Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mRNA vaccines drive the generation of affinity-matured B cell responses through germinal center (GC) reactions in vaccine draining lymph nodes. Herein, we describe a procedure for the acquisition of human lymph node samples via an ultrasound-guided fine needle aspiration-based approach. Additionally, we outline a suggested approach for the analysis of CD4 T helper cell subsets as well as antigen-specific GC B cells, memory B cells, and plasmablasts by high-parameter spectral flow cytometry.
For complete details on the use and execution of this protocol, please refer to Lederer et al. (2022).1
Subject areas: Flow Cytometry/Mass Cytometry, Health Sciences, Immunology
Graphical abstract
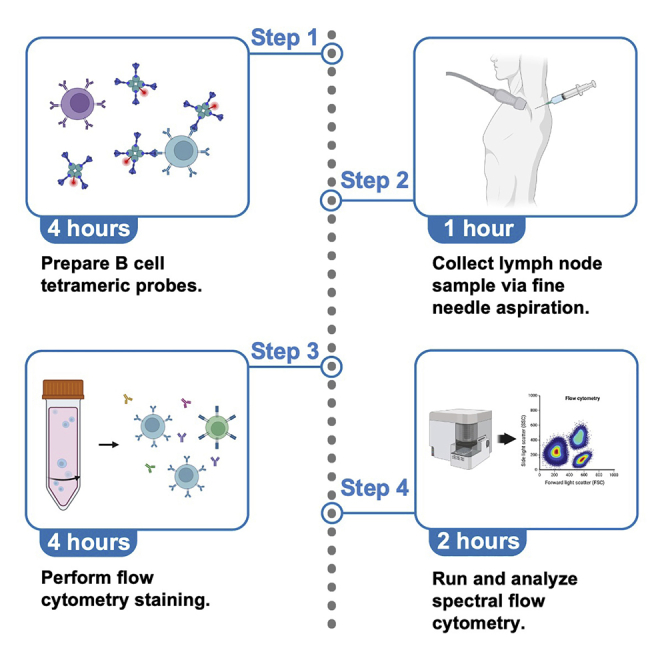
Highlights
-
•
Step-by-step procedure for SARS-CoV-2-specific B cell tetrameric probe generation
-
•
Detailed procedure for collecting fine needle aspirate (FNA) samples from lymph nodes
-
•
Long-term storage of FNA samples
-
•
Twenty-three-color panel for analysis of human germinal center responses by spectral flow cytometry
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Severe acute respiratory syndrome coronavirus 2 mRNA vaccines drive the generation of affinity-matured B cell responses through germinal center (GC) reactions in vaccine draining lymph nodes. Herein, we describe a procedure for the acquisition of human lymph node samples via an ultrasound-guided fine-needle-aspiration-based approach. Additionally, we outline a suggested approach for the analysis of CD4 T helper cell subsets as well as antigen-specific GC B cells, memory B cells, and plasmablasts by high-parameter, spectral flow cytometry.
Before you begin
This protocol describes both the collection and analysis of lymph node biopsy samples obtained via fine needle aspiration (FNA). The FNA procedure needs to be approved by the Institutional Review Board and performed by a trained, board-certified radiologist. Having the same radiologist/s performing all FNAs for a particular study can help to reduce the variability in FNA sample collection and cell recovery. Ideally, multiple FNAs will be scheduled to be collected on the same day, by the same radiologist. If this is not feasible, to limit sample-to-sample variability, it is important to ensure that all radiologists receive the same training and follow a standard operating protocol, such as the one outlined here.
Additionally, we have used tetrameric probes for the labeling of antigen-specific germinal center (GC) B cells, memory B cells, and plasmablasts. These probes must be prepared and titrated in advance, as described in detail below. We have found that the tetrameric probes described in this protocol are stable when stored at 4°C for up to six months. Thus, probes may be made days to months in advance. Antigen-specific T cell analysis is not discussed here but has been described by others.2
Institutional permissions
Patient samples should be collected in accordance with the Institutional Review Board rules, including appropriate recruitment and patient consent. Our FNA protocol was approved by the Institutional Review Board at the University of Pennsylvania (IRB# 844882).
Preparation of probes for labeling SARS-CoV-2-specific B cells
Timing: 4 h (for steps 1–6)
We use tetramerized-recombinant protein probes for the identification of antigen-specific GC B cells, memory B cells, and plasmablasts. We have found that using tetrameric probes, as opposed to monomeric probes, increases the quality of signal detection. Additionally, we typically use a dual probe approach, along with a decoy (irrelevant protein) probe, to ensure the specificity of the binding cells.
Specifically, for this protocol we used a recombinant biotinylated SARS-CoV-2 spike protein from R&D Systems. Additionally, we used a recombinant SARS-CoV-2 receptor binding domain (RBD) protein and a recombinant A/Puerto Rico/8/1934 (PR8, H1N1) influenza virus hemagglutinin (PR8-HA) protein, which were produced as described by others.3,4,5 The recombinant RBD and PR8-HA proteins were biotinylated using the EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit following the manufacturer’s instructions, with minor modifications, as described below.
CRITICAL: B cell tetrameric probes should be made with non-tandem dyes (i.e., PE, APC, etc). As discussed later, the use of tandem dyes (i.e., APC-Cy7, PerCP-Cy5.5, etc.) for the probes can make flow cytometry compensation challenging.
-
1.Biotinylation of recombinant RBD and PR8-HA:
-
a.Start with 20–200 micrograms (μg) of protein dissolved in 200–700 microliters (μL) of PBS. If protein is outside of this range, dilute it down to the appropriate range.
-
b.Calculate the millimoles (mmol) of Sulfo-NHS-LC-Biotin to add to the reaction for a 50-fold molar excess.V = volume (milliliters, mL).c = concentration (milligram (mg)/mL).MW = molecular weight (mmol/mg).n = mmol.
-
c.Remove the No-WeighTM Sulfo-NHS-LC-Biotin tube (included with the EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) containing 1 mg of biotin from the freezer.
-
i.Add 200 μL of molecular biology grade water to the tube.
-
ii.Mix by pipetting up and down.
-
iii.This creates a 9 mM Sulfo-NHS-LC-Biotin solution.
-
i.
-
d.Calculate the volume of 9 mM Sulfo-NHS-LC-Biotin to add to the reaction.557 mg = molecular weight of Sulfo-NHS-LC-Biotin.200 μL = volume of solvent used in step 1c to make a 9 mM Sulfo-NHS-LC-Biotin solution.V = volume (μL).n = mmol.
