Abstract
This paper aims to diagnose COVID-19 by using Chest X-Ray (CXR) scan images in a deep learning-based system. First of all, COVID-19 Chest X-Ray Dataset is used to segment the lung parts in CXR images semantically. DeepLabV3+ architecture is trained by using the masks of the lung parts in this dataset. The trained architecture is then fed with images in the COVID-19 Radiography Database. In order to improve the output images, some image preprocessing steps are applied. As a result, lung regions are successfully segmented from CXR images. The next step is feature extraction and classification. While features are extracted with modified AlexNet (mAlexNet), Support Vector Machine (SVM) is used for classification. As a result, 3-class data consisting of Normal, Viral Pneumonia and COVID-19 class are classified with 99.8% success. Classification results show that the proposed method is superior to previous state-of-the-art methods.
Keywords: AlexNet, Convolutional neural networks, COVID-19, DeepLabV3+, Support vector machine, Semantic segmentation
1. Introduction
The latest coronavirus infection, known as COVID-19, has spread rapidly from Wuhan to all of China and many other countries since December 2019. As of 22.09.2022, the number of cases exceeded 600 million and the number of deaths exceeded 6.5 million. Despite quarantine rules to limit its spread, COVID-19 infection has spread rapidly in countries worldwide. As a result of the rapid spread of COVID-19, a type of respiratory tract infection, worldwide, COVID-19 infection was declared as a global pandemic by the WHO (World Health Organization). Because of the lack of therapeutic treatment or vaccine for COVID-19 disease, immediate isolation of the infected person and early diagnosis of the disease is vital. The common method used to diagnose COVID-19 cases is mostly the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. However, RT-PCR testing is time-consuming, laborious and complex, and has little supply. Moreover, its sensitivity is highly variable [[1], [2], [3]].
Although the impact of COVID-19 has decreased over time, new cases are still being detected. There are also concerns that it may still increase the effect of the infection. For this reason, various studies and methods have been proposed for the detection of COVID-19 during the epidemic period and today. In general, the focus is on detecting COVID-19 early, thereby providing early treatment to the patient. Early detection also significantly limits the rate of transmission of the disease. Medical scan images are very useful for early detection. Magnetic Resonance Imaging (MRI), chest X-ray (CXR), Computed Tomography (CT), and Positron Emission Tomography (PET) are images that are frequently used for the early detection of COVID-19 [4]. Among these, it has been widely proven that CXR and CT scans can show pathological findings even in the early stages of the disease [[5], [6], [7]]. In addition, since the diagnoses provided by these scan images are more reliable than RT-PCR and provide easier diagnosis than RT-PCR, it has accelerated the COVID-19 detection studies based on medical scan images [8,9]. Particularly, in today's era of big data, the sharing of publicly available scan images of COVID-19 has enabled Artificial Intelligence (AI) researchers to make a major contribution to the diagnosis of COVID-19. Such AI studies aim to enable the automatic, rapid and high-accuracy diagnosis of COVID-19 through these medical scan images [10].
AI is crucial to developing solutions to support virus diagnosis. Therefore, AI methods are often preferred today to develop automated expert systems for COVID-19 diagnosis [11,12]. In recent years, Machine Learning (ML) and Deep Learning (DL) applications in the field of AI have achieved great success, especially in computer vision studies. Advances in computational capabilities and the availability of large labeled datasets have encouraged the use of DL techniques in the field of medical imaging. In particular, research papers using DL-based Convolutional Neural Networks (CNN) models have shown promising performance in identifying, classifying and measuring disease patterns in medical images such as CT and CXR. In DL applications, both feature extraction and classification/regression are provided through deep layers. The features extracted are of high level and have high separation ability. The fact that it automatically extracts powerful features makes DL superior to ML methods [[13], [14], [15]].
In a CNN architecture consisting of many layers, features are extracted through the first layers, and features are classified in the last layers. Classification success and extracted features depend on the designed architecture. In the last decade, many pre-trained and pre-designed CNN architectures such as AlexNet [16], VGG-Net [17], ResNet [18], Inceptionv3 [19], DenseNet [20] and GoogLeNet [21] have achieved successful results in pattern recognition and image classification. Many studies based on DL and transfer learning have previously been proposed using such pre-trained CNN models. Ardakani, Kanafi, Acharya, Khadem and Mohammadi [22] used the pre-trained CNN architectures to discriminate COVID-19 cases from non-COVID cases using CT images. Among all models, ResNet-101 and Xception achieved the best performance. Khan, Shah and Bhat [23] designed a new architecture, CoroNet, based on Xception model to diagnose COVID-19 infection using CT and X-ray images. As a result of the study, they provided 95% diagnostic accuracy for 3-class data. Asif and Wenhui [24] carried out a CNN-based study to automatically detect COVID-19 from CXR images. CXR images without preprocessing were given as input to Inception-V3 model with transfer learning method. At the end of the study, a detection rate of 96% was achieved. Narin, Kaya and Pamuk [25] proposed a method to detect COVID-19 from CXR images based on the InceptionV3, Inception-ResNetv2 and ResNet50 models by applying binary classification and 5-fold cross validation. With 98% accuracy, the ResNet50 model provided a very successful classification. In a different study, Chowdhury, Rahman, Khandakar, Mazhar, Kadir, Mahbub, Islam, Khan, Iqbal and Al-Emadi [26] performed a transfer learning-based benchmarking study for the detection of COVID-19. CheXNet, VGG19, Inceptionv3, MobileNetv2, ResNet18, SqueezeNet, ResNet101 and DenseNet201 CNN models were fed and tested with augmented CXR images. As a result of the comparison, DenseNet provided the most successful detection with 97.94%. Unlike these, while some studies extracted features with CNN models, they used different ML methods for more successful classification. Nour, Cömert and Polat [27] detected COVID-19 from CXR images with a CNN network they designed. The CNN network extracted the image features, and the classification was performed with optimized k-Nearest Neighbor (k-NN), Decision Tree and (DT) Support Vector Machine (SVM) algorithms. As a result, SVM showed a high classification success of 98.97%. Sethy and Behera [28] classified the features extracted from the CXR images via the ResNet50 model with 95.4% success using SVM.
