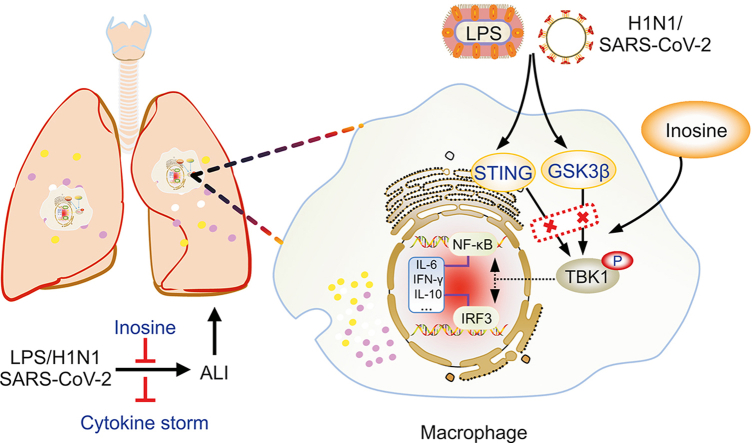
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced cytokine storms constitute the primary cause of coronavirus disease 19 (COVID-19) progression, severity, criticality, and death. Glucocorticoid and anti-cytokine therapies are frequently administered to treat COVID-19, but have limited clinical efficacy in severe and critical cases. Nevertheless, the weaknesses of these treatment modalities have prompted the development of anti-inflammatory therapy against this infection. We found that the broad-spectrum anti-inflammatory agent inosine downregulated proinflammatory interleukin (IL)-6, upregulated anti-inflammatory IL-10, and ameliorated acute inflammatory lung injury caused by multiple infectious agents. Inosine significantly improved survival in mice infected with SARS-CoV-2. It indirectly impeded TANK-binding kinase 1 (TBK1) phosphorylation by binding stimulator of interferon genes (STING) and glycogen synthase kinase-3β (GSK3β), inhibited the activation and nuclear translocation of the downstream transcription factors interferon regulatory factor (IRF3) and nuclear factor kappa B (NF-κB), and downregulated IL-6 in the sera and lung tissues of mice infected with lipopolysaccharide (LPS), H1N1, or SARS-CoV-2. Thus, inosine administration is feasible for clinical anti-inflammatory therapy against severe and critical COVID-19. Moreover, targeting TBK1 is a promising strategy for inhibiting cytokine storms and mitigating acute inflammatory lung injury induced by SARS-CoV-2 and other infectious agents.
Keywords: Cytokine storm, Interleukin 6 (IL-6), Inosine, SARS-CoV-2, TANK-binding kinase 1 (TBK1)
Graphical abstract
Highlights
-
•
Inosine significantly improved the survival rate of SARS-CoV-2-infected mice.
-
•
Inosine abrogated excessive IL-6 expression to ameliorate various infectious agents-induced acute inflammatory lung injury.
-
•
Inosine inhibited TBK1 phosphorylation via binding to STING and GSK3β, leading to decreases in IL-6 release.
1. Introduction
The coronavirus disease 19 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and has rapidly led to a global health crisis [1]. By the end of 2021, over 250 million people worldwide had been diagnosed with COVID-19. Ten to fifteen percent of these cases progressed to severe or critical status and over five million people died [2,3]. Newly developed antiviral drugs have demonstrated a certain degree of efficacy against mild COVID-19 [4]. Cases that progressed to severe or critical status were characterized by a pulmonary hyperinflammatory state known as a cytokine storm [5]. In these cases, only anti-inflammatory therapy is potentially clinically beneficial [6]. Although immunosuppressive glucocorticoids reduced the requirement for mechanical ventilation, they did not significantly lower mortality in severe and critical cases [7,8]. In fact, they sometimes worsened the condition by delaying viral clearance [9]. The ideal therapeutic agent for severe or critical COVID-19 controls cytokine release and prevents overactivation of the immune response.
Cytokine storms cause life-threatening systemic inflammation characterized by immune cell hyperactivation and the release of proinflammatory cytokines including interleukin-6 (IL-6), interferons (IFNs), and tumor necrosis factor-alpha (TNF-α) [10]. Our previous study and several recent articles indicate that a sharp increase in pleiotropic cytokine IL-6 content is associated with acute respiratory distress syndrome (ARDS), sepsis, and even death in patients with COVID-19 [11,12]. IL-6 receptor antagonists have been widely administered in human clinical trials. However, IL-6 signaling blockade provides no broad-based survival benefit in COVID-19 therapy. A possible explanation is that IL-6 is essential for activating innate/adaptive immunity and facilitating efficient pathogen clearance [13,14]. Therefore, it is necessary to develop a transcriptional regulation strategy wherein the SARS-CoV-2-induced rise in IL-6 content is controlled and the anti-inflammatory response initiated by IL-6 is partially conserved.