-
e.Add the appropriate volume of the biotin solution (step 1d) to the protein solution.
-
f.Incubate the reaction on ice (or at 4°C) for 2 h.
-
g.After the incubation, prepare the ZebaTM Spin Desalting Columns (included with the EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) by breaking off the bottom plug and placing in a 15 mL conical tube.
-
h.Spin at 1,000 × g for 2 min and discard the flow through.
-
i.Mark directly on the side of the column where the resin is slanted upward.
-
i.Place column back in the 15 mL conical tube.
-
ii.Place column in the centrifuge with the mark facing outwards for all remaining spins.
-
i.
-
j.Prepare BupH Phosphate Buffered Saline (PBS) by dissolving the provided packet (included with the EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) in 500 mL of deionized water.
-
k.Equilibrate the column by adding 1 mL of PBS.
-
i.Spin at 1,000 × g for 2 min.
-
ii.Discard the flow-through.
-
iii.Repeat step 1k 2×.
-
i.
-
l.Place column in a clean 15 mL conical tube.
-
i.Apply the protein sample directly to the resin bed.
-
ii.If the total volume is less than 400 μL, add 100 μL of molecular biology grade water on top of the resin after the sample is fully absorbed.
-
i.
-
m.Spin column at 1,000 × g for 2 min. Collect the flow-through which contains the biotinylated protein.
 Pause point: Biotinylated protein can be stored at 4°C before tetramerization. Theoretically, the protein should be stable for prolonged storage at 4°C, but we have not tested the storage for longer than 24 h.
Pause point: Biotinylated protein can be stored at 4°C before tetramerization. Theoretically, the protein should be stable for prolonged storage at 4°C, but we have not tested the storage for longer than 24 h.
-
a.
-
2.
Once the protein is biotinylated, follow the steps outlined below (steps 2–6) for tetramerizing RBD and PR8-HA proteins, similarly to what was reported in Taylor et al. (2012).6 First, calculate the mmol of protein. We typically assume no protein loss and negligible weight of the biotin.
V = volume (mL).
c = concentration (mg/mL).
MW = molecular weight (mmol/mg).
n = mmol.
-
3.
Calculate the molarity of the fluorochrome-labeled streptavidin (SA:fluorochrome) that will be conjugated to the biotinylated protein.
c = concentration (mg/mL).
MW = molecular weight (mmol/mg).
M = molarity (mmol/mL).
-
4.
Determine the volumes of biotinylated protein and SA:fluorochrome that will be needed for a 6:1 molar ratio (6 mol biotinylated protein to 1 mol SA:fluorochrome).
V = volume (mL).
n = mmol.
M = molarity (mmol/mL).
-
5.
Once the volume of SA:fluorochrome needed is determined, sequentially add 1/10 of the required volume of SA:fluorochrome to the biotinylated protein and mix gently while on ice (or at 4°C).
-
6.
Repeat step 5 every 10 min for a total of 10 additions (this will result in the total volume calculated in step 4 being added sequentially over 10 additions).
Note: The biotinylated-spike tetramers used for these studies were made as described in Goel et al. (2021).7 Biotinylated-spike protein was mixed with SA:fluorochrome at a 10:1 mass ratio during a single addition, and incubated on ice (or at 4°C) for at least 2 h before use.
Note: Please note that for the conjugation of recombinant proteins to phycoerythrin (PE), this method may not provide optimal results. PE is substantially larger than most other fluorochromes (∼240kDa), and when conjugating to smaller proteins, the volume of PE required to reach a 6:1 molar ratio may dilute out your protein more than is desirable. We recommend titrating to find the optimal amount of streptavidin-PE to conjugate your protein of interest.
Note: We have stored the RBD and PR8-HA tetramers described here at 4°C for up to six months without appreciable loss in staining quality. However, some proteins (e.g., SARS-CoV-2 full-length spike) may be less stable over time and/or at higher temperatures. Due to this, the shelf-life for all tetramer formulations should be assessed individually.
Titration of tetrameric probes for labeling SARS-CoV-2-specific B cells
Timing: 4–6 h (for steps 7–20)
Titration of tetrameric probes to label antigen-specific B cells should be performed every time that a new probe is made. For the titration, peripheral blood mononuclear cells (PBMC) from COVID-19 convalescent or SARS-CoV-2 vaccinated individuals, ideally within 3–4 months post infection or vaccination, can be used. For negative controls, use pre-pandemic PBMC if available.
In this protocol, tetrameric probes are titrated alongside a small antibody panel to follow the titration of probes directly on B cells. If comparing a new probe batch to a pre-existing one, also include a stained sample with the pre-existing probe at the appropriate concentration.
-
7.
Prepare FACS Buffer as described in the materials and equipment section.
-
8.
Prepare the eBioscienceTM Fixable Viability Dye eFluorTM 780 by diluting it 1:2,000 in FACS Buffer (for a final dilution of 1:4,000).
-
9.
Prepare the mix of antibodies to stain B cells (B Cell Mix) in a volume of 50 μL per sample/well in the viability dye prepared in step 8 to create a 2× B Cell Mix.
B Cell Mix
| Marker | Concentration (mg/mL; 2×) | Dilution (2×) |
|---|---|---|
| CD4 BUV563 | 0.00012 | 1:100 |
| CD8α PerCP-eFluor710 | 0.0005 | 1:100 |
| CD19 PE-Cy5.5 | 0.00025 | 1:100 |
-
10.Separately, prepare a 2×, 2-fold serial dilution of the probes (Probe Titration Solution) to be tested in FACS Buffer.
-
a.For a 2-fold serial dilution, start by adding the highest desired probe concentration to 100 μL of FACS Buffer.
-
b.Next, resuspend and transfer 50 μL of the probe mix to 50 μL of FACS Buffer. Repeat step as many times as necessary to reach the desired concentrations.
-
c.Recommended final dilutions (1×) are: 0, 1:50, 1:100, 1:200, 1:400, 1:800, 1:1,600.
-
a.
-
11.
Seed cells at approximately 1 × 106 cells per well. Ensure your plate has enough wells for all titrations and control samples.
-
12.
Spin cells at 400 × g for 2 min at 4°C.
-
13.
Aspirate supernatant and add 50 μL of the 2× B Cell Mix directly to each well.
-
14.