In addition to numerous studies with pre-trained CNN-based models, different DL architectures have also been proposed by different studies. Prominent studies among these use Recurrent Neural Networks (RNNs) layers in conjunction with CNN. Unlike CNN, RNN has the ability to process temporal information or sequence data. However, for long-term sequences, Long Short Term Memory (LSTM) and Bidirectional LSTM (BiLSTM) architectures outperform RNN. The LSTM and BiLSTM network has an internal memory that can learn from long-term states [29]. Successful studies have been carried out combining the features of both CNN and RNN-based architectures for COVID-19 detection. Islam, Islam and Asraf [30] conducted a DL study based on the combination of CNN and LSTM architectures to automatically detect COVID-19 from CXR images. Aslan, Unlersen, Sabanci and Durdu [1] first classified the CXR images with transfer learning including AlexNet (mAlexnet). Then, a classification was performed again by giving the features extracted with mAlexnet to the BiLSTM network. The results proved that the mAlexNEt-BiLSTM hybrid architecture provides better detection. In a different study, Islam, Islam, Asraf and Ding [31] proposed a combined architecture of CNN and RNN. First, features were extracted with pre-trained models such as VGG19, DenseNet121, InceptionV3 and InceptionResNetV2, and then RNN was used for classification.
In the light of the above information, it is seen that many DL-based studies have been conducted for the detection of COVID-19 with raw CXR and CT images. However, raw images contain irrelevant features that negatively affect the classification. Removing unnecessary features ensures that the classification is not affected by these irrelevant features, resulting in more reliable results. To achieve this, areas or pixels in the raw scan image that should not affect the classification result are removed. As a result, only useful areas or pixels remain in the processed image that should affect the classification. Image segmentation is a major component in many computer vision tasks. Segmentation has a wide range of applications, including medical image analysis, autonomous vehicles, video surveillance and augmented reality. Among these, the medical field that includes applications such as brain tumor segmentation [32], liver segmentation [33], breast image segmentation [34], etc. stands out. Numerous image segmentation algorithms have been proposed in the literature, such as thresholding [35], k-means clustering [36], watersheds [37], sparsity-based methods [38], etc. In recent segmentation applications, different DL architectures have come to the forefront by providing object-boundary segmentation in a human-like manner. DL-based segmentation applications provide segmentation of a particular object in the image by providing pixel-based classification. This segmentation, which is provided automatically, is called semantic segmentation [39]. Semantic segmentation-based applications are very useful. Because with this method, classification, detection, object size, object quantity and position of the object can be determined. Recently, successful applications of DL on semantic segmentation have increased and many models with different encoder-decoder architectures have been proposed. Examples of these models are Fully Convolutional Networks (FCN) [40], U-Net [41], SegNet [42], DeepLabv3 [43], DeepLabv3+ [44], etc. [45]. Many medical studies were carried out with these architectures. Khan, Yahya, Alsaih, Ali and Meriaudeau [46] evaluated and compared these FCN, SegNet, U-Net, and DeepLabV3+ models for segmentation of prostate T2W magnetic resonance imaging (MRI). At the end of the study, it was stated that the best segmentation was achieved with the DeepLabv3 + model.
While numerous AI applications have been developed to aid in the diagnosis of COVID-19 infection in clinical practice, there are unfortunately few studies on the semantic segmentation of CT and CXR images [47]. The main reason for this is that most of the COVID-19 datasets are for infection detection purposes only. However, over time, datasets such as [[48], [49], [50]] have been shared for segmentation purposes, these datasets provide label images in addition to medical images. Studies that perform segmentation using these datasets have started to increase recently. Oulefki, Agaian, Trongtirakul and Kassah Laouar [51] evaluated a modified Local Contrast Enhancement method for automatic COVID-19 lung infection segmentation using CT scan images. Yan, Wang, Gong, Luo, Zhao, Shen, Shi, Jin, Zhang and You [52] proposed a 3D tailored CNN (COVID-SegNet) for automatic segmentation of the COVID-19 infection regions from CT images. Saeedizadeh, Minaee, Kafieh, Yazdani and Sonka [53] carried out a semantic segmentation to detect chest regions in CT images using an architecture similar to U-Net model. Wang, Deng, Fu, Zhou, Feng, Ma, Liu and Zheng [54] performed lung segmentation in chest CT images using the U-net model to discriminate COVID-19 from non-COVID-19. The segmented 3D lung region was then fed into the 3D DL to detect the COVID-19 infectious. Chen, Wu, Zhang, Zhang, Gong, Zhao, Hu, Wang, Hu and Zheng [55] trained CT scans using U-Net++ to extract valid regions and predicted COVID-19 lesions. Ferdi, Benierbah and Ferdi [56] modified the U-Net model to segment chest CT scan images into ground-glass and consolidation. The encoder part of the U-Net model was replaced with the pre-trained EfficientNet-b0, EfficientNet-b1 and EfficientNet-b2 CNN models. As a result, the U-Net model with the EfficientNet-b2 encoder achieved 95.31% accuracy. Shamim, Awan, Mohd Zain, Naseem, Mohammed and Garcia-Zapirain [57] modified the U-Net model for diagnosing COVID-19 from CT images. They aimed to establish a connection between the encoder and decoder pipelines. In the developed model, they added complementary convolution layers to the encoder and decoder pipelines and named this model ConvUnet. At the end of that study, the authors stated that ConvUnet was more successful, achieving a diagnostic accuracy of 93.29%. Rajamani, Rani, Siebert, ElagiriRamalingam and Heinrich [58] aimed to diagnose COVID-19 from CT images by combining the semantic segmentation model with attention models. For this, the attention-augmented convolution network was integrated into the bottleneck of the U-Net encoder-decoder architecture. This new deep architecture was named Attention-augmented U-Net (AA-U-Net) and at the end of the study, AA-U-Net provided more successful classification than U-Net.
