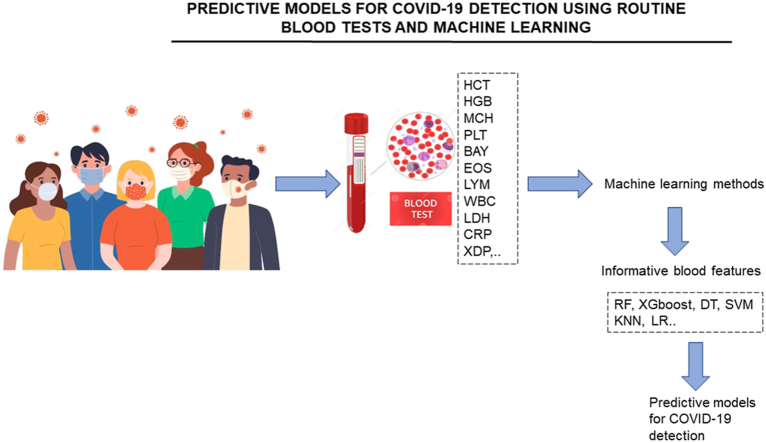
Abstract
The problem of accurate, fast, and inexpensive COVID-19 tests has been urgent till now. Standard COVID-19 tests need high-cost reagents and specialized laboratories with high safety requirements, are time-consuming. Data of routine blood tests as a base of SARS-CoV-2 invasion detection allows using the most practical medicine facilities. But blood tests give general information about a patient’s state, which is not directly associated with COVID-19. COVID-19-specific features should be selected from the list of standard blood characteristics, and decision-making software based on appropriate clinical data should be created. This review describes the abilities to develop predictive models for COVID-19 detection using routine blood tests and machine learning.
Keywords: COVID-19, Blood tests, Machine learning
Graphical abstract
COVID-19; Blood tests; Machine learning.
1. Introduction
The problem of accurate, fast, and inexpensive COVID-19 tests has been urgent till now. The key message of this review is that blood tests contain enough information to detect SARS-CoV-2 infected patients with acceptable accuracy. To establish the corresponding latent dependencies between blood test parameters and COVID-19 presence, a machine learning approach should be applied. The most effective prediction models with accuracy of COVID-19 diagnosis of about 85–95% are based on using Random forest, Gradient boosting, and their variations. Surprisingly, that these models use not more than only 8–10 blood parameters. A set of the most informative features includes mean corpuscular hemoglobin, lymphocytes, leukocytes, basophils, eosinophils, C-reactive protein, bilirubin, D-dimer.
Current “gold standard” for SARS-CoV-2 infection identification is based on a transcription-polymerase chain reaction (RT-PCR) and Enzyme-linked immunosorbent assay (ELISA) tests [1, 2]. RT-PCR has high specificity but low sensitivity [3, 4] and high misclassification in the early stage of disease [5]. The drawbacks are high-cost equipment and consumables, long time of analysis. Additional information can be achieved by computer tomography or X-ray visualization [6, 7, 8], but these methods require high-tech equipment and cannot be used often. A possible solution is to use more conventional clinical tests combined with sophisticated but powerful analytical methods from the field of Artificial Intelligence implemented as a Medical Decision Support System (MDS). The attractiveness of this idea is that Artificial Intelligence can compensate a nonspecificity of general clinical tests.
Since the COVID-19 outbreak at the end of 2019, several Medical Decision Support Systems (MDSSs) were developed for this disease diagnostic and severity prediction [9, 10, 11, 12, 13]. MDSSs can be based on data-driven, knowledge-driven, and hybrid (a combination of knowledge driven and data-based) data models. Data driven models (DDM) rely only on the observed data, while knowledge-driven (KDM) ones use human logic. The hybrid models take advantage of both approaches. For example, DDMs are superior in natural language processing and can extract knowledge from Electronic Health Records, and KDM can be used for the support of disease diagnostics. Usually, DDM MDSSs use machine learning (ML) algorithms like artificial neural networks; KDM MDSSs are based on an expert knowledge and human logic [14]. Most trustworthy are expert systems based on subjective evaluations described as medical treatment protocols. Such systems are the state-of-the-art for a well-known disease [15] but lack the knowledge with COVID-19. ML MDSSs are available to analyze latent relations in data and construct complex predictive models, but this is a purely data-driven approach and couldn’t handle data uncertainty.
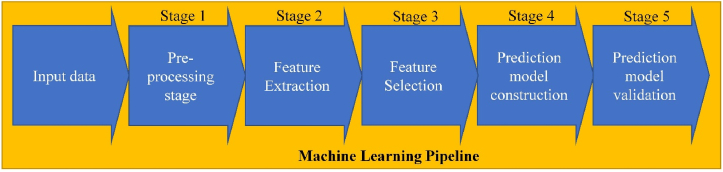
ML pipeline is a sequence of algorithms that includes preprocessing, feature extraction/selection, prediction model construction, and its validation (see Figure 1) [13]. A brief description of these steps is presented below.
Figure 1.
A general machine learning pipeline.
1.1. Preprocessing
The necessity of preprocessing depends on the quality of input data. Routine blood test results often have missing values, and imputation algorithms based on the statistical mean are used to fill it. Also, detection and removal of outliers, presenting data, significantly different from the major of the dataset, should be conducted. The statistical-based approaches like a parametric Z-score [4, 16], isolation forest [17], density-based spatial clustering of applications with noise [18] are considered to be the most effective.
1.2. Feature extraction
The feature extraction is based on a transformation of initial parameters to a feature vector of less dimension due to removing redundant features. Principal component analysis and multidimensional scaling are the most frequently used [19, 20].
1.3. Feature selection
Feature selection is based on uninformative parameters removing. There are univariate and multivariate methods of feature selection [21, 22]. Univariate methods analyze every feature impact on the predictive model accuracy independently. Multivariate methods do the same for several features jointly. Some classification methods, like Decision trees (DT) and Random forests (RF) can also estimate features' informativeness [23].
1.4. Prediction model construction
This step allows making a conclusion about a studied object belonging to one of the preliminarily determined classes. Supervised ML methods useful for creating prediction models in medicine are limited by the requirements of model transparency and a small volume of medical datasets [24]. DT, RF, Support vector machine (SVM), and gradient boosting are the most often used [24, 25].
1.5. Prediction model validation
The validation needs to estimate a prediction model accuracy. It should be conducted with a data set, which was not used for data model training. The latter most simple implementation is train-test split [19, 26]. Another option is to get a small standalone dataset from the other source and use it as a final performance measure [27]. K-fold cross-validation is often used [19].
Using routine blood tests as an additional diagnostic tool is very attractive because of its simplicity and availability in all medical institutions of practical healthcare. The routine blood tests issue is that they give general information about a patient’s state, which is not directly associated with COVID-19 presence or absence. COVID-19-specific features should be selected from the list of standard blood characteristics, and decision-making software based on appropriate clinical data should be created to overcome this issue. The latter can be implemented using ML.
This review aims to analyze routine blood test availability for COVID-19 detection and the ways of ML pipeline implementation using routine blood test data.
2. The blood test informative features selection
We analyzed about 50 articles published in journals in the field of medicine and machine learning and indexed in Scopus, Google Scholar, and Web of Science. The search included the following terms: COVID-19; Blood tests; Machine learning, Statistical analysis.
The features met in more than 10 papers and features considered by the authors as informative are presented in Table 1.
Table 1.