Immediately add 50 μL of the 2× Probe Titration Solution to the appropriate wells and pipette to mix.
-
15.
Incubate for 1 h at 4°C.
-
16.
Add 100 μL of FACS Buffer and spin at 400 × g for 2 min at 4°C.
-
17.
Aspirate supernatant and add 200 μL of a 1% paraformaldehyde solution (in FACS Buffer).
-
18.
Resuspend and incubate for 30 min at 4°C.
-
19.
Spin cells at 400 × g for 2 min at 4°C and discard supernatant.
-
20.
Resuspend cells in 200 μL of FACS Buffer and proceed to flow cytometry acquisition.
Note: For analysis, follow the gating strategy for CD19+ B cells (Figure 1). Within the B cell population, gate on the probe-positive cells as outlined in Figure 2. Compare the staining of probes in the positive samples to the negative samples (pre-pandemic sample, if available). Ascertain the optimal concentration for each probe, determined by the highest signal-to-noise ratio (calculated as the ratio between mean fluorescence intensity of the probe positive and probe negative B cells). Ensure that the negative control samples do not exhibit irrelevant probe binding to B cells at the desired concentration. Compare staining to a pre-validated batch of probe if applicable.
Note: If the probes have a high level of irrelevant binding in the negative controls, the probes should be remade with a new batch of protein. If persistent difficulty occurs with minimal probe staining or high background staining across batches, an alternative source of protein should be considered.
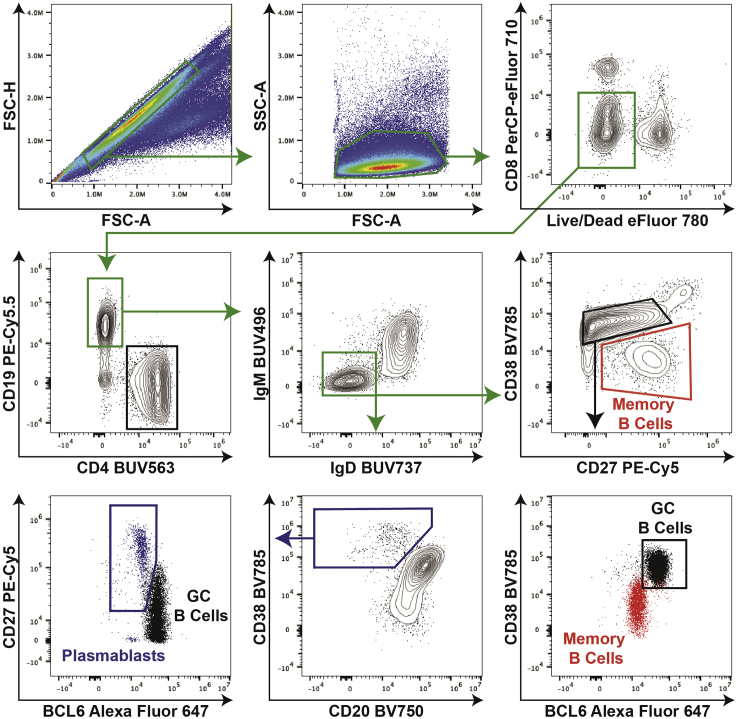
Figure 1.
GC B cell, memory B cell, and plasmablast gating strategy
For the B cell analysis, first gate on singlets and then on lymphocytes. Next, separate out live CD8- cells, and further gate on the CD19+CD4- population. Class-switched B cells (IgM-IgD-) are further selected to define: memory B cells (CD38-CD27+), GC B cells (CD38+CD27lo/intBCL6+), and plasmablasts (CD38+CD20lo/-CD27+BCL6-). Memory B cells, which are BCL6-, are overlaid in the plot showing CD38 versus BCL6 to guide the GC B cell gating (lower right panel). GC B cells, which are BCL6+, are overlaid in the plot showing CD38 versus BCL6 to guide the plasmablast gating (lower left panel). Memory B cells (CD38-CD27+) can also be analyzed in the IgM+ population, as some memory B cells are not class-switched. Plasmablasts can also be defined as BLIMP1+ cells as opposed to BCL6- cells. Since BLIMP1 and BCL6 antagonize each other, we have found that the definition of plasmablasts as CD38+CD20-CD27+BCL6- largely overlaps with the population of CD38+CD20-CD27+BLIMP1+ cells.
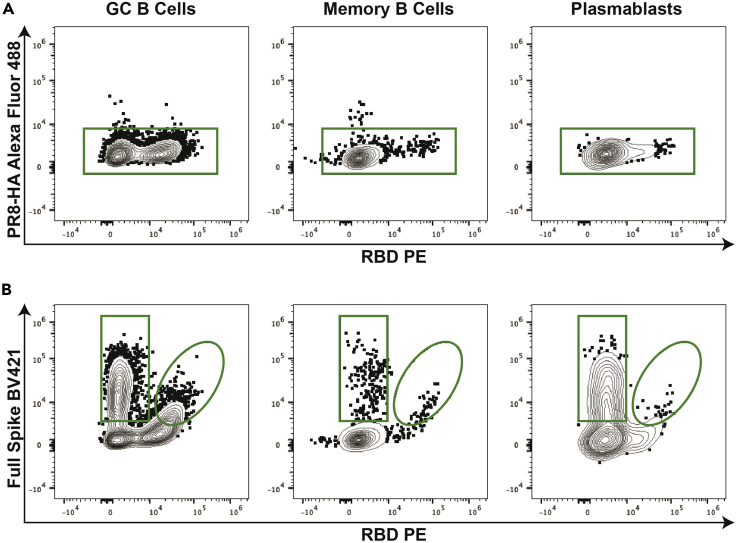
Figure 2.
Antigen-specific B cell gating strategy
Following the gating strategy from Figure 1, determine the antigen-specificity of the B cell subsets by using a combination of tetrameric probes.
(A) First, an irrelevant tetrameric probe (PR8-HA in this example) is used to gate out B cells potentially binding tetramers in an unspecific manner (i.e., streptavidin binders).
(B) Next, use a combination of SARS-CoV-2 full spike and RBD tetrameric probes to stratify antigen-specific GC B cells, memory B cells, and plasmablasts into spike-specific B cells binding the spike within or outside the RBD region.
Preparation for FNA collection
Timing: 1–5 min (for steps 21–23)
FNAs are collected into ice cold Culture Media and kept on ice (or at 4°C) until processing.