This study performs the diagnosis of COVID-19 from CXR images using CNN. Unlike most other studies, it does not directly give the raw images to the deep network for feature extraction, but instead gives the semantically segmented lung regions as input to the network. For the semantic segmentation of the lung regions, DeepLabV3 + is used, inspired by Ref. [46]. COVID-19 Chest X-Ray Dataset [50,[59], [60], [61]] is used for training the semantic segmentation network. After the training, the trained network is applied to viral, normal, and COVID-19 CXR images in the COVID-19 Radiography Database [26]. Then, image pre-processing is performed using the semantic lung masks and raw images. As a result, images containing only lung areas are obtained, and COVID-19 infection is detected with these images. Features for COVID-19 diagnosis are extracted with the modified AlexNet architecture and classification is performed with SVM. The classification success of the proposed method has been superior to previous studies in the literature. At the end of the study, the proposed method is compared with other successful studies. The contributions and differences of this study can be explained as follows:
-
1)
While CT images are generally used for segmentation in other studies, in this study lung segments are semantically segmented from CXR images.
-
2)
The suggested practice automatically segments the lungs on CXR images and provides an easy diagnosis. Since the diagnosis of infection was made only from lung fields, the results of the study are highly reliable.
-
3)
High classification success is achieved for the diagnosis of COVID-19.
The organization of the article is as follows. Section 2 introduces the public datasets used in the proposed method. The proposed methodology is explained in detail in section 3. The proposed method, results and discussion are shared in section 4. In addition, in this section, comparisons with previous studies are made. Finally, section 5 concludes the proposed method.
2. Datasets
In this section, information is given about the COVID-19 Chest X-Ray Dataset and the COVID-19 Radiography Database used in the proposed method.
2.1. COVID-19 chest X-Ray Dataset
This study uses COVID-19 Chest X-Ray Dataset [50,[59], [60], [61]] to train the DeepLAbV3 + network using CXR images. This dataset includes 6500 CXR images with pixel-level polygonal lung segmentations. Among them, there are 517 COVID-19 cases. There are two lung segmentation masks for each CXR image, including the posterior region behind the heart. Each of CXR images are in.JSON format and contain information such as viral, bacterial and healthy of the image. Images labeled with COVID-19 also contain information such as age, temperature, sex, intubation status, etc. The lung segmentations in this dataset also include most of the area of the heart. Since the sources of the images are different, their resolutions and orientations differ. Some CXR images and corresponding masks of this dataset are shown in Fig. 1 (a).
Fig. 1.
Sample images of the datasets used
(a) Sample COVID-19 Chest X-Ray Dataset CXR images and corresponding masks, (b) Sample COVID-19 Radiography Database scan images for each class (COVID-19, Normal, Viral Pneumonia).
2.2. COVID-19 Radiography Database
For the diagnosis of COVID-19, this proposed application uses a public database of CXR scan images [26]. The images in the dataset were created with the cooperation of different universities and many medical doctors. Three classes were specified for the CXR images in the dataset: Viral Pneumonia, Normal and COVID-19. The number of samples for each class in the dataset is given in Table 1 .
Table 1.
Sample numbers of classes in the COVID-19 Radiography Database.
| Class/Type | COVID-19 | Normal | Viral Pneumonia |
|---|---|---|---|
| Number of Samples | 219 | 1341 | 1345 |
| Total | 2905 | ||
The COVID-19 Radiology database was created by collecting samples from many different publications, as well as the Italian Society of Medical and Interventional Radiology (SIRM) COVID-19 Database [62] and Novel Corona Virus 2019 Dataset [63]. Fig. 1(b) shows a sample CXR image of each of the three classes in the COVID-19 Radiography dataset.
3. Proposed methodology
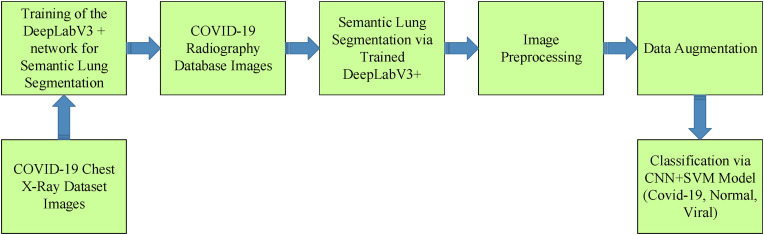
This section discusses the proposed methodology in detail. In this context, information is given about semantic segmentation of the lung, preprocessing to improve the semantic lung image, data augmentation and COVID-19 diagnosis applications. The flow diagram of the proposed method, including all the implementation steps, is given in Fig. 2 . According to Fig. 2, the proposed method can be briefly explained as a methodology as follows: DeepLabV3+ network is trained using CXR and binary mask images in the COVID-19 Chest X-Ray Dataset. The trained DeepLabV3+ is fed with CXR images from the COVID-19 Radiography Database. Segmentation results are enhanced by image preprocessing steps. Then data augmentation and CNN + SVM-based classification are performed.