Blood-test features associated with COVID-19.
| Abb. | Feature | Description | Articles: |
|
|---|---|---|---|---|
| features used | informative features | |||
| HCT | Hematocrit | The hematocrit is the calculated volume percentage of red blood cells (erythrocytes) in blood. HCT is a useful but low-specificity biomarker associated with number of diseases like thrombocythemia, thrombosis, hypoxia. | [28] [29, 30, 31] [32, 33, 34] [35, 36, 37] [38, 39, 40] [41, 42, 43] [44, 45, 46] [47] |
[35] |
| HGB | Hemoglobin | Hemoglobin is a conjugated protein molecule, which handles oxygen carrier to tissues. Therefore, it is the key component of the red blood cells. Both low/high hemoglobin levels can indicate certain diseases. |
[19, 26, 28] [29, 30, 31] [33, 34, 35] [36, 37, 38] [39, 40, 41] [42, 44, 45] [46, 47, 48] [49, 50, 51] |
[35, 51] |
| PLT | Platelets | Platelets are a component of blood whose function is to react to bleeding from blood vessel injury by clumping, that plays a major role in blood clotting. PLT reflects a pathology risk/severity, for example, cancer [87]. | [4, 23, 26] [28, 29, 30] [31, 32, 33] [35, 36, 37] [38, 39, 40] [42, 43, 44] [45, 46, 47] [50, 51, 52] |
[23, 28] [30, 38] [39, 51] |
| RBC | Red blood cells | Red blood cells are the most common type of blood cells that carry oxygen to the body tissues via blood flow through the circulatory system. Low/high RBC levels indicate health problems, for example liver disease. | [19, 26, 27] [28, 29, 30] [31, 33, 49] [35, 36, 37] [38, 42, 43] [46, 47, 53] |
[30, 35] |
| LYM | Lymphocytes | White blood cells are a cellular component of the adaptive (specific) immune response: B-Lymphocytes produce antibodies; T-lymphocytes destruct the infected cells and regulate other leukocytes. Their concentration is low in severe COVID-19. | [4, 19, 26] [28, 29, 30] [31, 33, 34] [35, 36, 38] [39, 40, 41] [42, 44, 45] [46, 47, 49] [50, 51, 52] [54, 55, 56] [57] |
[34, 39] [41, 44] [50, 51] [52, 55] [56, 57] |
| MCH | Mean Corpuscular Hemoglobin | The mean corpuscular hemoglobin is the average mass of HGB per a RBC in a blood sample. | [19, 26, 27] [28, 29, 30] [31, 32, 33] [34, 35, 36] [37, 46, 52], [53] |
|
| MCHC | Mean Corpuscular Hemoglobin Concentration | The mean corpuscular hemoglobin concentration characterizes an average concentration of HGB in a RBC. MCHC also reflects the amount of hemoglobin per unit volume in a single red blood cell | [19, 26, 28] [29, 30, 31] [34, 35, 36] [37, 42, 46] [52] |
[19] |
| WBC | Leukocytes | White blood cells include: granulocytes (neutrophils, eosinophils, basophils), monocytes, lymphocytes. WBC are the cells of the immune system involved in protecting the body against infectious disease. | [4, 19, 26] [27, 28, 29] [30, 31, 32] [33, 34, 35] [36, 37, 38] [39, 40, 41] [42, 43, 44] [45, 46, 47] [50, 51, 52] [53, 54, 55] [56, 57] |
[4, 26] [28, 30] [38, 39] [51, 52] [53, 56] [57] |
| BAY | Basophils | Basophils are a type of white blood cells. Basophils have granules containing biologically active substances, with the help of which they regulate immune responses. They handle detection of an antigen and its presentation to other cells, stimulating a response on an inflammation. | [4, 19, 23] [26, 27, 28] [29, 30, 31] [32, 33, 34] [36, 42, 43] [46, 47, 49] [50, 52, 53] |
[23, 34] [42, 50] [52] |
| EOS | Eosinophils | Eosinophils are a type of white blood cells, responsible for hypersensitive immune responses. They contain inflammatory mediators (prostaglandins, leukotrienes, platelet- activating factor, cytokines) and play a significant role in destroying foreign particles and removing toxic substances. |
[4, 19, 26] [27, 28, 29] [30, 31, 32] [33, 34, 36] [40, 42, 43] [46, 47, 49] [50, 52, 53] [57] |
[19, 27] [28, 30] [34, 42] [50, 52] |
| LDH | Lactate dehydrogenase | Lactate dehydrogenase is an important enzyme, which involves in cellular respiration and energy production. Also, LDH is a marker of cells and tissues damage. | [4, 23, 26] [28, 29, 30] [31, 32, 37] [39, 40, 41] [44, 45, 46] [49, 50, 51] [53, 57] |
[4, 23] [26, 29] [30, 32] [37, 39] [40, 41] [49, 50] [51, 53] [57] |
| MCV | Mean corpuscular volume | The average volume of an erythrocyte population | [19, 26, 27] [28, 29, 30] [31, 33, 34] [35, 36, 37] [41, 46, 52] [53] |
|
| RDW | Red blood cell distribution width | Red blood cell distribution width characterizes the variability RBC size. RDW reflects the extent of anisocytosis фтв is elevated in iron deficiency anemia. |
[19, 26, 27] [28, 29, 30] [31, 33, 35] [36, 37, 42] [46, 50, 52] |
|
| MONO | Monocytes | Monocytes are one of the largest among WBC, responsible for attacking and breaking down germs and bacteria that enter the body. Monocytes also influence adaptive immune responses and exert tissue repair functions. | [4, 19, 26] [28, 29, 30] [31, 32, 33] [34, 36, 39] [42, 43, 46] [47, 49, 50] [52, 53, 57] |
[30, 34] [43] |
| MPV | Mean platelet volume | Mean platelet volume is a measure of the average size of platelets in blood. MPV high value can be observed with increased destruction of platelets and sepsis. |
[19, 26, 28] [29, 30, 31] [33, 35, 36] [42, 46, 52] [53] |
|
| NEU | Neutrophils | Neutrophils are a type of white blood cells, being an immune system’s first line of defense against infections by ingesting microorganisms and releasing enzymes that kill them. | [4, 19, 26] [28, 29, 30] [31, 32, 34] [38, 39, 41] [42, 43, 44] [45, 46, 47] [49, 51, 56] [57] |
[41, 42] [43, 49] [56] |
| CRP | C-reactive protein | C-reactive protein is produced by the liver and is induced by various inflammatory mediators, such as interleukin-6. In a healthy person, it is absent or present in minimal amounts. Therefore, it is an early marker of an acute phase of inflammatory response. | [4, 19, 26] [28, 29, 30] [31, 32, 34] [35, 37, 39] [40, 43, 44] [45, 46, 47] [49, 50, 51] [53, 54, 57] [58] |
[4, 30] [34, 35] [37, 39] [40, 43], [40, 49] [51, 54] [57, 58] |
| CREAT | Creatinine | Creatinine is a chemical waste molecule resulting from muscle metabolism. It is formed mainly in the liver and excreted by the kidneys. | [19, 23, 26] [28, 29, 30] [31, 34, 40] [41, 44, 45] [46, 47, 51] [58] |
[23, 51] |
| UREA | Urea | Urea is a sub-product of the protein metabolism which is formed in the liver, excreted by the kidney, and is a standard biomarker of renal failure. | [26, 28, 29] [31, 34, 38] [41, 44, 46] [47, 53, 57] |
|
| K+ | Potassium | Potassium is an essential mineral that is needed by all tissues. It participates in the electrochemical processes in the cells, as well as in carbohydrate and protein metabolism, regulation of blood pressure. Potassium deficiency may be caused by increased renal excretion or its loss through the gastrointestinal tract. | [19, 23, 26] [27, 28, 29] [34, 38, 40] [41, 44, 45] [47, 48, 58] |
[23] |
| Na | Sodium | Sodium is the main electrolyte in the extracellular space, mainly responsible for osmotic pressure and electrolyte balance in the blood. | [19, 26, 27] [28, 29, 30] [34, 38, 40] [41, 43, 44] [45, 47, 48] [58] |
[48] |
| AST | Aspartate transaminase | Aspartate transaminase is an enzyme involved in amino acid metabolism. It is found mainly in the liver, heart, nervous tissue, skeletal muscles, and in smaller amounts in the kidneys, pancreas, spleen, and lung tissue. | [4, 19, 26] [27, 28, 29] [30, 31, 32] [37, 40, 41] [43, 44, 45] [47, 51, 53] [56] |
[4, 26] [27, 29] [30, 53] |
| ALT | Alanine transaminase | Alanine transaminase is an endogenous enzyme that commonly measured clinically as biomarker of liver injury. It is found in smallest amounts in the kidneys, heart, skeletal muscles, pancreas, therefore, it may indicate diseases of these organs. | [4, 19, 26] [28, 29, 30] [31, 34, 37] [40, 41, 43] [44, 45, 47] [51, 53] |
[4, 29] [37, 53] |
| BR | Bilirubin (Total) | Bilirubin is an intermediate product of hemoglobin metabolism, formed during the normal process of breaking down red blood cells. Higher than normal levels of bilirubin may be associated with liver, bile duct or gallbladder problems. | [19, 23, 26] [27, 29, 30] [31, 32, 34] [40, 43, 44] [45, 46, 51] [54, 59] |
[23, 29] [34, 51] |
| XDP | D-dimer | D-dimer is a plasmatic protein essential for dissolving blood clots. D-dimer is considered as a thrombosis marker, its high level indicates increased blood clotting. | [26, 30, 32] [37, 40, 44] [45, 46, 51] [57, 58] |
[26, 37] [51, 58] |
| Ferritin | Ferritin is a complex consisting of a protein shell (apoferritin) and iron hydroxide. The function of ferritin is to bind free iron ions, neutralising its toxic properties and increasing its solubility. It is used in clinical medicine as an indicator of iron stores. The highest concentrations of ferritin are usually found in hepatocytes and reticuloendothelial cells. | [29, 31, 32] [37, 40, 43] [45, 49, 50] [55] |
[29, 32] [37, 43] [49, 50] [55] |
|
The most often used features include: Lymphocytes (LYM), Leukocytes (WBC), Mean corpuscular hemoglobin (MCH), Basophils (BAY), Eosinophils (EOS), C-reactive protein (CRP), Bilirubin, D-dimer.
The oxygen desaturation is associated with severe respiratory failure in COVID-19 patients. SARS-CoV-2 surface glycoproteins bind to porphyrin on hemoglobin and inhibit heme metabolism. It is a possible mechanism of MCH and MCHC changing during COVID-19 [59].
Lymphopenia (temporary or persistent decrease in the level of lymphocytes in the blood) is a typical symptom for COVID-19 patients [60]. An essential decreasing CD8+ T cells, a type of lymphocytes, was established for SARS-CoV-2 infected patients compared to vaccinated subjects [61].
Eosinophils play an essential role in SARS-CoV-2 infection associated with high CD62L expression, a lung eosinophil marker [62]. The basophil level is correlated with humoral response to SARS-CoV-2 infection [62, 63].
CRP is usually produced by hepatocytes and, additionally, by macrophages in a place of inflammation [64]. The latter is strongly related to COVID-19 disease complications and mortality. A cytokine storm appearing at the most dangerous stage of COVID-19 pneumonia stimulates hepatocytes to produce CRP [65, 66]. CRP is considered an independent indicator of COVID-19 severity [64, 67, 68]. Aspartate transaminase is a statistically significant marker of pulmonary fibrosis caused by SarS-CoV-2 infection [69]. SARS-CoV-2-positive patients (positive test for SARS-CoV-2) showed a statistically significant decrease in calcium concentration compared to SARS-CoV-2-negative patients [70]. High levels of ferritin, as well as severe lymphopenia (decrease in T-, B – lymphocytes), also high levels of C-reactive protein, ferritin, D-dimer, ALT and AST, are signs of a cytokine storm [71]. High ferritin levels were found in autopsies of deceased COVID-19 patients [72].
A high bilirubin content was established in severe Covid-19 patients compared to milder forms (an averaged difference was in interval from 0.27 to 0.95 μmol/L) [73, 74, 75].
As mentioned above, D-dimer is a biomarker of thrombosis. A 3–4 fold D-dimer concentration increasing was associated with poor prognosis [76].
3. The COVID-19 prediction models construction using blood test data
The dataset description used by the authors, number of selected/extracted informative features, and created COVID-19 prediction model performance are shown in Table 2.
Table 2.
Description of used dataset size, predictive model construction methods, features, and model performance.
| Database |
Methods | Features | The best method | SP, % | SE, % | Accuracy, % | Refs. | |
|---|---|---|---|---|---|---|---|---|
| Total cases | Positive cases | |||||||
| 279 patients | ET, SVM, LR, NB, RF, KNN, TWRF, DT | Used: 12 Informative: 6 |
RF TWRF |
65 75 |
92 95 |
82 86 |
[4] | |
| 5352 patients | 131 positive cases | KNN, RF, SVM, XGBoost | All features: 117 Used: 35 Informative: 5 |
XGBoost | 97.9 ± 0.4 | 81.9 ± 6 | [19] | |
| 105 patients | RF | All features: 49 Used: 11 Informative: 11 |
94 | 100 | [23] | |||
| 1-First dataset used Data from San Raphael Hospital (OSR) with 72 features. 2-COVID-specific dataset (34 features) 3 - Complete Blood Count (CBC) dataset (21 features) |
positive cases | LR, NB, KNN, SVM, RF | All features: 72 Used: 69 Informative: 6 |
1) OSR: RF, SVM 2) COVID: KNN, SVM 3) CBC: KNN, RF |
1) OSR: 86, 89 2) COVID: 80, 83 3) CBC: 89, 82 |
1) OSR 91,87 2) COVID: 92, 89 3) CBC: 82, 84 |
1) OSR 88,88 2) COVID: 86,86 3) CBC: 86, 83 |
[26] |
| 12,183 patients | 2183 positive cases | XGBoost | All features: 29 Used: 15 Informative: 3 |
41,70 | 95,90 | [27] | ||
| 608 patients∗ | 84 positive cases | DTX, LR, XGBoost, RF, SVM SMOTE, MLP | All features: 111 Used: 23 Informative: 3 |
RF | 91 ± 2 | 66 ± 10 | 88 ± 2 | [28] |
| 5643 patients∗∗ | 557 positive cases | ANN, DT, PLS-DA, KNN | All features: 75 Used: 51 Informative: 10 |
ANN | 94 | 93 | 94 | [29] |
| First data set: 279 patients Second data set: 1624 patients Third data set: 600 patients |
177 positive cases 786 positive cases 80 positive cases |
LR, KNN, DT, SVM, NB, ET, RF, XGBoost, DNN, CNN, RNN, LSTM | Used: 52 Informative: 9 |
DNN | 84.56 93.02 95.27 |
96.14 93.27 77.05 | 92.11 93.16 93.33 |
[30] |
| 5644 patients | 558 positive cases | LR, RF, KNN, GNB, Ridge, Elastic net, DT, ET, AdaBoost, SMOTE, SV-LAR (ensemble based on LR, RF and AdaBoost) | Used: 48 | SV-LAR | 91 | 83 | 91 | [31] |
| 92,254 patients | 7335 positive cases | XGBoost | All features: 70 Used: 19 Informative: 4 |
86.8 | 82.4 | 86.4 | [32] | |
| 294 patients | SMOTE, DT, RF, KNN, SVC, GB, GNB, MLP, GP | All features: 111 Used: 33 Blood test: 13 |
GB | 98 | [33] | |||
| 405 patients | 212 positive cases | EBT | Used: 22 Informative: 6 |
76.65 | 85.85 | 81.79 | [34] | |
| The initial dataset had 9004 records. | RF, LR, ANN, SVM, KNN, Ensemble. | Used: 11 Informative: 4 |
[35] | |||||
| 5644 patients | KNN, RBF, NB, kStar, PART, RF, DT, OneR, SVM, MLP | Used: 14 | KNN, kStar, and RF | 100 (for the three best algorithms), 88 (test) | [36] | |||
| 143 patients | 88 positive cases | RF, SVM, NB | Used: 17 Informative: 6 |
SVM | 88 | [37] | ||
| 5644 patients | 558 positive cases | SVM, AdaBoost, RF, KNN, Ensemble | Used: 15 Informative: 4 |
The Ensemble Model | 65 | [38] | ||
| 3058 patients | 421 positive cases | CatBoost, SVM, LR, Radiologist + ML model | All features: 13 Used: 9 Informative: 5 |
ML 91.5–98.3 Radiologist + ML model 52.7–66.7 |
ML 55.5–77.8 Radiologist + ML model 92.3–92.5 |
ML 89.3–96.9 Radiologist + ML model 55.5–68.4 |
[39] | |
| 250 positive cases | SIMPLS | Used: 32 Informative: 3 |
[40] | |||||
| 2034 patients | DT, RF, GB, XGBoost, SVM, LGBM, KNN, ANN | Used: 59 Informative: 3 |
1) RF and GB 2) RF, LGBM 3) RF, LGBM, XGBoost |
1) 89, 89 2) 93, 91 3) 91, 93, 87 |
[41] | |||
| 1218 patients∗∗∗ | XGBoost, LR, SVM, RF, DT, Ensemble | Used: 15 Informative: 3 |
DT LR Ensample |
76 83 85 |
73 70 74 |
[42] | ||
| 137 patients | RF, SVM, KNN, Adaboost | Used: 30 Informative: 7 |
SVM | 81 | [43] | |||
| 422 patients | RF, NB, SVM, KNN, LR, ANN | Used: 38 Informative: 6 |
NB | 75.0 | 85.9 | [44] | ||
| 162 patients | CRT, RF,ANN | Used: 25 | CRT 92.7 RF 95.8 ANN 96.3 |
CRT 88.0 RF 75.0 ANN 59.0 |
CRT 92.0 RF 92.9 ANN 90.5 |
[45] | ||
| 5644 patients | 559 positive cases | MLP,SVM, RF, DT, Bayes Net, NB | All: 107 Used: 41 |
RF | 92.1 ± 1.2 | 93.6 ± 1.1 | 92.891 ± 0.851 | [46] |
| 5644 patients | CNN, DT, NB, kNN, | All features: 111 Used: 18 |
CNN | 80 | [47] | |||
| 51 positive cases | HHOSRL | Used: 20 Informative: 4 |
100.00 | 100.00 | 100.00 | [48] | ||
| 1455 patients | 182 positive cases | RF, LR, SVM, MLP, SGD, XGBoost, AdaBoost, Ensemble. | Used: 12 Informative: 3 |
64 (95 confidence interval 0.59–0.69) | 93 (95 confidence interval 0.84–0.98) | [49] | ||
| 127,115 patients | 1573 positive cases | SNN, KNN, LR, SVM, RF, XGBoost | All features: 100 Used: 19 Informative: 11 |
XGBoost RF |
[50] | |||
| 412 patients | 326 positive cases | RF, SVM | Used: 19 Informative: 11 |
SVM | 80 | 88 | 84 | [51] |
| 1500 patients | LR, EMB, RF, SVM, SHAP | Used: 16 Informative: 4 |
EBM RF |
(70–90) | [52] | |||
| 5644 patients | 559 positive cases | MLP,SVM, DT, Bayes Net, NB | Used: 24 Informative: 6 |
Bayes Net | 93.6 ± 1.1 | 96.8 ± 0.7 | 95.159 ± 0.693 | [53] |
| 88 positive cases | SVM, XGBoost, RF, GB, DT | Informative: 3 | Subsemble | 72 | 85 | [55] | ||
| 300 patients | 87 positive cases | LR, SVM, XGBoost | Used: 8 Informative: 6 |
XGBoost | 69 | 94 | 87 | [56] |
| 196 positive cases | MCDM | Used: 6 Informative: 3 |
82 | [57] | ||||
| 404 patients | XGBoost | Used: 20 Informative: 3 | 90 | [58] | ||||
| 212 patients | RF, MLP, SVM, GB, ET, AdaBoost | Used: 17 Informative: 5 |
RF | 74 | 73 | [59] | ||
| 250 patients | 126 positive cases | GB, MLP, KNN, DT, ET, LR, AdaBoost, SVM, RF. | All features: 100 | Subsemble | 98.6 | [77] | ||
| 117 positive cases | SVM-P, SVM-R, KNN, NNET, NB, RF | Informative: 6 | RF | 92 | 93 | 92 | [78] | |
| 4009 COVID-19 patients | 489 severe and 3520 non-severe cases | MLP, DT | Used 30 Informative: 6 |
MLP | 96.4 | 96.5 | [79] | |
Remarks and abbreviations.
- AdaBoost: Adaptive Boosting,
- ANN: Artificial Neural Networks,
- CatBoost: Categorical gradient boosting,
- CNN: Convolutional Neural network,
- CRT: Classification and Regression Decision Tree,
- DNN: Deep Neural Network,
- DT: Decision Trees,
- DTX: Decision Tree-based Explainer,
- EBT: Ensemble Bagged Tree model,
- EMB: Explainable Boosting Machine,
- ET: Extremely Randomized Trees,
- GB: Gradient Boost,
- GNB: Gaussian naive Bayes,
- GP: Gaussian process,
- HHO with specular reflection learning,
- HHOSRL: Harris hawks optimized extreme learning machine,
- KNN: K-Nearest Neighbors,
- LGBM: Light Gradient Boosting Machine,
- LR: Logistic Regression,
- LSTM: Long Short-term Memory,
- MCDM: Multi Criteria Decision Making,
- MLP: Multi-Layer Perceptron,
- NB: Naive Bayes,
- NNET: Neural Net,
- PLS-DA: Discriminant Analysis by Partial Least Squares,
- RBF: Radial Basis Functions,
- RF: Random Forest,
- RNN: Recurrent Neural Network,
- SGD: Stochastic Gradient Descent,
- SHAP: Shapley Additive Explanations,
- SIMPLS: inspired modification of partial least square,
- SNN: Self-normalizing neural network,
- SVC: Support Vector Classifier,
- SVM SMOTE: SVM-Synthetic Minority Over-sampling Technique,
- SVM: Support Vector Machine,
- SVM-P: Support Vector Machine Polynomial,
- SVM-R: Support Vector Machine Radial,
- TWRF: Three-Way Random Forest,
- XGBoost: Extreme Gradient Boosting machine.
The dataset is publicly available in www.kaggle.com/einsteindata4u/covid19, 2020. Online.
The dataset is publicly available in https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/discussion/139347.
The dataset is publicly available in https://zenodo.org/record/4686707.
Accuracy, sensitivity (SE), and specificity (SP) are defined as follows. Accuracy reflects how much the prediction is exact [36]:
| (1) |
The sensitivity is an ability of a method identifying true positives, and specificity (SP) is an ability to identify true negatives [44, 49]:
| (2) |
| (3) |
Here:
-
-
True Positive (TP): Participants with a target disease classified as belonging to the target group;
-
-
False Positive (FP): Healthy participants but classified as belonging to the target group;
-
-
True Negative (TN): Healthy participants classified as belonging to the control group;
-
-
False Negative (FN): Participants with the target disease but classified as belonging to the control group.
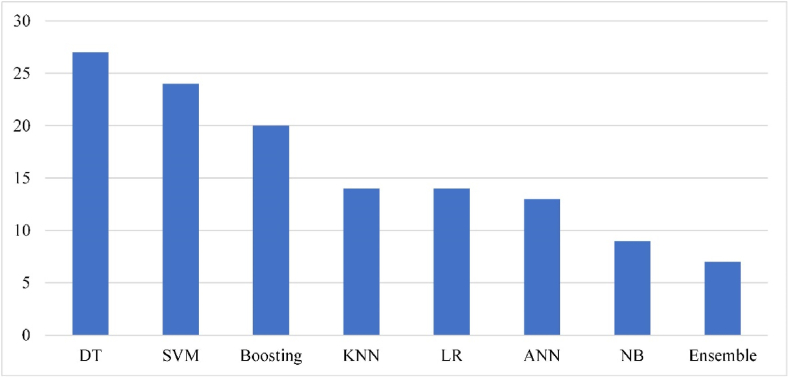
Machine learning methods met the most papers include Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Decision Tree (DT), Boosting, Artificial Neural Network (ANN), Logistic regression (LR) (see Figure 2). The popularity of DT-based methods, including Random Forest (RF), Regression DT, etc., is associated with the explainability of the created data model and excellent accuracy. In 9 cases, the DT-based methods demonstrated the best prediction accuracy. A RF-based demonstrated COVID-19 diagnosis accuracy of 92.9% [76].
Figure 2.
The frequency of various classification used methods.
The SVM generalization potential and good performance are the reasons of its frequent use [26, 31, 37, 43, 51]. The Boosting family is in the third place in popularity and in the second place of prediction accuracy (7 studies). The Logistic regression is effective for informative features' extraction and robust to overfitting. Researchers are still using KNN and Naïve Bayes (NB), which were among the early appeared supervised learning methods and do not show high results. An exception is the Bayesian network with diagnosis performance of 95.2% [76].
Researchers are still aware of ANN and Deep Neural Network (DNN) usage because the number of features in raw data is small (tens) compared to a number of samples (hundreds), which is not suitable for these methods.
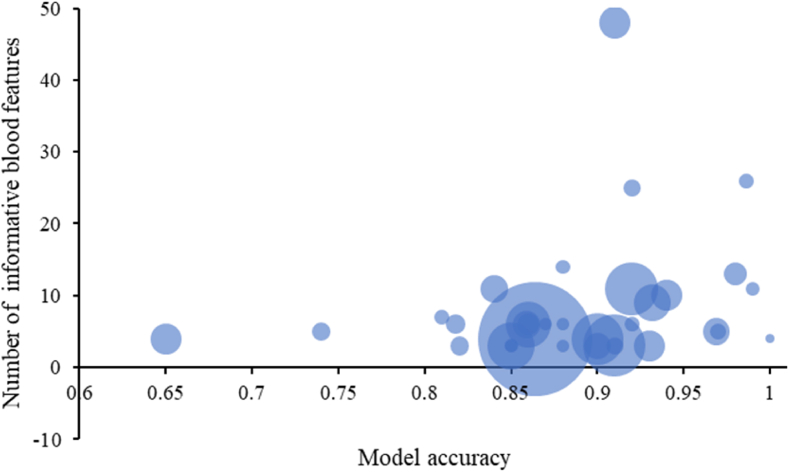
A predictive model accuracy depends on the data volume and quality. In the analyzed papers, the smallest dataset included 51 COVID-19 patients, the largest – 7335 ones [32]. The average number of COVID-19 patients was 919. The most large control group included 125,542 samples [50]. The average number of informative blood features was 8; the median was 6. The mean accuracy was 88%. The simplicity of the best predictive models is an argument that overfitting was avoided. A relation between the number of informative features and a predictive model accuracy is shown in Figure 3.
Figure 3.
Dataset and predictive model characteristics. The bubble size corresponds to the number of verified COVID-19 samples in the dataset; the bigger, the better. The vertical axis refers to the number of the used blood informative features. The horizontal axis describes model accuracy, taken from the papers. When it was not available, an approximation was used.
4. Discussion
Circulating blood contains the metabolic biomarkers of inflammatory processes in a human body, so the metabolic profile potentially can be used for COVID-19 diagnostics and severe prediction. It can be seen from Table 1 that the most full profile contains up to a hundred metabolites. Removing redundant ones can significantly improve the robustness of the predictive models on the one hand and reveal the most informative blood features for diagnostic purposes.
Preprocessing of input data includes missing data imputations based on statistics. Feature extraction was not usually used. The used machine learning systems provide the following result: there is a definite relation between mean corpuscular hemoglobin, lymphocytes, leukocytes, basophils, eosinophils, C-reactive protein, bilirubin, D-dimer, and COVID-19 (see Table 1). The most effective prediction models were based on using Random forest, Gradient boosting, and their variations. The predictive model achieved the highest accuracy score with a relatively small dataset size can be a subject of overfitting. Datasets with over 500 samples demonstrate an accuracy of about 85–95%, that can be considered as very good (see Table 2). Such results should be deemed positive, and specific blood metabolites-oriented diagnostics may be used in the clinical practice.
Significant progress in blood data analysis became available due to open datasets, also cited in this paper (see Table 2). Though merging the small datasets into large ones can benefit data scientists, it is still a problem from the ethical and medical points of view. Federative machine learning (for instance, OpenFL project by Intel Corporation ©) can solve a problem with data security, but the medical protocol unification issue is persisting. Different machine learning pipelines and the absence of the standard validation strategy that proves the quality of the predictive data model diminish the trust in such results. The open-source software, which allows the reproduction of achieved results, is a key to high-quality data models.
5. Conclusion
PCR test is still the best choice for diagnosing SARS-COV-2 infection [80] but, despite its usefulness, it requires expensive laboratory equipment, highly qualified laboratory personnel, and the time between sampling and the result is relatively long. Other clinical and immunodiagnostic tests should also be taken into account when interpreting the results of RT-PCR tests for a more reliable diagnosis [81]. It is necessary to improve RT-PCR tests to decrease false-negative and false-positive results [82]. COVID-19 pandemic force rapid development of a new method to study disease metabolic pathways.
Routine blood tests, including microfluidic devices, are minimally invasive, minimizing the sample volume to one drop. From this perspective, proposed predictive models allow us to understand the connections among human blood metabolites, COVID-19 diagnosis and prognosis of severe. Such knowledge is not a clinic-ready system, but a step before it. Once the verified set of metabolites is found, a simple expert knowledge-based system can be constructed to be used routinely.
Some ideas for the future development of COVID-19 blood tests are as follows. Classes imbalance can lead to a biased predictive model. Synthetic Minority Over-sampling Technique allows generating additional data samples through variations of each blood feature [28, 31, 33]. A game theory-based Shapley value method can provide a reliable, feature extraction [83, 84] An ensemble learning based on combining several classification algorithms and generating a final prediction, for instance, by the voting procedure allows improving a predictive model performance [38, 54, 85, 86, 87, 88].
Another achievement of such studies is the proof that machine learning methods coupled with thoroughly developed protocols and large datasets can provide rapid and reliable techniques to generate fundamental knowledge about socially significant diseases. We encourage scientific groups to share their datasets and develop machine learning models because humanity can overcome the pandemic by uniting efforts.
The essential proofs, which may lead to a clinical trial and further introduction of such methods into the clinical practice, are improvements and conclusions by physiologists found in many reviewed papers.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
Prof. Yury V. Kistenev was supported by the Tomsk State University Development Programme (Priority-2030).
Data availability statement
Data associated with this study has been deposited at www.kaggle.com/einsteindata4u/covid19, 2020. Online
https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/discussion/139347.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Dziąbowska K., Czaczyk E., Nidzworski D. Detection methods of human and animal influenza virus—current trends. Biosensors. 2018;8:94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George K.St. In: Virus Influenza, Kawaoka Y., Neumann G., editors. vol. 865. Humana Press; Totowa, NJ: 2012. Diagnosis of influenza virus; pp. 53–69. (Methods in Molecular Biology). [Google Scholar]
- 3.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinati D., Campagner A., Ferrari D., Locatelli M., Banfi G., Cabitza F. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J. Med. Syst. 2020;44:135. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin. Chem. Lab. Med.(CCLM) 2020;58:1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- 6.Mohammad-Rahimi H., Nadimi M., Ghalyanchi-Langeroudi A., Taheri M., Ghafouri-Fard S. Application of machine learning in diagnosis of COVID-19 through X-ray and CT images: a scoping review. Front. Cardiovasc. Med. 2021;8 doi: 10.3389/fcvm.2021.638011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zargari Khuzani A., Heidari M., Shariati S.A. COVID-Classifier: An automated machine learning model to assist in the diagnosis of COVID-19 infection in chest X-ray images. Sci. Rep. 2021;11:9887. doi: 10.1038/s41598-021-88807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain L., Nguyen T., Li H., Abbasi A.A., Lone K.J., Zhao Z., Zaib M., Chen A., Duong T.Q. Machine-learning classification of texture features of portable chest X-ray accurately classifies COVID-19 lung infection. Biomed. Eng. Online. 2020;19:88. doi: 10.1186/s12938-020-00831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Сallejon-Leblic M.A., Moreno-Luna R., Del Cuvillo A., Reyes-Tejero I.M., Garcia-Villaran M.A., Santos-Peña M., Maza-Solano J.M., Martín-Jimenez D.I., Palacios-Garcia J.M., Fernandez-Velez C., et al. Loss of smell and taste can accurately predict COVID-19 infection: a machine-learning approach. J. Clin. Med. 2021;10:570. doi: 10.3390/jcm10040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arpaci I., Huang S., Al-Emran M., Al-Kabi M.N., Peng M. Predicting the COVID-19 infection with fourteen clinical features using machine learning classification algorithms. Multimed. Tool. Appl. 2021;80:11943–11957. doi: 10.1007/s11042-020-10340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan Y., Zhou H., Zhang X. An interpretation architecture for Deep learning models with the application of COVID-19 diagnosis. Entropy. 2021;23:204. doi: 10.3390/e23020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imran A., Posokhova I., Qureshi H.N., Masood U., Riaz M.S., Ali K., John C.N., Hussain M.I., Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthikeyan A., Garg A., Vinod P.K., Priyakumar U.D. Machine learning based clinical decision support system for early COVID-19 mortality prediction. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.626697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed F., Hossain M.S., Islam R.U., Andersson K. An evolutionary belief rule-based clinical decision support system to predict COVID-19 severity under uncertainty. Appl. Sci. 2021;11:5810. [Google Scholar]
- 15.Ayo F.E., Awotunde J.B., Ogundokun R.O., Folorunso S.O., Adekunle A.O. A decision support system for multi-target disease diagnosis: a bioinformatics approach. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cousineau D., Chartier S. Outliers detection and treatment: a review. Int. J. Psychol. Res. 2010;3:58–67. [Google Scholar]
- 17.Liu F.T., Ting K.M., Zhou Z.-H. Proceedings of the 2008 Eighth IEEE International Conference on Data Mining. IEEE; Pisa, Italy: 2008. Isolation forest; pp. 413–422. [Google Scholar]
- 18.Celik M., Dadaser-Celik F., Dokuz A.S. Proceedings of the 2011 International Symposium on Innovations in Intelligent Systems and Applications. IEEE; Istanbul, Turkey: 2011. Anomaly detection in temperature data using DBSCAN algorithm; pp. 91–95. [Google Scholar]
- 19.Kukar M., Gunčar G., Vovko T., Podnar S., Černelč P., Brvar M., Zalaznik M., Notar M., Moškon S., Notar M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenenbaum J.B., Silva V. de, Langford J.C. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 21.Hsing T., Attoor S., Dougherty E. 2003. Relation between Permutation-Test P Values and Classfier Error Estimates. [Google Scholar]
- 22.Kistenev Y.V., Borisov A.V., Vrazhnov D.A. SPIE; 2021. Medical Applications of Laser Molecular Imaging and Machine Learning. [Google Scholar]
- 23.Wu J., Zhang P., Zhang L., Meng W., Li J., Tong C., Li Y., Cai J., Yang Z., Zhu J., et al. 2020. Rapid and Accurate Identification of COVID-19 Infection through Machine Learning Based on Clinical Available Blood Test Results. medRxiv. [Google Scholar]
- 24.Aktar S., Ahamad M.M., Rashed-Al-Mahfuz M., Azad A., Uddin S., Kamal A., Alyami S.A., Lin P.-I., Islam S.M.S., Quinn J.M., et al. Machine learning approach to predicting COVID-19 disease severity based on clinical blood test data: statistical analysis and model development. JMIR Med. Inf. 2021;9 doi: 10.2196/25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knapič S., Malhi A., Saluja R., Främling K. Explainable artificial intelligence for human decision support system in the medical domain. Mach. Learn. Knowl. Extr. 2021;3:740–770. [Google Scholar]
- 26.Cabitza F., Campagner A., Ferrari D., Di Resta C., Ceriotti D., Sabetta E., Colombini A., De Vecchi E., Banfi G., Locatelli M., et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med.(CCLM) 2021;59:421–431. doi: 10.1515/cclm-2020-1294. [DOI] [PubMed] [Google Scholar]
- 27.Plante T.B., Blau A.M., Berg A.N., Weinberg A.S., Jun I.C., Tapson V.F., Kanigan T.S., Adib A.B. Development and external validation of a machine learning tool to rule out COVID-19 among adults in the emergency department using routine blood tests: a large, multicenter, real-world study. J. Med. Internet Res. 2020;22:1–12. doi: 10.2196/24048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alves M.A., Castro G.Z., Oliveira B.A.S., Ferreira L.A., Ramírez J.A., Silva R., Guimarães F.G. Explaining machine learning based diagnosis of COVID-19 from routine blood tests with decision trees and Criteria graphs. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cobre A. de F., Stremel D.P., Noleto G.R., Fachi M.M., Surek M., Wiens A., Tonin F.S., Pontarolo R. Diagnosis and prediction of COVID-19 severity: can biochemical tests and machine learning Be used as prognostic indicators? Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babaei Rikan S., Sorayaie Azar A., Ghafari A., Bagherzadeh Mohasefi J., Pirnejad H. COVID-19 diagnosis from routine blood tests using artificial intelligence techniques. Biomed. Signal Process Control. 2022;72 doi: 10.1016/j.bspc.2021.103263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darapaneni N., Gupta M., Paduri A.R., Agrawal R., Padasali S., Kumari A., Purushothaman P. 2021. A novel machine learning based screening method for high-risk covid-19 patients based on simple blood exams. (2021 IEEE International IOT, Electronics and Mechatronics Conference, IEMTRONICS 2021 - Proceedings). [Google Scholar]
- 32.Bayat V., Phelps S., Ryono R., Lee C., Parekh H., Mewton J., Sedghi F., Etminani P., Holodniy M. A severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) prediction model from standard laboratory tests. Clin. Infect. Dis. 2021;73:E2901–E2907. doi: 10.1093/cid/ciaa1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gök E.C., Olgun M.O. SMOTE-NC and gradient boosting imputation based random forest classifier for predicting severity level of covid-19 patients with blood samples. Neural Comput. Appl. 2021;33:15693–15707. doi: 10.1007/s00521-021-06189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baktash V., Hosack T., Rule R., Patel N., Kho J., Sekhar R., Mandal A.K.J., Missouris C.G. Development. Evaluation and validation of machine learning algorithms to detect atypical and asymptomatic presentations of Covid-19 in hospital practice. QJM. 2021;114:496–501. doi: 10.1093/qjmed/hcab172. [DOI] [PubMed] [Google Scholar]
- 35.Eid M.M., Ibrahim A. vol. 2. 2021. Anemia Estimation for COVID-19 Patients using A Machine Learning Model; pp. 1–7. [Google Scholar]
- 36.Akhtar A., Akhtar S., Bakhtawar B., Kashif A.A., Aziz N., Javeid M.S. COVID-19 detection from CBC using machine learning techniques. Int. J. Technol., Innov. Manag. (IJTIM) 2021;1:65–78. [Google Scholar]
- 37.Hany N., Atef N., Mostafa N., Mohamed S., Elsahhar M., Abdelraouf A. 2021. Detection COVID-19 using machine learning from blood tests; pp. 229–234. (2021 International Mobile, Intelligent, and Ubiquitous Computing Conference, MIUCC 2021). [Google Scholar]
- 38.Almansoor M., Hewahi N.M. 2020. Exploring the relation between blood tests and Covid-19 using machine learning. (2020 International Conference on Data Analytics for Business and Industry: Way towards a Sustainable Economy, ICDABI 2020). [Google Scholar]
- 39.Du R., Tsougenis E.D., Ho J.W.K., Chan J.K.Y., Chiu K.W.H., Fang B.X.H., Ng M.Y., Leung S.T., Lo C.S.Y., Wong H.Y.F., et al. Machine learning application for the prediction of SARS-CoV-2 infection using blood tests and chest radiograph. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-93719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banoei M.M., Dinparastisaleh R., Zadeh A.V., Mirsaeidi M. Machine-learning-based COVID-19 mortality prediction model and identification of patients at low and high risk of dying. Crit. Care. 2021;25:1–14. doi: 10.1186/s13054-021-03749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aktar S., Ahamad Md.M., Rashed-Al-Mahfuz Md., Azad A., Uddin S., Kamal A.H.M., Alyami S.A., Lin P.-I., Islam S.M.S., Quinn J.M.W., et al. 2020. Predicting Patient COVID-19 Disease Severity by Means of Statistical and Machine Learning Analysis of Blood Cell Transcriptome Data. [Google Scholar]
- 42.Famiglini L., Bini G., Carobene A., Campagner A., Cabitza F. Prediction of ICU admission for COVID-19 patients: a machine learning approach based on complete blood count data. Comput. Base Med. Syst. 2021;2021:160–165. [Google Scholar]
- 43.Yao H., Zhang N., Zhang R., Duan M., Xie T., Pan J., Peng E., Huang J., Zhang Y., Xu X., et al. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front. Cell Dev. Biol. 2020;8:1–10. doi: 10.3389/fcell.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R. kun, Xiao Q., Zhu S. lang, Lin H. yan, Tang M. Using different machine learning models to classify patients into mild and severe cases of COVID-19 based on multivariate blood testing. J. Med. Virol. 2022;94:357–365. doi: 10.1002/jmv.27352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Assaf D., Gutman Y., Neuman Y., Segal G., Amit S., Gefen-Halevi S., Shilo N., Epstein A., Mor-Cohen R., Biber A., et al. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern. Emerg. Med. 2020;15:1435–1443. doi: 10.1007/s11739-020-02475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbosa V.A. de F., Gomes J.C., de Santana M.A., de Lima C.L., Calado R.B., Bertoldo Júnior C.R., Albuquerque J.E. de A., de Souza R.G., de Araújo R.J.E., Mattos Júnior L.A.R., et al. Covid-19 rapid test by combining a random forest-based Web system and blood tests. J. Biomol. Struct. Dyn. 2021:1–20. doi: 10.1080/07391102.2021.1966509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turabieh H., Ben Abdessalem Karaa W. Proceedings of the 2021 International Conference of Women in Data Science at Taif University (WiDSTaif) IEEE; Taif, Saudi Arabia: 2021. Predicting the existence of COVID-19 using machine learning based on laboratory findings; pp. 1–7. [Google Scholar]
- 48.Hu J. Detection of COVID-19 severity using blood gas analysis parameters and Harris hawks optimized extreme learning machine. Comput. Biol. Med. 2022:14. doi: 10.1016/j.compbiomed.2021.105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman-Meza D., Rudas A., Chiang J.N., Adamson P.C., Ebinger J., Sun N., Botting P., Fulcher J.A., Saab F.G., Brook R., et al. A machine learning algorithm to increase COVID-19 inpatient diagnostic capacity. PLoS One. 2020;15:1–10. doi: 10.1371/journal.pone.0239474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roland T., Boeck C., Tschoellitsch T., Maletzky A., Hochreiter S., Meier J., Klambauer G. 2021. Machine Learning Based COVID-19 Diagnosis from Blood Tests with Robustness to Domain Shifts. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao F.S., He Y., Liu J., Chen Y., Li Q., Zhang C.R., Han L., Zhu B., Ge Y., Chen S., et al. 2020. Triaging Moderate COVID-19 and other Viral Pneumonias from Routine Blood Tests; pp. 1–18. [Google Scholar]
- 52.Thimoteo L.M., Vellasco M.M., Amaral J., Figueiredo K., Yokoyama C.L., Marques E. Explainable artificial intelligence for COVID-19 diagnosis through blood test variables. J. Control, Autom. Electr. Syst. 2022 [Google Scholar]
- 53.Barbosa V.A. de F., Gomes J.C., Santana M.A. de, Albuquerque J.E. de A., Souza R.G. de, Souza R.E. de, Santos W.P. dos. 2020. Heg.IA: An Intelligent System to Support Diagnosis of Covid-19 Based on Blood Tests. [Google Scholar]
- 54.Faria S.P., Carpinteiro C., Pinto V., Rodrigues S.M., Alves J., Marques F., Lourenço M., Santos P.H., Ramos A., Cardoso M.J., et al. Forecasting covid-19 severity by intelligent optical fingerprinting of blood samples. Diagnostics. 2021;11:1–16. doi: 10.3390/diagnostics11081309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousif A.Y., Younis S.M., Hussein S.A., Al-Saidi N.M.G. An intelligent computing for diagnosing covid-19 using available blood tests. Int. J. Innov. Comput., Inf. Control. 2022;18:57–72. [Google Scholar]
- 56.Luo J., Zhou L., Feng Y., Li B., Guo S. The selection of indicators from initial blood routine test results to improve the accuracy of early prediction of COVID-19 severity. PLoS One. 2021;16:1–18. doi: 10.1371/journal.pone.0253329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan L., Zhang H.T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jin L., Zhang M., et al. 2020. A Machine Learning-Based Model for Survival Prediction in Patients with Severe COVID-19 Infection. medRxiv. [Google Scholar]
- 58.Patel D., Kher V., Desai B., Lei X., Cen S., Nanda N., Gholamrezanezhad A., Duddalwar V., Varghese B., Oberai A.A. Machine learning based predictors for COVID-19 disease severity. Sci. Rep. 2021;11:1–7. doi: 10.1038/s41598-021-83967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., Xia B., Ma X., Yang T., Yu F., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J., Zhong M., Zhang E., Hong K., Yang Q., Zhou D., Xia J., Chen Y.-Q., Sun M., Zhao B., et al. Broad phenotypic alterations and potential dysfunction of lymphocytes in individuals clinically recovered from COVID-19. J. Mol. Cell Biol. 2021;13:197–209. doi: 10.1093/jmcb/mjab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimaldi V., Benincasa G., Moccia G., Sansone A., Signoriello G., Napoli C. Evaluation of circulating leucocyte populations both in subjects with previous SARS-COV-2 infection and in Healthy subjects after vaccination. J. Immunol. Methods. 2022;502 doi: 10.1016/j.jim.2022.113230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez L., Pekkarinen P.T., Lakshmikanth T., Tan Z., Consiglio C.R., Pou C., Chen Y., Mugabo C.H., Nguyen N.A., Nowlan K., et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denzel A., Maus U.A., Gomez M.R., Moll C., Niedermeier M., Winter C., Maus R., Hollingshead S., Briles D.E., Kunz-Schughart L.A., et al. Basophils enhance immunological memory responses. Nat. Immunol. 2008;9:733–742. doi: 10.1038/ni.1621. [DOI] [PubMed] [Google Scholar]
- 64.Luan Y., Yin C., Yao Y. Update advances on C-reactive protein in COVID-19 and other viral infections. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.720363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo X., Zhou W., Yan X., Guo T., Wang B., Xia H., Ye L., Xiong J., Jiang Z., Liu Y., et al. Prognostic value of C-reactive protein in patients with coronavirus 2019. Clin. Infect. Dis. 2020;71:2174–2179. doi: 10.1093/cid/ciaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bisoendial R.J., Kastelein J.J.P., Levels J.H.M., Zwaginga J.J., van den Bogaard B., Reitsma P.H., Meijers J.C.M., Hartman D., Levi M., Stroes E.S.G. Activation of inflammation and coagulation after infusion of C-reactive protein in humans. Circ. Res. 2005;96:714–716. doi: 10.1161/01.RES.0000163015.67711.AB. [DOI] [PubMed] [Google Scholar]
- 69.Zou J.-N., Sun L., Wang B.-R., Zou Y., Xu S., Ding Y.-J., Shen L.-J., Huang W.-C., Jiang X.-J., Chen S.-M. The characteristics and evolution of pulmonary fibrosis in COVID-19 patients as assessed by AI-assisted chest HRCT. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X., Chen D., Wang L., Zhao Y., Wei L., Chen Z., Yang B. Low serum calcium: a new, important indicator of COVID-19 patients from mild/moderate to severe/critical. Biosci. Rep. 2020:40. doi: 10.1042/BSR20202690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vargas-Vargas M., Cortés-Rojo C. Ferritin levels and COVID-19. Rev. Panam. Salud Públic. 2020;44:1. doi: 10.26633/RPSP.2020.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fox S.E., Akmatbekov A., Harbert J.L., Li G., Quincy Brown J., Vander Heide R.S. Pulmonary and cardiac pathology in african American patients with COVID-19: an autopsy series from new orleans. Lancet Respir. Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paliogiannis P., Zinellu A. Bilirubin levels in patients with mild and severe covid-19: a pooled analysis. Liver Int. 2020;40:1787–1788. doi: 10.1111/liv.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qian Z.P., Mei X., Zhang Y.Y., Zou Y., Zhang Z.G., Zhu H., Guo H.Y., Liu Y., Ling Y., Zhang X.Y., Wang J.F., Lu H. Analysis of baseline liver biochemical parameters in 324 cases with novel coronavirus pneumonia in shanghai area. Zhonghua Gan Zang Bing Za Zhi. 2020;28:229–233. doi: 10.3760/cma.j.cn501113-20200229-00076. [DOI] [PubMed] [Google Scholar]
- 75.Zhang X., Cai H., Hu J., Lian J., Gu J., Zhang S., Ye C., Lu Y., Jin C., Yu G., et al. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int. J. Infect. Dis. 2020;94:81–87. doi: 10.1016/j.ijid.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rostami M., Mansouritorghabeh H. D-dimer level in COVID-19 infection: a systematic review. Expet Rev. Hematol. 2020;13:1265–1275. doi: 10.1080/17474086.2020.1831383. [DOI] [PubMed] [Google Scholar]
- 77.Chen D. Analysis of machine learning methods for COVID-19 detection using serum Raman spectroscopy. Appl. Artif. Intell. 2021;35:1147–1168. [Google Scholar]
- 78.Lazari L.C., de Rose Ghilardi F., Rosa-Fernandes L., Assis D.M., Nicolau J.C., Santiago V.F., Dalçóquio T.F., Angeli C.B., Bertolin A.J., Marinho C.R.F., et al. Prognostic accuracy of MALDI-TOF mass spectrometric analysis of plasma in COVID-19. Life Sci. Alliance. 2021;4:1–12. doi: 10.26508/lsa.202000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pulgar-Sánchez M., Chamorro K., Fors M., Mora F.X., Ramírez H., Fernandez-Moreira E., Ballaz S.J. Biomarkers of severe COVID-19 pneumonia on admission using data-mining powered by common laboratory blood tests-datasets. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jarrom D., Elston L., Washington J., Prettyjohns M., Cann K., Myles S., Groves P. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ EBM. 2022;27:33–45. doi: 10.1136/bmjebm-2020-111511. [DOI] [PubMed] [Google Scholar]
- 82.Teymouri M., Mollazadeh S., Mortazavi H., Naderi Ghale-noie Z., Keyvani V., Aghababaei F., Hamblin M.R., Abbaszadeh-Goudarzi G., Pourghadamyari H., Hashemian S.M.R., et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021;221 doi: 10.1016/j.prp.2021.153443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith M., Alvarez F. Identifying mortality factors from machine learning using Shapley values – a case of COVID19. Expert Syst. Appl. 2021;176 doi: 10.1016/j.eswa.2021.114832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davazdahemami, B. An Explanatory Machine Learning Framework for Studying Pandemics: the Case of COVID-19 Emergency Department Readmissions. 13. [DOI] [PMC free article] [PubMed]
- 85.Doewes R.I., Nair R., Sharma T. Diagnosis of COVID-19 through blood sample using ensemble genetic algorithms and machine learning classifier. World J. Eng. 2021 [Google Scholar]
- 86.Raihan M.M.S., Khan M.M.U., Akter L., Shams A.B. 2021. Development of Risk-free COVID-19 Screening Algorithm from Routine Blood Test Using Ensemble Machine Learning. arXiv:2108.05660 [cs, q-bio, stat] [Google Scholar]
- 87.AlJame M. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked. 2020;10 doi: 10.1016/j.imu.2020.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Navarro R.J.M., Gallego P.J., Borrás R.F., Calpena R.R., Ruiz M.J.A., Morcillo R.M.Á. Is it possible to predict the presence of colorectal cancer in a blood test? A probabilistic approach method. Rev. Esp. Enferm. Dig. 2017;109(10):694–703. doi: 10.17235/reed.2017.4645/2016. PMID: 28929777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at www.kaggle.com/einsteindata4u/covid19, 2020. Online
https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/discussion/139347.