-
21.
Prepare Culture Media as described in the materials and equipment section.
-
22.
Add 20 mL of Culture Media into a 50 mL conical tube.
-
23.
Store the tube on ice (or at 4°C) and keep cold throughout the entire FNA procedure.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CXCR5 BUV395 (final dilution 1:80) | BD Biosciences | Cat# 740266; RRID: AB_2740008 |
| Anti-human CCR4 PE-CF594 (final dilution 1:40) | BD Biosciences | Cat# 565391; RRID: AB_2739215 |
| Anti-human CCR6 BV480 (final dilution 1:66) | BD Biosciences | Cat# 566130; RRID: AB_2739529 |
| Anti-human CXCR3 BV510 (final dilution 1:40) | Biolegend | Cat# 353726; RRID: AB_2563642 |
| Anti-human IgM BUV496 (final dilution 1:200) | BD Biosciences | Cat# 750366; RRID: AB_2874541 |
| Anti-human CD4 BUV563 (final dilution 1:200) | BD Biosciences | Cat# 612912; RRID: AB_2739451 |
| Anti-human CD11c BUV661 (final dilution 1:100) | BD Biosciences | Cat# 612967, RRID: AB_2870241 |
| Anti-human IgD BUV737 (final dilution 1:200) | BD Biosciences | Cat# 612798; RRID: AB_2738894 |
| Anti-human CD45RA BUV805 (final dilution 1:200) | BD Biosciences | Cat# 742020; RRID: AB_2871317 |
| Anti-human CD138 BV605 (final dilution 1:50) | Biolegend | Cat# 356520; RRID: AB_2562862 |
| Anti-human ICOS BV650 (final dilution 1:50) | Biolegend | Cat# 313550; RRID: AB_2749929 |
| Anti-human CD20 BV750 (final dilution 1:200) | BD Biosciences | Cat# 747062; RRID: AB_2871819 |
| Anti-human CD38 BV785 (final dilution 1:100) | Biolegend | Cat# 303530; RRID: AB_2565893 |
| Anti-human CD8α PerCP-eFluor710 (final dilution 1:200) | Invitrogen | Cat# 46-0086-42; RRID: AB_2848331 |
| Anti-human CD27 PE-Cy5 (final dilution 1:50) | Invitrogen | Cat# 15-0279-42; RRID: AB_10717249 |
| Anti-human CD19 PE-Cy5.5 (final dilution 1:200) | Invitrogen | Cat# 35-0198-42; RRID: AB_11218903 |
| Anti-human CD11b Alexa Fluor 700 (final dilution 1:100) | Biolegend | Cat# 301356; RRID: AB_2750075 |
| Anti-human PD-1 PE-Cy7 (final dilution 1:50) | Biolegend | Cat# 329918; RRID: AB_2159324 |
| Anti-human/mouse BCL6 Alexa Fluor 647 (final dilution 1:50) | BD Biosciences | Cat# 561525; RRID: AB_10898007 |
| Biological samples | ||
| Human FNA | Perelman Center for Advanced Medicine | N/A |
| Human PBMC | Perelman Center for Advanced Medicine | N/A |
| Human pediatric tonsils | Children’s Hospital of Philadelphia | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fixable viability dye eFluor 780 | Invitrogen | Cat# 65086518 |
| Recombinant RBD | Norbert Pardi | N/A |
| Recombinant Spike-Biotin | R&D Systems | Cat# BT10549-050 |
| Recombinant PR8 HA | Norbert Pardi | N/A |
| Streptavidin BV421 | Biolegend | Cat# 405225 |
| Streptavidin Alexa Fluor 488 | Biolegend | Cat# 405235 |
| Streptavidin Alexa Fluor 647 | Biolegend | Cat# 405237 |
| EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit | Thermo Fisher Scientific | Cat# 21935 |
| EZ-Link™ Sulfo-NHS-LC-Biotin, No-Weigh™ format (included in EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) | Thermo Fisher Scientific | Cat# A39257 |
| ZebaTM spin desalting columns (included in EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) | Thermo Fisher Scientific | Cat# 89889 |
| BupH phosphate-buffered saline (included in EZ-LinkTM Micro Sulfo-NHS-LC-Biotinylation Kit) | Thermo Fisher Scientific | Cat# 28372 |
| FoxP3/transcription factor staining buffer set | ThermoFisher Scientific | Cat# 00-5523-00 |
| ACK lysis buffer | Gibco | Cat# A1049201 |
| DMSO | Sigma | Cat# D2650 |
| FBS | Corning | Cat# 35-010-CV |
| EDTA | Invitrogen | Cat# 15575-038 |
| PBS | Corning | Cat# 21-031-CV |
| Glutamax | Gibco | Cat# 35050-061 |
| Penicillin-streptomycin | Gibco | Cat# 15140-122 |
| RPMI | Gibco | Cat# 22400-089 |
| Brilliant Stain Buffer | BD BioSciences | Cat# 563794 |
| 0.9% Buffered lidocaine | Hospital of the University of Pennsylvania | N/A |
| 4% Chlorhexidine gluconate | SC Johnson | Cat# 134439 |
| 16% Paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| Molecular biology grade water | Corning | Cat# 46-000-Cl |
| Software and algorithms | ||
| FlowJo v10 | FlowJo LLC | RRID: SCR_008520 |
| Other | ||
| UltraComp eBeadsTM compensation beads | Thermo Fisher | Cat# 01-2222-42 |
| Spectral cytometer | Cytek | Aurora spectral analyzer (5 lasers, 64 detectors) |
| Ultrasound machine | Phillips | EPIQ ELITE or IU222 |
| Ultrasound probes | Phillips | L15-7io or C8-5 |
Materials and equipment
Culture Media
| Component | Final concentration | Volume |
|---|---|---|
| FBS | 10% | 50 mL |
| Glutamax | 1× (Equivalent to 2 mM L-alanyl-L-glutamine dipeptide) | 5 mL |
| Penicillin-Streptomycin | 1× (Equivalent to 100 units/mL of penicillin and 100 μg/mL of streptomycin) | 5 mL |
| RPMI | 1× | 440 mL |
Store at 4°C for up to 1 month.
FACS Buffer
| Component | Final concentration | Volume |
|---|---|---|
| FBS | 2% | 10 mL |
| EDTA | 5 mM | 5 mL |
| PBS | 1× | 485 mL |
Store at 4°C for up to 1 month.