Viral genes are detected by classical Toll-like receptors (TLRs) as well as other specific pattern recognition receptors (PRRs) that initiate the host innate immune response. Viral RNA is detected by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) while viral DNA is sensed by cyclic guanosine monophosphate-adenosine monophosphate (GMP-AMP) synthase (cGAS) [15]. In turn, RIG-I and cGAS activate particular signaling cascades and induce IL-6 and other inflammatory cytokines via mitochondrial antiviral signaling (MAVS) protein and stimulator of interferon genes (STING), respectively [16,17]. The RNA virus SARS-CoV-2 can evade RLR sensors and inhibit the MAVS downstream adaptor [18]. By contrast, host self-DNA in the cytoplasms of SARS-CoV-2-induced syncytia triggers the antiviral innate immune response via the cGAS-STING pathway [15], activates interferon regulatory factor (IRF3) and nuclear factor kappa B (NF-κB), induces IFNs, and upregulates proinflammatory cytokines [19]. Glycogen synthase kinase-3-beta (GSK3β) is activated in SARS-CoV-2-infected cells and is required for viral replication [20]. Activated GSK3β phosphorylates NF-κB and provokes systemic inflammation [21]. Hence, multiple signaling pathway-induced NF-κB responses may control IL-6 transcription and release in patients with COVID-19. Recent evidence demonstrates that TANK-binding kinase 1 (TBK1) is a vital factor in the foregoing innate immune signaling pathways and mediating upstream stimuli and downstream NF-κB or IRF3 signaling [22]. For these reasons, suppressing TBK1 activation can modulate the host innate immune response following SARS-CoV-2 infection.
Inosine, a type of purine nucleoside, is a deamination metabolite of adenosine and is also formed by binding hypoxanthine with ribose. Endogenous inosine is a key intermediate in purine biosynthesis and regulates RNA replication and translation [23]. Exogenous inosine supplementation protects against lipopolysaccharide (LPS)-induced acute lung injury and endotoxin-induced septic shock by suppressing the release of the proinflammatory cytokines IL-6, IL-1β, TNF-α, and so on [24,25]. Inosine may also modulate Th1 cell differentiation and/or activation and improve immunotherapy efficacy via the T cell-specific adenosine A2A receptor (A2AR) and STING signaling pathways [26,27]. Inosine was detected in certain Chinese material medica and medical formulae such as Lumbricus terrestris and Pinellia ternata (Thunb.) that are administered for the treatment of pneumonia and COVID-19 [28,29]. Hence, inosine might control SARS-CoV-2-induced cytokine storms.
Here, we found that inosine functioned as a broad-spectrum anti-inflammatory agent. It attenuated acute lung inflammatory injury triggered by various stimuli and markedly improved survival in mice infected with SARS-CoV-2. Inosine demonstrated strong efficacy against cytokine storms as it inhibited innate immune signaling-mediated TBK1 phosphorylation by multiple routes. The findings of this work showed that inosine could serve as an adjuvant for the treatment of severe or critical COVID-19 and suggested that TBK1 is a potential therapeutic target.
2. Materials and methods
2.1. Cell lines and viruses
All cell lines used in the present study were purchased from the National Infrastructure of Cell Line Resource (Beijing, China). Mouse leukemic monocyte/macrophages (RAW264.7) and human embryonic kidney 293 (HEK293) cells were maintained in a humidified incubator (37 °C, 5% CO2) in Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher Scientific Inc., Waltham, MA, USA) containing 10% (V/V) fetal bovine serum (FBS; Tianhang Biotechnology, Hangzhou, China) and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin) (Sigma-Aldrich Corp., St. Louis, MO, USA). Influenza A (H1N1) virus Beijing/501/2009 strain (BJ501) was isolated from a patient in Beijing confirmed to be infected in 2009. The genome sequence of this strain is deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession No. GQ223415. The mouse-adapted SARS-CoV-2 C57MA14 strain was isolated from a COVID-19 patient in Wuhan, China and obtained by continuous passage in C57BL/6N mice. The genome sequence of this strain is deposited in the GenBank database under accession No. OL913104.1. All experiments involving live H1N1 virus and SARS-CoV-2 were performed in a biosafety level three (BSL-3) laboratory at Changchun Veterinary Research Institute of the Chinese Academy of Agricultural Sciences and in strict accordance with biosafety standard operating procedures.
