Abstract
This study aims to fully exploit the natural compound; bee venom (BV) as a substance that can kill and inhibit the growth of microbes and viruses. For this target, BV was loaded onto a safe, natural, and economically inexpensive polymer; chitosan (Ch) in its nano-size form prepared using ionic gelation method in the presence of chemical crosslinking agent (sodium tripolyphosphate; TPP). The findings illustrated that chitosan nanoparticles (ChNPs) were prepared thru this method and exhibited spherical shape and average hydrodynamic size of 202 nm with a polydispersity index (PDI = 0.44). However, the size was increased to 221 nm with PDI (0.37) when chitosan nanoparticles were loaded with BV (ChNC). In addition, the particles of BV appeared as a core and chitosan nanoparticles as a shell implying the successful preparation of nanocomposite (ChNC). Encapsulation of BV into ChNPs with significantly small size distribution and good stability that protect these formed nanocomposites from agglomeration. The cytopathic effect (CPE) inhibition assay was used to identify potential antivirals for Middle East respiratory syndrome coronavirus (MERS-CoV). The response of the dose study was designed to influence the range of effectiveness for the chosen antiviral, i.e., the 50 % inhibitory concentration (IC50), as well as the range of cytotoxicity (CC50). However, our results indicated that crude BV had mild anti-MERS-COV with selective index (SI = 4.6), followed by ChNPs that exhibited moderate anti-MERS-COV with SI = 8.6. Meanwhile. The nanocomposite of ChNC displayed a promising anti-MERS-COV with SI = 12.1. Additionally, the synthesized nanocomposite (ChNC) had greater antimicrobial activity against both Gram-positive and Gram-negative bacteria when compared with ChNPs, BV or the utilized model drug.
Keywords: Chitosan nanoparticles, Bee venom, MERS, MDR bacteria
1. Introduction
Nanotechnology refers to the study, production, and synthesis of different components thru the manipulation of the material at the nanoscale scale [1]. Drug formulations of nanoparticles have a very important role in drug delivery [2]. Modern science in the delivery of a drug is focused on polymers and their kinds as clear biological functions. Chitosan nanoparticles (ChNPs) are one of the expectations for this design, depending on the kind of nanoparticles used in treatment [3]. However, particles of colloidal that can be trapped, dissolved, absorbed, or encapsulated, into polymer matrix might be used in drug carriers or vaccines [4].
Nanomedicine is new approach and very important role in preventing and treating coronavirus [5], [6]. Birds, some mammals, and humans are all primarily affected by gastrointestinal and respiratory infections brought on by coronaviruses. But a lot of species can lead to serious conditions like neurological, hepatitis, or peritonitis disease. Seven coronaviruses can infect people, and four of them the human coronaviruses including HcoV-NL63, HcoV-OC43, HcoV HKU1, and 229E can cause minor upper and lower respiratory tract infections. SARS-CoV-2 and SARS-CoV, as well as MERS-CoV (Middle East respiratory syndrome coronavirus), are 3 more zoonotic coronaviruses that are linked to severe, life-threatening organ dysfunction and respiratory problems. [7], [8], [9]. Similar to SARS-CoV-2, MERS-CoV also includes several bat viruses. Bind the MERS-COV instance, bats are thought to have served as the initial reservoir [10], but, the intermediate host of MERS-COV is defined as a camel [11]. The human respiratory disease caused by MERS-CoV ranges in severity from mild to potentially deadly acute respiratory failure. [12], [13], [14]. There have been >2000 instances of the MERS-CoV infection documented to date, with a death rate of 35 %.
Microbes acquired the capability to create biofilms set in an extracellular matrix that are more resistive to and more difficult for antibiotics to penetrate as a result of the inappropriate and excessive use of antibiotics. The development of drugs resistant to the microbial effect stimulates the search for new alternatives with unique action modes. Both chitosan (Ch) as linear natural polysaccharide (Fig. S1a) and bee venom (BV) and have shown very promising and powerful properties as alternatives to antibiotics. Bee venom has been reported to have multiple effects as such s anti-inflammation, antibacterial, and antivirus in various types of cells. BV is a complex combination of active peptides, amines, and enzymes (Fig. S1b). Chitosan has fundamental biological features, including such antioxidant, anti-inflammatory, and antimicrobial activities. The biocompatibility, biodegradability, and its non-toxicity of chitosan [17], [18], [19] facilitate its application in the nanoform as a carrier for BV. The most acceptable antimicrobial mechanism of BV is to form pores in the cell membrane which resulting increases in cell permeability [24], while in chitosan attributes to their ionic interactions of charged groups located onto the polymer molecules with the walls of bacteria that causes hydrolysis of the peptidoglycans and seepage of intracellular electrolytes, and finally, leading to the microorganism death [25].
The aim of the work was designed to prepare nanocomposite (ChNC) based on the encapsulation of bee venom (BV) into the environmentally biodegradable chitosan nanoparticles (ChNPs) thru the ionic gelation method using sodium tripolyphosphate as crosslinking agent. The whole nanocomposite was assessed using TEM, DLS, zeta potential and FTIR analysis. The work was extended to evaluate the effect of nanocomposite as antiviral (anti-MERS-COV) and antimicrobial (multi-drug resistance bacteria) agents.
2. Material and methods
2.1. Materials
Sodium tripolyphosphate (TPP) as crosslinking agent and chitosan (low molecular weight, purity >99 %) were purchased from Sigma Aldrich Co. USA. Bee venom (BV) was obtained from the plant protection Dep., Faculty of Agriculture, Al-azhar University, Cairo, Egypt. Middle East respiratory syndrome coronavirus (MERS-CoV) antivirals were donated by Nawah-Scientific in Egypt. Both Gram-negative bacteria (Escherichia coli; ATCC 8739 and Pseudomonas aeruginosa; ATCC 9027) and Gram-positive bacteria (Bacillus subtilis; ATCC 6633 and Staphylococcus aureus; ATCC7984) were used for nanocomposite evaluation.