Fig. 2.
Block diagram of the proposed algorithm.
3.1. Training of DeepLabV3+ network for semantic lung segmentation
Segmentation of desired regions in the image is vital for AI. Because noises in the image, or areas that should not affect the diagnosis, may lead to incorrect diagnosis. These unwanted pixels affect the AI's decision and negatively affect prediction reliability. Undoubtedly, the most successful methods for image segmentation are those that can perform segmentation semantically. In this way, the desired region is segmented based on pixels starting from its edges. In particular, CT and CXR scan images should be denoised or segmented before being fed to the AI algorithm. For example, a raw CXR image in the COVID-19 Radiography Database contains various noises during scanning, as seen in Fig. 3 . This study segments the lung region semantically so that the prediction algorithm is not affected by these unnecessary noises and makes predictions only on the diseased area.
Fig. 3.
Example CXR Images with unnecessary pixels or patterns.
For detection of COVID-19 from a CXR image, the lung area included in the raw image is taken into account. Therefore, the DL network, which is fed with a scan image containing only the lung area, creates a more reliable system. The areas and positions of the lungs in the CXR images in the COVID-19 Radiography Database are not fixed or specific. Therefore, each CXR image must be viewed individually to segment the lung area. However, there are 2905 images in the COVID-19 Radiography Database. Therefore, automatic segmentation of lung regions will provide great convenience. To achieve this, the DeepLabV3+, which is state-of-the-art algorithm in the semantic segmentation field, is used.
As a result of stride and pooling operations, there is a reduction in output size in traditional CNN networks. However, in semantic segmentation, the output size must be the same as the input. Therefore, traditional CNN models cannot generate semantic segmentation output. To overcome this limitation, DL models for semantic segmentation adopt the encoder-decoder structure. In this context, architectures such as U-Net, SegNet and DeepLabV3+ have been developed. U-Net adopts the technique of combining feature maps in encoder and decoder architectures. SegNet, on the other hand, keeps pooling indices in its encoder architecture. DeepLabV3+ [44], introduced by Google, applies parallel atrous convolutions (Atrous Spatial Pyramid Pooling (ASPP)) at different rates to capture information at different scales [64]. Fig. 4 shows the design of the DeepLabV3 + architecture. DeepLabV3 + has started to be used in the medical field as it proved that it is more successful than its other varieties (DeepLabV1, DeepLabV2 and DeepLabV3) and PSPNet [65] for semantic segmentation.
Fig. 4.
DeepLabv3+ structure used for CXR image segmentation [44].
Since the architecture used is in a deep structure, a lot of data is needed for training. Since COVID-19 Chest X-Ray Dataset contains 6500 raw CXR images and masks in total, it is suitable for DL training. In the encoder part of the DeepLabV3+ structure, basic features are extracted from the CXR images. In the decoder stage, an appropriate size output is created according to the features extracted in the encoder. DeppLabV3+ supports network backbones such as Xception, ResNet, MobileNetv2. Our study uses ResNet18 as a backbone. After ResNet18, an ASPP module is used to aggregate multiscale contextual information. The decoder combines ResNet18 low-level features with upsampled deep level multiscale features extracted from ASPP. Lastly, it upsamples the combined feature maps to generate the final semantic segmentation results [66].
In practice, 6000 of 6500 images in the COVID-19 Chest X-Ray Dataset are reserved for training and the remainder for testing to train the DeepLabV3+ network. Since the dimensions of the raw images are different, all images are set to 224 × 224 × 3. The raw images that have been resized are applied as an input to the DeepLabV3 + network for training. Table 2 shows the hyperparameter values set during the training phase. According to Table 2, the Mini Batch value is 60 and the optimization algorithm is Stochastic Gradient Descent with Momentum (SGDM).
Table 2.
Training parameters of the DeepLabV3+ network.
| Training Hyperparameters | ||||
|---|---|---|---|---|
| Optimization Algorithm | Maximum Epoch | Mini Batch Size | Learning Rate () | Momentum () |
| SGDM | 100 | 32 | 0.001 | 0.95 |
The parameter updating equation used for the SGDM optimization algorithm is shown in Eq. (1). The aim is to update these weights based on the error value called loss function () and to reduce the error as the number of iterations increases. Therefore, in each iteration, the error value is sought in the negative gradient direction of the loss function with the learning rate steps. Meanwhile, the weights are updated with the backpropagation algorithm at every iteration. Momentum () coefficient represents the relationship of the current weight value with the weight value in previous iteration.
| (1) |
The graphic obtained as a result of the training performed with the parameters in Table 2 and the SGDM optimization algorithm is shown in Fig. 5 . According to Fig. 5, training and testing accuracies are calculated as 98.1% and 95.82%, respectively. Since the trained network will be used for a different dataset, the number of training is much higher than the number of tests. In this way, a more robust network is created. After the training is completed, the mask images obtained as a result of applying some test images in COVID-19 Chest X-Ray Dataset to the trained DeepLabV3+ network are given in Fig. 6 . Masks predicted as a result of semantic segmentation have been classified quite successfully. However, because of pixel-based classification, some pixels are classified incorrectly. Defective pixels are removed with the image preprocessing steps described in the next section.
