Abstract
The COVID-19 pandemic has caused severe health problems worldwide and unprecedented decimation of the global economy. Moreover, after more than 2 years, many populations are still under pressure of infection. Thus, a broader perspective in developing antiviral strategies is still of great importance. Inspired by the observed multiple benefits of heparin in the treatment of thrombosis, the potential of low molecular weight heparin (LMWH) for the treatment of COVID-19 have been explored. Clinical applications found that LMWH decreased the level of inflammatory cytokines in COVID-19 patients, accordingly reducing lethality. Furthermore, several in vitro studies have demonstrated the important roles of heparan sulfate in SARS-CoV-2 infection and the inhibitory effects of heparin and heparin mimetics in viral infection. These clinical observations and designed studies argue for the potential to develop heparin mimetics as anti-SARS-CoV-2 drug candidates. In this review, we summarize the properties of heparin as an anticoagulant and the pharmaceutical possibilities for the treatment of virus infection, focusing on the perspectives of developing heparin mimetics via chemical synthesis, chemoenzymatic synthesis, and bioengineered production by microbial cell factories. The ultimate goal is to pave the eminent need for exploring novel compounds to treat coronavirus infection-caused diseases.
Keywords: Heparin, Biosynthesis, Biological activities, Bio-production
1. Introduction
Coronavirus is a family of RNA viruses that infect the respiratory tract system. The ongoing COVID-19 (coronavirus disease 2019) pandemic has caused more than 500 million human infections, including at least 6 million deaths worldwide (as of Apr. 15, 2022), and led to unprecedented decimation of the global economy [1]. COVID-19 is caused by the highly transmissible and pathogenic SARS-CoV-2 RNA virus species with a high fatality rate [2]. The phylogenetic network of SARS-CoV-2 genomes sampled worldwide revealed closely related evolutionary selection in their human hosts [3]. It should be noted that, within about 2 years, SARS-CoV-2 has already resulted in several evolutionary variants different from the original isolate spreading rapidly over the global areas [4], from Alpha to Delta to the latest globally spreading Omicron. Unfortunately, it is highly possible that Omicron is not the last variant of SARS-CoV-2 [5,6].
In advanced stage of COVID-19, vascular leakage, abnormal coagulopathy and excessive inflammation are the major complications leading to rapid deterioration and death of COVID-19 patients. Furthermore, a progressive alteration of some inflammatory and coagulative parameters was observed in this stage, leading to an increased risk of both arterial and venous thrombosis [[7], [8], [9]]. Thus, anticoagulation has been proposed as prognostically significant for reducing mortality in COVID-19 treatment [10]. Heparin or low molecular weight heparins (LMWHs) are the mostly used anticoagulant for the prevention and treatment of thrombosis, therefore have been recommended by WHO to apply to moderate-severe COVID-19 patients [[10], [11], [12], [13], [14], [15]]. In addition, heparin also shows ‘side’ effects, including attenuation of inflammatory reaction and acting as a “trap” to bind the virus [16]. Though these non-anticoagulant effects of heparin are beneficial to patients, a dosage of heparin must be tightly regulated to avoid the risk of bleeding complications. To overcome this potential adverse effect, heparin mimetics with a weakened anticoagulant effect but more potent anti-inflammatory and antiviral activity are attracting attention [17]. To pave the rapid progress in developing non-anticoagulant heparin mimetics, a number of approaches are in the process.
This review summarizes the functional properties of heparin in treating thrombosis and COVID-19 patients and the underlying mechanisms of heparin's beneficial effects in addition to its anticoagulant activity. Production of heparin and heparin mimetics via chemical synthesis, chemoenzymatic synthesis, and the current understandings of the biosynthesis of heparin as well as efforts toward the bioengineered production by microbial cell factories are discussed. Finally, synthetic biology approaches are highlighted as the most efficient strategy for the future supply of heparins.
2. Biological activities of heparin
2.1. Anticoagulation
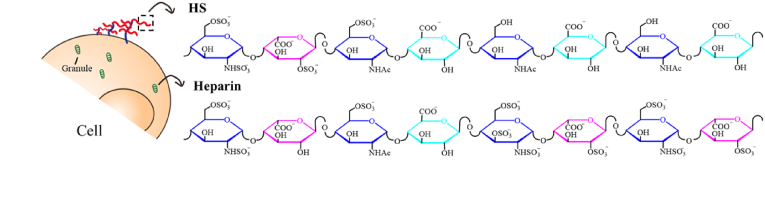
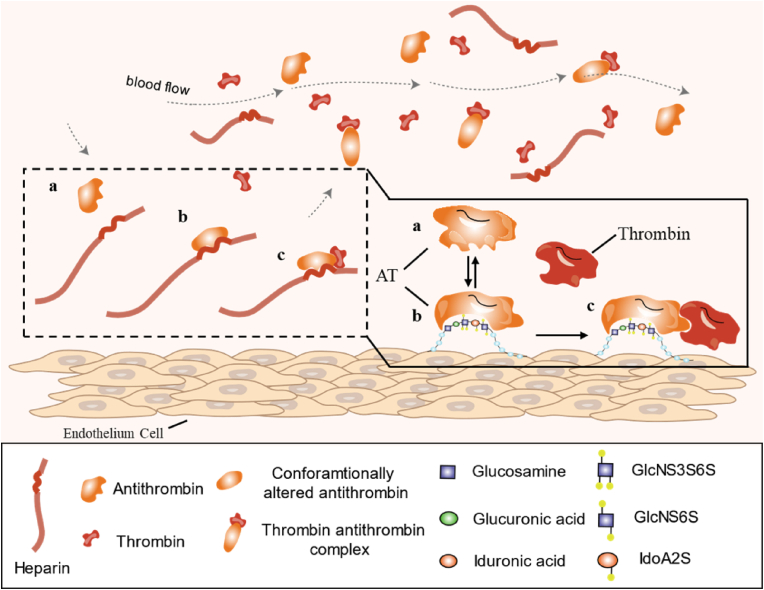
Heparin was the first glycosaminoglycans (GAGs) molecule discovered, originally isolated from dog liver [18]. A few decades after this discovery, the clinical application of heparin as an anticoagulant was introduced. However, the knowledge regarding the molecular structure and functional mechanism of heparin's anticoagulation activity was utterly lacking. The breakthrough came from 1970 to 1980, when the unique pentasaccharide sequence in heparin chains was identified (Fig. 1). Since then, it has been clarified that the anticoagulation activity of heparin is through a specific interaction between the pentasaccharide sequence and antithrombin (AT), an endogenous anticoagulant protein inhibiting several coagulation factors [19,20] (Fig. 2). This finding significantly promoted the application of heparin and the generation of low molecular weight heparin (LMWH).
Fig. 1.
Illustration of heparin and heparan sulfate (HS) structure.
Fig. 2.
The anticoagulation activity of heparin. Antithrombin (a) has low affinity to coagulation enzymes. The conformation of AT (b) is changed after specific binding to the pentasaccharide in heparin chains, which significantly increased the affinity of AT to the coagulation enzymes (c).
It should be pointed out that heparin is currently isolated from animal tissues, mainly porcine intestinal mucosa or bovine lung. This animal-derived medicine is highly heterogenous and has an average molecular weight of 14–15 kDa, leading to poor pharmacokinetics and clinical inconvenience in dosage. Moreover, the macromolecular property of heparin tends to be immunogenic, causing severe side effects, e.g., heparin-induced thrombocytopenia (HIT) [21]. LMWH is generated by partial degradation (enzymatic or chemical) of heparin, resulting in 4–5 kDa fragments. LMWH has several advantages over full-length heparin, including better pharmacokinetics, easier dosage and higher safety. Regardless of the drawbacks of animal resources, more than 100 years after its discovery, this potent anticoagulant remains one of the most widely used clinical drugs. The primary clinical indication of heparin is for prophylactic treatment of postoperative thrombosis and acute venous thrombosis [22]. However, it has been observed that the application of heparin has exhibited multiple beneficial effects other than anticoagulation, e.g., anti-cancer and anti-inflammation properties [23]. In addition, experimental and retrospective studies showed that LMWH had anti-neoplastic effects [24,25]. These encouraging observations support the beneficial impact of heparin in treating cancer patients and chronic inflammatory diseases complicated by thrombosis.
2.2. Heparin and heparin mimetics contributions to the treatment of COVID-19 patients
One of the significant pathological symptoms of severe COVID-19 patients is hyper-coagulopathy-associated thrombosis, a leading cause of high mortality. To prevent thrombosis, WHO recommended heparin to be included in the treatment regime for hospitalized COVID-19 patients. Numerous clinical observations evidenced that treatment with heparin, primarily LMWH, reduced lethality [26,27]. As the hyper-coagulopathy is triggered by an acute immunological reaction (“cytokine storm”) towards SARS-CoV-2 infection, it is assumed that heparin, apart from anticoagulation, may have also exhibited anti-inflammatory effects. Retrospective studies revealed reduced inflammatory cytokine levels were shown in COVID-19 patients treated with LMWH [28]. These findings inspire and warrant a more profound and broader investigation to explore the multiple-target effects of heparin, e.g. anticoagulation, anti-inflammation and antiviral infection, in the treatment of COVID-19 and other coronavirus infections [29]. However, it is to be noted that heparin is a potent anticoagulant, and its dosage has to be tightly regulated to avoid the risk of bleeding complications. Thus, interest is increasing in designing heparin mimetics with low or no anticoagulation activity but with high anti-inflammatory and anti-viral activity [30,31].
Heparin mimetics are highly sulfated synthetic and semi-synthetic glycosaminoglycans with different structures. In vitro studies from different labs demonstrated the antiviral effect of heparin mimetics [32,33]. Many heparin-like drugs, commonly used to treat other diseases, also show good anti-SARS-CoV2 activity. For example, Pixatimod, a synthetic HS mimetic, which is in clinical trials for cancer, was found to have anti-SARS-CoV-2 activity (including those variants of concern) through disruption of the interaction between spike protein and ACE2 [34]; Pentosan polysulfate, a semisynthetic heparin-like glucosaminoglycan, which is an established drug for the oral treatment of interstitial cystitis, exhibits weaker anticoagulant effects and more potent SARS-CoV-2 inhibition than heparin in Vero cell model [35]; Mucopolysaccharide polysulfate, a heparinoid which is a clinic drug for antithrombotic, exhibited effectively antiviral activities against wild-type or Delta S-proteins of SARS-CoV-2 than heparin in an in vitro cell-based assay [36]. Sulodexide, composed of a mixture of fast-moving heparin and dermatan sulfate, has been used clinically for the prevention and treatment of vascular diseases, improving the efficacy of COVID-19 patients' clinical outcomes [37]. However, discrepancies in the antiviral potencies of heparin analogs are also indicated. For instance, Gasbarri et al. revealed that SARS-CoV-2 does not use heparan sulfate for infection and the inhibition against SARS-CoV-2 was found to be simply reversible [38]. The search for novel low/non-anticoagulant heparin mimetics with the increased binding ability and fewer side effects remains a research subject.
3. Biological functions of heparan sulfate
3.1. Expression and functions
Heparan sulfate (HS) was initially discovered as a ‘contaminant’ or ‘by-product’ of heparin production from animal tissues [[39], [40], [41]]. The important biological functions of HS were recognized gradually when the biochemical and biological techniques were advanced. The studies during the past 40+ years have established multiple indispensable functions of HS in animal development and homeostasis, which are nicely described in several reviews [42,43]. The most direct shreds of evidence are from transgenic animal models, showing that HS is essential for animal development. Eliminating the enzymes involved in HS biosynthesis led to the production of distorted HS structure, resulting in different degrees of the developmental defect and embryonic lethality in the animals [[44], [45], [46], [47]]. Due to its ubiquitous expression and structural diversity, HS interacts with diverse protein ligands, including enzymes, growth factors/morphogens and their receptors, cytokines and ECM components [48,49], enabling its participation in various biological activities, covering coordinating cellular signaling to cell-cell, cell-matrix communications. Recent studies have increasingly implicated HS in pathological processes of various diseases, such as diabetic nephropathy [50], inflammation [51,52], amyloid deposition [53,54] and tumor biology [55]. These findings imply that the molecular structure of HS may be altered under pathological conditions, resulting in abnormal biological functions.
3.2. HS as co-receptor for virus infection
HS is attached to a variety of cell surfaces as heparan sulfate proteoglycans (HSPGs), composed of a core protein and covalently linked to GAG chains. One of the well-recognized functions of HS is as co-receptors for many morphogens and growth factors, regulating key signaling activities [44]. Accumulated evidence shows that HS on cell surfaces also serves as a receptor for viral attachment and cell entry [56]. The sulfated GAG chains exhibit global negative charges that can interact electrostatically with a surface of viral or the basic residues of viral capsid proteins that are not enveloped in the virus. Viruses use these weak ionic interactions to increase their concentration at the cell surface and augment the chances of binding to more specific receptors [57,58].
Several virus species like Herpes Simplex Virus (HSV), dengue virus (DENV) [59,60], and human papillomavirus (HPV) [61,62] have the natural dependence of binding to HSPG for their attachment to the host cells. In addition, some highly prevalent viruses could gradually adapt to HSPG after widely passaging in cells, such as foot-and-mouth disease virus (FMDV) [63], Coxsackie virus B3 (CV–B3) [64], or the Rhinoviruses-C15 (RV-C15), RV-A8, and RV-A89 [[65], [66], [67]]. Interestingly, other viruses such as enterovirus 71 (EV-A71) [68,69] and Cunningham polyomavirus (JCV) [70] can even obtain HSPG-binding ability in vivo after adapting to the host. However, for respiratory syncytial virus (RSV) [71,72] and Zika Virus (ZIKV) [[73], [74], [75]], the literature contains contradictory findings showing that their binding to HSPG remains controversial [58].
One solid evidence for the essential roles of HS in virus infection is Herpes Simplex Virus (HSV) infection, where it is demonstrated that a rare modification of HS described as 3-O-sulfation can interact with the envelope protein of HSV to trigger viral penetration into cells [76]. Yet, most oncogenic viruses' entry into the host is initiated by attachment to HSPG followed by important conformational changes of viral proteins and then fuses with cellular membranes. For example, Kaposi's sarcoma-associated herpes virus (KSHV) attaches primarily to HSPGs on the surface of host cells via gB and gpK8.1A [77,78], subsequently changes the conformation of viral glycoproteins to allow access to specific entry receptors. It also has been demonstrated that HSPGs expressed on the endothelial cell surface act as a receptor for HIV-1 Tat [79,80], and their binding promotes HIV neurovirulence by allowing the infection of endothelial cells that do not express CD4 and facilitating the crossing of the blood-brain barrier [81]. Furthermore, using a mouse embryonic fibroblast cell line defective in HS production, an important role of HS on Sindbis virus infection has been demonstrated through the direct interaction of envelope protein (E2) with HS [82].
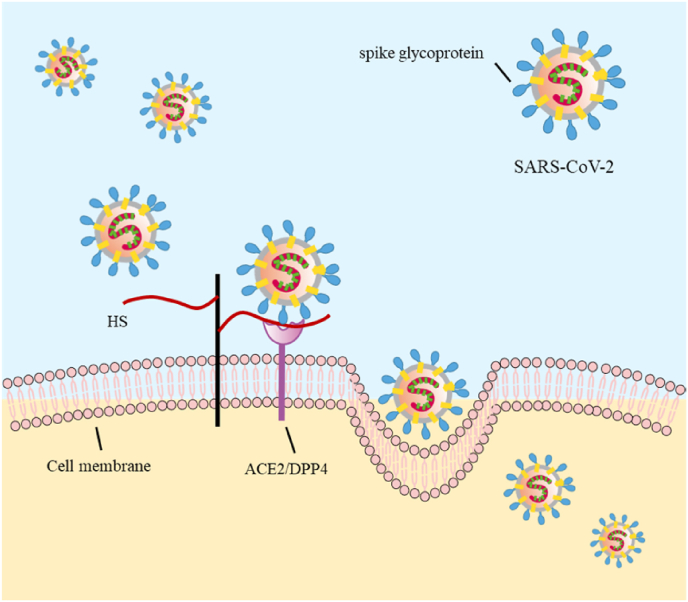
Most interestingly, recent studies have provided ample evidence for the critical role of HS on SARS-CoV-2 infection [83]. Numerous associated findings support this milestone report. The study shows that human Cov-NL63 utilizes HSPG for attachment to target cells [84]. Treatment of HEK293E/ACE2-Myc cells with heparinase (degrading cell surface HS) or exogenous heparin (competing with cell surface HS) prevents viral spike protein binding to host cells. This treatment also impedes SARS pseudovirus infection, supporting the receptor roles of HS for SARS-CoV-2 attachment at the early phase of invasion [85]. Indeed, heparin and heparin mimetics are demonstrated to bind to the S1 protein receptor binding domain (RBD), leading to a conformational change that blocks the viral attachment and/or entry, subsequently, the cellular invasion by SARS-CoV-2 (Fig. 3). Further, the molecular structure of heparin to interact with the RBD has been investigated, showing a preference in 2-O and 6-O sulfation of the polysaccharide [86].
Fig. 3.
Heparan sulfate serves as a receptor for viral attachment. Heparan sulfate interacts with the receptor-binding domain of spike glycoprotein, adjacent specific entry receptor, shifting the spike structure to an open conformation to facilitate ACE2 binding.
More and more evidence suggest that HSPGs are required for virus binding to host cells, while the HS moieties on HSPGs play the key role as virus receptors. Thus, HS or HSPG could serve as an excellent broad-spectrum antiviral target. Theoretically, these also provide a hint that exogenous sulfated polysaccharides may have a broad inhibition effect on virus infection.
4. Synthesis of heparin mimetics
As a front-line anticoagulant drug, the pharmaceutical production of heparin currently exceeds 100 tons/year using porcine slaughter by-products intestinal mucosa as starting material. However, animal sources bring up major concerns in aspects of safety, environmental and supply stress. Therefore, there is an urgent need to develop non-animal source heparins to satisfy the essential need as an anticoagulant and meet the perspective for further widening the applications.
4.1. Biosynthetic enzymes for heparin and heparan sulfate
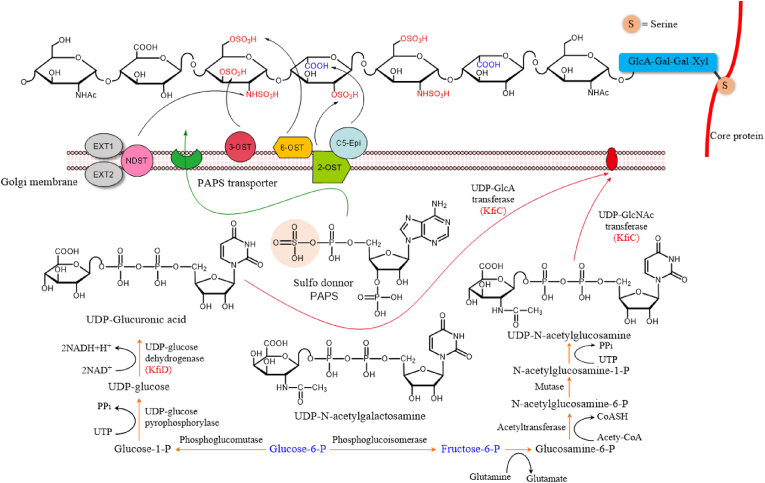
Heparin and HS are composed of alternating hexuronic acid (d-glucuronic acid (GlcA)/l-iduronic acid (IdoA)) and d-glucosamine (GlcN) that are sulfated at various positions. Heparin is exclusively found in connective tissue mast cells, while HS is expressed in all cell types. They share a common biosynthesis pathway that involves at least 11 different enzymes (Table 1). The biosynthetic process is described as two stages: 1) assembly of repeating –GlcA–GlcNAc– disaccharide units to produce the backbone of [GlcA–GlcNAc]n structure that is accomplished by 6 different glycosyltransferases; and 2) modification of the polymers (heparosan) by five reactions catalyzed by distinct enzymes [43]. The first modification is N-deacetylation and N-sulfation of the GlcNAc units catalyzed by N-deacetylase/N-sulfotransferase (Ndst), creating substrate for the C5-epimerase (Glce) to convert GlcA to IdoA. Then the polymer serves as an optimal substrate for 2-O-sulfotransferase (Hs2st) that prefers to transfer sulfate group to the C2 of IdoA than to GlcA residues. The 6-O-sulfotransferase (Hs6st) transfers sulfate group to the C6 of GlcN, which seems less selective and can occur even before the N-sulfation (Fig. 4). The 3-O-sulfotransferase (Hs3st) adds sulfates at C3 of GlcN units, which rarely occurs in HS [87], but is critical for the anticoagulation activity in heparin. While a single gene codes Glce and Hs2st in mammalian cells, Ndst, Hs6st, and Hs3st are expressed in several isoforms [43].
Table 1.
The enzymes involved in the biosynthesis of heparin and heparan sulfate.
| Enzyme | Abbreviation | Gene | Function | Reference |
|---|---|---|---|---|
| Xylosyltransferase | Xyl-T | XylT | Transfer xylose to core protein | [126] |
| Galactosyltransferase 1 | Gal-T1 | B4galT7 | Transfer 1st galactose | [127] |
| Galactosyltransferase 2 | Gal-T2 | B3galT6 | Transfer 2nd galactose | [127] |
| Glucuronytransferase 1 | GlcA-T1 | B3gat3 | Transfer first glucoronic acid | [128] |
| N-acetylated glucusamyltransferase 1 | EXTL2 | Extl2 | Transfer first N-acetyl glucosamine | [129] |
| Polymerase | EXT | Extl1/2 | Transfer alternate GlcA and GlcNAc | [130] |
| N-deacetylase/N-sulfotransferase | NDST | Ndst1-4 | N-deacetylation and sulfation of GlcNAc | [131] |
| Glucuorynyl C5-epimerase | Hsepi | Glce | Epimerization of GlcA to IdoA | [132] |
| Hexuronyl 2-O-sulfotransferase | HS2OST | Hs2st | Sulfation of IdoA and GlcA at C2 | [133] |
| Glucosaminyl 6-O-sulfotransferase | HS6OST | Hs6st1-3 | Sulfation of GlcN at C6 | [134] |
| Glucosaminyl 3-O-sulfotransferase | HS3OST | Hs3st1-6 | Sulfation of GlcN at C3 | [22] |
Fig. 4.
Biosynthesis of heparin and heparan sulfate.
4.2. Chemical synthesis
On average, the natural heparin isolated from porcine intestinal mucosa is composed of 60 mono-sugars and the size of LMWH is around 20 sugar units, which makes the chemical synthesis of these complicated carbohydrate molecules practically impossible. Nonetheless, the finding of specific binding of the pentasaccharide with antithrombin facilitated the chemical synthesis of anticoagulant oligosaccharides [88]. So far, the pentasaccharide fonduparinux is still the only synthetic oligosaccharide drug that inhibits coagulation enzymes through binding to antithrombin [89]. Although chemical synthesis of polysaccharides remains a major challenge, recent progress has been made to synthesize heparin-like oligosaccharides [[90], [91], [92], [93]]. Instead of directly synthesizing an oligosaccharide, one strategy is to prepare ‘building blocks’, e.g. disaccharides that are linked to form a longer chain [94]. This approach was further elaborated using the ‘building blocks’ derived from a natural polysaccharide, e.g., heparin or HS [95]. In this way, one may build up ‘tailored’ oligosaccharides.
Chemically synthesized compounds are more homogeneous than those isolated from natural sources. However, restricted by the chemical nature of sugars, synthesis difficulty increases with increased chain length concerning the protection of the hydroxyl group for each step of the reaction; therefore, there is still a bottleneck to produce sufficient quantities of oligosaccharides for pharmaceutical use. An alternative is to produce non-sugar polymers. One of the successive approaches is RAFT (Reversible Addition-Fragmentation chain Transfer) strategy to synthesize heparin-mimic polymers, generating homopolymers with molecular weights ranging from 5 to 50 kDa [96]. Among these synthetic products, the polymer with sodium 4-styrene sulfonate as the monomer and the copolymer composed of the sulfonated monomer and acrylic acid at a ratio of 1:1 show better anticoagulant activity [96].
4.3. Chemoenzymatic synthesis
Chemical synthesis has the drawbacks of multiple protection/synthesis steps, low yield and high cost, which greatly limit its practice. To avoid this, strategies for the chemoenzymatic synthesis process have been explored [36,97]. The capsular polysaccharide produced by E. coli K5 strain has the repeating disaccharide (GlcAβ1–4GlcNAcα1-4)n structure (denoted as K5 polysaccharide) and was used as a precursor for heparin/HS. K5 polysaccharides can be modified to ‘neoheparin’ by chemoenzymatic synthesis [98]. The polysaccharide, often denoted as heparosan, was chemically deacetylated and N-sulfated, then incubated with C5-epimerase to generate IdoA residues, followed by O-sulfating of various hydroxyl groups on both HexA and GlcNS residues [99,100]. The products displayed anticoagulant activity to a similar level to heparin. Unfortunately, due to the non-selective chemical O-sulfation, the reaction generated an artificial structure of non-desired sulfation at C3 of HexA that is not found in natural heparin and HS. The above procedure is advanced by an alternative strategy using O-sulfotransferases, which significantly improved selective O-sulfation reactions [[101], [102], [103]]. One interesting achievement is the mutational expression of one 6-O-sulfotransferase (6-OST), 6-OST Mt-4, to induce specific sulfation of the non-reducing terminal glucosamine residues [104]. In addition, N-deacetylation by chemical reaction, and enzymatic modification on human 3-O-sulfotransferase-1 (3-OST-1), successively led to the single-site sulfated heparosan, 2-deacetyl-3-O-sulfo-heparosan, which exhibits certain anti-tumor activity in vitro [105]. Although the progress in chemoenzymatic synthesis are encouraging, there is still a long way to achieve industrial-scale production.
5. Bio-production of heparin and heparin mimetics
Synthetic biology emerged at the beginning of the 21st Century and has proved to be an effective technological approach to designing and engineering biological systems for diverse applications, e.g., bioproduction of medicines, biofuels, and biomaterials. The significant progress made in synthetic biology over the last decade will continue to accelerate as design and testing cycles rely less on traditional molecular cloning tools [106]. This is an ideal strategy to produce heparin (as well as heparin mimetics) owing to its capacity and sustainability.
The biosynthesis process of heparin/HS taking place in the Golgi apparatus is believed to be highly efficient through the concerted action of each enzyme [107]. The idea is to transfer this process to a microorganism to produce polysaccharides. As discussed above, heparosan, the precursor of heparin and HS, can be produced by E. coli K5. To increase the production of heparosan and develop an alternative source of heparosan from a non-pathogenic strain, many strategies have been used, including co-expression of the key genes (such as elmA, pgcA, gtaB and tuaD) involved in heparosan synthesis, use of different chassis cells as bioreactor and optimization of culture conditions (compositions of medium and culture phase) [[108], [109], [110], [111], [112]]. Bacillus megaterium does not appear to be a very efficient chassis; with a modular approach for UDP-precursor production and pmHS1 heparosan synthesis in B. megaterium, the titer of heparosan was 237.6 mg/L [112]. By up-regulating a single precursor pathway gene UDP and expressing kfiC and kfiA glycosyltransferase genes from E. coli K5, the maximum concentration of heparosan produced by engineering B. megaterium in the bioreactor increased to 394 mg/L, further through fed-batch fermentation, the heparosan titer was increased to 1.32 g/L [113]. The non-pathogenic E. coli BL21 was also used to express heparosan; by inducing the four essential heparosan biosynthesis genes pKfiA-pKfiD in it and optimizing the cultivation process, heparosan titer reached 1.88 g/L [109], and when co-express the eliminase gene elmA, heparosan polysaccharide (about 60–100 kDa) can be cleaved into smaller oligosaccharide [114]. The most effective chassis may be B. subtilis (a recognized safe strain for industrial production), with heparosan titers reaching 5.82 g/L through up-regulation of the tauD gene (encoding the UDPGDH enzyme) and fermentation optimization [110,115]. However, it is still far from using E. coli K5, which has four essential genes (pKfiA-pKfiD) organized in one operon, heparosan titer up to 15 g/L in bioreactor [116,117].
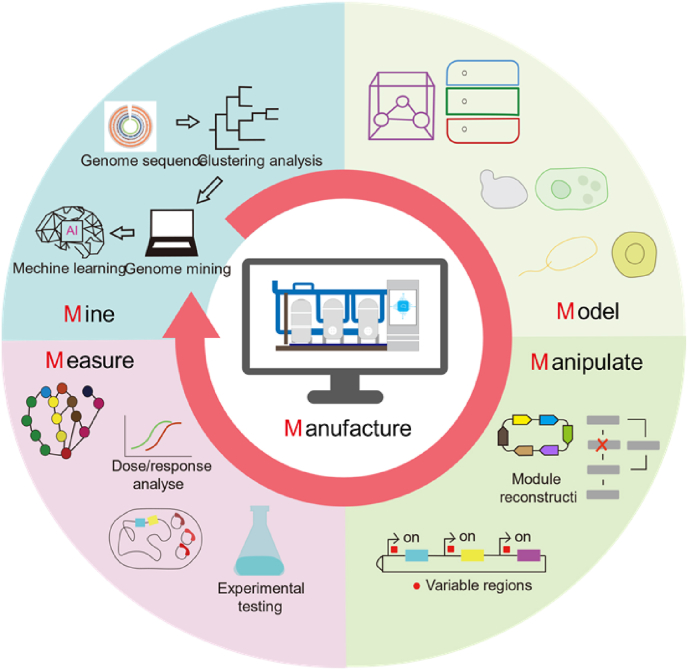
These efforts are still trying to produce heparin precursors in the microorganisms. The most challenging strategy is to engineer the microorganisms to make heparin! One key point is that these post-modified enzymes acting on heparosan to produce heparin have extremely high stereoselectivity and specificity, and it's challenging to find natural enzymes that meet such conditions from microorganisms [118]. It is possible that the enzymes with high specificity or stereoselectivity can be obtained by artificially creating proteins like using “de novo design” to produce “mini proteins” (only the active domain is retained) or by rational design strategy such as the biopharmaceutical company Optimvia, using amino acid sequences to build entirely new proteins to produce heparin recently [119]. This is a step closer to the microorganism production of heparin. Nevertheless, to meet the demand for larger quantities of heparin and the potential of heparin mimetics in drug development (global market = > 100 metric tons/year) [120], we still need to work on several aspects: 1) optimizing the substrate specificity and stereoselectivity of the enzyme; 2) screening a non-toxic and genetically manipulated microbial chassis suitable for high-yielding heparin; 3) using Scale-up technology to optimize fermentation process to achieve high yield of heparin. To reach the above achievements for heparin overproduction through the desired pathways, the 5Ms strategy (including Mine, Model, Manipulation, Measure & Manufacture) could be applied (Fig. 5) [121,122]. Firstly, ‘Mine’ is used to clarify the internal connections in big data sets, including various omics, to generate hypotheses for ‘Model’. The subsequent ‘Manipulation’ is applied to test the above hypotheses and figure out the feasible solution. Then ‘Measure’ is to uncover the phenotype of genome-modified microorganisms. These four procedures formed iterative cycles of experimentation and computation, which could be put forward for the application of ‘Manufacture’ [123,124]. With this 5Ms strategy, heparin and analogs could be manufactured and scaled up from the tube scale to industrial production. This system has been used for the successful overproduction of avermectins [121] and integrated multi-scale data-driven engineering to improve the production of natural products [125].