Freezing Media
| Component | Final concentration | Volume |
|---|---|---|
| FBS | 90% | 900 μL |
| DMSO | 10% | 100 μL |
Make fresh and store at 4°C until use.
1% Paraformaldehyde Solution
| Component | Final concentration | Volume |
|---|---|---|
| 16% Paraformaldehyde | 1% | 3.125 mL |
| FACS Buffer | 1× | 46.875 mL |
Store at 4°C for up to 1 month.
Step-by-step method details
Fine needle aspiration
Timing: 30–60 min (for steps 1–6)
In this step of the protocol, ultrasound is used to identify superficial lateral axillary lymph nodes in donors. The lymph node to biopsy is selected based on accessibility, larger size/cortical thickness, and relative distance to the surface of the skin. If no lateral lymph node is accessible, a central lymph node may be biopsied instead, however, posterior and/or apical lymph nodes are never sampled. Once a suitable lymph node is identified, ultrasound guidance is used to insert the tip of the needle into the lymph node and gently aspirate lymphoid cells from the cortex. After aspiration, the specimen is expelled from the syringe into ice cold media to collect the cells. FNAs are performed similar to those reported in Havenar-Daughton et al. (2020),8 with a few minor differences highlighted below.
-
1.
Prior to the FNA procedure, prepare the collection syringes by attaching a 25-gauge needle to a 10 mL syringe and withdraw the plunger to pull 1 mL of air into the syringe.
Note: This helps to eject the specimen. A separate sterile needle/syringe is prepared for each pass (5 total).
Note: In the original protocol,8 a 22-, 25-, or 27-gauge needle along with a 3 mL or 5 mL syringe were all successfully used. We opt to consistently use a 25-gauge needle with a 10 mL syringe.
-
2.
Use an ultrasound instrument (Philips EPIQ ELITE or PHILIPS IU222) to visualize superficial axillary draining lymph nodes. Select the most accessible and best visualized lymph node for biopsy.
-
3.
Clean the skin around the lymph node with 4% chlorhexidine gluconate solution, or another antiseptic solution. Anesthetize this area by injecting 2–6 mL of 0.9% buffered lidocaine solution.
-
4.Under ultrasound guidance, place the tip of the 25-gauge needle into the cortex of the lymph node and move back and forth for 10–20 s to collect cells.
-
a.Eject the sample into the prepared tube of ice-cold Culture Media.
-
b.This step is considered as a single “pass”. See Methods video S1.
-
a.
Note: Negative pressure (light suction) is usually applied to the syringe in step 4 to increase the cell yield and is recommended in the original protocol.8 Suction should be released prior to removing the needle from the lymph node to avoid aspirating subcutaneous fat.
-
5.
Rinse the needle with 1–2 mL of media to collect residual cells before disposal.
-
6.
Perform a total of five passes (as defined in step 4).
Note: In the original protocol,8 four lymph nodes, with a total of four passes each, were probed per donor. In our recent study, we opted for a single lymph node per donor with a total of five passes.1 In some donors, it can be difficult to visualize multiple lymph nodes that are accessible for FNA, thus a single lymph node per donor tends to be more feasible for collection.
Flow cytometry
Timing: 4–6 h (for steps 7–28)
After the FNA is performed, cells are immediately processed and stained with fluorescent antibodies to identify B cell and T cell populations by spectral flow cytometry.
-
7.Prepare single-color compensation controls.
-
a.In a V-bottom 96-well plate, set up an unstained control and a compensation control for the viability dye by adding 1 × 106 cells per well into two separate wells.Note: Since FNA cell recovery can be limiting, fresh PBMC or other human cells (e.g., tonsillar cells) can be used for these compensation controls.
-
b.Bring the volume of the well to 200 μL with FACS Buffer, then spin the plate at 400 × g for 2 min at 4°C.
-
c.Aspirate the supernatant and resuspend the cells with 100 μL of FACS Buffer (unstained control) or eBioscienceTM Fixable Viability Dye eFluorTM 780 diluted 1:4,000 in FACS Buffer (viability dye control). Keep the plate at 4°C (or on ice) until step 7g.
-
d.For the other single-color compensation controls, add 15 μL per well of UltraComp eBeadsTM Compensation Beads to the plate. One well is needed for each fluorochrome included in the panel.
-
e.The total volume of each well is brought to 100 μL by adding 85 μL of FACS Buffer.
-
f.Add 1 μL (or more) of each individual antibody to the appropriate wells.
-
g.Incubate all controls at 4°C for 30 min.
 CRITICAL: It is critical that, for each fluorochrome, the single-color compensation controls are as bright, or brighter, than the stained sample. For this reason, the concentration of the antibody used to stain the samples (see Stain Mix 1 and Stain Mix 2 tables for the suggested concentrations), is an appropriate starting point for staining the compensation beads. From there, the concentration may need to be adjusted depending on signal brightness (use lower concentration if the signal is too bright, or higher concentration if the signal is too dim).Note: The fluorescently labeled tetrameric B cell probes do not bind to these compensation beads. To set up compensation controls for the tetrameric probes, we instead stain compensation beads with an antibody conjugated to the same fluorochrome as the one conjugated to the probes (i.e., we use an anti-CD4-PE antibody as the single-color compensation control for the RBD-PE tetrameric probe). Importantly, if possible, tandem dyes should be avoided for making tetrameric probes as tandem dyes have a much higher variability from lot-to-lot (due to the dissolution of the tandem fluorochromes) and cannot be accurately compensated in this manner.
CRITICAL: It is critical that, for each fluorochrome, the single-color compensation controls are as bright, or brighter, than the stained sample. For this reason, the concentration of the antibody used to stain the samples (see Stain Mix 1 and Stain Mix 2 tables for the suggested concentrations), is an appropriate starting point for staining the compensation beads. From there, the concentration may need to be adjusted depending on signal brightness (use lower concentration if the signal is too bright, or higher concentration if the signal is too dim).Note: The fluorescently labeled tetrameric B cell probes do not bind to these compensation beads. To set up compensation controls for the tetrameric probes, we instead stain compensation beads with an antibody conjugated to the same fluorochrome as the one conjugated to the probes (i.e., we use an anti-CD4-PE antibody as the single-color compensation control for the RBD-PE tetrameric probe). Importantly, if possible, tandem dyes should be avoided for making tetrameric probes as tandem dyes have a much higher variability from lot-to-lot (due to the dissolution of the tandem fluorochromes) and cannot be accurately compensated in this manner. -
h.After incubation, add 100 μL of FACS Buffer to each well and spin the plate at 400 × g for 2 min at 4°C.
