;11:338. doi: 10.1186/1471-2458-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T.H., Hong K.J., Shin S.D., Park G.J., Kim S., Hong N. Forecasting respiratory infectious outbreaks using ED-based syndromic surveillance for febrile ED visits in a Metropolitan City. Am J Emerg Med. 2019 Feb;37(2):183–188. doi: 10.1016/j.ajem.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buda S., Tolksdorf K., Schuler E., Kuhlen R., Haas W. Establishing an ICD-10 code based SARI-surveillance in Germany - description of the system and first results from five recent influenza seasons. BMC Public Health. 2017 Jun 30;17(1):612. doi: 10.1186/s12889-017-4515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Asten L., Luna Pinzon A., de Lange D.W., de Jonge E., Dijkstra F., Marbus S., et al. Estimating severity of influenza epidemics from severe acute respiratory infections (SARI) in intensive care units. Crit Care. 2018 Dec 19;22(1):351. doi: 10.1186/s13054-018-2274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruzda G., Rawlins F., Sumpter C., Garner H.R. Evaluating disease outbreaks with syndromic surveillance using medical student clinical rotation patient encounter logs. J Osteopath Med. 2021 Feb 1;121(2):211–220. doi: 10.1515/jom-2020-0129. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization (WHO) 2020. Emergency use ICD codes for COVID-19 disease outbreak.https://www.who.int/standards/classifications/classification-of-diseases/emergency-use-icd-codes-for-covid-19-disease-outbreak [Internet] [cited 2022 Apr 21]. Available from: [Google Scholar]
- 22.World Health Organization (WHO) 2021. Updates 3 & 4 in relation to COVID-19 coding in ICD-10.https://cdn.who.int/media/docs/default-source/classification/icd/covid-19/covid-19-coding-updates-3-4-combined.pdf [Internet] [cited 2022 Mar 18]. Available from: [Google Scholar]
- 23.Hamilton M.A., Calzavara A., Emerson S.D., Djebli M., Sundaram M.E., Chan A.K., et al. Validating International Classification of Disease 10th Revision algorithms for identifying influenza and respiratory syncytial virus hospitalizations. PLoS One. 2021 Jan 7;16(1) doi: 10.1371/journal.pone.0244746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention (CDC) ICD-10-CM Official Coding and Reporting Guidelines April 1, 2020 through September 30, 2020. https://www.cdc.gov/nchs/data/icd/COVID-19-guidelines-final.pdf [Internet]. [cited 2022 Mar 18]. Available from:
- 25.National Data Catalogue, National Datasets, ISD Scotland. General Acute Inpatient and Day Case - Scottish Morbidity Record (SMR01) https://www.ndc.scot.nhs.uk/National-Datasets/data.asp?SubID=5 [Internet]. [cited 2022 Feb 9]. Available from:
- 26.National Data Catalogue, National Datasets, ISD Scotland. Rapid Preliminary Inpatient Data (RAPID) https://www.ndc.scot.nhs.uk/National-Datasets/data.asp?SubID=37 [Internet]. [cited 2022 Feb 9]. Available from:
- 27.Yu J., Xie Z., Zhang T., Lu Y., Fan H., Yang D., et al. Comparison of the prevalence of respiratory viruses in patients with acute respiratory infections at different hospital settings in North China, 2012-2015. BMC Infect Dis. 2018 Feb 8;18(1):72. doi: 10.1186/s12879-018-2982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair H., Nokes D.J., Gessner B.D., Dherani M., Madhi S.A., Singleton R.J., et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010 May 1;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain S., Self W.H., Wunderink R.G., Fakhran S., Balk R., Bramley A.M., et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015 Jul 30;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.RStudio Team . RStudio, PBC; Boston, MA: 2020. RStudio: integrated development for R.http://www.rstudio.com/ [Internet] [cited 2022 Apr 6]. Available from: [Google Scholar]
- 31.Mulholland R.H., Wood R., Stagg H.R., Fischbacher C., Villacampa J., Simpson C.R., et al. Impact of COVID-19 on accident and emergency attendances and emergency and planned hospital admissions in Scotland: an interrupted time-series analysis. J R Soc Med. 2020 Nov;113(11):444–453. doi: 10.1177/0141076820962447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moynihan R., Sanders S., Michaleff Z.A., Scott A.M., Clark J., To E.J., et al. Impact of COVID-19 pandemic on utilisation of healthcare services: a systematic review. BMJ Open. 2021 Mar 16;11(3) doi: 10.1136/bmjopen-2020-045343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scottish Government . 2020. NHS Scotland placed on emergency footing.https://www.gov.scot/news/nhs-scotland-placed-on-emergency-footing/ [Internet] [cited 2022 Apr 21]. Available from: [Google Scholar]
- 34.COVID-19: First case of community transmission in Scotland - gov.scot. https://www.gov.scot/news/covid-19-first-case-of-community-transmission-in-scotland/ [Internet]. [cited 2022 Aug 30]. Available from:
- 35.Epiconcept Epiconcept. https://www.epiconcept.fr/en/ [Internet]. [cited 2022 Apr 21]. Available from: