Abstract
SARS-CoV-2 is a coronavirus that has created a global pandemic. The virus contains a spike protein which has been shown to bind to the ACE2 receptor on the surface of human cells. Vaccines have been developed that recognize elements of the SARS-CoV-2 spike protein and they have been successful in preventing infection. Recently, the Omicron variant of the SARS-CoV-2 virus was reported and quickly became a variant of concern due to its transmissibility. This variant contained an unusually large number (32) of point mutations, of which 15 of those mutations are in the receptor binding domain of the spike protein. While several computational and experimental investigations comparing the binding of the Omicron and wild type RBD to the human ACE2 receptor have been conducted, many of these report contradictory findings. In order to assess the differential binding ability, we conducted 2 μs of classical molecular dynamics (cMD) simulation to estimate the binding affinities and behaviors. Based upon MM-GBSA binding affinity, per-residue energy decomposition analysis, center of mass distance measurements, ensemble clustering, pairwise residue decomposition and hydrogen bonding analysis, our results suggest that a single point mutation is responsible for the enhanced binding of the Omicron mutant relative to the WT. While the 15-point mutations in the receptor binding domain contribute positively and negatively to the affinity of the spike protein for the human ACE2 receptor, it is the point mutation Q493R that confers enhanced binding while the Q493K mutation results in similar binding. The MM-GBSA binding estimations over a 2 μs trajectory, suggest that the wild type binds to ACE2 with a value of −29.69 kcal/mol while the Q493K and Q493R Omicron mutants bind with energy values of −26.67 and −34.56 kcal/mol, respectively. These values are significantly different, given the error estimates associated with the MM-GBSA method. In general, while some mutations increase binding, more mutations diminish binding, leading to an overall similar picture of binding for Q493K and enhanced binding for Q493R.
Keywords: SARS-CoV-2, COVID-19, Omicron, Residue mutations, Spike protein, Receptor binding domain, Human ACE2 receptor, Molecular dynamics, MM-GBSA
Graphical abstract
1. Introduction
Coronavirus disease-2019 (COVID-19) is a global pandemic caused by infection with the severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) [[1], [2], [3]]. The virus has been circulating globally since late 2019, and has evolved genetically [4]. In the first 11 months of the pandemic, viral mutations were occurring at a relatively slow rate; however, since late 2020 mutational variants of concern (VOC) have been identified (Table 1 ). VOCs such as the Alpha, Beta, Delta and Omicron may be associated with enhanced viral fitness and typically display increased transmissibility and infectivity characteristics [[4], [5], [6]].
Table 1.
Summary and timeline of SARS-CoV-2 Mutational Variants with a particular focus on the Spike RBD [4,15,16]. The sequence of the wild type Wuhan Hu-1 Spike RBD [17] against which early vaccines were prepared is shown in Fig. S1. The ∗ indicates conflicting reports regarding which mutation is present.
| WHO Designation | Detected | S-RBD Mutation(s) | Comments |
|---|---|---|---|
| Alpha | Sept 2020 | N501Y | Increased transmission [18] |
| Beta | Oct 2020 | K417 N, E484K, N501Y | Binds ACE2 receptor with 4.62 times greater affinity than Wuhan Hu-1 spike RBD [8,19,20]. Increased transmission rates [21] |
| Delta | late 2020 | K417 N, L452R, T478K | Increased transmission |
| Gamma | Jan 2021 | L452R, T478K | Increased transmission |
| Omicron B.1.1.529 | Nov 2021 | G339D, S371L, S373P, S375F, K417 N, N440K, G446S, S477 N, T478K, E484A, Q493 K/R∗, G496S, Q498R, N501Y, Y505H | Concerning number (15) of mutations in the Spike protein RBD |
The SARS-CoV-2 virus encodes for a spike protein that contains a receptor binding domain (RBD) that binds favorably to the ACE2 receptor present on the surface of human throat cells, and lung epithelial cells [7,8]. This binding creates a fusion between the human cell membrane and the spike protein, allowing the viral genetic material to be replicated within the human host cell [9].
In December 2020, the United States Food and Drug Administration issued emergency use authorization (EUA) for mRNA vaccines developed by Pfizer-BioNTech and Moderna, followed quickly by EUAs for the adenovirus (viral vector) vaccines developed by Johnson & Johnson and Astra-Zeneca [10]. These vaccines produced multiple antibodies to various regions of the SARS-CoV-2 spike protein including antibodies that targeted the RBD of the spike protein, thereby preventing the virus from anchoring to the human ACE2 receptor and preventing host cell entry [11,12]. The vaccines were extremely effective; reducing hospitalizations and deaths among the vaccinated [13,14].
The vaccines continued to provide protection from mutated forms of the virus [4]. Breakthrough infections were known, but hospitalizations and deaths among the vaccinated remained low. In November 2021, a mutant variant was detected in a variety of locations around the world. This mutant, labeled as Omicron by the World Health Organization, contained a much larger number of mutations than had previously been observed in the SARS-CoV-2 virus and initial reports displayed a concerning rate of transmission [22,23].
There is concern that mutations in the RBD region of the spike protein may increase the ability of the spike to bind to the human ACE2 receptor or allow it to evade polyclonal antibody recognition (Fig. 1 ). A number of spike protein residues in the wild type (WT) Wuhan Hu-1 RBD (Lys417; Glu484, Gln493, Gln498, Gly496, Asn501, Tyr505 and Gly446) that were mutated in the Omicron variant have previously shown persistent interactions with the hACE2 receptor [8,24]. Mutations at positions 498 and 501 are particularly worrisome as in-vitro evolution studies suggest that such mutations increase the binding between the spike protein and hACE2 [25]. Tyr449 may also be an important residue for interaction with ACE2 [24] while the N439K spike protein variant may increase ACE2 affinity by the formation of a new salt bridge at the RBD-ACE2 interface with Glu329 of hACE2 [26].
Fig. 1.
Omicron SARS-CoV-2 RBD - hACE2 interaction. Omicron SARS-CoV-2 RBD is displayed in blue and hACE2 is displayed in a lighter blue. In green are the Omicron SARS-CoV-2 RBD residue mutations: G339D, S371L, S373P, S375F, K417 N, N440K, G446S, S477 N, T478K, E484A, Q493K, G496S, Q498R, N501Y, and Y505H. The binding site for the RBD - hACE2 interaction and the residues K417 N, G446S, S477 N, T478K, E484A, Q493K, G496S, N501Y, and Y505H are shown in the inset.
While several studies comparing ACE2 binding of the Omicron and wild type RBD have been conducted, results of these studies appear to be largely contradictory. In a study by Han et al., a flow cytometry assay and surface plasmon resonance (SPR) were used to analyze the binding affinity of multiple RBD variants (Alpha, Beta, Delta, Gamma, and Omicron) with hACE2-expressing BHK-21 cells [27]. These results suggest that the Omicron RBD binds with similar affinity to the WT RBD. However, Geng et al. also used SPR to study the RBD binding affinity and found a 3.9 fold increase in Omicron RBD binding relative to WT [28]. Geng et al. noted that the Han SPR study used a CM5 chip coated with random ACE2 orientations, whereas their study used a protein A chip coated with fixed ACE2 orientations to ensure that the binding surface of ACE2 was fully accessible. Using a CM5 chip coated with random orientations similar to the Han study, Geng et al. found a 1.7-fold increase favoring Omicron. Cui et al. used SPR to demonstrate that the Omicron RBD binds to hACE2 with a 2.8-fold increase relative to the WT RBD [29]. Schubert et al. analyzed ACE2 - RBD binding using titration enzyme-linked immunoassay (ELISA) on RBDs from the WT and variant strains produced in insect cells [30]. The ELISA results indicate that the Omicron RBD binds less tightly to ACE2 (EC50 = 150 ng/mL, 5.6 nM) compared to the WT (EC50 = 120 ng/mL, 4.6 nM), while other variants such as Beta and Delta bind more tightly. The same study also used microscale thermophoresis (MST) and found that the Omicron binding affinity was slightly lower than for WT, Beta and Delta.
In an accelerated molecular dynamics (aMD) study, da Costa et al. used MM-GBSA, principal component analysis (PCA), and per-residue decomposition energy to suggest that the Omicron RBD binds to hACE2 with an enhanced binding affinity [31]. They found that the MM-GBSA binding energy for WT was −59.7 kcal/mol and, after manually mutating a WT SARS-CoV-2 RBD - hACE2 structure (PDB 6M0J) to match the Omicron RBD mutations, Omicron was −75.4 kcal/mol. The binding free energies reported in the da Costa et al. study are larger than the ensemble average as MM-GBSA was only performed on selected frames representing the most stable structures determined by PCA. In a cMD study conducted by Kumar et al. on a WT SARS-CoV-2 RBD - hACE2 structure (PDB 7A91) mutated to match the Omicron RBD mutations, MM-GBSA binding free energy analysis was conducted over frames corresponding to the last 10 ns of their 100 ns trajectories [32]. For the WT and Omicron RBD respectively, binding free energies of −32.43 and −41.00 kcal/mol were computed (standard deviations were not reported). Both studies utilized the ff14SB protein and TIP3P water/ion force fields.