Fig. 5.
Training and testing (validation) graphics of DeepLabV3+ network.
Fig. 6.
Some COVID-19 Chest X-Ray Dataset CXR images, real lung masks and predicted lung masks via DeepLabV3+.
3.2. Preprocessing of semantically segmented lung parts in the COVID-19 Radiography Database
In Fig. 6, as a result of applying some COVID-19 Chest X-Ray Dataset CXR test images to the DeepLabV3 + network, it is seen that some pixels are misclassified in the estimated mask images. This situation is the same for the mask to be created for COVID-19 Radiography Database CXR images, as seen in Fig. 7 (a). As a result of applying the trained DeepLabV3 + network to these CXR images, some lung image masks are perfectly predicted and some are predicted defectively.
Fig. 7.
Some CXR mask images segmented with DeepLabV3+ and preprocessing steps applied to these mask images
(a) Some defective and perfect mask samples as a result of DeepLabV3+ semantic segmentation, (b) Preprocessing steps applied to a sample defective segmented mask.
For a successful diagnosis of COVID-19 with COVID-19 Radiography Database images, these defective pixels must be removed by image preprocessing. In addition, for the diagnosis of COVID-19, lung images obtained using mask images should be given to the feature extraction network. For this, the image preprocessing steps applied to the COVID-19 Radiography Database CXR images after semantic segmentation are shown in Fig. 7(b). As seen in Fig. 7(b), binary lung masks are generated from raw CXR images as a result of semantic segmentation. Preprocessing steps are applied to these mask images. Errors in the outputs of the DeepLabV3+ network are of two types: blobs in areas outside the lung and insufficient blobs inside the lung. The most common image processing steps can be used for these binary pixel errors. These steps are morphology, filtering and thresholding. After these steps, the blobs outside the lung are removed and the black pixels inside the lung are converted to white. As a result, noise in binary mask images is removed. In the last step, the raw image and the thresholding image are multiplied, so that only the raw lung parts are formed in the final image. This process is applied to all 3-class COVID-19 Radiography Database images and feature extraction and classification is performed using the obtained lung parts.
3.3. Data augmentation
It is necessary to have a large dataset for a successful prediction in DL applications. But, a large amount of data is not easily accessible for many fields. Therefore, many applications artificially augment data with a computer for robust classification. In this application, image rotation technique between −30 and + 30 is applied (see Fig. 8 ). Because there are CXR images at different angles in raw images. Rotation is used to the COVID-19 class scan images that have fewer samples than others (see Table 1). As a result of the data augmentation, the number of samples in the COVID-19 class becomes 1225.
Fig. 8.
Rotation operation.
3.4. Deep feature extraction and classification
Fig. 9 summarizes the steps described in this section. The lung images obtained after the semantic segmentation and preprocessing steps performed in the previous sections are given to the modified AlexNet (mAlexNet) architecture for feature extraction. Prior to this, images are resized according to the AlexNet architecture (227 × 227 × 3). The input images given to the model are processed through various layers such as convolution, pooling (ex. Max-pooling), Rectified Linear Unit (ReLU) and features are extracted. The last fully connected layer, fc8, has been modified. fc8 layer has 100 output sizes. In this case, instead of 1000, 100 features are extracted from each semantically segmented image. Output layer size is set to three and the mAlexNet network is trained. In total, 85% of 3911 (1341 + 1345 + 1225) images are used as the training and the remainder as a test. Training parameters are shown in Table 3 . At the end of the training, the accuracy rate is calculated as 97.26%. Then the features extracted through the fc8 layer are given to the SVM for a more successful classification.
Fig. 9.
Block diagram of deep feature extraction and classification steps.
Table 3.
Training parameters of the mAlexNet network.
| Training Hyperparameters | ||||
|---|---|---|---|---|
| Optimization Algorithm | Maximum Epoch | Mini Batch Size | Learning Rate () | Momentum () |
| SGDM | 30 | 166 | 0.001 | 0.95 |
4. Results and discussion
Semantically segmented lung images are classified using features extracted via mAlexNet network. In order to test the performance of the proposed method, the classification results of the lung images determined as tests are analyzed in this section. Intel Core i7-7700HG CPU, NVIDIA GeForce GTX 1050, 16 GB RAM, 4 GB laptop are used in the experimental studies. The total training time is 760 s and the number of iterations is 600. However, this study only uses mAlexNet for feature extraction. SVM is used for a high classification accuracy. As a result, a success of 99.8% is achieved. The confusion matrix obtained as a result of the classification is shown in Fig. 10 (a). According to Fig. 10(a), all of the samples with Normal and COVID-19 classes are classified correctly with the proposed method. Only one sample belonging to the Viral class was estimated as the Normal class. As a result, the average accuracy for Viral is 99.5%, while the diagnostic accuracy for Normal and COVID-19 classes is 100%.
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
Fig. 10.
Performance of the proposed method
(a) Confusion matrix, (b) ROC curves for Normal, Viral and COVID-19 class.