2.2. Methods
2.2.1. Preparation of bee venom-loaded chitosan nanoparticles (ChNC)
Chitosan nanoparticles (ChNPs) were primarily created via the ionic gelation process, which involved mixing sodium tripolyphosphate (TPP) anions with chitosan cations. To make 50 mL of chitosan solution, chitosan (1.5 mg/mL) was dissolved in an aqueous solution of acetic acid (0.5 mL and 49.5 mL H2O) and maintained for 120 min at room temperature under mechanical stirring. On the other hand, sodium tripolyphosphate (TPP, 0.75 mg/mL; 20 mL) was dissolved in water and agitated for 30 min before adding it to chitosan solution. To chitosan solution, TPP solution (10 mL) was then added. Rapid mixing with high-speed ultrasound at room temperature for 60 min caused nanoparticles to develop on their own, allowing the nanoparticulate system to be completely stabilized and an opalescent suspension to be produced. The nanoparticles were then lyophilized at −80 °C and kept in the refrigerator after being separated by a centrifuge at 12,000 rpm and 10 °C for 120 min. Bee venom (BV) at various concentrations (1, 2, and 4 mg/mL) were added to TPP solution and then, added to chitosan solution in order to prepare BV loaded chitosan nanoparticles (ChNC).
2.2.2. Characterization of ChNPs and ChNC
The particle shapes of chitosan nanoparticles (ChNPs) and BV loaded ChNPs (ChNC) were assessed using Transmission electron microscopy (TEM, JEOL, Japan). The produced nanoparticle solution was first dropped onto a carbon-coated grid, dyed with phospho-tungamic acid, let to air dry, and then put into TEM instrument for analysis. Based on the dynamic light scattering (DLS) method, the average diameter, and polydispersity index (PDI) of ChNPs and ChNC were assessed by Zeta sizer (Malvern Instruments, UK). The same device was used to test the zeta potential of freshly produced nanoparticles. The chemical structure of BV, Ch, ChNPs, and ChNC were conducted using FTIR (PerkinElmer, Germany) Spectrophotometry at wavenumbers of 400–4000 cm−1, and their FTIR spectra were plotted.
2.2.3. Anti-MERS-COV activity
The cytopathic effect (CPE) inhibition assay was utilized to identify the potential for MERS-CoV. The goal of the dosage response study was to adjust the IC50 (50 percent inhibitory concentration) range for the selected antiviral as well as the cytotoxicity range (CC50). This test is essential for assessing the effectiveness of antivirals in cell culture systems.
MERS-CoV antivirals were provided by the Egyptian company Nawah-Scientific, and Vero E6 cells were grown in Dulbecco's Modified Eagle (DMEM) Medium with fetal bovine serum (10 %) and antimicrobials solution (0.1 %). Gibco BRL provided the trypsin-EDTA, fetal bovine serum, antimycotic solution and antimicrobial, and DMEM medium (Grand Island, NY, USA). The recently revealed cytopathic inhibitory effect was used to evaluate antiviral activity and cytotoxicity experiments utilizing the crystal violet method [26]. In brief, Vero E6 cells were seeded into well culture plate (96) at a density for cells/well equals 2 × 104 one day before infection. The next day, the cells were rinsed with phosphate-buffered saline after the culture media was withdrawn. The crystal violet method, which tracked CPE and allowed the percentage of cell viability to be measured, was used to assess the infectivity of the MERS-CoV virus. Mammalian cells were infected with diluted MERS-CoV viral suspension (0.1 mL) containing CCID50 (1.0 × 104) virus stock. This dose was chosen so that two days after infection, the appropriate CPEs would be produced. To treat the cells with a drug, 0.01 mL of media having the required compound concentration was added. The antiviral potency of each test sample was assessed at doses that were two times diluted, starting at 1000 g/mL. The virus controls were nondrug-treated cells, and virus-infected. Meanwhile, the cell controls were nondrug treated cells, and non-infected, and. For 72 h, culture plates were incubated at 37 °C in CO2 (5 %). The development of the cytopathic effect was monitored by light microscopy. Following a PBS wash, the cell monolayers were fixed and stained with a 0.03 % crystal violet solution in ethanol (2 %) and formalin (10 %). After washing and drying the optical density of individual wells was quantified spectrophotometrically at 570/630 nm. According to Pauwels et al. [27] the percentages of antiviral activity of the substances tested were determined by using the following equation:
Considering the above results, CPE inhibitory dosage (IC50) of 50 % was assessed. Before this assay, we assessed the cytotoxicity according to Vanicha and Kanyawim [4]. In a 96-well culture plate, cells were planted at a density of 2 × 104 cells/well. The next day, the cells were cultured in growth media containing serially diluted samples for 72 h before being removed and rinsed with buffer (PBS). The subsequent steps were carried out in the same sequence as those for the antiviral activity assay. The findings of CC50 and IC50 were calculated using Graph-Pad Software, San Diego (USA).
2.2.4. Evaluation of antibacterial affectivity
The antibacterial measurements were performed using two methods.
2.2.4.1. Agar-well diffusion method for inhibition zone (IZ) determination
The agar-well diffusion test described by Hsouna et al. [28] was used with a slightly modified technique. In brief, about 20 mL of Mueller Hinton agar (MHA) medium at 45 °C were poured into 10 mL petri plates and then seeded with a 24 h culture of the bacterial strains. About 100 μL of a bacterial suspension with approximately 108 CFU/mL was inoculated spreading on the entire surface of agar utilizing a sterile spreader, once the plates were dried aseptically, wells with diameter (6 mm) were cut into the agar and 50 μL of BV, ChNPs and ChNc were tested. A standard drug (tetracycline 500 μg/mL) was used for comparison of the antibacterial behavior. The plates were left at +4 °C for 2 h to assist the diffusion of the tested samples in the agar [29]. For 24 h, incubation was carried out at 37 °C. The evaluation of bactericidal property was based on the diameter of the inhibition zone (IZ) which formed around the well and the mean value was taken for three replicate experiments.