Taken together, the previously reported experimental and computational studies of SARS-CoV-2 RBD binding to human ACE2 suggest that this is a complex system, and while it appears that the Omicron RBD may bind more strongly than the WT, results seem to depend on the experimental or computational design, and the quantitative and atomistic details remain elusive.
In this report, we utilize cMD simulations, structural analysis of a mutated WT structure (PDB 6LZG) [33] and MM-GBSA binding estimations to compare the binding properties of the Wuhan Hu-1 and Omicron RBD spike protein to the hACE2 receptor. We compare the binding of a WT RBD structure (PDB 6LZG) and two mutated models (both containing all Omicron mutations but differing at residue 493) using cMD simulation and MM-GBSA binding free energy estimation. Molecular simulations play a critically important role in understanding the atomistic nature of protein dynamics and protein-ligand binding affinities [[34], [35], [36], [37]]. Since the pandemic began, there have been multiple reports describing the use of molecular dynamics and MM-GBSA to understand SARS-CoV-2 behavior. MM-GBSA has been used to estimate hACE2 binding with a.) the WT SARS-CoV-2 spike protein RBD [38], b.) SARS-CoV and SARS-CoV-2 [24,39,40], c.) in vitro single point spike mutations [41], d.) Alpha, Kappa and Delta mutants [42], and e.) RBD hotspot mutations [43]. MM-GBSA has also been used for an evaluation of the binding affinity between the SARS-CoV-2 spike RBD and the ACE2 receptor from various mammals (human, monkey, hamster, ferret, dog, cat) [44].
In this study, we analyze how the Omicron mutations affect the binding of the spike protein to the hACE2 receptor by performing molecular dynamics and binding free energy computations on the wild type (WT) and Omicron mutant versions. We find that in spite of the significant number of mutations in the RBD of the spike protein, the binding affinity between the WT virus and an Omicron model containing mutation Q493K are similar. We do see structural differences, and these are described in some detail; however, these do not lead to an overall increase in hACE2 affinity. However, with an Omicron model containing the Q493R mutation, we find that Omicron binds more tightly. Given the variability of information described above regarding RBD binding, this work seeks to systematically provide an atomistic evaluation of such binding.
2. Methods
2.1. Protein retrieval and preparation
An hACE2 - SARS-CoV-2 RBD crystal structure was obtained from the Protein Data Bank (PDB Code 6LZG) [51]. Chains A (hACE2) and B (SARS-CoV-2 RBD) were selected from 6LZG, and all waters were removed. The Omicron structure was computationally constructed according to the 15 RBD point mutations shown in red in Fig. S2. Schrödinger's Protein Preparation Wizard was used to add missing hydrogen atoms, assign bond orders according to the CCD database, fill missing side chains using Prime, predict side chain protonation states using Epik with a pH range of 7 ± 2, and optimize H-bonds using PROPKA at a pH of 7 [[52], [53], [54]]. Schrodinger's Protein Preparation Wizard was used to sample side chain conformers of amino acid residues Asn, Gln, His, Asp, and Glu during the optimization of H-bonding. Restrained minimization was then performed using the OPLS3e force field [55].
2.2. Molecular dynamics simulations
Unrestrained classical molecular dynamics (cMD) was performed on the binary complex of SARS-CoV-2 RBD with hACE2 using the GPU-accelerated pmemd code of AMBER18 [[56], [57], [58]]. The ff14SB and Glycam06j force fields were used to model standard amino acids, and glycosylated amino acids/glycans, respectively [59,60]. All models were neutralized with Na+ ions and explicitly solvated in a TIP3P [61] unit cell using the program tleap [56]. Further details describing the MD protocol can be found in the Supplemental Information. Initially, 20 100 ns trajectories were generated, each using different seeds to speed surface coverage. We concatenated the 10 seeds from each of the different spike RBD structures to obtain 1 μs ensembles for wild type and both mutant binary complexes, initiated using the 6LZG experimental structure. We then extended these seeds by another 100 ns (200 ns total) and produced concatenated 2 μs ensembles for the wild type and mutant complexes.
2.3. Molecular dynamics analyses
Trajectory visualization was conducted using UCSF Chimera and UCSF ChimeraX [62,63]. From this analysis we confirm that amino acid side chains sampled all possible rotamers as part of the cMD simulation. This is in addition to the rotamer analysis and screening that was performed as part of our initial protein preparation. Using the AmberTools MMPBSA.py package, MM-GBSA binding free energies and per-residue decomposition energies were calculated for every frame, and pairwise decomposition energies were obtained for frames at a 1 ns interval [64]. Hydrogen bonding, center-of-mass distance (COM), root-mean-squared deviation (RMSD), root-mean-squared fluctuation (RMSF), secondary structure, backbone atom RMSD-based clustering, and non-hydrogen atom pairwise distance-based clustering analyses were conducted using the AmberTools cpptraj module [56]. Each clustering method resulted in 10 families per model.
3. Results and discussion
Our overarching goal is to compare the hACE2 binding behavior of the RBD in the Hu-1 spike protein (Wild Type (WT)) to the mutated Omicron RBD spike protein. When this work was first initiated in November 2021, there were no experimental structures of the Omicron mutant RBD in either apo form, or bound to hACE2. Therefore, we built an Omicron spike RBD by making in silico mutations of the RBD using the WT-ACE2 experimental structure (PDB: 6LZG). Our approach assumed that the 15 mutations present in the Omicron variant do not change significantly the conformation of the spike RBD, and that atomic relaxation via local minimization is enough to stabilize our in silico mutated structure.
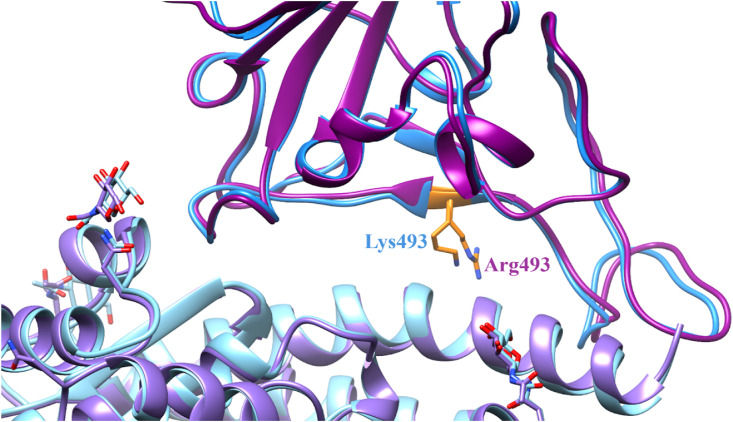
While our current study was under review, experimental structures of the Omicron SARS-CoV-2 RBD complexed with the hACE2 receptor have become available, including the cryo-EM Omicron B.1.1.529 variant structure published by Guo et al. (PDB: 7WSA) [65]. Our detailed residue comparison of the 6LZG and 7WSA structure can be found in the Supporting Information (S1 Note). A superimposition of 7WSA with our mutated 6LZG Omicron model shows that the model is structurally true to the experimental structure (Fig. 2 and S3). There are additional residues on both the 6LZG and 7WSA structures but they are well removed from the binding site. From this comparison, we believe that our results using 6LZG are representative of the results that would be obtained using the 7WSA structure, i.e. these structures have a similar capacity for use in determining the binding ability of the Omicron RBD to the hACE2 receptor (Table 2 ).
Fig. 2.
Comparison of the 6LZG and 7WSA binding sites. The 6LZG structure is in blue with the RBD depicted in dark blue and the hACE2 receptor depicted in light blue. The 7WSA structure is in purple with the RBD depicted in dark purple and the hACE2 receptor depicted in light purple. The Omicron mutation of the residue 493 is highlighted in orange on both structures. On the 6LZG WT structure residue 493 is a lysine and on the 7WSA Omicron structure this residue is an arginine. Both mutated residues have similar lengths and placement of the side chain conformers.
Table 2.