: True Positive : True Negative : False Positive : False Negative
Apart from accuracy, different metrics are often used to interpret classification results. This study also calculates the recall (or sensitivity), specificity, precision, F1-score, kappa and Matthews Correlation Coefficient (MCC) metrics of the proposed method. The formula for each metric is shared between Eqs (2), (3), (4), (5), (6), (7)). These metrics show the quality of the classification results, the success and weakness of the model over a particular category. The results for each metric are shown in Table 4 . The results obtained are 0.9983,0.9991,0.9983,0.9983 and 0.9975, respectively. These results prove successful and unbiased classification of semantic lung images with the proposed mAlexNet-SVM model. Apart from these, the results of this study are also expressed by Receiver operating characteristic (ROC) curves. ROC analysis is very popular for evaluating the accuracy of medical diagnostic systems. A ROC curve is constructed by plotting the true positive rate () against the false positive rate (). The ROC curve shows the balance between sensitivity and recall. Consequently, the ROC curve must be perpendicular along the diagonal for successful classification. The curve approaching the 45-degree angle indicates poor performance of the classifier [67]. Therefore, as a result of a classification, it is desirable that the Area under curve (AUC) be close to 1. Fig. 10(b) shows the ROC curves for each class. Curves are perpendicular along the diagonal and have AUC values of 1. As a result, the proposed method provides reliable results.
Table 4.
Performance metrics of proposed method.
| Model | Accuracy | Recall | Specificity | Precision | F1_score | MCC |
|---|---|---|---|---|---|---|
| mAlexNet + SVM | 0.9983 | 0.9983 | 0.9991 | 0.9983 | 0.9983 | 0.9975 |
As seen in Table 5 , the accuracy of the proposed method is compared with different studies. According to this table, the proposed method is superior to previous state-of-the-art studies. All of the previous studies in the table are based on deep learning and have achieved very high accuracy rates. In general, previous studies have adopted the Transfer Learning (TL) structure. Apart from this, there are also studies using RNN-based LSTM and BiLSTM structures. In addition, some studies have used the CNN + SVM structure as in this study. Previous studies have generally made modifications to the classifier rather than the network input images. However, the most important difference of our study is the preprocessing and feeding the semantically segmented lung images to the deep network. The next implemented CNN + SVM structure also classified the input images in the best way.
Table 5.
Comparison of the proposed method with previous state-of-the-art studies.
| Previous Study | Method | Accuracy (%) |
|---|---|---|
| Wang et al. [68] | DL | 92.30 |
| Gupta et al. [69] | InstaCovNet-19 | 99.08 |
| Afshar et al. [70] | Capsule Network | 95.70 |
| Islam et al. [30] | LSTM & CNN | 99.40 |
| Chowdhury et al. [26] | TL | 97.94 |
| Sethy et al. [28] | ResNet50 - SVM | 95.40 |
| Farooq et al. [71] | TL | 96.20 |
| Das et al. [72] | Xception | 97.40 |
| Ucar et al. [73] | Bayes - SqueezeNet | 98.30 |
| Ismael et al. [74] | DL & SVM | 94.70 |
| Apostolopoulos et al. [75] | TL | 93.48 |
| Hemdan et al. [76] | VGG19 | 90.00 |
| Xu et al. [77] | ResNet - Location Attention | 86.70 |
| Brunese et al. [78] | TL | 97.00 |
| Ozturk et al. [79] | DarkCovidNet | 87.02 |
| Narin et al. [25] | TL | 98.00 |
| Rahimzadeh et al. [80] | Xception & ResNet50V2 | 91.40 |
| Asif et al. [24] | TL | 96.00 |
| Nour et al. [27] | DL & SVM | 98.97 |
| Khan et al. [23] | TL | 95.00 |
| Wang et al. [81]. | ResNet - Feature Pyramid Network | 94.00 |
| Proposed Method | DL & SVM | 99.80 |
Although the proposed method provides high accuracy rates, some improvements are required to increase the success and reliability of the results. The number of data and the data diversity significantly affect accuracy and reliability. It is desirable that the proposed method achieves similar success on all CXR images. However, this is not usually possible. One reason is that the datasets are not uniform. Another reason is that the parameter settings of the networks provide less success for different datasets. Another reason is the data augmentation steps applied to CXR images. Although the data augmentation steps applied in this study increased the data diversity, each augmented image is now artificial. As a result, the performance of the network is determined by many images that do not have real clinical data. This creates doubts about the results. In addition, the number and data diversity used in the study give important clues about the generalization capacity of the method. Considering these limitations, in future studies, data augmentation steps will not be used and the proposed method will be tested by mixing different datasets. In addition, different hyperparameter optimization techniques will be used to provide the optimum result for different datasets of the proposed deep network.
5. Conclusion
Early detection of the COVID-19 pandemic is crucial to ensuring early treatment and preventing the spread of the disease. Unlike studies that directly use raw CXR images, this study first semantically segments the lung from the raw images and then applies preprocessing steps to the segmented images. Semantic segmentation aims to segment the lungs in CXR images. As a result of feeding these images to the designed mAlexNet + SVM network, three-class CXR images are distinguished with 99.8% success. The study owes its success to successful lung segmentation. Previous studies often use raw images for feature extraction. The results show that this study outperforms previous state-of-the-art studies. The proposed application automatically classifies lung images for COVID-19 diagnosis after the semantic segmentation stage, without the need for any handcrafted feature extraction technique. Prediction with only lung images enables the feature extraction algorithm to obtain more reliable features. Because the features are extracted from the area where the disease is effective. Ultimately, the proposed method is fast and stable and helps expert radiologists as a decision support system. In this way, the workload of radiologists is reduced, misdiagnosis is prevented and early infection detection is provided.
CRediT authorship contribution statement
Muhammet Fatih Aslan: Investigation, Methodology, Software, Writing – review & editing.
Declaration of competing interest
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
The data used is public and there is citation for access within the article. The code can be requested from the author.
References
- 1.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. Applied Soft Computing; 2020. CNN-based Transfer Learning-BiLSTM Network: A Novel Approach for COVID-19 Infection Detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. 2020. Deep-covid: Predicting Covid-19 from Chest X-Ray Images Using Deep Transfer Learning. arXiv preprint arXiv:2004.09363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horry M.J., Chakraborty S., Paul M., Ulhaq A., Pradhan B., Saha M., Shukla N. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) Statpearls [internet] [PubMed] [Google Scholar]
- 6.ACR . 2020. ACR Recommendations for the Use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection. [Google Scholar]
- 7.Tartaglione E., Barbano C.A., Berzovini C., Calandri M., Grangetto M. 2020. Unveiling COVID-19 from Chest X-Ray with Deep Learning: a Hurdles Race with Small Data. arXiv preprint arXiv:2004.05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh Y., Park S., Ye J.C. IEEE Transactions on Medical Imaging; 2020. Deep Learning Covid-19 Features on Cxr Using Limited Training Data Sets. [DOI] [PubMed] [Google Scholar]
- 10.Aslan M.F., Sabanci K., Ropelewska E. A new approach to COVID-19 detection: an ANN proposal optimized through tree-seed algorithm. Symmetry. 2022;14:1310. [Google Scholar]
- 11.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Waisy A.S., Al-Fahdawi S., Mohammed M.A., Abdulkareem K.H., Mostafa S.A., Maashi M.S., Arif M., Garcia-Zapirain B. COVID-CheXNet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020:1–16. doi: 10.1007/s00500-020-05424-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks, Chaos. Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moura J.D., García L.R., Vidal P.F.L., Cruz M., López L.A., Lopez E.C., Novo J., Ortega M. Deep convolutional approaches for the analysis of COVID-19 using chest X-ray images from portable devices. IEEE Access. 2020;8:195594–195607. doi: 10.1109/ACCESS.2020.3033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J., Zhang D., Cheng G., Liu N., Xu D. Advanced deep-learning techniques for salient and category-specific object detection: a survey. IEEE Signal Process. Mag. 2018;35:84–100. [Google Scholar]
- 17.Simonyan K., Zisserman A. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 18.Yu S., Xie L., Liu L., Xia D. Learning long-term temporal features with deep neural networks for human action recognition. IEEE Access. 2019;8:1840–1850. [Google Scholar]
- 19.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 20.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 21.Al-Dhamari A., Sudirman R., Mahmood N.H. Transfer deep learning along with binary support vector machine for abnormal behavior detection. IEEE Access. 2020;8:61085–61095. [Google Scholar]
- 22.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asif S., Wenhui Y. medRxiv; 2020. Automatic Detection of COVID-19 Using X-Ray Images with Deep Convolutional Neural Networks and Machine Learning. [Google Scholar]
- 25.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N. 2020. Can AI Help in Screening Viral and COVID-19 Pneumonia? arXiv preprint arXiv:2003.13145. [Google Scholar]
- 27.Nour M., Cömert Z., Polat K. Applied Soft Computing; 2020. A Novel Medical Diagnosis Model for COVID-19 Infection Detection Based on Deep Features and Bayesian Optimization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethy P.K., Behera S.K. 2020. Detection of Coronavirus Disease (Covid-19) Based on Deep Features; p. 2020. Preprints, 2020030300. [Google Scholar]
- 29.Aslan M.F., Durdu A., Yusefi A., Yilmaz A. Neural Networks; 2022. HVIOnet: A Deep Learning Based Hybrid Visual-Inertial Odometry Approach for Unmanned Aerial System Position Estimation. [DOI] [PubMed] [Google Scholar]
- 30.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam M.M., Islam M.Z., Asraf A., Ding W. medRxiv; 2020. Diagnosis of COVID-19 from X-Rays Using Combined CNN-RNN Architecture with Transfer Learning. [Google Scholar]
- 32.Zhang D., Huang G., Zhang Q., Han J., Han J., Yu Y. Cross-modality deep feature learning for brain tumor segmentation. Pattern Recogn. 2020;110 [Google Scholar]
- 33.Gloger O., Tönnies K. Subject-Specific prior shape knowledge in feature-oriented probability maps for fully automatized liver segmentation in MR volume data. Pattern Recogn. 2018;84:288–300. [Google Scholar]
- 34.Xian M., Zhang Y., Cheng H.-D., Xu F., Zhang B., Ding J. Automatic breast ultrasound image segmentation: a survey. Pattern Recogn. 2018;79:340–355. [Google Scholar]
- 35.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979;9:62–66. [Google Scholar]
- 36.Wang L., Pan C. Robust level set image segmentation via a local correntropy-based K-means clustering. Pattern Recogn. 2014;47:1917–1925. [Google Scholar]
- 37.De Smet P. Optimized high speed pixel sorting and its application in watershed based image segmentation. Pattern Recogn. 2010;43:2359–2366. [Google Scholar]
- 38.Minaee S., Wang Y. An ADMM approach to masked signal decomposition using subspace representation. IEEE Trans. Image Process. 2019;28:3192–3204. doi: 10.1109/TIP.2019.2894966. [DOI] [PubMed] [Google Scholar]
- 39.Minaee S., Boykov Y., Porikli F., Plaza A., Kehtarnavaz N., Terzopoulos D. 2020. Image Segmentation Using Deep Learning: A Survey. arXiv preprint arXiv:2001.05566. [DOI] [PubMed] [Google Scholar]
- 40.Long J., Shelhamer E., Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 41.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 42.Badrinarayanan V., Kendall A., Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 43.Chen L.-C., Papandreou G., Schroff F., Adam H. 2017. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv preprint arXiv:1706.05587. [Google Scholar]