2.2.4.2. Macro-dilution method
To determining, the MIC of the BV, ChNPs and ChNc, standard broth dilution method (Clinical and Laboratory Standards Institute, CLSI M07-A9) was used by estimating the apparent growth of the bacteria in Mueller Hinton broth (MHB) medium. Firstly, serial two-fold, dilutions of BV, ChNPs and ChNc in concentrations ranging from 500 to 7.8 μg/mL in a liquid growth medium distributed in tubes with about 2 mL (macro-dilution). Then, each tube is inoculated with a bacterial inoculum adjusted to (108 CFU/mL, 0.5 McFarland's standard), and the bacterial suspension was prepared in the same medium. One tube has only inoculated a broth is considered as control tube and incubated for 24 h at 37 °C. The minimum inhibitory concentration (MIC) endpoint is the lowest concentration of the tested specimens where no visible growth is seen in the tubes [30]. After broth macro-dilution, the minimum bactericidal concentration (MBC) can be determined by sub-culturing samples from tubes that gave a negative bacterial growth on the surface of MHA plates for 24 h incubation and then counting the number of viable cells (CFU/mL). The bactericidal (MBC) is endpoint described as the lowest concentration; at which the final bacterial inoculum is killed by about 99.9 % [30]. After MIC determination for each sample, the fractional inhibitory concentration index (FICI) can be calculated by using the following formula:
In which, .
According to Bassolé and Juliani [31], synergism if FICI ≤0.5; antagonism if FICI >4.0; interpreted if FICI 0.5–1; indifference if FICI 1–4.0.
3. Results and discussion
3.1. Characterization of chitosan nanoparticles loaded with bee venom (ChNC)
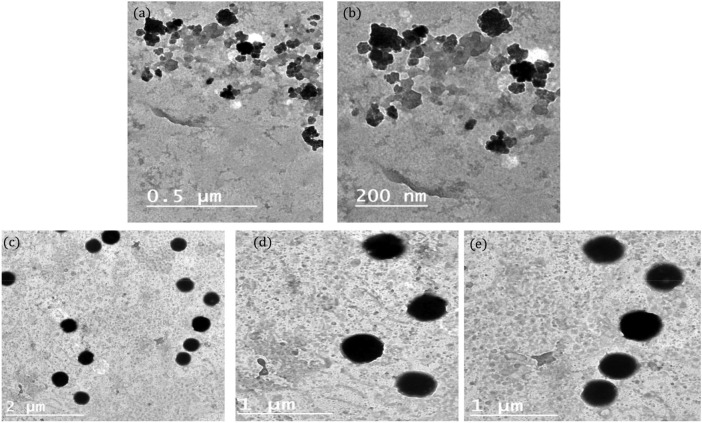
Based on the cytotoxic data, chitosan nanoparticles that loaded with low concentration of BV (1 mg/mL) were selected for further characterization and application. Using a transmission electron microscope (TEM), ChNPs and ChNC were created and analyzed at different magnifications. ChNPs exhibited spherical shapes and smooth surfaces but with no clear edges (Fig. 1 a, b). On the other hand, after BV loading, TEM demonstrated the produced nanocomposite's core shell. The black spots are associated with the encapsulated BV, while the faint particles could be ChNPs (Fig. 1c, d). The majority of the nanoconjugate particles seemed to be spherical, with smooth surfaces and homogeneous distribution.
Fig. 1.
TEM at different magnifications of (a, b) ChNPs, and (c, d) ChNC.
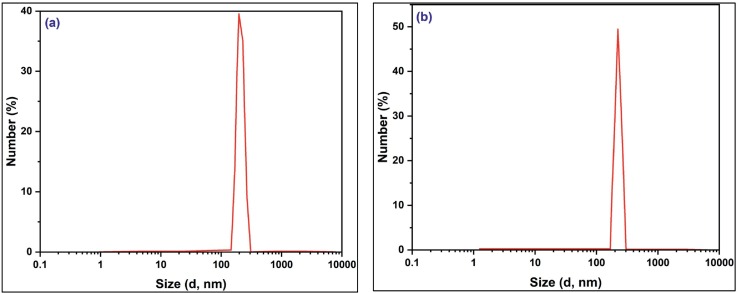
The finding indicated that ChNPs have 202 nm in size, with a PDI of 0.44, as shown in Fig. 2a. As observed, the PDI of ChNPs was found to be <0.5, confirming the homogeneity of the produced nanoparticles. Meanwhile. The average hydrodynamic size of ChNC was 221 nm, with a PDI of 0.37 (Fig. 2b). It was noted that the encapsulation of BV increased the average size while improving PDI. The obtained data from DLS about the synthesized ChNPs and ChNC showed that BV loaded ChNPs had larger sizes than ChNPs; this may be because ChNC have more molecular weight and a composited structure after being encapsulated with BV, which may increase their size. As known, zeta potential value tells us about the stability and homogenoty of the produced nanoparticles. Via measuring the surface charge of ChNPs and ChNC, it is slightly decreased from 36 mV to 34.7 mV, suggesting that the impact of the TPP crosslinking agent and the entire process of ionic gelation is what makes the formed nanoparticles of chitosan and chitosan loaded with BV stable and protected from agglomeration. By and large, the production of nanocomposites with a size around 200 nm, and above 30 mV (regarding zeta potential; surface charge), is suitable for the effective biomedical applications since the small particles may reach any portion of the body.
Fig. 2.
Average hydrodynamic size using dynamic light scattering (DLS) for (a) ChNPs, and (b) ChNC.
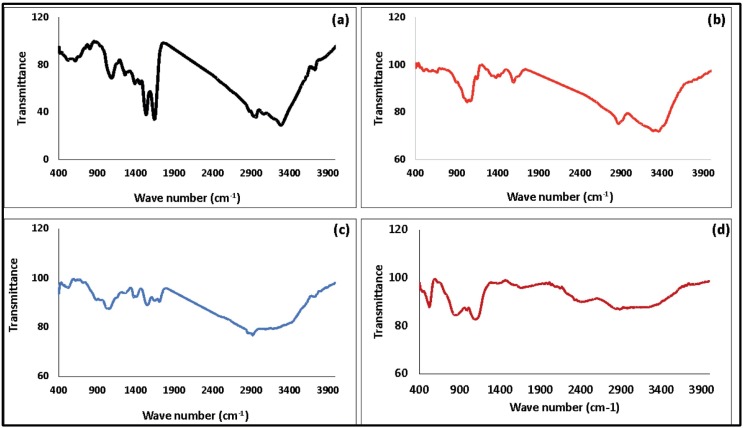
Using FTIR to analyze BV-ChNPs interactions, the ability of the ionic gelation procedure to create ChNC was evaluated. Fig. 3 displays the FTIR spectra of BV, Ch, ChNPs, and ChNC. The detected peak in the absorbance range of 3250–3450 cm−1 in the BV spectrum (Fig. 3a) denotes the free vibrations of N—H stretching. The distinctive amide bands, including amide I (1645 cm−1), amide II (1534 cm−1), and the bands assigned at 1104 cm−1 and 1240 cm−1 that denote unsystematic coil conformation, were also revealed in the FTIR spectrum of BV.