Experimental and Computational Binding Data for Complexes of SARS-CoV-2 Variants with hACE2.
| KD (nM) | Ref. | |
|---|---|---|
| WT | 60 ± 1.4 | [45] |
| 13.20 | [46] | |
| 24.63 ± 5.00 | [27] | |
| 16.6 ± 8.4 | [47] | |
| 22.0 | [48] | |
| 68.3 | [29] | |
| Omicron |
25.3 ± 1.2 | [45] |
| 8.85 | [46] | |
| 31.40 ± 11.62 | [27] | |
| 270.27 ± 3403.94 | [47] | |
| 24.4 | [29] | |
| KAFF (L/mol) |
Ref. |
|
| WT | 6.01 ± 3.02 x 107 | [49] |
| Omicron |
0.37 ± 4.66 x 107 | [49] |
| Computational ΔG estimation (kcal/mol) |
Ref. |
|
| WT | −33.13 ± 3.26 | [49] |
| −18.32 ± 1.62 | [50] | |
| −32.43 | [32] | |
| −59.7 | [31] | |
| Omicron | −29.43 ± 3.01 | [49] |
| −30.21 ± 4.48 | [50] | |
| −41 | [32] | |
| −75.4 | [31] |
Using the initial WT structure (6LZG), as well as the computationally mutated Omicron-6LZG structure, we first performed 100 ns of molecular dynamics simulation using 10 different, randomly selected, initial seeds. This generated 1 μs ensembles for each molecular system. We then extended these 10 seeds by another 100 ns each (for a total of 200 ns per seed) and conducted the same analyses on the resultant 2 μs ensembles. No significant differences in ensemble analyses were detected, and the MM-GBSA binding free energy from the 1 and 2 μs ensembles were similar (Table 3 ). As such, all further analyses were conducted on the 1 μs ensembles for computational efficiency, except where specified.
Table 3.
MM-GBSA Binding Energy (kcal/mol) for the 1 and 2 μs ensembles of WT and mutants SARS-CoV-2 RBD with hACE2.
|
Model |
1 μs |
2 μs |
||||
|---|---|---|---|---|---|---|
| MM-GBSA Avg. | Std. Dev. | Std Error Mean | MM-GBSA Avg. | Std. Dev. | Std Error Mean | |
| WT | −28.45 | 11.33 | 0.11 | −29.69 | 10.61 | 0.07 |
| Omicron (K493) | −25.61 | 7.57 | 0.08 | −26.67 | 7.31 | 0.05 |
| Omicron (R493) | −34.82 | 8.44 | 0.08 | −34.56 | 8.24 | 0.06 |
To assess conformational dynamics and simulation convergence, we computed the RMSD of each ensemble conformation, relative to the corresponding initial structure. Apart from a brief increase in RMSD between 20 and 40 ns of WT seed 1 (Fig. S4 top), increased RMSD from 30 ns onwards in seed 5 of the Omicron Q493K model (Fig. S4 middle), and a brief increase in RMSD between 100 and 150 ns in seed 4 of the Omicron Q493R model (Fig. S4 bottom), RMSD analysis shows that the WT and both Omicron mutant complexes are relatively well converged across each seed, with RMSD values ranging from 2 to 3.5 Å. There are no significantly notable differences in the RMSD behavior of the WT and Omicron models. This suggests thorough sampling of the dynamics of each complex and that the ensembles we are using to estimate binding free energy are conformationally converged.
We also performed root mean square fluctuation (RMSF) per residue analysis on each 1 μs ensemble (Fig. 3 ). This analysis indicates that the fluctuations of individual residues are also well conserved between seeds. Comparison of the RMSF trajectories for the WT and both Omicron models suggest relatively similar residue movements with the only areas of marked difference occurring between spike residues 358 to 376 and 384 to 390. The region 358 to 376 contains the point mutations S371L, S373P, and S375F, while the region 384 to 390 does not contain any mutations and is relatively distant from spike residues responsible for hACE2 binding, but is notably 10 residues away from the proline point mutation S373P. While all models show differences in fluctuation in the region 384 to 390, only the Q493R model shows increased fluctuation in the region 358 to 376. Secondary structure analysis for the region 358 to 376 indicates the occurrence of alpha and 3–10 helices between residues 365 to 371 with a frequency of 95.29, 94.26, and 87.71% for the WT, Q493K, and Q493R models, respectively. The decreased helical character of Q493R may explain the increase in RMSF. Secondary structure analysis for the region 384–390 indicates that the decrease in RMSF for Omicron Q493K results from the formation of a 3–10 helix involving residues L387, N388, and D389. Within the Omicron Q493K ensemble, this helix occurs with a frequency of 69.26% while the corresponding region of the WT complex shows helical secondary structure in only 4.25% of the RBD ensemble (Fig. 3). While the Q493R ensemble shows this helix occurring for 33.78%, increased RMSF in this region may be due to both decreased helical character compared to the Q493K model and increased turn and bend frequencies compared to the WT and Q493K models. For the WT, Q493K, and Q493R models respectively, combined turn and bend frequencies for residue 387 are 15.21, 14.33, and 22.23%. Likewise, these frequencies are 60.83, 26.96, and 53.34% for residue 388 and 37.06, 22.14, and 41.62% for residue 389. Secondary structure plots over time are shown in Figs. S5–S10.
Fig. 3.
Wild Type and Omicron SARS-CoV-2 RBD and hACE2 Root Mean Square Fluctuation Graphs. [A] RMSF graph of the WT and Omicron RBD concatenated 1 μs trajectories. Notably, differences in RMSF in RBD residues 358–376 and 384–390 are highlighted in the black box. [B] The Omicron Q493K RBD with the WT RBD superimposed. The most prevalent average structures of both models are shown. The Omicron Q493K RBD is displayed in blue and hACE2 in lighter blue, while the WT RBD and hACE2 are displayed in the same colors with a transparency effect applied. Amino acids highlighted in green are mutated residues that are notably close to the residues highlighted in red. In orange are WT RBD residues 384–390.
Early reports (Nov–Dec 2021) described the Omicron variant as containing mutation Q493K [30] whereas the 7WSA structure contains the Q493R mutation [27]. Arg and Lys are both positively charged amino acid residues, with similar side chains - if there is an effect - previous systematic mutational studies using the SARS-CoV-2 RBD would suggest that a Lys to Arg mutation at residue 493 would decrease the RBD binding affinity [66]. To assess this difference, we ran 2 μs of cMD simulation on 6LZG with Q493R, for comparison to the Q493K and WT behavior, and computed MM-GBSA binding estimations (Table 3) and per-residue decomposition analysis (Table 4 ) for all ensembles. These results suggest similar hACE2 binding behavior between the WT and Omicron Q493K but significantly enhanced affinity for the Q493R mutant.
Table 4.
Determining the Effect of Omicron Lys vs Arg Mutation at Residue 493. Per-residue Decomposition Energy (kcal/mol) for the 2 μs ensembles of Q493K and Q493R mutant SARS-CoV-2 RBD with hACE2.
| Model | Per-residue Decomp. | Std. Dev. | Std Error Mean |
|---|---|---|---|
| WT (Q493) | −4.60 | 1.90 | 0.01 |
| Omicron (K493) | −5.05 | 2.62 | 0.02 |
| Omicron (R493) | −9.86 | 3.49 | 0.02 |
From this we see that residue 493 plays a significant, perhaps outsized role in the binding of RBD to hACE2; a single point mutation causes Omicron RBD to bind significantly more tightly to hACE2 than the WT. This may explain the conflicting results in the literature, i.e. if previous computational or experimental studies utilized the Q493R mutant they will show enhanced Omicron binding but if they used the Q493K mutant binding between Omicron and WT will be similar. For instance, the Schubert et al. ELISA study used the originally available Q493K mutation and they report slightly decreased binding of the Omicron RBD. This is consistent with our findings for the Q493K mutant [30].
The computational studies described in the introduction utilized the Q493R mutation so our results are in agreement with these earlier findings [31,32]. However, unlike these studies, we conducted our MM-GBSA analysis over all frames of our concatenated trajectory while da Costa et al. and Kumar at al. conducted MM-GBSA analysis on only the most stable structure and the last 10 ns of their trajectory, respectively. As such, our report provides long time scale information and a more detailed atomistic picture of binding.
Per-residue decomposition energies were calculated for both the WT and Omicron 2 μs ensemble. Table S1 displays the values that provide a more negative contribution than −1 kcal/mol to the binding energy while Fig. 4 highlights the 15 residues that undergo mutation from WT (blue) to Omicron Q493K (orange) and Omicron Q493R (green). There are 8 unmutated residues that are significant for binding of both the WT and Omicron (Q493K and Q493R) RBD: F486F, F456F, Y489Y, L455L, G502G, N487 N, A475A, and T500T. There are 4 mutated residues that are important for binding of both WT and Omicron (Q493K and Q493R): Q493 K/R, N501Y, Y505H and G496S. Residues Y449 and Q498 were significant only for WT, and the 477 N mutant was significant only for the Omicron models. While residue 449 remains a tyrosine in both WT and Omicron mutants, there is a sizable decrease in the per-residue contribution (WT: -1.35 ± 1.00; Omicron Q493K: -0.11 ± 0.32; and Omicron Q493R −0.15 ± 0.41 kcal/mol). As Fig. 4 shows, the mutations S477 N, Q493 K/R, G496S, and Q498R are significant for both the WT and Omicron RBD interactions with hACE2; however, there is a notable decrease for the mutations Q498R (WT: -2.98 ± 3.21; Omicron Q493K: -0.16 ± 1.26; and Omicron Q493R: -0.30 ± 1.62 kcal/mol) and Y505H (WT: -4.92 ± 0.98; Omicron Q493K: -3.09 ± 1.32; and Omicron Q493R −3.33 ± 1.61 kcal/mol), and a notable increase for the mutation S477 N (WT: -0.31 ± 0.82; Omicron Q493K: -1.10 ± 1.15; and Omicron Q493R: -1.02 ± 1.17 kcal/mol). There is a significantly favorable free energy contribution for the mutant N501Y (WT: -2.05 ± 1.19; Omicron Q493K: -6.69 ± 1.17; and Omicron Q493R −6.46 ± 1.15 kcal/mol). For both Omicron models, the per-residue decomposition energies of the mutations are consistent; however, there is a notable per-residue contribution difference between the Q493K and Q493R mutations (Table 4).
Fig. 4.
WT and Omicron Per-Residue Decomposition of SARS-CoV-2 RBD Mutated Residues. This bar graph demonstrates the per-residue decomposition energies for the 15 WT and Omicron (Q493K and Q493R) mutated RBD residues. The energies depicted are WT (blue), Omicron Q493K (orange) and Omicron Q493R (green). Standard deviations are reported in black.
A previous cMD report by Kumar et al., focusing on the last 10 ns of a 100 ns trajectory using the Q493R mutant, suggested that Omicron residues 475–477, 489, 493 and 501 contribute significantly to enhancing the RBD - hACE2 interaction [32]. As can be seen in Fig. 4 and S1, our results averaged over longer (2000 ns) trajectories agree that the N501Y and S477 N mutations confer enhanced binding affinity for Omicron relative to WT. Our MM-GBSA and per-residue decomposition results also strongly suggest that the Q493R mutation enhances Omicron binding affinity; however, the Q493K mutation shows similar binding between Omicron and WT. Our per-residue results suggest that the other residues identified in the Kumar study either do not contribute (476) or contribute significantly in WT and mutant (475, 489, Q493K; Table S1). To further understand the binding affinity of the Omicron (Q493K and Q493R) and WT RBD we conducted center-of-mass, pairwise decomposition and hydrogen bonding analyses described below.
As previously reported, molecular dynamics was used by Ali and Vijayan to characterize residues responsible for the affinity between the WT RBD of the spike protein and hACE2 [24]. To better understand the Omicron RBD – hACE2 interactions and the relative proximity of each monomer, we used the 15 residue contacts of Ali and Vijayan (listed in Table S2) to perform an averaged center of mass distance analysis between WT and Omicron RBD and hACE2. Our data suggests few differences between WT and Omicron interactions except for the RBD Q493 K/R – hACE2 K31 contact which shows significant increase in both α carbon (WT: 8.53 ± 0.49; Omicron Q493K: 9.32 ± 0.51; and Omicron Q493R: 9.46 ± 0.49 Å) and end-to-end distances (WT: 4.00 ± 0.99; Omicron Q493K: 6.88 ± 1.11; and Omicron Q493R: 6.45 ± 0.86 Å), and the RBD K417 N – hACE2 D30 contact which shows significant all-atom (WT: 8.29 ± 0.68; Omicron Q493K: 9.75 ± 0.74; and Omicron Q493R: 9.81 ± 0.75 Å) and end-to-end distance increases (WT: 4.04 ± 1.30; Omicron Q493K: 7.85 ± 1.30; and Omicron Q493R: 8.41 ± 1.42 Å) (Table S3). These distance increases corresponding with the Q493 K/R and K417 N mutations are visualized in the graphs of Table S4. These COM increases make sense as in the first case (RBD Q493 K/R – hACE2 K31) it is a neutral residue (Q493) in the WT interacting with a positively charged residue (K31), and that is replaced with a positively charged residue (493K), leading to an unfavorable electrostatically repulsive interaction. In the second case (RBD K417 N – hACE2 D30), a positively charged residue in the WT (K417) interacting with a negatively charged hACE2 residue (D30) is replaced in the mutant with a neutral residue (417 N) reducing what was an electrostatically favorable interaction. We also see that, within the large standard deviations shown for the WT contacts RBD Q498R - hACE2 D38 and RBD Q498R - hACE2 K353, the end-to-end distances increase in the mutant (WT: 6.88 ± 2.38; Omicron Q493K: 8.21 ± 1.38; Omicron Q493R: 7.71 ± 1.20 Å, and WT: 5.51 ± 2.25; Omicron Q493K: 8.73 ± 1.29; Omicron Q493R: 9.05 ± 1.01 Å, respectively) while the end-to-end distance for RBD K417 N – hACE2 H34 decreases in the Omicron mutant (WT: 7.93 ± 1.78; Omicron Q493K: 6.66 ± 0.87; and Omicron Q493R: 6.70 ± 0.80 Å). In the case of Q493K and Q498R, the mutations replace a neutral glutamine residue with a charged lysine or arginine, whereas for K417 N, the mutation replaces a charged lysine with an uncharged asparagine. This further suggests that the Omicron mutations produce both increased favorable and unfavorable interactions relative to the WT. The graphs in Table S4 suggest that a ∼4 Å end-to-end distance for contacts between RBD Q498R – hACE2 D38, and RBD Q498R – hACE2 K353, rarely occur with Omicron. Notably, despite not being subject to mutation, the RDB Y449 – hACE2 D38 interaction also shows an increased average distance in the mutant (WT: 4.50 ± 2.53; Omicron Q493K: 6.85 ± 1.58; and Omicron Q493R: 7.80 ± 3.37 Å) and little occurrence of a ∼4 Å end-to-end distance contact in the Omicron trajectories.
Pairwise decomposition analysis is largely consistent with our MM-GBSA results. A simple summation of each favorable interacting residue pair (Table 5 ) yields total energies of −54.68, −59.63, and −57.73 kcal/mol for the WT and Omicron Q493K and Q493R models respectively. Likewise, a summation of all pairwise interactions (both favorable and unfavorable) yields total energies of −117.53 ± 0.05, −110.83 ± 0.06, −116.30 ± 0.06 kcal/mol, for the WT, Q493K and Q493R structures, respectively. This further suggests little difference in hACE2 binding affinity for the WT and Omicron Q493K mutant, within the limits of the molecular mechanics energy model. Our data suggests that favorable WT interactions lost as a result of mutation appear to be offset by roughly equally favorable new interactions in the Q493 K/R mutants, and vice versa.
Table 5.
WT and Omicron SARS-CoV-2 RBD - hACE2 Average Pairwise Decomposition Energies (average ± standard deviation; kcal/mol). Pairwise decomposition energies that are more favorable (less) than −2.00 kcal/mol are listed. The pairwise decomposition energies are calculated from the full 1 μs ensemble and are reported with the corresponding SARS-CoV-2 RBD and hACE2 residues. Spike mutant residues are shown in bold.