-
i.Aspirate supernatant and wash the single-color compensation controls one additional time by adding 200 μL of FACS Buffer to each well and spin as above.
-
j.Resuspend the single-color compensation controls in 100–200 μL of FACS Buffer and use to set the compensation matrix on the spectral flow cytometer.Note: Single-color compensation controls can be prepared during any incubation steps.Note: Spectral flow cytometers, such as the Cytek Aurora, tend to be quite stable over time. For this reason, we have had success with acquiring single-color compensation controls once and reusing the same controls across multiple weeks to set the compensation matrix.
 CRITICAL: If adopting the above strategy and staining fresh FNA samples on different days, the same selected frozen human sample should be used as an internal control for every new FNA sample acquisition. This internal control is used to validate the accuracy of the compensation matrix over time and ensure the consistency of the gating strategy between batches. A tonsil sample is preferred for this purpose, as it contains the cell populations of interest. However, PBMC could also be used if human tonsils are not available, keeping in mind that certain markers (e.g., BCL6) are not be expressed, or expressed at low levels in cell populations from blood.
CRITICAL: If adopting the above strategy and staining fresh FNA samples on different days, the same selected frozen human sample should be used as an internal control for every new FNA sample acquisition. This internal control is used to validate the accuracy of the compensation matrix over time and ensure the consistency of the gating strategy between batches. A tonsil sample is preferred for this purpose, as it contains the cell populations of interest. However, PBMC could also be used if human tonsils are not available, keeping in mind that certain markers (e.g., BCL6) are not be expressed, or expressed at low levels in cell populations from blood.
-
a.
-
8.
After obtaining the FNA sample, spin at 300 × g for 10 min at 4°C.
-
9.
Remove supernatant by aspiration, leaving 100–200 μL of residual volume. If red blood cells appear abundant in the pellet, proceed to step 10. Otherwise, proceed to step 13.
-
10.
To lyse red blood cells, resuspend cells in 2 mL of ACK Lysis Buffer and incubate on ice (or at 4°C) for 3–5 min.
Note: Longer incubation times in ACK Lysis Buffer can decrease cell viability. We tend to keep this step as short as possible by ending the incubation as soon as the solution clears (becomes transparent).
-
11.
Stop the lysis reaction by adding 20 mL of ice-cold PBS and spin cells at 300 × g for 10 min at 4°C.
-
12.
Remove supernatant by aspiration.
-
13.
Resuspend cell pellet in 500 μL of Culture Media. Count cells on a hemocytometer. Cells should be kept at 4°C (or on ice) throughout processing, unless otherwise denoted.
-
14.Prepare staining mixes. It is recommended to prepare a two-sample excess of these mixes.Note: The concentrations listed below are 2× concentrations, as Stain Mix 1 and Stain Mix 2 are ultimately combined 1:1 (see step 19).
-
a.Stain Mix 1: Prepare by mixing the appropriate volume of each antibody in FACS Buffer as described in the table below. 50 μL of mix is required per sample.Stain Mix 1
Marker Concentration (mg/mL; 2×) Dilution (2×) CXCR5 BUV395 0.005 1:40 CCR4 PE-CF594 0.01 1:20 CCR6 BV480 0.003 1:33 CXCR3 BV510 0.005 1:20 -
b.Stain Mix 2:
-
i.First prepare the eBioscienceTM Fixable Viability Dye eFluorTM 780 by diluting it 1:1,000 in FACS Buffer (for a final dilution of 1:4,000).
-
ii.Then combine diluted viability dye 1:1 with Brilliant Stain Buffer to achieve a total volume of 50 μL of mix per sample (mix 25 μL of diluted viability dye with 25 μL of Brilliant Stain Buffer per sample).
-
iii.Add the appropriate volume of each antibody/tetramer as described in the table below to obtain a 2× dilution.Stain Mix 2
Marker Concentration (mg/mL; 2×) Dilution (2×) IgM BUV496 0.002 1:100 CD4 BUV563 0.00012 1:100 CD11c BUV661 0.004 1:50 IgD BUV737 0.001 1:100 CD45RA BUV805 0.002 1:100 CD138 BV605 0.004 1:25 ICOS BV650 0.004 1:25 CD20 BV750 0.002 1:100 CD38 BV785 0.003 1:50 CD8α PerCP-eFluor710 0.0005 1:100 CD27 PE-Cy5 0.002 1:25 CD19 PE-Cy5.5 0.00025 1:100 CD11b Alexa Fluor 700 0.004 1:50 PD-1 PE-Cy7 0.004 1:25 Full S BV421 Varies Based on titration RBD PE Varies Based on titration PR8-HA Alexa Fluor 488 Varies Based on titration
-
i.
-
a.
-
15.
Add the appropriate number/volume of cells to a V-bottom 96-well plate. 0.5–1 × 106 cells per staining are recommended, but lower numbers can be stained according to the FNA yield.
Note: If larger numbers of cells have been collected, they can be cryopreserved as described in the FNA cryopreservation section.
-
16.
Spin cells at 400 × g for 2 min at 4°C.
-
17.
Aspirate supernatant and resuspend cells in 50 μL of Stain Mix 1.
-
18.
Incubate plate for 10 min at 37°C.
CRITICAL: Chemokine staining with Stain Mix 1 must be performed at 37°C. Staining for chemokine receptors at 4°C yields poor separation of positive and negative signals, while staining at 37°C markedly improves separation.
-
19.
After incubation, add 50 μL of Stain Mix 2 directly to each well (without washing) and gently resuspend by pipetting.
-
20.
Incubate for 1 h at 4°C. During this incubation, prepare the solutions for the fixation/permeabilization step (see step 22a).
-
21.
After the incubation is complete, add 100 μL of ice-cold FACS Buffer to each sample and spin cells at 400 × g for 2 min at 4°C.
-
22.Fix and permeabilize the cells using the FoxP3 / Transcription Factor Staining Buffer Set:
-
a.During the incubation in step 20, prepare 200 μL per sample of the Fixation/Permeabilization Solution by diluting the 4× Fixation/Permeabilization Concentrate in the Fixation/Permeabilization Diluent (for 200 μL total: mix 50 μL Concentrate with 150 μL Diluent).