Fig. 5.
Intelligent bio-production of heparin with 5Ms strategy.
6. Concluding remarks and future perspectives
Heparin has been extensively used in research to elucidate the interactions between HS and protein ligands. As a result, more than 100 HS-binding proteins, including cytokines, growth factors and viral proteins have been identified. Thus, it is believed that heparin can modulate each of the functions of HS. Considering the significant multi-effects of heparin used to treat COVID-19, well-designed heparin mimetics could possess multi-target biological activities, including inhibition of viral infection and replication, modulation of inflammatory responses, and prevention of hyper-coagulopathy and lung damage [27,28,41].
Thus, apart from the important clinical application for the prevention and treatment of thrombosis, the accumulated experimental and clinical findings potential opens a broader venue for heparin mimetics in pharmaceutical development. However, the issue of biosafety and raw material resource limitation remains as far as heparin is exclusively extracted from animal tissues. To meet the requirements in safety, quantity and cost, bio-engineering production in microorganisms should be the most optimal approach to produce heparin. Thousands of bacterial and plant genomes revealed a tremendous breadth of previously unknown secondary metabolite biosynthetic genes. Integrated strategies of genome mining, synthetic biology, and synergistic bioactivity screening will facilitate designing a better gene circuit for an efficient bio-manufacturing of heparin in a selected microorganism.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We gratefully acknowledge the financial support from the National Key Research and Development Program of China (2021YFF0600700, 2020YFA090032 and 2019YFA0906201), the Swedish research council (2020–05759), and the National Natural Science Foundation of China (31720103901, 31961133004, 21907031, 21977029, 22108266).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Lan Jiang, Email: Jianglan@ecust.edu.cn.
Kan Ding, Email: dingkan@simm.ac.cn.
Xueting Liu, Email: liuxueting@ecust.edu.cn.
References
- 1.WHO Coronavirus (COVID-19) Dashboard . 2022. WHO coronavirus (COVID-19) dashboard with vaccination data.https://covid19.who.int/ [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr Biol. 2020;30(15):849–857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X., Hong W., Pan X., Lu G., Wei X. SARS-CoV-2 Omicron variant: characteristics and prevention. MedComm. 2020;2(4):838–845. doi: 10.1002/mco2.110. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scudellari M. How the coronavirus infects cells - and why Delta is so dangerous. Nature. 2021;595(7869):640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 7.Kollias A., Kyriakoulis K.G., Lagou S., Kontopantelis E., Stergiou G.S., Syrigos K. Venous thromboembolism in COVID-19: a systematic review and meta-analysis. Vasc Med. 2021;26(4):415–425. doi: 10.1177/1358863X21995566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang K., Fu Y., Kang Y., Shao H., Yang J., Cui M., et al. Clinical features of COVID-19 patients with venous thromboembolism. Clin Appl Thromb Hemost. 2021;27 doi: 10.1177/10760296211013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali M.A.M., Spinler S.A. COVID-19 and thrombosis: from bench to bedside. Trends Cardiovasc Med. 2021;31(3):143–160. doi: 10.1016/j.tcm.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billett H.H., Reyes-Gil M., Szymanski J., Ikemura K., Stahl L.R., Lo Y., et al. Anticoagulation in COVID-19: effect of enoxaparin, heparin, and apixaban on mortality. Thromb Haemostasis. 2020;120(12):1691–1699. doi: 10.1055/s-0040-1720978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl U., Li J.P. Heparin - an old drug with multiple potential targets in Covid-19 therapy. J Thromb Haemostasis. 2020;18(9):2422–2424. doi: 10.1111/jth.14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindahl U., Li J.P. Heparanase - discovery and targets. Adv Exp Med Biol. 2020;1221:61–69. doi: 10.1007/978-3-030-34521-1_2. [DOI] [PubMed] [Google Scholar]
- 13.Shi C., Wang C., Wang H., Yang C., Cai F., Zeng F., et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: a retrospective cohort study. Clin Transl Sci. 2020;13(6):1087–1095. doi: 10.1111/cts.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie M., Li J.P. Heparan sulfate proteoglycan - a common receptor for diverse cytokines. Cell Signal. 2019;54:115–121. doi: 10.1016/j.cellsig.2018.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Yu M., Zhang T., Zhang W., Sun Q., Li H., Li J.P. Elucidating the interactions between heparin/heparan sulfate and SARS-CoV-2-related proteins-an important strategy for developing novel therapeutics for the COVID-19 pandemic. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.628551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020;181 doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohamed S., Coombe D. Heparin mimetics: their therapeutic potential. Pharmaceuticals. 2017;10:78. doi: 10.3390/ph10040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mclean J. The thromboplastic action of heparin. J Am J Physiol. 1916:41. [Google Scholar]
- 19.Lindahl U., Backstrom G., Hook M., Thunberg L., Fransson L.A., Linker A. Structure of the antithrombin-binding site in heparin. Proc Natl Acad Sci USA. 1979;76(7):3198–3202. doi: 10.1073/pnas.76.7.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg R.D., Jordan R.E., Favreau L.V., Lam L.H. Highly active heparin species with multiple binding sites for antithrombin. Biochem Biophys Res Commun. 1979;86(4):1319–1324. doi: 10.1016/0006-291x(79)90260-2. [DOI] [PubMed] [Google Scholar]
- 21.Castelli R., Cassinerio E., Cappellini M., Porro F., Graziadei G., Fabris F. Heparin induced thrombocytopenia: pathogenetic, clinical, diagnostic and therapeutic aspects. Cardiovasc Hematol Disord: Drug Targets. 2007;7:153–162. doi: 10.2174/187152907781745251. [DOI] [PubMed] [Google Scholar]
- 22.Petitou M., Casu B., Lindahl U. 1976-1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochimie. 2003;85(1–2):83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
- 23.Sarah M., Mandana M., Tina K., MJAiPe Maryam. 2015. Anti-inflammatory effects of heparin and its derivatives: a systematic review; pp. 1–14. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakkar A.K., Levine M.N. Thrombosis and cancer: implications beyond trousseau. J Thromb Haemostasis. 2004;2(8):1261–1262. doi: 10.1111/j.1538-7836.2004.00867.x. [DOI] [PubMed] [Google Scholar]
- 25.Lyman G.H., Khorana A.A., Kuderer N.M., Lee A.Y., Arcelus J.I., Balaban E.P., et al. Venous thromboembolism prophylaxis and treatment in patients with cancer: american society of clinical oncology clinical practice guideline update. J Clin Oncol. 2013;31(17):2189–2204. doi: 10.1200/JCO.2013.49.1118. [DOI] [PubMed] [Google Scholar]