Fig. 3.
FTIR of (a) BV, (b) Ch, (c) ChNPs, and (d) ChNC.
The prominent peak appeared in 3200–3411 cm−1 region in the chitosan spectra (Fig. 3b) corresponds to coupled peaks of O—H stretching and intermolecular hydrogen bonding. In the same area, the NH extending from primary amines overlapped. The secondary amide's carbonyl (C=O) stretching absorption band (amide I band) has a peak at 1027 cm−1, which is associated with the C-O-C stretching. The spectra of ChNPs and ChNC were different. The peak of 3400 cm−1 in ChNPs (Fig. 3c) widened and the relative intensity rose, suggesting an improvement in hydrogen bonding. Additionally, the NH2 bending vibration's 1610 cm−1 peak changed to 1563 cm−1. A P O peak assigned 1164 cm−1 on the cross-linked chitosan further demonstrated the cross-linkage with TPP. By comparing with the spectrum of pure chitosan nanoparticles, the spectrum of ChNC (Fig. 3d) did not exhibit any differences.
3.2. Anti-MERS-COV activity
Several studies have proven the efficacy of different natural products and nanoparticles as therapy strategies for severe illnesses. However, only a few classes have been actively employed for antiviral therapeutic targeting. Based on our previous work on the conjugation of bee venom as a natural product and ChNPs, a reasonable hypothesis would be to start investigating the features and benefits of BV and ChNPs as antiviral, as well as chitosan as a nano-carrier for antiviral drug delivery in the fight against the Middle East respiratory syndrome coronavirus (MERS-CoV).
It is well understood that when the IC50 concentration decreases than the CC50 concentration, the virus is destroyed before causing any harm to host cells.
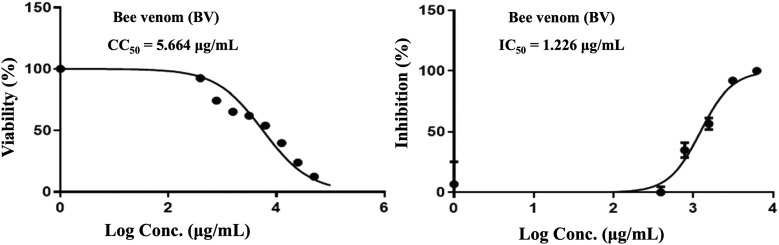
Our results indicated that, crude BV has mild antiviral activity against MERS-CoV, with an IC50 (1.226 g/mL) higher than the CC50 (5.664 g/mL) and SI of 4.6 (Table 1 and Fig. 4 ) [32]. Melittin has the ability to pierce virus protective membrane envelopes, particularly those protecting the HIV [33].
Table 1.
Antiviral activity of bee venom (BV), ChNPs and ChNC against human coronavirus MERS-CoV.
| Treatment compounds | Virus | CC50 (μg/mL) | IC50 (μg/mL) | Selective index (*SI) |
|---|---|---|---|---|
| BV | MERS-CoV | 5.664 | 1.226 | 4.6 |
| ChNPs | 11.921 | 1.382 | 8.6 | |
| ChNC | 14.106 | 1.165 | 12.1 |
Fig. 4.
Effect of bee venom (BV) on coronavirus MERS-CoV.
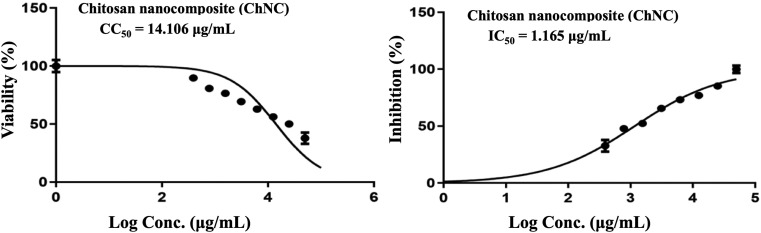
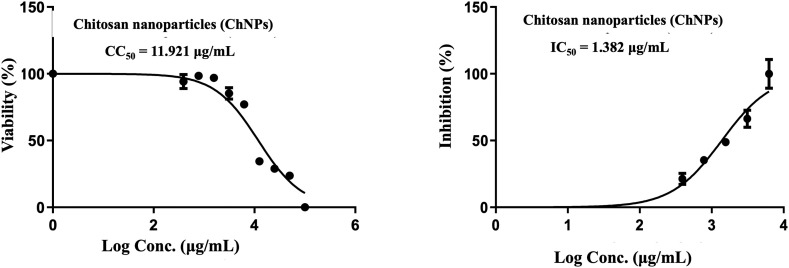
Chitosan nanoparticles (ChNPs) had a CC50 of 11.921 g/mL, an IC50 of 1.382 g/mL, and SI of 8.6 (Table 1 and Fig. 5 ). This indicates that it has moderate antiviral action against MERS-COV coronavirus, and more research may be done to increase its anti-coronavirus activity. So we suggest that the utilization of ChNPs as a carrier for bee venom led to enhance the properties of BV which, in turn, increases the antiviral activities against MERS-COV. This idea is in line with other studies that shown how ChNPs enhance the qualities of natural products and deliver them directly to the lungs. Thus, our results revealed that ChNC had a cytotoxic concentration (CC50) of 14.106 g/mL, an IC 50 of 1.165 g/mL, and a selective index (SI) of 12.1 (Table 1 and Fig. 6) which affirmed the importnace role of ChNPs as natural carrier for BV. It was also depicted that ChNC provides a large surface area, which is a benefit for them and enables the use of ChNPs as a carrier for BV for treating MERS [3], [35] (Fig. 7).