2.2. Reagents
Lipopolysaccharide (LPS; purity ≥98%) and dexamethasone sodium phosphate (DXM; purity ≥99%) were obtained from Solarbio Science & Technology (Beijing, China). Poly (I:C) (purity ≥99%) was purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). High-quality specific agonists or antagonists of STING, TBK1, GSK3β, and A2AR were purchased from Topscience Co., Ltd. (Shanghai, China). Primary anti-A2AR and anti-hemagglutinin (HA) antibodies were obtained from Proteintech Co., Ltd. (Wuhan, China). Anti-biotin antibody was obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Anti-phospho-GSK3β (p-GSK3β), anti-GSK3β, anti-phospho-STING (p-STING), anti-STING, anti-phospho-TBK1 (p-TBK1), anti-TBK1, anti-phospho-IRF3 (p-IRF3), anti-IRF3, anti-phospho-NF-κB (p-NF-κB), anti-NF-κB, anti-IL-6, anti-β-actin, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Alexa Fluor secondary antibodies were purchased from Servicebio (Wuhan, China).
2.3. Biosafety facility and ethics statement
All work involving live SARS-CoV-2 viruses was conducted in a BSL-3 laboratory at Changchun Veterinary Research Institute of the Chinese Academy of Agricultural Sciences. The BALB/c mice were housed and maintained in accordance with the Guidelines for the Welfare and Ethics of Laboratory Animals of China. All animal studies were approved by the Animal Welfare and Ethics Committee of Changchun Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (No. IACUC-AMMS-11-2020-020).
2.4. Mouse husbandry and treatment
C57BL/6 N mice aged 7–10 weeks and weighing 20–22 g and BALB/c mice aged 9 months and weighing 25–30 g were obtained from Beijing Charles River Laboratory Animal Co., Ltd. (Beijing, China). The mice were maintained under specific pathogen-free (SPF) conditions and ∼12 h light/12 h dark cycles. The SARS-CoV-2 challenge was performed on BALB/c mice. They were intranasally infected with 50 μL of a 103 median tissue culture infective dose (TCID50) SARS-CoV-2 strain C57MA14. The C57BL/6 N mice were exposed to H1N1 virus or LPS by intranasal inoculation or intratracheal instillation, respectively. Mice exposed to SARS-CoV-2, H1N1 virus, or LPS were randomly assigned either to an inosine treatment group or a vehicle group. The animals in the control group were administered equivalent volumes of sterile saline solution by intranasal inoculation or intratracheal instillation. Mouse survival and body weight changes were monitored daily. All experimental protocols were approved by the Ethics Committee of Animal Experiments of Beijing Institute of Radiation Medicine (No. IACUC-DWZX-2020-762).
2.5. Viral RNA extraction and reverse-transcription quantitative real-time polymerase chain reaction (RT-qPCR)
The mouse tissues were mixed with DMEM and homogenized (homogenizer; Thermo Fisher Scientific Inc.) for 5 min. The suspension was centrifuged at 12,000 g and 4 °C for 10 min and the supernatant was conserved. The RNA viruses were extracted with a QIAamp Viral RNA Mini Kit (QIAGEN, Dusseldorf, Germany). The viral RNA was quantified by RT-qPCR with Premix Ex Taq (Takara, Beijing, China) targeting the SARS-CoV-2 N gene. The primer and probe sequences were as follows: NF (5′-GGGGAACTTCTCCTGCTAGAAT-3′); NR (5′-CAGACATTTTGCTCTCAAGCTG-3′); and NP (5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′).
2.6. SARS-CoV-2 load quantification by TCID50
The supernatants of the lung and turbinate bone tissue homogenates were diluted with DMEM and applied to Vero E6 cells cultured on 96-well plates (Corning Inc., Corning, NY, USA). The Vero E6 cells were incubated for 1 h and the DMEM was replenished. After 72 h incubation, the TCID50 was determined based on the cytopathic effect.
2.7. Serum cytokine/chemokine measurements
Mice were euthanized at 1 day or 3 day post-infection/treatment and their sera were collected. Serum cytokines were measured with a ProcartaPlex Mouse Cytokine Panel (Thermo Fisher Scientific Inc.). The assays were conducted in 96-well filter plates and in a flow-based instrument (Luminex® 200™; Thermo Fisher Scientific Inc.) according to the manufacturer's instructions. Serum IL-6 levels were confirmed with a Sandwich enzyme-linked immunosorbent assay (ELISA) kit (Meimian industrial Co., Ltd, Yancheng, China). All data were calculated by deducting the background value.
2.8. Histological analysis and immunofluorescence
Mice were sacrificed at 1 day or 3 day post-infection/treatment and their lungs were harvested and fixed in 10% (V/V) neutral buffered formalin, embedded in paraffin, cut into 3.5-μm sections (Leica, Weztlar, Germany), and subjected to hematoxylin-eosin (H&E) staining. For the lung tissue immunofluorescence assay, the sections were sealed with bovine serum albumin (BSA) at 25 °C for 30 min and incubated at 4 °C overnight with the indicated primary antibodies. The slides were then incubated with fluorescein isothiocyanate (FITC) anti-rabbit secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) containing anti-fluorescence quenching agent. The lung sections were then visualized under a whole-slide imaging microscope fitted with Pannoramic Scanner software (Pannoramic DESK, Budapest, Hungary). RAW264.7 cells were fixed with immunostaining fixing solution (Beyotime Biotechnology, Shanghai, China) and blocked with immunostaining blocking buffer (Beyotime Biotechnology). The corresponding primary antibody was incubated at 4 °C overnight. The cells were then washed with phosphate-buffered saline (PBS) and incubated with fluorescently-coupled secondary antibodies and DAPI. Fluorescence images were visualized under a confocal laser scanning microscope (CLSM; Carl Zeiss AG, Oberkochen, Germany). Primary antibodies were used at the following dilutions: anti-p-GSK3β (1:300), anti-p-STING (1:400), anti-p-TBK1 (1:100), anti-p-IRF3 (1:200), anti-p-NF-κB (1:1000), and anti-IL-6 (1:200), and Alexa Fluor secondary antibodies were used at 1:1000 dilution.
2.9. Western blotting
Proteins were extracted from the cells or lung tissue with cold radioimmunoprecipitation assay (RIPA) buffer and quantified by bicinchoninic acid (BCA) assay. The proteins were separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel (PAGE) (Epizyme Biomedical Technology Co., Ltd, Shanghai, China) and transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). The latter were sealed with 5% skim milk and successively incubated with the indicated primary (1:1000) and secondary (1:3000) antibodies. The bands were incubated with SuperSignal West Pico PLUS Detection Reagent (Thermo Fisher Scientific Inc.) and visualized with ImageQuant LAS 500 (GE Healthcare BioSciences AB, Chicago, IL, USA).
2.10. Co-immunoprecipitation (Co-IP) assay
The TBK1 cDNA sequence was cloned into a pCDNA3.1 expression plasmid and fused with HA-tag at the C-terminal. HEK293 cells were transfected with 2 μg TBK1-HA expression plasmid using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cells were collected and resuspended in lysis buffer containing protease and phosphatase inhibitors. The lysates were mixed with rabbit monoclonal anti-HA at 4 °C overnight to form the immune complex. Prewashed Pierce™ classic magnetic beads (Thermo Fisher Scientific Inc.) were added to the lysate samples and the mixture was incubated at room temperature for 1 h. The beads were harvested on a magnetic rack, washed thrice with cold rinse buffer, and eluted with eluent. The proteins were eluted from the beads by boiling with loading buffers and then subjected to immunoblot analysis.
2.11. Biotin-tagged inosine synthesis
To identify binding between inosine and its targets, the former was labeled with biotin at its hydroxyl moiety and biotinylated inosine (bio-inosine) was synthesized. One equivalent biotin was dissolved in dry dimethyl sulfoxide (DMSO), 1.2 eq N,N,N′,N′-tetramethyl-O-(N-succinimidyl)uronium tetrafluoroborate (TSTU) and 0.1 mL of triethylamine were added, and the reaction was conducted at room temperature for 4 h. Then diethyl ether was added to precipitate the biotin succinimidyl ester (SE). Then 1 eq biotin SE was dissolved in 2 mL of N,N-dimethylformamide (DMF) and dripped into DMF containing 1 eq inosine through a constant-pressure dropping funnel for 4 h. The reaction continued for another 4 h and the solvent was evaporated at low pressure to prepare the liquid phase for purification. Bio-inosine structure and purity were determined by nuclear magnetic resonance (Bruker AVANCE NEO, karlsruhe, Germany) and liquid chromatography-mass spectrometry-ion trap-time-of-flight (LC/MS-IT/TOF; Shimadzu, Kyoto, Japan) (Fig. S1). The inosine successfully bound the biotin and the target compound was relatively pure.