- 26.Tang N., Bai H., Chen X., Gong J.L., Li D.J., Sun Z.Y. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemostasis. 2020;18(5):1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayerbe L., Risco C., Ayis S. The association between treatment with heparin and survival in patients with Covid-19. J Thromb Thrombolysis. 2020;50(2):298–301. doi: 10.1007/s11239-020-02162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S., Wang C., Wang H., Yang C., Cai F., Zeng F., et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: a retrospective clinical study. Clin Transl Sci. 2020;13(6):1087–1095. doi: 10.1111/cts.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S., Wang T., Li J.P., Sullivan M.A., Wang C., Wang H., et al. Comprehensive landscape of heparin therapy for COVID-19. Carbohydr Polym. 2021 doi: 10.1016/j.carbpol.2020.117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindahl U., Li J.P. Heparin - an old drug with multiple potential targets in Covid-19 therapy. J Thromb Haemostasis. 2020;18(9):2422–2424. doi: 10.1111/jth.14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thachil J. Clinical differentiation of anticoagulant and non-anticoagulant properties of heparin. J Thromb Haemostasis. 2020;18(9):2424–2425. doi: 10.1111/jth.14933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S.Y., Jin W.H., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020:181. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2020;95(3) doi: 10.1128/JVI.01987-20. e01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guimond S.E., Mycroft-West C.J., Gandhi N.S., Tree J.A., Le T.T., Spalluto C.M., et al. Synthetic heparan sulfate mimetic Pixatimod (PG545) potently inhibits SARS-CoV-2 by disrupting the spike-ace2 interaction. ACS Cent Sci. 2022;8(5):527–545. doi: 10.1021/acscentsci.1c01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertini S., Alekseeva A., Elli S., Pagani I., Zanzoni S., Eisele G., et al. Pentosan polysulfate inhibits attachment and infection by SARS-CoV-2 in vitro: insights into structural requirements for binding. Thromb Haemostasis. 2022;122(6):984–997. doi: 10.1055/a-1807-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Lin L., Huang H., Linhardt R.J. Chemoenzymatic synthesis of glycosaminoglycans. Acc Chem Res. 2020;53(2):335–346. doi: 10.1021/acs.accounts.9b00420. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Ochoa A.J., Raffetto J.D., Hernández A.G., Zavala N., Gutiérrez O., Vargas A., et al. Sulodexide in the treatment of patients with early stages of COVID-19: a randomized controlled trial. Thromb Haemostasis. 2021;121(7):944–954. doi: 10.1055/a-1414-5216. [DOI] [PubMed] [Google Scholar]
- 38.Gasbarri M., V'Kovski P., Torriani G., Thiel V., Stellacci F., Tapparel C., et al. SARS-CoV-2 inhibition by sulfonated compounds. Microorganisms. 2020;8(12):1894. doi: 10.3390/microorganisms8121894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorpes J.E., Gardell S. On heparin monosulfuric acid. J Biol Chem. 1948;176(1):267–276. [PubMed] [Google Scholar]
- 40.Cifonelli J.A., Dorfman A.J.B., Communications B.R. The uronic acid of heparin. 1962;7(1):41–45. doi: 10.1016/0006-291x(62)90141-9. [DOI] [PubMed] [Google Scholar]
- 41.Hippensteel J.A., LaRiviere W.B., Colbert J.F., Langouet-Astrie C.J., Schmidt E.P. Heparin as a therapy for COVID-19: current evidence and future possibilities. Am J Physiol Lung Cell Mol Physiol. 2020;319(2):211–217. doi: 10.1152/ajplung.00199.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop J.R., Schuksz M., Esko J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446(7139):1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Kusche Gullberg M. Heparan sulfate: biosynthesis, structure, and function. Int Rev Cell Mol Biol. 2016;325:215–273. doi: 10.1016/bs.ircmb.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Lin X., Wei G., Shi Z., Dryer L., Esko J.D., Wells D.E., et al. vol. 224. 2000. pp. 299–311. (Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice). 2. [DOI] [PubMed] [Google Scholar]
- 45.Bullock S., Fletcher J., Heyningen V.V., Beddington R., Wilson V. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulphate 2-sulphotransferase. J Genetics Research. 1998;12(12):1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringvall M., Ledin J., Holmborn K., Kuppevelt T.V., Forsberg E.J. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J Biol Chem. 2000;275(34):25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- 47.Li J.P., Gong F., Hagner-Mcwhirter A., Forsberg E., Abrink M., Kisilevsky R., et al. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking l-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278(31):28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 48.Lindahl U., Li J.P. Interactions between heparan sulfate and proteins—design and functional implications. Int Rev Cell Mol Biol. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- 49.Lindahl U., Kusche-Gullberg M., Kjellén L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273(39):24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 50.Raats C.J., Van Den Born J., Berden J.H. Glomerular heparan sulfate alterations: mechanisms and relevance for proteinuria. Kidney Int. 2000;57(2):385–400. doi: 10.1046/j.1523-1755.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Fuster M., Sriramarao P., Esko J.D. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6(9):902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X., Wang B., O'Callaghan P., Hjertström E., Jia J., Gong F., et al. Heparanase overexpression impairs inflammatory response and macrophage-mediated clearance of amyloid-β in murine brain. Acta Neuropathol. 2012;124(4):465–478. doi: 10.1007/s00401-012-0997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noborn F., O'Callaghan P., Hermansson E., Zhang X., Ancsin J.B., Damas A.M., et al. Heparan sulfate/heparin promotes transthyretin fibrillization through selective binding to a basic motif in the protein. Proc Natl Acad Sci U S A. 2011;108(14):5584–5589. doi: 10.1073/pnas.1101194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jendresen C.B., Cui H., Zhang X., Vlodavsky I., Nilsson L.N.G., Li J.P. Overexpression of heparanase lowers the amyloid burden in amyloid-β precursor protein transgenic mice. J Biol Chem. 2015;290(8):5053–5064. doi: 10.1074/jbc.M114.600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Escobar Galvis M.L., Jia J., Zhang X., Jastrebova N., Spillmann D., Gottfridsson E., et al. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 2007;3(12):773. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 56.Agelidis A., Shukla D. Heparanase, heparan sulfate and viral infection. Adv Exp Med Biol. 2020;1221:759–770. doi: 10.1007/978-3-030-34521-1_32. [DOI] [PubMed] [Google Scholar]
- 57.Rusnati M., Vicenzi E., Donalisio M., Oreste P., Landolfo S., Lembo D. Sulfated K5 Escherichia coli polysaccharide derivatives: a novel class of candidate antiviral microbicides. Pharmacol Ther. 2009;123(3):310–322. doi: 10.1016/j.pharmthera.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Cagno V., Tseligka E.D., Jones S.T., Tapparel C. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11(7):596. doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalrymple N., Mackow E.R. Productive dengue virus infection of human endothelial cells is directed by heparan sulfate-containing proteoglycan receptors. J Virol. 2011;85(18):9478–9485. doi: 10.1128/JVI.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donalisio M., Rusnati M., Civra A., Bugatti A., Allemand D., Pirri G., et al. Identification of a dendrimeric heparan sulfate-binding peptide that inhibits infectivity of genital types of human papillomaviruses. Antimicrob Agents Chemother. 2010;54(10):4290–4299. doi: 10.1128/AAC.00471-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cagno V., Donalisio M., Bugatti A., Civra A., Cavalli R., Ranucci E., et al. The agmatine-containing poly(amidoamine) polymer AGMA1 binds cell surface heparan sulfates and prevents attachment of mucosal human papillomaviruses. Antimicrob Agents Chemother. 2015;59(9):5250–5259. doi: 10.1128/AAC.00443-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sa-Carvalho D., Rieder E., Baxt B., Rodarte R., Tanuri A., Mason P.W. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J Virol. 1997;71(7):5115–5123. doi: 10.1128/jvi.71.7.5115-5123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zautner A.E., Körner U., Henke A., Badorff C., Schmidtke M. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection. J Virol. 2003;77(18):10071–10077. doi: 10.1128/JVI.77.18.10071-10077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kherad O., Kaiser L., Bridevaux P.O., Sarasin F., Thomas Y., Janssens J.P., et al. Upper-respiratory viral infection, biomarkers, and COPD exacerbations. Chest. 2010;138(4):896–904. doi: 10.1378/chest.09-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards M.R., Bartlett N.W., Hussell T., Openshaw P., Johnston S.L. The microbiology of asthma. Nat Rev Microbiol. 