-
b.Aspirate supernatant and resuspend in 200 μL of Fixation/Permeabilization Solution per well.
-
c.Incubate for 1 h at 4°C. During this time, complete steps 23 and 24.
-
a.
-
23.
Prepare a 1× Permeabilization Buffer by diluting the Permeabilization Buffer (10×) in milliQ water (for 1 mL total: mix 100 μL of 10× Buffer in 900 μL of water).
-
24.
Prepare 100 μL of BCL6 staining mix per sample by diluting the BCL6 antibody 1:50 in 1× Permeabilization Buffer.
-
25.
After incubation is complete, spin cells at 400 × g for 2 min at 4°C. Then, aspirate supernatant and resuspend samples in 100 μL of BCL6 staining mix.
-
26.
Incubate for 1 h at 4°C.
-
27.
After incubation, add 100 μL of 1× Permeabilization Buffer to each sample and spin cells at 400 × g for 2 min at 4°C.
-
28.
Aspirate supernatant and resuspend in 200 μL of FACS Buffer and proceed to flow cytometry sample acquisition.
CRITICAL: To ensure an adequate number of events for analysis, the entire sample volume should be acquired on the flow cytometer.
Note: Because of the intracellular BCL6 staining, samples are best acquired as soon as possible on the flow cytometer. In extreme circumstances they can be left up to 16 h before acquiring, but this will decrease the intensity of BCL6 signal.
FNA cryopreservation
Timing: 24 h (for steps 29–31)
If enough cells are recovered from the FNA procedure, excess sample can be cryopreserved for future use. In this section, we describe how to cryopreserve these samples for long term storage.
-
29.
If freezing cells, after spinning at 300 × g for 10 min at 4°C, gently resuspend the desired number of cells in the appropriate volume (2–10 × 106 cells/mL) of Freezing Media and rapidly transfer to a cryogenic vial.
-
30.
Immediately transfer vials to a pre-chilled isopropanol freezing container, such as a Nalgene® Mr.Frosty, and place in a −80°C freezer.
-
31.
The following day (approximately 24 h following step 30), transfer the vials into cryoboxes and place in vapor phase liquid nitrogen (below −135°C) for long-term storage.
Note: Performing the flow analysis on frozen samples, instead of fresh, is possible if enough cells are recovered from the FNA procedure to allow for cryopreservation. FNA cell recovery will depend on the time point of collection post-vaccination and on whether the subjects undergoing the FNA collection are immunocompetent or immunocompromised. In our published study,1 we opted to perform spectral flow cytometry analysis of fresh FNA samples because of the variability in cell yield at certain time points for immunocompromised individuals. It is also worth mentioning that while the concurrent analysis of multiple cryopreserved samples might reduce variability introduced by performing the staining on different days and/or the cytometer set up, cryopreservation itself can affect the quality of detection for certain markers. For instance, the signal intensity of some chemokine receptors might be reduced after cryopreservation.
Expected outcomes
The yield of FNA samples collected 14 days (+/- 2) after the 1st dose (V2) and 8 days (+/-2) after the 2nd dose (V3) of SARS-CoV-2 mRNA vaccines is variable and can range from 0.3 × 106–40 × 106 cells in immunocompetent donors. In our study,1 we found that GC B cells and Tfh cells were present at high frequencies at V2, and that these frequencies typically increased at V3. The same finding held true for antigen-specific GC B cells, antigen-specific memory B cells, and both total and antigen-specific plasmablasts. The expected ranges of cell frequencies at V2 and V3 are outlined in Table 1.
Table 1.
Expected frequencies of specific cell types from FNAs
| Cell type | V2 (% of lymphocytes) | V3 (% of lymphocytes) |
|---|---|---|
| GC Tfh cells | 0.04–1.58 | 0.34–3.08 |
| GC B cells | 0.01–4.03 | 0.26–8.22 |
| S+ RBD- GC B cells | 0.00–0.87 | 0.03–1.87 |
| S+ RBD+ GC B cells | 0.00–0.19 | 0.00–1.17 |
| S+ RBD- MBCs | 0.00–0.04 | 0.01–0.05 |
| S+ RBD+ MBCs | 0.00–0.01 | 0.00–0.03 |
| Plasmablasts | 0.01–0.11 | 0.05–1.00 |
| S+RBD- Plasmablasts | 0.00–0.01 | 0.00–0.03 |
| S+ RBD+ Plasmablasts | 0.00–0.00 | 0.00–0.01 |
Expected frequencies of the denoted cell types as measured from immunocompetent donors at V2 and V3 as detailed in Lederer et al. (2022).1
Quantification and statistical analysis
FlowJo v10 is used for the analysis of flow cytometry files. Compensation should be run and applied on your spectral flow cytometer. Before beginning analysis, the accuracy of the compensation matrix should be validated.
Gating strategies for GC B cells, memory B cells and plasmablasts (Figures 1 and 2), GC Tfh cells (Figure 3), and functional polarization of CD4 T cells (Figure 4), are outlined below.
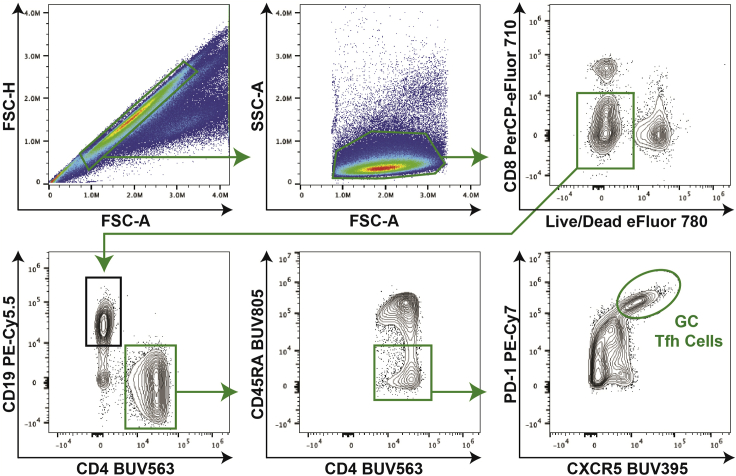
Figure 3.
Tfh cell gating strategy
For Tfh cell analysis, first gate on singlets and then on lymphocytes. Next, separate out live CD8- cells, and further gate on the CD19-CD4+ population. Antigen-experienced CD4 T cells, defined as CD45RA- cells, are selected for further analysis. GC Tfh cells are then defined as PD-1hiCXCR5hi cells.