| Wild Type |
Omicron Q493K |
Omicron Q493R |
||||||
|---|---|---|---|---|---|---|---|---|
| Residue | ACE2 Residue | Pairwise Decomp. | Residue | ACE2 Residue | Pairwise Decomp. | Residue | ACE2 Residue | Pairwise Decomp. |
| Lys417 | Asp30 | −5.92 ± 3.34 | Lys493 | Glu35 | −10.25 ± 3.21 | Arg493 | Glu35 | −12.65 ± 4.42 |
| Thr500 | Asp355 | −5.14 ± 2.38 | Lys493 | Asp38 | −8.09 ± 3.61 | Arg493 | Glu38 | −9.29 ± 5.22 |
| Tyr505 | Lys353 | −5.06 ± 0.54 | Thr500 | Asp355 | −6.44 ± 2.08 | Tyr501 | Lys355 | −6.31 ± 0.94 |
| Asn501 | Lys353 | −5.01 ± 1.47 | Tyr501 | Lys353 | −6.38 + 1.00 | Thr500 | Asp355 | −5.95 ± 2.24 |
| Gln493 | Glu35 | −4.84 ± 1.62 | His505 | Lys353 | −4.96 ± 1.45 | His505 | Lys353 | −5.17 ± 1.41 |
| Gln493 | Lys31 | −4.10 ± 2.04 | Asn477 | Ser19 | −3.37 ± 2.68 | Asn487 | Gln24 | −2.95 ± 0.93 |
| Tyr449 | Asp38 | −3.26 ± 2.17 | Asn487 | Tyr83 | −2.98 ± 0.89 | Asn487 | Tyr83 | −2.93 ± 0.88 |
| Gly496 | Lys353 | −3.07 ± 1.74 | Arg498 | Tyr41 | −2.76 ± 0.70 | Asn477 | Ser19 | −2.86 ± 2.71 |
| Asn487 | Tyr83 | −3.02 ± 0.92 | Asn487 | Gln24 | −2.75 ± 0.89 | Ser496 | Asp38 | −2.79 ± 2.41 |
| Gln498 | Lys353 | −3.00 ± 3.33 | Lys493 | His34 | −2.61 ± 1.41 | Arg498 | Tyr41 | −2.66 ± 0.71 |
| Asn487 | Gln24 | −2.99 ± 0.90 | Ala475 | Ser19 | −2.46 ± 1.54 | Arg493 | His34 | −2.47 ± 1.67 |
| Tyr505 | Glu37 | −2.59 ± 2.37 | Phe486 | Met82 | −2.35 ± 0.79 | Phe486 | Met82 | −2.35 ± 0.75 |
| Phe486 | Met82 | −2.50 ± 0.81 | Tyr489 | Lys31 | −2.16 ± 0.67 | Tyr489 | Lys31 | −2.21 ± 0.75 |
| Asn501 | Tyr41 | −2.13 ± 1.21 | Ser496 | Asp38 | −2.05 ± 2.40 | |||
| Gln493 | His34 | −2.05 ± 1.34 | ||||||
Several pairwise residue interaction energies change upon mutation from WT to Omicron. Most notably, the Q493 R/K mutation significantly increases binding, enhancing the strength of interaction with hACE2 residues His 34 and Glu 35, and providing a new interaction with hACE2 Asp38. While this disagrees with Geng et al., who reported that the Q493R mutation significantly reduced binding, several other studies agree with our findings [28,29,31,32]. In the WT, neutral Gln493 contributes −4.84 and −4.10 kcal/mol when bound to hACE2 Glu35 and Lys31, respectively. In the Omicron Q493K and Q493R RBD however, the mutation to a positively charged Lys or Arg creates very favorable interactions with hACE2 Glu35 (Q493K: -10.25; Q493R: -12.65 kcal/mol), and with hACE2 Asp38 (Q493K: -8.09; Q493R: -9.29 kcal/mol), while the interaction with hACE2 Lys31 is expectedly lost. In the WT, S477 is not involved in a significant interaction, however in Omicron the S477 N mutation creates an interaction with a binding energy contribution of −3.37 (Q493K) or −2.86 (Q493R) kcal/mol with hACE2 Ser19. Conversely, one of the strongest WT interactions between RBD Lys417 and hACE2 Asp 30 (−5.92 kcal/mol) is eliminated by the K417 N mutation in Omicron. Similarly, the G496S and Q498R mutations result in lost interactions with hACE2 Lys353 (−3.07 and −3.00 kcal/mol respectively). Despite not being a subject of mutation, WT interaction RBD Tyr449 – hACE2 Asp38 is also diminished in both Omicron models (Tables 5, S5 - S7). Many of these changes are supported by COM distance analysis (Tables S3–S4).
Interactions unaffected or preserved by mutation are also observed. The Q493 K/R mutation does not disrupt the favorable interaction with His34 in the WT. The mutation Y505H does not seem to affect binding. In both the WT and Omicron mutants, residue 505 interacts similarly with hACE2 residue Lys353 (WT: -5.06; Omicron Q493K: -4.96; and Omicron Q493R: -5.17 kcal/mol), while the interaction with Glu37 does not change significantly with respect to standard deviation. Important interactions not subject to mutation, RBD Thr500 – hACE2 Asp355, RBD Asn487 – hACE2 Tyr83, RBD Asn487 - hACE2 Gln24, and RBD Ala475 – hACE2 Ser19, are unchanged between the WT and Omicron models (Table 5, S5 - S7).
Based on hydrogen bonding analysis, there is a decrease in the number of significant (defined as occurring for more than 5% of the ensemble) hydrogen bonding interactions between RBD and hACE2 residues upon Omicron mutation (Table 6, Table 7 ). One interaction that is present in both WT and Omicron is the hydrogen bond with the residue Q493 K/R to the hACE2 residue Glu35 (WT: 68.16%, −4.84 ± 1.62; Omicron Q493K: 66.05%, −10.25 ± 3.21; and Omicron Q493R 99.6%, −12.65 ± 4.42 kcal/mol). Kumar et al. [32] reported this interaction as important but occurring to a lesser extent (WT: 38.20% and Omicron Q493R: 11.10%). In the WT, 7 more hydrogen bonds between RBD and hACE2 occur than in the mutants. (Table 6). Notably, these interactions are between non-mutated RBD residues for both WT and Omicron. The interaction RBD Thr500 – hACE2 Tyr41 appears to be significant for both WT and Omicron but there is a notable difference in hydrogen bonding occurrence (WT: 29.74%; Omicron Q493K: 10.26%; and Omicron Q493R: 12.10%). In comparison, Kumar et al. reports that the RBD Thr500 – hACE2 Tyr41 interaction is only significant for the WT, occurring 18.40%. Also, Table 6 shows that the interactions RBD Tyr489 - hACE2 Tyr83, and RBD Tyr495 – hACE2 Lys353 are significant hydrogen bonding interactions for the WT but are not significant interactions for the Omicron models. The interaction RBD Tyr449 - hACE2 Asp38 appears to be significant for WT and the Omicron Q493R mutant, but there is a significant decrease in hydrogen bonding occurrence. Similarly, Kumar et al. saw a significant decrease in hydrogen bonding occurrence for RBD Tyr449 - hACE2 Asp38 (WT: 70.50% and Omicron Q493R: 17.10%); however, the RBD Tyr489 - hACE2 Tyr83 and RBD Tyr495 – hACE2 Lys353 were not significant interactions for their WT model. These discrepancies are likely due to significant differences in simulation time, i.e. analysis conducted over 1 μs in this study versus sampling hydrogen-bonding over only 100 ns in the Kumar et al. report [32].
Table 6.
WT and Omicron SARS-CoV-2 RBD - hACE2 Hydrogen Bonding Occurrences. Individual hydrogen bonding percentages (Tables S8–S10) were combined for interactions between the same residues and rotationally equivalent atoms on each residue. For example, the hydrogen bonding interactions between RBD Lys 493 and ACE2 Asp 38 (62.53%) are comprised of individual interactions between OD1 and OD2 with N–H1, N–H2 and N–H3 (Table S5 rows 10 (11.66%); 15 (10.65%); 18 (10.30%); 20 (10.14%) 21 (10.05%) and 22 (9.73%). Unless specified, these consist of sidechain - sidechain interactions. Hydrogen bonding percentages that are greater than 5% are listed. Spike mutant residues are shown in bold.
|
RBD WT Residue |
hACE2 Residue | H-Bond % Occur. (Avg.) | RBD Omicron Q493K Residue | hACE2 Residue | H-Bond % Occur. (Avg.) | RBD Omicron Q493R Residue | hACE2 Residue | H-Bond % Occur. (Avg.) |
|---|---|---|---|---|---|---|---|---|
| Asn487 | Tyr83 | 83.27 | Asn487 | Tyr83 | 72.75 | Asn487 | Tyr83 | 61.16 |
| Gln24 | 23.27 | Gln24 | 14.15 | Gln24 | 14.60 | |||
| Gly502 | Lys353 | 77.58 | Thr500 | Asp355 | 67.49 | Thr500 | Asp355 | 52.58 |
| Gln493 | Glu35 | 68.07 | Tyr41 | 10.26 | Tyr41 | 12.10 | ||
| His34 | 7.66 | Lys493 | Glu35 | 66.05 | Arg493 | Asp38 | 74.21 | |
| Lys31 | 42.66 | Asp38 | 62.53 | Glu35 Sidechain | 66.44 | |||
| Glu35 Sidechain | 33.29 | |||||||
| Tyr449 | Asp38 | 62.55 | Gly502 | Lys353 | 57.00 | |||
| Lys417 | Asp30 | 62.26 | Ala475 | Ser19 Sidechain | 38.27 | Ala475 | Ser19 Sidechain | 32.79 |
| Ser19 Backbone | 16.28 | Ser19 Backbone | 13.98 | |||||
| Thr500 | Asp355 | 58.69 | Gln24 | 5.28 | Gly502 | Lys353 | 51.59 | |
| Tyr41 | 29.75 | Ser496 | Asp38 | 38.20 | Ser496 | Asp38 | 41.38 | |
| Tyr505 | Glu37 | 49.41 | Asn477 | Ser19 Backbone | 28.22 | Asn477 | Ser19 Backbone | 22.90 |
| Ser19 Sidechain | 11.42 | Ser19 Sidechain | 9.88 | |||||
| Ala386 | 7.21 | Tyr453 | His34 | 19.68 | Tyr453 | His34 | 20.52 | |
| Gln498 | Lys353 | 35.95 | Arg498 | Gln42 | 12.77 | Arg498 | Gln42 | 13.57 |
| Asp38 | 24.17 | Asp38 | 5.81 | |||||
| Ala475 | Ser19 Sidechain | 31.34 | Tyr449 | Asp38 | 10.25 | |||
| Ser19 Backbone | 5.85 | |||||||
| Gln24 | 7.60 | |||||||
| Gly496 | Lys353 | 30.70 | ||||||
| Tyr453 | His34 | 24.79 | ||||||
| Tyr489 | Tyr83 | 8.29 | ||||||
| Tyr495 | Lys353 | 9.32 | ||||||
| Gly446 | Gln42 | 8.17 |
Table 7.