Fig. 6.
Effect of ChNC on coronavirus MERS-CoV.
Fig. 5.
Effect of chitosan nanoparticles (ChNPs) on coronavirus MERS-CoV.
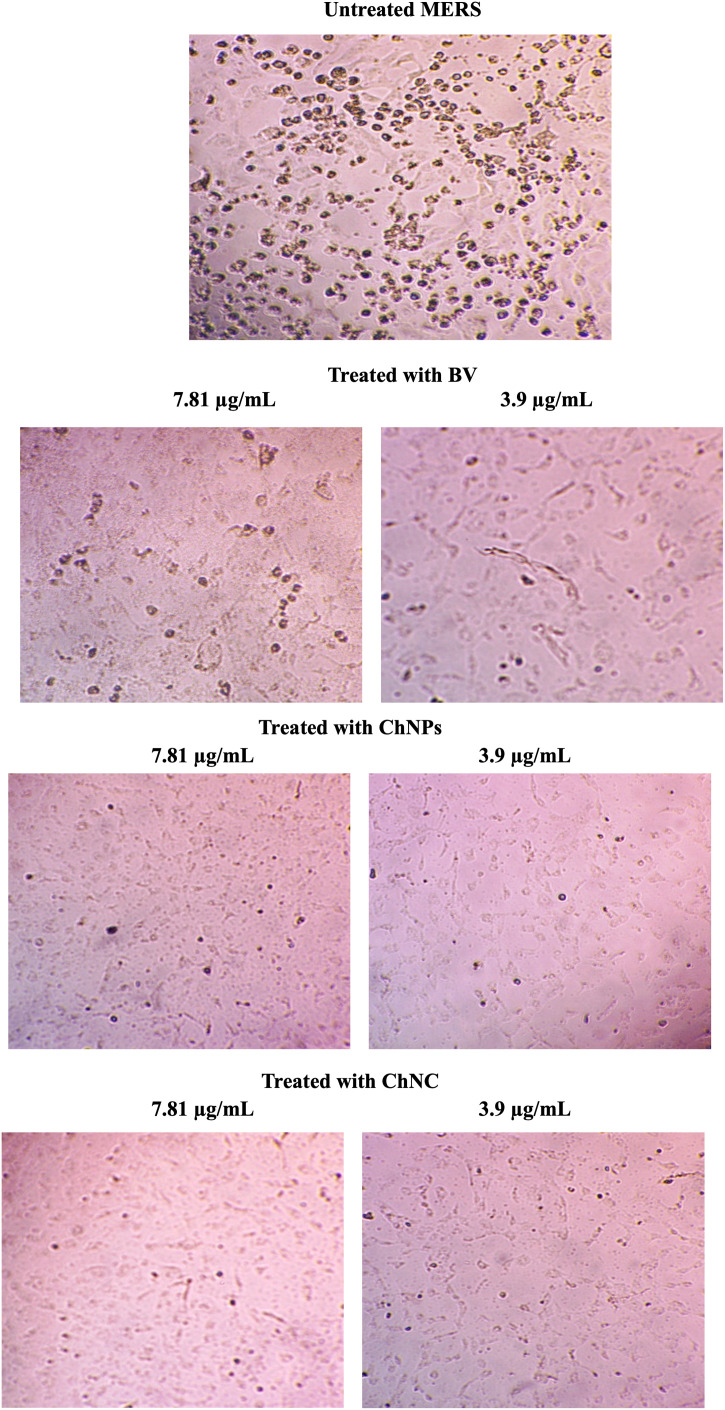
Fig. 7.
The antiviral effect CPE of Vero E6 cells infected with untreated or treated (MNTD of BV, ChNPs or ChNC) MERS-CoV.
3.3. Bactericidal and antiviral activities of BV, ChNPs and ChNC
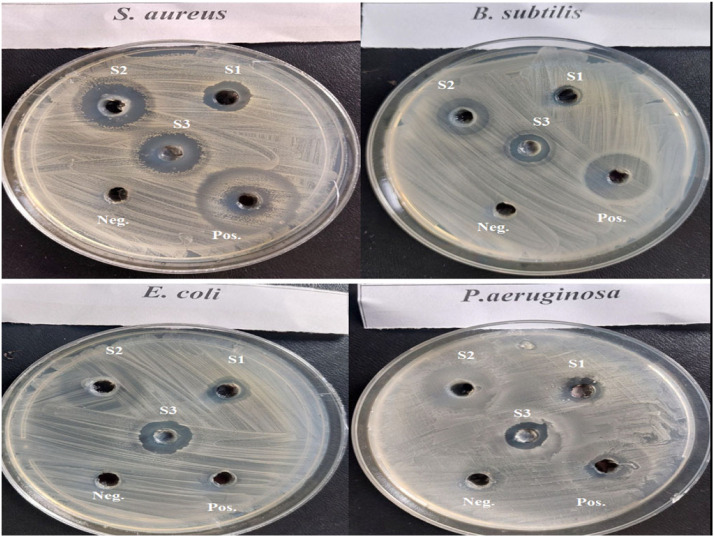
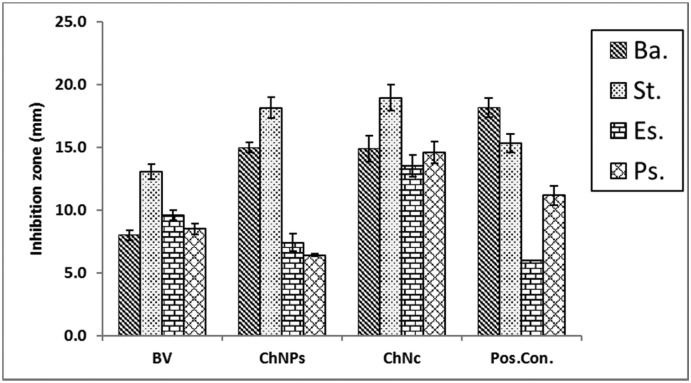
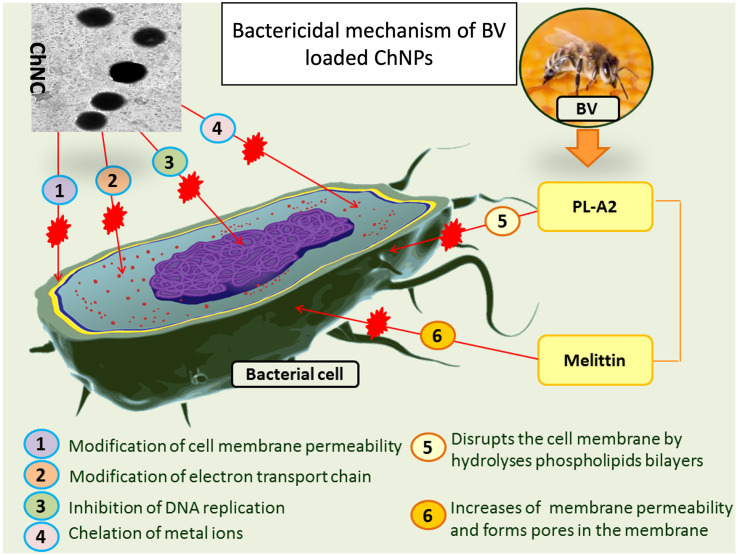
The bacterial behavior of BV, ChNPs and ChNC were estimated against the mentioned bacteria using the well diffusion approach. The sensitivity of the bacteria was qualitatively determined by measuring the inhibition zone (IZ) which are depicted in Fig. 8, Fig. 9 and reveal that, the strong effect of BV is obviously for Gram positive strain (S. aureus) which had significant higher IZ compared to other strains, and this is due to melittin in honey bee venom, which has greater efficacy towards Gram-positive bacteria than Gram-negative ones. Ortel and Markwardt [36] found that, low concentrations of BV have higher effect on Gram positive bacteria. The elevating values of MIC for BV (125 to 500) is agreed with El-Bahnasy et al. [37] which reported that, elevating the levels of BV appeared to be has very efficiency against both of Gram negative and positive organisms. Also, the ATP level of E. coli was significantly decreased when subjected to about 500 μg/mL of BV [38]. The two main components that give BV its antibacterial properties are melittin and phospholipase A2 (PLA2), where both of them affecting cell membrane permeability (Fig. 10). Melittin integrates into the phospholipid bilayers and forming pores in the membrane, this results in the breakdown of phospholipid groups or the release of Ca++ [39], while PLA 2 hydrolyses phospholipids, so indirectly damages the bacterial cell membrane [40].
Fig. 8.
Antimicrobial activity of BV, ChNPs and ChNC compared to reference drug against both Gram-positive and Gram-negative strains.
*S1; BV, S2; ChNPs, S3; ChNC, positive control; tetracycline and negative control; saline (0.85 %).
Fig. 9.
Antimicrobial activity of BV, ChNPs, and ChNC then compared to reference drug against both of Gram positive and Gram negative strains.
*Ba; Bacillus subtilis, St; Staphylococcus aureus, Es; Escherichia coli, Ps; Pseudomonas aeruginosa, BV; bee venom, ChNPs; chitosan nanoparticles, ChNC; nanocomposite, positive control; tetracycline.
Fig. 10.
Mechanism suggestion of BV loaded ChNPs as anti-microbial agent.