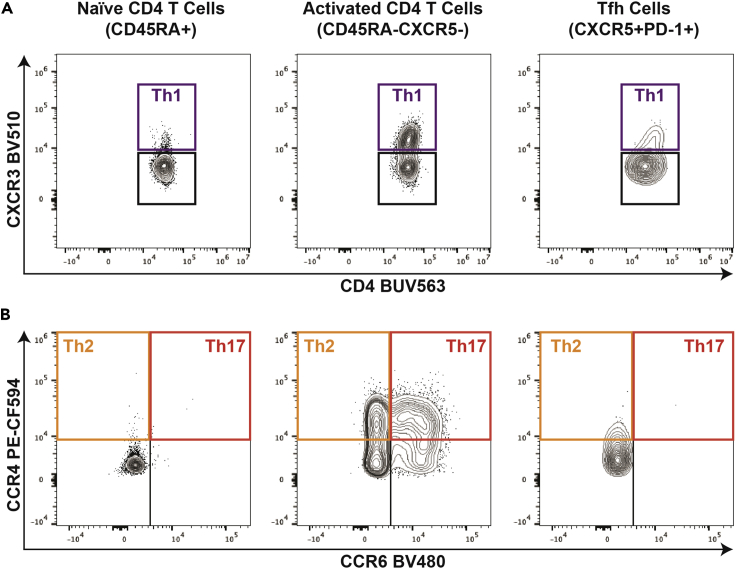
Figure 4.
Evaluation of CD4 T cell functional polarization
The initial analysis of live CD8-CD19-CD4+ cells is shown in Figure 3. The live CD8-CD19-CD4+ cell population is further subdivided into naïve CD4 T cells (CD45RA+), serving as a negative control to define chemokine receptor gating, antigen-experienced non-Tfh CD4 T cells (CD45RA-CXCR5-) or Tfh cells (CXCR5hiPD-1hi, gating shown in Figure 3).
(A and B) These 3 cell populations are further subdivided into (A) Th1 (CXCR3+) polarized cells and (B) Th2 (CXCR3-CCR4+CCR6-) or Th17 (CXCR3-CCR4+CCR6+) polarized cells.
Limitations
This procedure outlines a fine needle aspiration-based method to access cells from vaccine draining lymph nodes in humans. This approach, however, is limited in that it does not preserve any information about the location of recovered cells or the architecture of the lymph node. The cells recovered using this method are in a single-cell suspension, and therefore most suited to analysis applications such as flow cytometry. If spatial information about the draining lymph nodes is required for a particular study, then a different approach will need to be deployed.
Additionally, while FNAs are usually successful, there have been occasions where the FNA procedure does not result in sufficient viable cell recovery for staining. Due to this, we plan for about a 10% failure rate in FNAs.
Troubleshooting
Problem 1
Low cell yield and/or low viability of FNA biopsy.
Potential solution
-
•
If it is difficult to identify a lymph node (step 2), it may be helpful to switch between high frequency linear transducers (L15-7io or L12-4) and lower frequency small footprint transducers (C8-5), as well as optimizing other scanning parameters such as focal zone.
-
•
While performing the biopsy (step 4), it is important to ensure that the tip of the needle remains in the cortex and not in the adjacent fat or in the lymph node hilum and to stay within the lymph node an adequate amount of time (at least 10 s).
-
•
Selecting a lymph node with a thicker cortex in step 2 allows for a greater needle excursion and lowers the likelihood of inadvertently coming out of the lymph node cortex.
-
•
Ensure media tube is kept at 4°C or on ice at all times during the procedure through processing until after the fixation/permeabilization (step 22) is complete. This approach will help to limit cell death.
-
•
If initial cell pellet after centrifugation in step 8 is small, consider decreasing the time of ACK lysis to preserve viability.
Problem 2
Little or no cells binding probe staining.
Potential solution
In steps 7–20 of Titration of tetrameric probes for labeling SARS-CoV-2-specific B cells, validate tetrameric probe staining using a sample with known positive, probe-binding cells. PBMCs from individuals recently vaccinated with a SARS-CoV-2 mRNA vaccine or COVID-19 convalescent subjects will suffice for validation purposes if human lymph node biopsies are not readily available. If no cells from the positive control sample bind the probes, follow the steps outlined in this protocol to remake tetramers. Biotinylated RBD can be purchased from R&D Systems to eliminate a step in the process of making tetramers for an additional cost.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michela Locci (michela.locci@pennmedicine.upenn.edu).
Materials availability
All unique reagents generated in this study will be available from the lead contact upon reasonable request.
Acknowledgments
M.L. was supported by NIH NIAID grant R01 AI168312. E.B. was supported by the NIH fellowship T32 AI055400. We thank Dr. Norbert Pardi for kindly providing the RBD and HA proteins and Ali Naji and Vijay Bhoj for assistance in organizing the initial study.1 We thank Dr. Florin Tuluc and Jennifer Murray of the CHOP flow cytometry core facility for technical assistance. The graphical abstract was created with BioRender.com.
Author contributions
M.L. and K.L. adapted the protocol. K.L. and E.B. developed the FNA analysis strategy. E.B. and M.L. wrote the manuscript with help from K.L., H.S., and L.J. M.K. performed all clinical coordination. M.L. supervised the generation of this protocol.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101840.
Data and code availability
All raw data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Additional supplemental information are available from Mendeley Data at https://doi.org/10.17632/4kh76pv4sj.1.
References
- 1.Lederer K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castaño D., Goel R.R., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185:1008–1024.e15. doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185:603–613.e15. doi: 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margine I., Palese P., Krammer F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp. 2013:e51112. doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor J.J., Martinez R.J., Titcombe P.J., Barsness L.O., Thomas S.R., Zhang N., Katzman S.D., Jenkins M.K., Mueller D.L. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J. Exp. Med. 2012;209:2065–2077. doi: 10.1084/jem.20112272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havenar-Daughton C., Newton I.G., Zare S.Y., Reiss S.M., Schwan B., Suh M.J., Hasteh F., Levi G., Crotty S. Normal human lymph node T follicular helper cells and germinal center B cells accessed via fine needle aspirations. J. Immunol. Methods. 2020;479:112746. doi: 10.1016/j.jim.2020.112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Additional supplemental information are available from Mendeley Data at https://doi.org/10.17632/4kh76pv4sj.1.