A comparison of the hydrogen bonding percent occurrence and pairwise decomposition energies (average value ± standard deviation; kcal/mol) for WT and Q493 K/R SARS-CoV-2 RBD mutations. For Omicron and WT each residue is listed with the corresponding hydrogen bonding hACE2 residue(s) and the pairwise decomposition energy for the RBD - hACE2 interaction. All percent occurrences are calculated from the 1 μs ensemble. Shown in bold are the most significant interactions. [A] The RBD - hACE2 interaction type is indicated. S corresponds to the side chain and B corresponds to backbone, with the first letter representing that of the RBD and the second representing hACE2. The interaction information distinguishes the various hydrogen bond percent occurrences listed.
| Wild Type |
Omicron (Q493K) |
Omicron (Q493R) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residue | hACE2 Residue | Interaction Type[A] | Ave. H-Bond % Occurrence | Pairwise Decomp. | Residue | hACE2 Residue |
Interaction Type[A] | Ave. H-Bond % Occurrence | Pairwise Decomp. | Residue | hACE2 Residue |
Interaction Type[A] | Ave. H-Bond % Occurrence | Pairwise Decomp. |
| Gly339 | Asp339 | Asp339 | ||||||||||||
| Ser371 | Leu371 | Leu371 | ||||||||||||
| Ser373 | Pro373 | Pro373 | ||||||||||||
| Ser375 | Phe375 | Phe375 | ||||||||||||
| Lys417 | Asp30 | S–S | 37.05 | −5.92 ± 3.34 | Asn417 | Asp30 | S–S | 0.06 | −0.12 ± 0.27 | Asn417 | ||||
| S–S | 25.21 | |||||||||||||
| S–B | 0.01 | |||||||||||||
| NAG -Asn90 | S–B | 0.06 | −0.002 ± 0.18 | His34 | S–S | 0.03 | −0.32 ± 0.25 | His34 | S–S | 0.07 | −0.28 ± 0.28 | |||
| Asn440 | Lys440 | Gln325 | S–S | 0.02 | −0.015 ± 0.028 | Lys440 | ||||||||
| Glu329 | S–S | 0.01 | −0.075 ± 0.092 | |||||||||||
| Gly446 | Gln42 | S–B | 8.68 | −0.66 ± 0.92 | Ser446 | Gln42 | S–S | 0.02 | −0.11 ± 0.16 | Ser446 | Gln42 | S–S | 0.27 | −0.17 ± 0.32 |
| B–S | 0.01 | B–S | 0.02 | |||||||||||
| Tyr41 | B–S | 0.18 | −0.17 ± 0.01 | |||||||||||
| B–S | 0.01 | |||||||||||||
| Ser477 | Ser19 | S–B | 1.22 | −0.39 ± 0.80 | Asn477 | Ser19 | S–B | 11.42 | −3.37 ± 2.68 | Asn477 | Ser19 | S–B | 9.88 | −2.86 ± 2.71 |
| S–B | 0.11 | S–B | 28.22 | S–B | 22.90 | |||||||||
| S–S | 0.10 | S–S | 0.18 | S–S | 0.05 | |||||||||
| S–B | 0.05 | S–S | 0.06 | B–S | 0.04 | |||||||||
| B–S | 0.02 | S–S | 0.02 | |||||||||||
| S–B | 0.01 | |||||||||||||
| Gln24 | S–S | 0.93 | −0.40 ± 0.53 | Gln24 | S–S | 2.30 | −0.80 ± 0.76 | Gln24 | S–S | 1.48 | −0.71 ± 0.74 | |||
| B–S | 0.14 | S–S | 0.08 | B–S | 0.09 | |||||||||
| B–S | 0.09 | B–S | 0.07 | S–S | 0.07 | |||||||||
| S–S | 0.06 | S–S | 0.07 | |||||||||||
| B–S | 0.02 | B–S | 0.01 | |||||||||||
| S–S | 0.01 | |||||||||||||
| Thr20 | B–S | 0.32 | −0.09 ± 0.34 | Thr20 | S–S | 0.47 | −0.13 ± 0.38 | Thr20 | S–S | 0.04 | −0.10 ± 0.24 | |||
| S–S | 0.09 | S–S | 0.13 | S–S | 0.01 | |||||||||
| Thr478 | Gln24 | S–S | 1.68 | −0.25 ± 0.70 | Lys478 | Lys478 | ||||||||
| B–S | 0.55 | |||||||||||||
| S–S | 0.16 | |||||||||||||
| Ser19 | S–B | 0.04 | −0.03 ± 0.18 | |||||||||||
| S–S | 0.01 | |||||||||||||
| Glu484 | Lys31 | S–S | 1.04 | −0.65 ± 1.26 | Ala484 | Ala484 | ||||||||
| Gln493 | Glu35 | S–S | 68.16 | −4.84 ± 1.62 | Lys493 | Glu35 | S–S | 66.05 | −10.25 ± 3.21 | Arg493 | Glu35 | S–S | 66.44 | −12.65 ± 4.42 |
| S–B | 0.01 | S–S | 33.29 | |||||||||||
| Asp38 | S–S | 0.06 | −0.27 ± 0.31 | Asp38 | S–S | 62.53 | −8.09 ± 3.61 | Asp38 | S–S | 74.21 | −9.29 ± 5.22 | |||
| S–S | 0.20 | |||||||||||||
| His34 | S–S | 4.42 | −2.05 ± 1.34 | His34 | S–B | 3.98 | −2.61 ± 1.41 | His34 | S–B | 1.80 | −2.47 ± 1.67 | |||
| S–B | 3.24 | S–S | 0.21 | S–S | 1.20 | |||||||||
| S–S | 0.81 | |||||||||||||
| S–B | 0.31 | |||||||||||||
| S–S | 0.22 | |||||||||||||
| S–B | 0.07 | |||||||||||||
| Lys31 | S–S | 42.66 | −4.10 ± 2.04 | Lys31 | S–B | 0.01 | 0.36 ± 0.53 | Lys31 | S–B | 0.02 | 0.49 ± 0.66 | |||
| S–B | 0.06 | |||||||||||||
| Glu37 | S–S | 0.14 | −0.09 ± 0.58 | Glu37 | S–S | 0.63 | −0.18 ± 1.00 | |||||||
| Gly496 | Asp38 | B–S | 0.42 | −0.86 ± 0.85 | Ser496 | Asp38 | S–S | 38.20 | −2.05 ± 2.40 | Ser496 | Asp38 | S–S | 41.38 | −2.79 ± 2.41 |
| Lys353 | B–S | 30.70 | −3.07 ± 1.74 | Lys353 | S–S | 2.18 | −1.86 ± 1.36 | Lys353 | S–S B–S |
1.86 0.04 |
−1.82 ± 1.45 | |||
| Gln498 | Asp38 | S–S | 25.34 | −1.84 ± 2.41 | Arg498 | Asp38 | S–S | 1.14 | −0.71 ± 1.54 | Arg498 | Asp38 | S–S | 5.81 | −1.52 ± 3.44 |
| S–B | 0.01 | S–S | 0.26 | S–S | 0.24 | |||||||||
| S–B | 0.05 | S–B | 0.08 | |||||||||||
| Gln42 | S–S | 2.33 | −0.41 ± 1.03 | Gln42 | S–S | 12.77 | −1.52 ± 1.93 | Gln42 | S–S | 13.57 | −1.80 ± 2.26 | |||
| S–S | 2.94 | S–S | 0.04 | S–S | 0.25 | |||||||||
| S–S | 0.51 | S–S | 0.02 | S–S | 0.04 | |||||||||
| Tyr41 | S–S | 0.21 | −1.97 ± 0.64 | Tyr41 | S–S | 0.12 | −2.76 ± 0.70 | Tyr41 | S–S | 0.14 | −2.66 ± 0.71 | |||
| S–S | 0.08 | |||||||||||||
| Lys353 | S–S | 37.05 | −3.00 ± 3.33 | |||||||||||
| Asp355 | S–S | 0.06 | −0.003 ± 0.001 | |||||||||||
| Asn501 | Gln325 | B–S | 0.02 | −0.01 ± 0.12 | Tyr501 | Gln325 | B–S | 4.95 | −0.32 ± 1.05 | Tyr501 | Gln325 | B–S | 2.84 | −0.23 ± 0.83 |
| Lys353 | S–S | 1.37 | −5.01 ± 1.47 | Asp38 | S–S | 2.29 | −0.05 ± 0.79 | Asp38 | S–S | 1.90 | 0.05 ± 0.72 | |||
| S–B | 0.14 | |||||||||||||
| Tyr41 | S–S | 1.58 | −2.13 ± 1.21 | Lys353 | S–S | 0.60 | −6.38 ± 1.00 | Lys353 | S–S | 0.78 | −6.31 ± 0.94 | |||
| S–S | 0.40 | |||||||||||||
| Asp355 | S–S | 0.03 | −1.84 ± 0.60 | |||||||||||
| Tyr505 | Glu37 | S–S | 49.41 | −2.59 ± 2.37 | His505 | Glu37 | S–S | 3.30 | −0.62 ± 1.18 | His505 | Glu37 | S–S | 0.49 | −0.39 ± 0.68 |
| Ala386 | S–B | 7.21 | −0.43 ± 0.68 | Ala386 | S–B | 0.25 | −0.19 ± 0.23 | Ala386 | S–B | 0.02 | −0.16 ± 0.15 | |||
| Arg393 | S–S | 1.47 | −1.46 ± 1.12 | Lys353 | S–S | 0.15 | −4.96 ± 1.45 | Lys353 | S–S | 0.24 | −5.17 ± 1.41 | |||
| Ala387 | S–B | 0.02 | −0.13 ± 0.28 | |||||||||||
In Table 5, Table 6 we have presented pairwise decomposition and hydrogen bonding analysis of the WT and Omicron 1 μs ensembles rank ordered according to favorability and percent occurrence, respectively. In Table 7 we present pairwise and hydrogen-bonding data for all RBD mutated residues regardless of their values. In addition, we specify whether the hydrogen bonding occurs between side chain (S) or backbone (B) atoms. Significant hydrogen bonding interactions of the WT RBD with hACE2 (RBD Lys417 – hACE2 Asp30, RBD Gly446 – hACE2 Gln42, RBD Gln493 – hACE2 Lys31, RBD Gly496 – hACE2 Lys353, RBD Gln498 – hACE2 Asp38, RBD Gln498 – hACE2 Lys353, RBD Tyr505 – hACE2 Glu37, and RBD Tyr505 – hACE2 Ala386) appear to be destroyed or diminished upon residue mutation in the Omicron variant. Notably, such residue mutations do not seem to introduce many new hydrogen bonding interactions, and as such, the data in Table 7 suggests that, relative to WT, Omicron mutations reduce hydrogen bonding occurrences more than they increase it.