Additionally, compared to Gram-negative strains, Gram-positive strains were more sensitive to ChNPs. while all the tested strains were sensitive to ChNC. Several authors reported the same finding [41], while in some published papers, the researchers reported that, unmodified chitosan mostly more effective on Gram negative than Gram positive strains [42]. The mechanisms by which ChNPs affecting bacterial cell are summarized in Fig. 9. The antimicrobial activity of ChNC was higher than standard drug for all strains except for S. aureus. Our data also revealed that, the minimum inhibitory concentration (MIC) for BV ranged from 125 to 500, ChNPs from 15.6 to 500 and ChNC from 15.6 to 125 μg/mL, and also, S. aureus recorded the lowest MIC values for all the tested samples (Table 2, Table 4 ). The minimum bactericidal concentration (MBC) for BV ranged from 125 to 500 μg/mL and from 62.5 to 500 for both of ChNPs and ChNc (Table 3, Table 4 ). Standard drug displayed that MIC and MBC were around 500 μg/mL. The combination between BV and ChNPs was expressed as fractional inhibitory concentration index (FICI) and fractional bactericidal concentration index (FBCI) which recorded in Table 4. By calculating the fractional inhibitory concentration index (FICI), it is clear that, FICI was 2.5, 4.51, 1.0 and 0.5 for B. subtilis, S. aureus, E. coli and P. aeruginosa respectively, and according to Bassolé and Juliani [31], FICI showed synergism effect on P. aeruginosa, antagonism effect on S. aureus and indifference effect on both of E. coli and B. subtilis.
Table 2.
Minimum inhibitory concentration (MIC), turbidity for different concentrations of BV, ChNPs, ChNC and tetracycline after 24 h.
| Strains | 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 |
|---|---|---|---|---|---|---|---|
| BV concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| E. coli | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| P. aeruginosa | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| ChNPs concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | Pos. |
| E. coli | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| P. aeruginosa | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| ChNC concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Neg. | Neg. | Neg. | Neg. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | Pos. |
| E. coli | Neg. | Neg. | Neg. | Neg. | Neg. | Pos. | Pos. |
| P. aeruginosa | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| Standard drug (tetracycline μg/mL) | |||||||
| B. subtilis | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| S. aureus | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| E. coli | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| P. aeruginosa | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
BV; bee venom, ChNPs; chitosan nanoparticles, ChNC; nanocomposite, negative (Neg.) no turbidity indicating absence of growth and positive (Pos.) turbidity indicating growth;
Table 4.