- 44.Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. Proceedings of the European Conference on Computer Vision. ECCV); 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 801–818. [Google Scholar]
- 45.Hu J., Li L., Lin Y., Wu F., Zhao J. Springer; 2019. A Comparison and Strategy of Semantic Segmentation on Remote Sensing Images, the International Conference on Natural Computation, Fuzzy Systems and Knowledge Discovery; pp. 21–29. [Google Scholar]
- 46.Khan Z., Yahya N., Alsaih K., Ali S.S.A., Meriaudeau F. Evaluation of deep neural networks for semantic segmentation of prostate in T2W MRI. Sensors. 2020;20:3183. doi: 10.3390/s20113183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J., Zhang Y., He X., Xie P. 2020. COVID-CT-Dataset: a CT Scan Dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 49.medicalsegmentation . 2020. COVID-19 CT Segmentation Dataset. [Google Scholar]
- 50.COVID-19 Xray Dataset. 2020. [Google Scholar]
- 51.Oulefki A., Agaian S., Trongtirakul T., Kassah Laouar A. Pattern Recognition; 2020. Automatic COVID-19 Lung Infected Region Segmentation and Measurement Using CT-scans Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Q., Wang B., Gong D., Luo C., Zhao W., Shen J., Shi Q., Jin S., Zhang L., You Z. 2020. COVID-19 Chest CT Image Segmentation--A Deep Convolutional Neural Network Solution. arXiv preprint arXiv:2004.10987. [Google Scholar]
- 53.Saeedizadeh N., Minaee S., Kafieh R., Yazdani S., Sonka M. 2020. Covid Tv-Unet: Segmenting Covid-19 Chest Ct Images Using Connectivity Imposed U-Net. arXiv preprint arXiv:2007.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. IEEE Transactions on Medical Imaging; 2020. A Weakly-Supervised Framework for COVID-19 Classification and Lesion Localization from Chest CT. [DOI] [PubMed] [Google Scholar]
- 55.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B. MedRxiv; 2020. Deep Learning-Based Model for Detecting 2019 Novel Coronavirus Pneumonia on High-Resolution Computed Tomography: a Prospective Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferdi A., Benierbah S., Ferdi Y. 2022 7th International Conference on Image and Signal Processing and Their Applications (ISPA) IEEE; 2022. U-Net-based covid-19 CT image semantic segmentation: a transfer learning approach; pp. 1–5. [Google Scholar]
- 57.Shamim S., Awan M.J., Mohd Zain A., Naseem U., Mohammed M.A., Garcia-Zapirain B. Automatic COVID-19 lung infection segmentation through modified unet model. J. Healthc. Eng. 2022;2022 doi: 10.1155/2022/6566982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Rajamani K.T., Rani P., Siebert H., ElagiriRamalingam R., Heinrich M.P. Attention-augmented U-Net (AA-U-Net) for semantic segmentation. Signal. Image.Video Process. 2022 doi: 10.1007/s11760-022-02302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. Covid-19 Image Data Collection: Prospective Predictions Are the Future. arXiv preprint arXiv:2006.11988. [Google Scholar]
- 60.Kermany D., Zhang K., Goldbaum M. Mendeley data; 2018. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification; p. 2. [Google Scholar]
- 61.Darwin . 2020. COVID-19 Chest X-Ray Dataset. [Google Scholar]
- 62.SIRM . 2020. COVID-19 DATABASE. [Google Scholar]
- 63.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 Image Data Collection. ArXiv, abs/2003. [Google Scholar]
- 64.Roy Choudhury A., Vanguri R., Jambawalikar S.R., Kumar P. Springer International Publishing; Cham: 2019. Segmentation of Brain Tumors Using DeepLabv3+ pp. 154–167. [Google Scholar]
- 65.Zhao H., Shi J., Qi X., Wang X., Jia J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Pyramid scene parsing network; pp. 2881–2890. [Google Scholar]
- 66.Zhang S., Ma Z., Zhang G., Lei T., Zhang R., Cui Y. Semantic image segmentation with deep convolutional neural networks and quick shift. Symmetry. 2020;12:427. [Google Scholar]
- 67.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Intern. Med. 2013;4:627. [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Wong A. 2020. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta A., Gupta S., Katarya R. Applied Soft Computing; 2020. InstaCovNet-19: A Deep Learning Classification Model for the Detection of COVID-19 Patients Using Chest X-Ray. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. Covid-caps: A Capsule Network-Based Framework for Identification of Covid-19 Cases from X-Ray Images. arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farooq M., Hafeez A. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 72.Das N.N., Kumar N., Kaur M., Kumar V., Singh D. Irbm; 2020. Automated Deep Transfer Learning-Based Approach for Detection of COVID-19 Infection in Chest X-Rays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ucar F., Korkmaz D. Medical Hypotheses; 2020. COVIDiagnosis-Net: Deep Bayes-SqueezeNet Based Diagnostic of the Coronavirus Disease 2019 (COVID-19) from X-Ray Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2020;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 77.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Progr. Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahimzadeh M., Attar A. 2020. A New Modified Deep Convolutional Neural Network for Detecting COVID-19 from X-Ray Images. arXiv preprint arXiv:2004.08052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recogn. 2020;110 doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used is public and there is citation for access within the article. The code can be requested from the author.