2012;10(7):459–471. doi: 10.1038/nrmicro2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tapparel C., Cordey S., Junier T., Farinelli L., Van Belle S., Soccal P.M., et al. Rhinovirus genome variation during chronic upper and lower respiratory tract infections. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bochkov Y.A., Watters K., Ashraf S., Griggs T.F., Devries M.K., Jackson D.J., et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112(17):5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tseligka E.D., Sobo K., Stoppini L., Cagno V., Abdul F., Piuz I., et al. A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeCaprio J.A., Garcea R.L. A cornucopia of human polyomaviruses. Nat Rev Microbiol. 2013;11(4):264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crim R.L., Audet S.A., Feldman S.A., Mostowski H.S., Beeler J.A. Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J Virol. 2007;81(1):261–271. doi: 10.1128/JVI.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilman M.S., Moin S.M., Mas V., Chen M., Patel N.K., Kramer K., et al. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog. 2015;11(7) doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghezzi S., Cooper L., Rubio A., Pagani I., Capobianchi M.R., Ippolito G., et al. Heparin prevents Zika virus induced-cytopathic effects in human neural progenitor cells. Antivir Res. 2017;140:13–17. doi: 10.1016/j.antiviral.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pierson T.C., Diamond M.S. The emergence of Zika virus and its new clinical syndromes. Nature. 2018;560(7720):573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 75.Kim S.Y., Koetzner C.A., Payne A.F., Nierode G.J., Yu Y., Wang R., et al. Glycosaminoglycan compositional analysis of relevant tissues in Zika virus pathogenesis and in vitro evaluation of heparin as an antiviral against zika virus infection. Biochemistry. 2019;58(8):1155–1166. doi: 10.1021/acs.biochem.8b01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shukla D., Liu J., Blaiklock P., Shworak N.W., Bai X., Esko J.D., et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99(1):13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 77.Birkmann A., Mahr K., Ensser A., Yağuboğlu S., Titgemeyer F., Fleckenstein B., et al. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J Virol. 2001;75(23):11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang F.Z., Akula S.M., Sharma-Walia N., Zeng L., Chandran B. Human herpesvirus 8 envelope glycoprotein B mediates cell adhesion via its RGD sequence. J Virol. 2003;77(5):3131–3147. doi: 10.1128/JVI.77.5.3131-3147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tyagi M., Rusnati M., Presta M., Giacca M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem. 2001;276(5):3254–3261. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- 80.Urbinati C., Nicoli S., Giacca M., David G., Fiorentini S., Caruso A., et al. HIV-1 Tat and heparan sulfate proteoglycan interaction: a novel mechanism of lymphocyte adhesion and migration across the endothelium. Blood. 2009;114(15):3335–3342. doi: 10.1182/blood-2009-01-198945. [DOI] [PubMed] [Google Scholar]
- 81.Bobardt M.D., Salmon P., Wang L., Esko J.D., Gabuzda D., Fiala M., et al. Contribution of proteoglycans to human immunodeficiency virus type 1 brain invasion. J Virol. 2004;78(12):6567–6584. doi: 10.1128/JVI.78.12.6567-6584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu W., Fu S., He Y., Li J., Liang G. Amino acid substitutions in the E2 glycoprotein of Sindbis-like virus XJ-160 confer the ability to undergo heparan sulfate-dependent infection of mouse embryonic fibroblasts. Virol J. 2010;7:225. doi: 10.1186/1743-422X-7-225. 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88(22):13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., et al. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Guimond S.E., et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. Thromb Haemostasis. 2020;120(12):1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Agostini A.I., Dong J.C., de Vantéry Arrighi C., Ramus M.A., Dentand-Quadri I., Thalmann S., et al. Human follicular fluid heparan sulfate contains abundant 3-O-sulfated chains with anticoagulant activity. J Biol Chem. 2008;283(42):28115–28124. doi: 10.1074/jbc.M805338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Petitou M., Hérault J.P., Bernat A., Driguez P.A., Herbert J.M.J.N. Synthesis of thrombin-inhibiting heparin mimetics without side effects. Nature. 1999;398(6726):417–422. doi: 10.1038/18877. [DOI] [PubMed] [Google Scholar]
- 89.Petitou M., Duchaussoy P., Herbert J.M., Duc G., El Hajji M., Branellec J.F.O., et al. The synthetic pentasaccharide fondaparinux: first in the class of antithrombotic agents that selectively inhibit coagulation factor Xa. Semin Thromb Hemost. 2002;28(4):393–402. doi: 10.1055/s-2002-34309. [DOI] [PubMed] [Google Scholar]
- 90.Barath M., Hansen S.U., Dalton C.E., Jayson G.C., Miller G.J., Gardiner J.M. Modular synthesis of heparin-related tetra-, hexa- and octasaccharides with differential O-6 protections: programming for regiodefined 6-O-modifications. Molecules. 2015;20(4):6167–6180. doi: 10.3390/molecules20046167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hansen S.U., Miller G.J., Cliff M.J., Jaysonc G.C., Gardiner J.M. Making the longest sugars: a chemical synthesis of heparin-related [4](n) oligosaccharides from 16-mer to 40-mer. Chem Sci. 2015;6(11):6158–6164. doi: 10.1039/c5sc02091c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy S., El Hadri A., Richard S., Denis F., Holte K., Duffner J., et al. Synthesis and biological evaluation of a unique heparin mimetic hexasaccharide for structure–activity relationship studies. J Med Chem. 2014;57(11):4511–4520. doi: 10.1021/jm4016069. [DOI] [PubMed] [Google Scholar]
- 93.Baytas S.N., Linhardt R.J. Advances in the preparation and synthesis of heparin and related products. Drug Discov Today. 2020;25(12):2095–2109. doi: 10.1016/j.drudis.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu L.D., Shie C.R., Kulkarni S.S., Pan G.R., Lu X.A., Hung S.C. Synthesis of 48 disaccharide building blocks for the assembly of a heparin and heparan sulfate oligosaccharide library. Org Lett. 2006;8(26):5995–5998. doi: 10.1021/ol062464t. [DOI] [PubMed] [Google Scholar]
- 95.Pawar N.J., Wang L., Higo T., Bhattacharya C., Hsieh-Wilson L.C. Expedient synthesis of core disaccharide building blocks from natural polysaccharides for heparan sulfate oligosaccharide assembly. Angew Chem Int Ed Engl. 2019;58(51):18577–18583. doi: 10.1002/anie.201908805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nahain A.A., Ignjatovic V., Monagle P., Tsanaktsidis J., Vamvounis G., Ferro V. Sulfonated RAFT copolymers as heparin mimetics: synthesis, reactivity ratios, and anticoagulant activity. Macromol Biosci. 2020;20(9) doi: 10.1002/mabi.202000110. [DOI] [PubMed] [Google Scholar]
- 97.Gottschalk J., Elling L. Current state on the enzymatic synthesis of glycosaminoglycans. Curr Opin Chem Biol. 2021;61:71–80. doi: 10.1016/j.cbpa.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 98.Lindahl U., Li J.P., Kusche-Gullberg M., Salmivirta M., Alaranta S., Veromaa T., et al. Generation of "neoheparin" from E. coli K5 capsular polysaccharide. J Med Chem. 2005;48(2):349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 99.Casu B., Grazioli G., Razi N., Guerrini M., Naggi A., Torri G., et al. Heparin-like compounds prepared by chemical modification of capsular polysaccharide from E. coli K5. Carbohydr Res. 1994;263(2):271–284. doi: 10.1016/0008-6215(94)00172-3. [DOI] [PubMed] [Google Scholar]
- 100.Datta P., Yan L., Awofiranye A., Dordick J.S., Linhardt R.J. Heparosan chain characterization: sequential depolymerization of E. coli K5 heparosan by a bacterial eliminase heparin lyase iii and a bacterial hydrolase heparanase Bp to prepare defined oligomers. Biotechnol J. 2021;16(3) doi: 10.1002/biot.202000336. [DOI] [PubMed] [Google Scholar]