However, the mutated RBD residues Asn477, Lys493/Arg493, Ser496, and Arg498 participate in significant hydrogen bonding interactions: RBD Asn477 – hACE2 Ser19, RBD Lys493/Arg493 – hACE2 Asp38, RBD Ser496 – hACE2 Asp38, and RBD Arg498 – hACE2 Gln42. The WT interaction RBD Gly496 – hACE2 Asp38 and Omicron interaction RBD Ser496 – hACE2 Asp38, both have relatively high pairwise decomposition values (WT: -0.86 ± 0.85; Omicron Q493K: -2.05 ± 2.40; and Omicron Q493R: -2.79 ± 2.41 kcal/mol). The Omicron Q493K hydrogen bonding interactions RBD Ser496 – hACE2 Lys353, RBD Arg498 – hACE3 Asp38, and RBD His505 – hACE2 Glu37, appear to have low hydrogen bonding occurrences compared to the equivalent WT interactions (with Omicron percent occurrences less than 5%); however, the pairwise decomposition values of these interactions are significant (Table 7). The Omicron Q493R hydrogen bonding occurrences for the previously mentioned interactions are similar to the Omicron Q493K model.
Overall, MM-GBSA estimations, per-residue and pairwise decomposition energies, hydrogen-bonding interactions and center-of-mass distance measurements indicate 3 very important Omicron mutations: Q493 K/R, N501Y and S477 N. Strikingly, for Q493K, hACE2 binding to the WT and Omicron are similar, whereas for the Q493R mutant, Omicron binds more tightly. This is in agreement with previous experimental reports, using the Q493K mutant, that suggest Omicron binds similar or less tightly than WT [27,30], and also in agreement with computational and experimental reports that suggest Omicron binds more tightly [28,31,32]. It is remarkable that a single point mutation can confer such differences in binding and it is imperative to experimentally test this computational prediction. The overall atomistic picture that emerges from our detailed study is that some of the mutations in the Spike-RBD hACE2 interface enhance binding and that some of those enhancements are balanced by mutations that disfavor binding. The mutations that have the most significant effect on binding are shown in Fig. 5 , where favorable and unfavorable point mutations are shown in green and red, respectively.
Fig. 5.
Omicron Q493K SARS-CoV-2 RBD - hACE2 interactions. Highlighted are residues that most significantly affect binding based upon hydrogen bonding occurrence and pairwise residue decomposition changes in the WT and Omicron variant. Shown in red are mutated residues that display less favorable or less significant interaction. Shown in green are the mutated residues that became more significant for RBD - hACE2 interactions. See Fig. S11 for a complete visualization of all residues that play a significant role in WT and Omicron binding to hACE2.
3.1. Average structures
Clustering analysis was conducted using cpptraj to output 10 families for the WT and Omicron Q493K mutant models. Clustering was not performed on the Q493R model. As the Q493R model differs by only a single residue, it could be reasonably expected to form similar clusters to the Q493K model. This is supported by similar pairwise COM distances between both models (Tables S3 and S4). Families were produced using both backbone atom RMSD and non-hydrogen atom pairwise distance clustering. Representative average structures of each family are displayed in Fig. 6, S12-S17. The most populated of the families are displayed in Table 8 . Notably, both RMSD and pairwise distance clustering reports a single dominant family for the Omicron Q493K ensemble with respective occurrences of 77.0 and 80.2%. For the WT, RMSD-based clustering reports four families ranging between ∼15 and 30% occurrences, while pairwise distance-based clustering reports a single dominant family with an occurrence of 89.6%. Ultimately, RMSD comparisons of representative structures of each family to each other suggests few structural differences (Table 9 ). These representative structures are freely available at https://github.com/Parish-Lab/spikeace2.
Fig. 6.
Comparison of the WT and Omicron Q493K Cluster Families. [A] Representative structures from WT and Omicron Q493K clustering using backbone-atom RMSD. The 4 different WT cluster families are represented in shades of blue and the two different Omicron cluster families are depicted in shades of red. Individual images of representative structures from each cluster family are shown in Figs. S12 and S13. [B] Representative structures from WT and Omicron Q493K clustering using pairwise distance-based clustering. The WT cluster family is displayed in green and the two different Omicron cluster families are depicted in shades of orange. Individual images of the representative structures from each cluster family are shown in Figs. S15 and S16.
Table 8.
Percent occurrence of WT and Omicron Q493K families obtained from clustering analysis. Only families contributing occurrences of greater than 10% are included.
| Family # | RMSD Based % |
Pairwise Distance % |
||
|---|---|---|---|---|
| WT | Q493K | WT | Q493K | |
| 1 | 30.4 | 77.0 | 89.6 | 80.2 |
| 2 | 23.3 | 11.0 | – | 11.7 |
| 3 | 19.2 | – | – | – |
| 4 | 14.8 | – | – | – |
Table 9.
RMSD comparison of representative structures of each clustering family. The RMSD (Å) of each family compared with each other is reported. Families labeled with “Pair” were generated from pairwise distance-based clustering, while all other families were generated with RMSD-based clustering.
| Cluster | WT 1 | WT 2 | WT 3 | WT 4 | O 1 | O 2 | Pair WT 1 | Pair O 1 |
Pair O 2 |
|---|---|---|---|---|---|---|---|---|---|
| WT 1 | 0 | 2.314 | 1.848 | 2.406 | 2.034 | 2.845 | 2.327 | 2.983 | 2.262 |
| WT 2 | 0 | 2.168 | 1.926 | 2.169 | 1.819 | 2.767 | 1.982 | 2.048 | |
| WT 3 | 0 | 2.565 | 2.211 | 2.838 | 2.665 | 2.867 | 2.241 | ||
| WT 4 | 0 | 2.504 | 1.996 | 2.184 | 2.126 | 2.376 | |||
| O 1 | 0 | 2.292 | 2.793 | 2.601 | 1.493 | ||||
| O 2 | 0 | 2.737 | 1.902 | 2.045 | |||||
|
Pair WT 1 |
0 | 2.937 | 2.953 | ||||||
|
Pair O 1 |
0 | 2.265 | |||||||
|
Pair O 2 |
0 |
4. Conclusion
The SARS-CoV-2 virus encodes for a spike protein that contains a receptor binding domain that binds favorably to the ACE2 receptor present on the surface of human cells. This binding allows the virus to enter the cell and to begin host infection. The Omicron variant of the SARS-CoV-2 virus possesses 32 point mutations, including 15 in the receptor binding domain of the spike protein. Current vaccines produce polyclonal antibodies that target the RBD of the spike protein and prevent the virus from gaining access to human cells. We have utilized classical molecular dynamics to sample the binding behavior of the wild type and Omicron spike protein RBD with hACE2 as well as MM-GBSA to compare their binding affinities for hACE2. Remarkably, while we find that the binding affinity between the hACE2 receptor and the WT and Omicron Q493K mutant spike protein RBD are similar and within the limits of error of the MM-GBSA binding estimation, the binding for the Q493R mutant is significantly enhanced. A detailed analysis of the per-residue interaction energies, pairwise decomposition energies, hydrogen-bonding interactions, center-of-mass distance measurements and clustering suggests that while the Omicron RBD mutations disrupt some favorable wild type residue interactions, such mutations also produce new favorable interactions. For instance, for the Q493K mutant, pairwise analysis shows that the Q493K and S477 N mutations significantly increase binding whereas K417 N, G496S and Q498R reduce binding. For the Q493R mutant, per-residue decomposition analysis indicates that the point mutation confers a significantly larger contribution to favorable binding. For both mutants, and in agreement with previous studies, this detailed atomistic analysis points to the importance of 3 residues: Q493 K/R, N501Y and S477 N.