The fractional inhibitory concentration index (FICI) and the fractional bactericidal concentration index (FBCI).
| Strains | MIC (A) | MIC (B) | MIC (C) | MIC (D) | FIC (A) | FIC (B) | FIC index |
|---|---|---|---|---|---|---|---|
| B. subtilis | 250.0 | 62.5 | 31.3 | 500.0 | 0.50 | 2.00 | 2.50 |
| S. aureus | 125.0 | 15.6 | 15.6 | 500.0 | 0.50 | 4.01 | 4.51 |
| E. coli | 500.0 | 500.0 | 31.3 | 500.0 | 0.50 | 0.50 | 1.00 |
| P. aeruginosa | 500.0 | 500.0 | 125.0 | 500.0 | 0.25 | 0.25 | 0.50 |
| Strains | MBC (A) | MBC (B) | MBC (C) | MBC (D) | FBC (A) | FBC (B) | FBC index |
|---|---|---|---|---|---|---|---|
| B. subtilis | 500.0 | 125.0 | 125.0 | 500.0 | 0.25 | 1.00 | 1.25 |
| S. aureus | 125.0 | 62.5 | 62.5 | 500.0 | 0.50 | 1.00 | 1.50 |
| E. coli | 500.0 | 500.0 | 125.0 | 500.0 | 0.25 | 0.25 | 0.50 |
| P. aeruginosa | 500.0 | 500.0 | 500.0 | 500.0 | 1.00 | 1.00 | 2.00 |
MIC; minimum inhibitory concentration, FIC; fractional inhibitory concentration, MBC; minimum bactericidal concentration, FIC; fractional bactericidal concentration, A; bee venom, B; chitosan nanoparticles, C; nanocomposite.
Table 3.
Minimum bactericidal concentration (MBC) of BV, ChNPs and ChNc after 24 h.
| Strains | 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 |
|---|---|---|---|---|---|---|---|
| BV concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| E. coli | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| P. aeruginosa | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| ChNPs concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. |
| E. coli | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| P. aeruginosa | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| ChNC concentrations (μg/mL) | |||||||
| B. subtilis | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| S. aureus | Neg. | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. |
| E. coli | Neg. | Neg. | Neg. | Pos. | Pos. | Pos. | Pos. |
| P. aeruginosa | Neg. | Pos. | Pos. | Pos. | Pos. | Pos. | Pos. |
| Standard drug (tetracycline μg/mL) | |||||||
| B. subtilis | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| S. aureus | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| E. coli | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
| P. aeruginosa | Pos. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. |
Positive (Pos.): Indicating growth; Negative (Neg.): Indicating absence of growth.
4. Conclusion
It can be concluded that chitosan as a promising natural carbohydrate polymer can be used as a carrier for many drugs and extracts. In order to increase chitosan efficiency, ionic gelation method was used to prepared chitosan nanoparticles (ChNPs). After that, the as-prepared ChNPs was used as a carrier for encapsulating bee venom (BV). The data depicted that the formed nanocomposite of ChNPs loaded with BV (ChNC) exhibited spherical size with well homogeneity. The finding illustrated that ChNPs exhibited moderate antiviral activity while ChNC showed promising antiviral activity against Human coronavirus MERS-CoV with selective index (SI) = estimated CC50/estimated IC50 = 8.5 for ChNP while SI = 12.1 for ChNC. On other hand the bee venom (BV) showed low antiviral activity. The results affirmed that ChNC is a good candidate for further experiments as anti-MERS-CoV than ChNPs. The experimental data from the current study concluded that, nanocomposite (ChNC) had greater antimicrobial activity against both Gram-positive and Gram-negative bacteria compared to the standard drug (tetracycline), bee venom (BV) and chitosan nanoparticles (ChNPs).
The following is the supplementary data related to this article.
Chemical structure of (a) chitosan and (b) bee venom (BV).
CRediT authorship contribution statement
Mohamed E. Elnosary: Conceptualization, Methodology, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Hesham A. Aboelmagd: Methodology, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Manal A. Habaka: Methodology, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Salem R. Salem: Methodology, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Mehrez E. El-Naggar: Supervision, Software, Conceptualization, Data curation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Rytting E., Nguyen J., Wang X., Kissel T. Biodegradable polymeric nanocarriers for pulmonary drug delivery. Expert Opin. Drug Deliv. 2008;5:629–639. doi: 10.1517/17425247.5.6.629. [DOI] [PubMed] [Google Scholar]
- 2.Karuppusamy C., Venkatesan P. Role of Nanoparticles in Drug Delivery System : A Comprehensive review. J. Pharm. Sci. & Res. 2017;9(3):318–325. [Google Scholar]
- 3.Safer A.M., Leporatti S., Leporatti J., Leporatti MS. Conjugation of EGCG and Chitosan NPs as a novel nano-drug delivery system. Int J Nanomedicine. 2019;14:8033–8804. doi: 10.2147/IJN.S217898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanicha V., Kanyawim K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 5.Nel AE. Nano-enabled COVID-19 vaccines: meeting the challenges of durable antibody plus cellular immunity and immune escape. 2021;15(4):5793–5818. ACS Nano. 2021;15:5793–5818. doi: 10.1021/acsnano.1c01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang D. Application of nanotechnology in the COVID-19 pandemic. Int J Nanomedicine. 2021;2021:623–649. doi: 10.2147/IJN.S296383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knipe DM., Howley P., Cohen J., Griffin D., Lamb R., Martin M., Racaniello V., Roizman B. Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed). Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. 2013 [Google Scholar]
- 8.de Groot RJ., Baker SC., Baric RS., Brown RS., Brown CS., Drosten C., Enjuanes L., Fouchier RA., Galiano M., Gorbalenya AE., Memish ZA., Perlman S., Poon LL., Snijder EJ., Stephens GM., Woo PC., Zaki AM., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao GF., Tan W. China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM., ColomIthetebi NL., Richards LR., Schoeman MC., Preiser W., Drosten C., Drexler JF. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Shen L., Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci Rep. 2016;6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackay IM., Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. J Virol. 2015;12:222. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne-Price S., Miazgowicz KL., Munster VJ. The emergence of the Middle East respiratory syndrome coronavirus. Pathog Dis. 2014;71:121–136. doi: 10.1111/2049-632X.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan RW., Hemida MG., Kayali G., Chu DK., Poon LL., Alnaeem A., Ali MA., Tao KP., Ng HY., Chan MC., Guan Y., Nicholls JM., Peiris JS. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med. 2014;2:813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehman K., Fiayyaz F., Khurshid M., Sabir S., Akash M.S.H. Elsevier; in: 2020. Antibiotics and antimicrobial resistance: temporal and global trends in the environment. In Antibiotics and Antimicrobial Resistance Genes in the Environment; pp. 7–27. [Google Scholar]