- 101.Xu Y., Masuko S., Takieddin M., Xu H., Liu R., Jing J., et al. Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science. 2011;334(6055):498–501. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu J., Linhardt R.J. Chemoenzymatic synthesis of heparan sulfate and heparin. Nat Prod Rep. 2014;31(12):1676–1685. doi: 10.1039/c4np00076e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu Y., Pempe E.H., Liu J. Chemoenzymatic synthesis of heparin oligosaccharides with both anti-factor Xa and anti-factor IIa activities. J Biol Chem. 2012;287(34):29054–29061. doi: 10.1074/jbc.M112.358523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yi L., Xu Y., Kaminski A.M., Chang X., Pagadala V., Horton M., et al. Using engineered 6-O-sulfotransferase to improve the synthesis of anticoagulant heparin. Org Biomol Chem. 2020;18(40):8094–8102. doi: 10.1039/d0ob01736a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zha Z., Liu Y., Miao Y., Liao S., Wang S.Y., Tang H., et al. Preparation and characterization of 2-deacetyl-3-O-sulfo-heparosan and its antitumor effects via the fibroblast growth factor receptor pathw ay. Int J Biol Macromol. 2022;201:47–58. doi: 10.1016/j.ijbiomac.2021.12.098. [DOI] [PubMed] [Google Scholar]
- 106.Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12(5):381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 107.Jacobsson I., Lindahl U. Biosynthesis of heparin. Concerted action of late polymer-modification reactions. J Biol Chem. 1980;255(11):5094–5100. [PubMed] [Google Scholar]
- 108.Lohse D.L., Linhardt R.J. Purification and characterization of heparin lyases from Flavobacterium heparinum. J Biol Chem. 1992;267(34):24347–24355. [PubMed] [Google Scholar]
- 109.Zhang C., Liu L., Teng L., Chen J., Liu J., Li J., et al. Metabolic engineering of Escherichia coli BL21 for biosynthesis of heparosan, a bioengineered heparin precursor. Metab Eng. 2012;14(5):521–527. doi: 10.1016/j.ymben.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 110.Jin P., Zhang L., Yuan P., Kang Z., Du G., Chen J. Efficient biosynthesis of polysaccharides chondroitin and heparosan by metabolically engineered Bacillus subtilis. Carbohydr Polym. 2016;140:424–432. doi: 10.1016/j.carbpol.2015.12.065. [DOI] [PubMed] [Google Scholar]
- 111.Sarnaik A., Abernathy M.H., Han X., Ouyang Y., Xia K., Chen Y., et al. Metabolic engineering of cyanobacteria for photoautotrophic production of heparosan, a pharmaceutical precursor of heparin. Algal Res. 2019;37:57–63. [Google Scholar]
- 112.Williams A., Gedeon K.S., Vaidyanathan D., Yu Y., Collins C.H., Dordick J.S., et al. Metabolic engineering of Bacillus megaterium for heparosan biosynthesis using Pasteurella multocida heparosan synthase, PmHS2. Microb Cell Fact. 2019;18(1):132. doi: 10.1186/s12934-019-1187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nehru G., Tadi S.R.R., Limaye A.M., Sivaprakasam S. Production and characterization of low molecular weight heparosan in Bacillus megaterium using Escherichia coli K5 glycosyltransferases. Int J Biol Macromol. 2020;160:69–76. doi: 10.1016/j.ijbiomac.2020.05.159. [DOI] [PubMed] [Google Scholar]
- 114.Roy A., Miyai Y., Rossi A., Paraswar K., Desai U.R., Saijoh Y., et al. Metabolic engineering of non-pathogenic Escherichia coli strains for the controlled production of low molecular weight heparosan and size-specific heparosan oligosaccharides. Biochim Biophys Acta Gen Subj. 2021;1865(1) doi: 10.1016/j.bbagen.2020.129765. [DOI] [PubMed] [Google Scholar]
- 115.Van Dijl J.M., Hecker M. Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Factories. 2013;12:3. doi: 10.1186/1475-2859-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Z., Dordick J.S., Linhardt R.J. Escherichia coli K5 heparosan fermentation and improvement by genetic engineering. Bioeng Bugs. 2011;2(1):63–67. doi: 10.4161/bbug.2.1.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Z., Ly M., Zhang F., Zhong W., Suen A., Hickey A.M., et al. E. coli K5 fermentation and the preparation of heparosan, a bioengineered heparin precursor. Biotechnol Bioeng. 2010;107(6):964–973. doi: 10.1002/bit.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oduah E.I., Linhardt R.J., Sharfstein S.T. Heparin: past, present, and future. Pharmaceuticals. 2016;9(3):38. doi: 10.3390/ph9030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bioworks Ginkgo. 2022. Optimvia and ginkgo bioworks announce partnership to manufacture biosynthetic heparin.https://www.prnewswire.com/news-releases/optimvia-and-ginkgo-bioworks-announce-partnership-to-manufacture-biosynthetic-heparin-301455332.html [Google Scholar]
- 120.Digital Journal . 2022. Heparin market growth to remain stable during the projection period 2019- 2027.https://www.digitaljournal.com/pr/heparin-market-growth-to-remain-stable-during-the-projection-period-2019-2027 [Google Scholar]
- 121.Gao Q., Tan G.-Y., Xia X., Zhang L. Learn from microbial intelligence for avermectins overproduction. Curr Opin Biotechnol. 2017;48:251–257. doi: 10.1016/j.copbio.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 122.Li S., Wang J., Li X., Yin S., Wang W., Yang K. Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microb Cell Factories. 2015;14:172. doi: 10.1186/s12934-015-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen J., Liu M., Liu X., Miao J., Fu C., Gao H., et al. Interrogation of Streptomyces avermitilis for efficient product ion of avermectins. Synth Syst Biotechnol. 2016;1(1):7–16. doi: 10.1016/j.synbio.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tadmor B., Tidor B. Interdisciplinary research and education at the biology-engineering-co mputer science interface: a perspective. Drug Discov Today. 2005;10(17):1183–1189. doi: 10.1016/S1359-6446(05)03540-3. [DOI] [PubMed] [Google Scholar]
- 125.Cao Z., Yu J., Wang W., Lu H., Xia X., Xu H., et al. Multi-scale data-driven engineering for biosynthetic titer improvement. Curr Opin Biotechnol. 2020;65:205–212. doi: 10.1016/j.copbio.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 126.Wilson I.B. The never-ending story of peptide O-xylosyltransferase. Cell Mol Life Sci. 2004;61(7–8):794–809. doi: 10.1007/s00018-003-3278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Almeida R., Levery S.B., Mandel U., Kresse H., Schwientek T., Bennett E.P., et al. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J Biol Chem. 1999;274(37):26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 128.Bai X., Zhou D., Brown J.R., Crawford B.E., Hennet T., Esko J.D. Biosynthesis of the linkage region of glycosaminoglycans: cloning and activity of galactosyltransferase II, the sixth member of the beta 1,3-galactosyltransferase family (beta 3GalT6) J Biol Chem. 2001;276(51):48189–48195. doi: 10.1074/jbc.M107339200. [DOI] [PubMed] [Google Scholar]
- 129.Kitagawa H., Shimakawa H., Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J Biol Chem. 1999;274(20):13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- 130.Zak B.M., Crawford B.E., Esko J.D. Hereditary multiple exostoses and heparan sulfate polymerization. Biochim Biophys Acta. 2002;1573(3):346–355. doi: 10.1016/s0304-4165(02)00402-6. [DOI] [PubMed] [Google Scholar]
- 131.Lindahl U., Li J.P. Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- 132.Jacobsson I., Lindahl U., Jensen J.W., Rodén L., Prihar H., Feingold D.S. Biosynthesis of heparin. Substrate specificity of heparosan N-sulfate D-glucuronosyl 5-epimerase. J Biol Chem. 1984;259(2):1056–1063. [PubMed] [Google Scholar]
- 133.Rong J., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry. 2001;40(18):5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 134.Habuchi H., Miyake G., Nogami K., Kuroiwa A., Matsuda Y., Kusche-Gullberg M., et al. Biosynthesis of heparan sulphate with diverse structures and functions: two alternatively spliced forms of human heparan sulphate 6-O-sulphotransferase-2 having different expression patterns and properties. Biochem J. 2003;371(Pt 1):131–142. doi: 10.1042/BJ20021259. [DOI] [PMC free article] [PubMed] [Google Scholar]