Author contributions
The study was conceived by CC, JA and CP; CC and JA ran the simulations and performed the analyses. CC, JA and CP wrote the main manuscript and SI. CC, JA and CP reviewed all documents.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by funding from the National Science Foundation (Grant CHE-18800014) and the Floyd D. and Elisabeth S. Gottwald Endowment. J.A. and C.C. were recipients of a summer fellowship from the Puryear-Topham-Pierce-Gupton endowment from the Department of Chemistry at the University of Richmond, and from the University of Richmond Integrated and Inclusive Science program, and the Arts and Sciences Undergraduate Research Committee.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2022.108360.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data is publicly available on our github site described in the paper.
References
- 1.Berlin D.A., Gulick R.M., Martinez F.J. Severe covid-19. New England Journal of Medicine. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 2.Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F., et al. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nature Reviews Microbiology. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 6.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. The Lancet Infectious Diseases. 2021;21:1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidry J.P.D., Laestadius L.I., Vraga E.K., Miller C.A., Perrin P.B., Burton C.W., et al. Willingness to get the COVID-19 vaccine with and without emergency use authorization. American Journal of Infection Control. 2021;49:137–142. doi: 10.1016/j.ajic.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 12.Mascellino M.T., Di Timoteo F., De Angelis M., Oliva A. Overview of the main anti-SARS-CoV-2 vaccines: mechanism of action, efficacy and safety. Infection and drug resistance. 2021;14:3459–3476. doi: 10.2147/IDR.S315727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruxvoort K.J., Sy L.S., Qian L., Ackerson B.K., Luo Y., Lee G.S., et al. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ. 2021;375 doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenforde M.W., Self W.H., Adams K., Gaglani M., Ginde A.A., McNeal T., et al. Association between mRNA vaccination and COVID-19 hospitalization and disease severity. JAMA. 2021;326:2043–2054. doi: 10.1001/jama.2021.19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramesh S., Govindarajulu M., Parise R.S., Neel L., Shankar T., Patel S., et al. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines. 2021;9:1195. doi: 10.3390/vaccines9101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alkhatib M., Svicher V., Salpini R., Ambrosio F.A., Bellocchi M.C., Carioti L., et al. SARS-CoV-2 variants and their relevant mutational profiles: update summer 2021. Microbiology Spectrum. 2021;9 doi: 10.1128/Spectrum.01096-21. e01096-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 19.Fiorentini S., Messali S., Zani A., Caccuri F., Giovanetti M., Ciccozzi M., et al. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. The Lancet Infectious Diseases. 2021;21:e147. doi: 10.1016/S1473-3099(21)00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 21.Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., et al. bioRxiv; 2021. Neutralization of Viruses with European, South African, and United States SARS-CoV-2 Variant Spike Proteins by Convalescent Sera and BNT162b2 mRNA Vaccine-Elicited Antibodies. [Google Scholar]
- 22.He X., Hong W., Pan X., Lu G., Wei X. SARS-CoV-2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. The Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Scientific Reports. 2020;10 doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nature Microbiology. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W., Xu C., Wang P., Luo M., Xu Z., Cheng R., et al. N439K variant in spike protein alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. Frontiers in cell and developmental biology. 2021;9 doi: 10.3389/fcell.2021.697035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han P., Li L., Liu S., Wang Q., Zhang D., Xu Z., et al. Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and delta SARS-CoV-2. Cell. 2022;185:630–640. doi: 10.1016/j.cell.2022.01.001. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng Q., Shi K., Ye G., Zhang W., Aihara H., Li F., et al. Structural basis for human receptor recognition by SARS-CoV-2 Omicron variant BA.1. Journal of Virology. 2022;96 doi: 10.1128/jvi.00249-22. e00249-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871. doi: 10.1016/j.cell.2022.01.019. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert M., Bertoglio F., Steinke S., Heine P.A., Ynga-Durand M.A., Maass H., et al. Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant. BMC Medicine. 2022;20:102. doi: 10.1186/s12916-022-02312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Costa C.H.S., de Freitas C.A.B., Alves C.N., Lameira J. Assessment of mutations on RBD in the spike protein of SARS-CoV-2 alpha, delta and Omicron variants. Scientific Reports. 2022;12:8540. doi: 10.1038/s41598-022-12479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar R., Murugan N.A., Srivastava V. Improved binding affinity of omicron's spike protein for the human angiotensin-converting enzyme 2 receptor is the key behind its increased virulence. International Journal of Molecular Sciences. 2022;23:3409. doi: 10.3390/ijms23063409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert opinion on drug discovery. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuccinardi T. What is the current value of MM/PBSA and MM/GBSA methods in drug discovery? Expert Opinion on Drug Discovery. 2021;16:1233–1237. doi: 10.1080/17460441.2021.1942836. [DOI] [PubMed] [Google Scholar]
- 37.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z.H., et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chemical Reviews. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 38.Forouzesh N., Mishra N. An effective MM/GBSA protocol for absolute binding free energy calculations: a case study on SARS-CoV-2 spike protein and the human ACE2 receptor. Molecules. 2021:26. doi: 10.3390/molecules26082383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Andrade J., Gonçalves P.F.B., Netz P.A. Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. ChemBioChem. 2021;22:865–875. doi: 10.1002/cbic.202000455. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen H.L., Lan P.D., Thai N.Q., Nissley D.A., O'Brien E.P., Li M.S. Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J Phys Chem B. 2020;124:7336–7347. doi: 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aljindan R.Y., Al-Subaie A.M., Al-Ohali A.I., Kumar D.T., Doss C.G., Kamaraj B. Investigation of nonsynonymous mutations in the spike protein of SARS-CoV-2 and its interaction with the ACE2 receptor by molecular docking and MM/GBSA approach. Comput Biol Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar V., Singh J., Hasnain S.E., Sundar D. Possible link between higher transmissibility of alpha, Kappa and delta variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. International Journal of Molecular Sciences. 2021;22:9131. doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma J., Subbarao N. Insilico study on the effect of SARS-CoV-2 RBD hotspot mutants' interaction with ACE2 to understand the binding affinity and stability. Virology. 2021;561:107–116. doi: 10.1016/j.virol.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piplani S., Singh P.K., Winkler D.A., Petrovsky N. In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Scientific Reports. 2021;11 doi: 10.1038/s41598-021-92388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Wu S., Wu B., Yang Q., Chen A., Li Y., et al. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduction and Targeted Therapy. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tareq A.M., Emran T.B., Dhama K., Dhawan M., Tallei T.E. Impact of SARS-CoV-2 delta variant (B.1.617.2) in surging second wave of COVID-19 and efficacy of vaccines in tackling the ongoing pandemic. Human Vaccines & Immunotherapeutics. 2021;17:4126–4127. doi: 10.1080/21645515.2021.1963601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., et al. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L., Zhou L., Mo M., Liu T., Wu C., Gong C., et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduction and Targeted Therapy. 2022;7:8. doi: 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen H.L., Thai N.Q., Nguyen P.H., Li M.S. SARS-CoV-2 Omicron variant binds to human cells more strongly than the wild type: evidence from molecular dynamics simulation. The Journal of Physical Chemistry B. 2022;126:4669–4678. doi: 10.1021/acs.jpcb.2c01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrodinger Release 2019-3. Schrodinger, LLC; New York, NY: 2019. [Google Scholar]
- 53.Olsson M.H.M., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. Journal of Chemical Theory and Computation. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 54.Søndergaard C.R., Olsson M.H.M., Rostkowski M., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. Journal of Chemical Theory and Computation. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 55.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory and Computation. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 56.Case D.A., Belfon K., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., T E Cheatham I., et al. University of California; San Francisco: 2020. AMBER20. [Google Scholar]
- 57.Götz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. Journal of chemical theory and computation. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. Journal of Chemical Theory and Computation. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 59.Kirschner K.N., Yongye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L., et al. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. Journal of Computational Chemistry. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen W.L., Madura J.D. Quantum and statistical mechanical studies of liquids. 25. Solvation and conformation of methanol in water. Journal of the American Chemical Society. 1983;105:1407–1413. [Google Scholar]
- 62.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Science. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 65.Guo H., Gao Y., Li T., Li T., Lu Y., Zheng L., et al. Structures of Omicron spike complexes and implications for neutralizing antibody development. Cell Reports. 2022;39 doi: 10.1016/j.celrep.2022.110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is publicly available on our github site described in the paper.