- 16.Memariani H., Memariani M., Shahidi-Dadras M., Nasiri S., Akhavan M.M., Moravvej H. Melittin: from honeybees to superbugs. Appl. Microbiol. Biotechnol. 2019;103:3265–3276. doi: 10.1007/s00253-019-09698-y. [DOI] [PubMed] [Google Scholar]
- 17.Ilium L. Chitosan and its use as a pharmaceutical excipient. Pharm. Res. 1998;15(9):1326–1331. doi: 10.1023/a:1011929016601. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed S.A., Mohamed R.R., Elella M.H.A., Sabaa M.W. Synthesis, characterization and applications of N-quaternized chitosan/poly (vinyl alcohol) hydrogels. Int. J. Biol. Macromol. 2015;80:149–161. doi: 10.1016/j.ijbiomac.2015.06.041. [DOI] [PubMed] [Google Scholar]
- 19.Goda E.S., Abu Elella M.H., Hong S.E., Pandit, B., Yoon K.R., Gamal H. Smart flame retardant coating containing carboxymethyl chitosan nanoparticles decorated graphene for obtaining multifunctional textiles. Cellulose. 2021;28:5087–5105. [Google Scholar]
- 24.Socarras K.M., Theophilus P.A., Torres J.P., Gupta K, Sapi E. Antimicrobial activity of bee venom and melittin against Borrelia burgdorferi. Antibiotics. 2017;6:31. doi: 10.3390/antibiotics6040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goy R.C., Morais S.T.B., Assis O.B.G. Evaluation of the antimicrobial activity of chitosan and its quaternized derivative on Ecoli and S. aureus growth. Rev. Bras. Farmacogn. 2016;26:122–127. [Google Scholar]
- 26.Donalisio M., Huguette M., Rosalie A., Donatien G., Alembert T., Roberta R., Valeria C., Cecilia C., Fabrice F., Patrizia R., Carlo B., David L. Donalisio M, et al. In vitro anti-Herpes simplex virus activity of crude extract of the roots of Nauclea latifolia Smith (Rubiaceae) ECAM Comp & Alternative Med. 2013;13:266. doi: 10.1186/1472-6882-13-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herdewijn R., Balzarini J., Baba M., Snoeck R., Schols D., Herdewijn P., Desmyter J., De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods. 1988;20(4):309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 28.Hsouna A.B., Trigui M., Mansour R.B., Jarraya R.M., Damak M., Jaoua S. Chemical composition, cytotoxicity effect and antimicrobial activity of Ceratonia siliqua essential oil with preservative effects against Listeria inoculated in minced beef meat. Int. J. Food Microbiol. 2011;148(1):66–72. doi: 10.1016/j.ijfoodmicro.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 29.Trigui M., Hsouna A.B., Tounsi S., Jaoua S. Chemical composition and evaluation of antioxidant and antimicrobial activities of Tunisian Thymelaea hirsuta with special reference to its mode of action. Ind Crops Prod. 2013;41:150–157. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute (CLSI). Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7th ed., CLSI document M02-A11. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA. 2012 [Google Scholar]
- 31.Bassolé I.H.N., Juliani H.R. Essential oils in combination and their antimicrobial properties. Molecules. 2012;17:3989–4006. doi: 10.3390/molecules17043989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uddin MB., Lee B-H., Nikapitiya C., Kim J-H., Kim T-H., Lee H-C., Choul G-K., Jong S-L., Kim C-J. Inhibitory effects of bee venom and its components against viruses in vitro and in vivo.J Microbiol.(2016) 54:853–853. J Microbiol. 2016;54:853–866. doi: 10.1007/s12275-016-6376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawood A.A., Rifaat H.M., Menazea A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020;35:100673. doi: 10.1016/j.nmni.2020.100673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safarzadeh M., Sadeghi S., Azizi M., Rastegari-Pouyani M., Pouriran R., Haji M.C. Chitin and chitosan as tools to combat COVID-19: a triple approach. Int J Biol Macromol. 2021;18:235–244. doi: 10.1016/j.ijbiomac.2021.04.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortel V., Markwardt F. Studies on the antibacterial properties of bee venom. Die Pharmazie. 1955;10:743–746. [PubMed] [Google Scholar]
- 37.El-Bahnasy, S.A., Mahfouz H.M., ElBassiony M., El-Shibiny A., El-Bolok D.M. ANTIBACTERIAL ACTIVITIES OF BEE VENOM PRODUCED BY TWO HONEYBEE, Apis mellifera L., HYBRIDS. SINJAS. 2022;11:257–270. [Google Scholar]
- 38.Haktanir I., Masoura M., Mantzouridou F.T., Gkatzionis K. Mechanism of antimicrobial activity of honeybee (Apis mellifera) venom on Gram-negative bacteria: Escherichia coli and Pseudomonas spp. AMB Express. 2021;11(1):1–11. doi: 10.1186/s13568-021-01214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X., Singh A.K., Wu X., Lyu Y., Bhunia A.K., Narsimhan G. Characterization of antimicrobial activity against Listeria and cytotoxicity of native melittin and its mutant variants. Colloids Surf. B. 2016;143:194–205. doi: 10.1016/j.colsurfb.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 40.Banks B.E., Shipolini R.A. Chemistry and pharmacology of honey-bee venom. Venoms of the hymenoptera. Biochemical, pharmacological and behavioural aspects. 1986:329–416. [Google Scholar]
- 41.Eaton P., Fernandes J.C., Pereira E., Pintado M.E., Malcata F.X. Atomic force microscopy study of the antibacterial effects of chitosans on Escherichia coli and Staphylococcus aureus. Ultramicroscopy. 2008;108:1128–1134. doi: 10.1016/j.ultramic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Silva L.P.D., de Britto D., Seleghim M.H.R., Assis O.B. In vitro activity of water-soluble quaternary chitosan chloride salt against E. coli. World J. Microbiol. Biotechnol. 2010;26:2089–2092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chemical structure of (a) chitosan and (b) bee venom (BV).