2.12. Biotin pulldown assay
Streptavidin isolated from Streptomyces avidinii was covalently bound to agarose to perform pull-down experiments. Five hundred microliters of 500 μM of bio-inosine or biotin was added to 60 μL of streptavidin-agarose beads (Sigma-Aldrich Corp.) and incubated at 4 °C overnight. The cell lysates were then added to the streptavidin-agarose beads containing bio-inosine or biotin. The mixture was placed in a blender, incubated at 4 °C overnight, and washed thrice with PBS. The bead-bound proteins were boiled in 2× loading buffer and subjected to immunoblot analysis on SDS-PAGE.
2.13. Computational docking and molecular simulation
Docking was performed using the Glide module in Schrödinger 2020-2 software (https://www.schrodinger.com/releases/release-2020-2). The protein structures used in the docking studies included human GSK3β and STING. Their pdb codes were 1Q5K and 6UKZ, respectively. All crystal structures were prepared according to the recommended procedure in the Protein Preparation Wizard (https://www.schrodinger.com/science-articles/protein-preparation-wizard). The ligands and solvent molecules were removed. Polar hydrogen atoms were added for each protein structure and the atomic charges were assigned based on the OPLS3e force field. Residues within 20 Å of the active ligand binding sites in the crystal structure were defined as the binding sites at which the docking grids were created. The ligand structures were prepared with the LigPrep module (https://www.schrodinger.com/products/ligprep) to define the protonation state and the atomic charges (reference pH = 7.0 ± 2.0; OPLS3e force field). The native ligand was removed and docking was conducted in the standard docking mode with the receptor. The default input parameters were no scaling factor for the VdW radii of the nonpolar protein atoms, and 0.8 scaling factor for the nonpolar ligand atoms. Both were used in all computations. Inosine was docked and scored in Glide standard precision (SP) mode and the optimal inosine pose was selected with Glide Score. The binding interaction modes of inosine with human GSK3β and STING were analyzed with PyMOL (https://pymol.org/2/).
2.14. Statistical analysis
Statistical analyses were conducted in GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA). The data were means ± standard error of the mean (SEM). One-way analysis of variance (ANOVA) followed by a least significant difference (LSD) post-hoc test compared multiple treatment means. A log-rank (Mantel-Cox) test was used to determine mouse survival rates. P < 0.05 indicated statistical significance.
3. Results
3.1. Inosine alleviated LPS- and H1N1 virus-induced acute inflammatory lung injury by regulating cytokine secretion
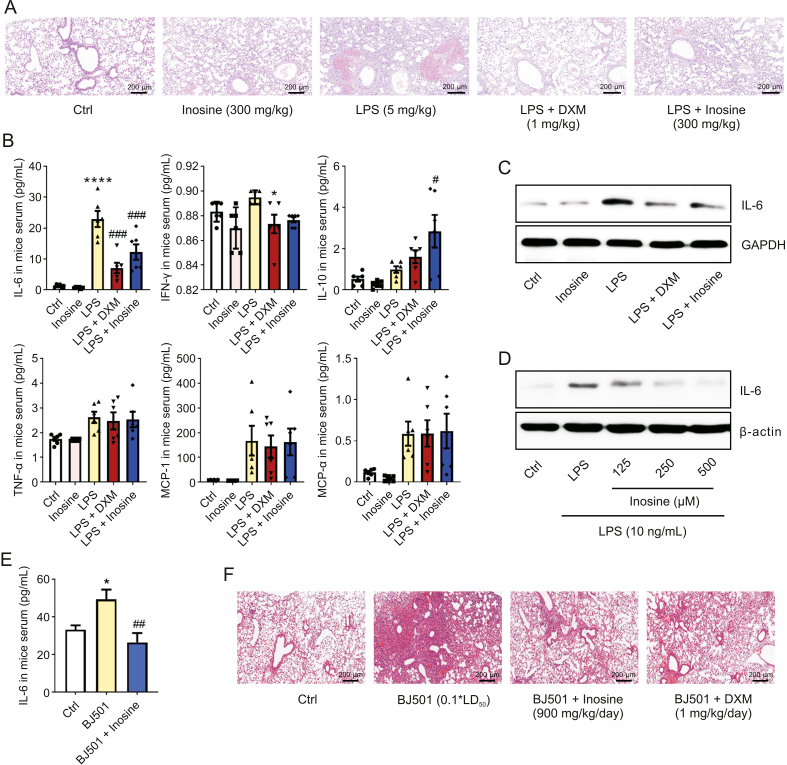
We used LPS and H1N1 virus to establish acute inflammatory lung injury models in mice and evaluate the anti-inflammatory effect of inosine. H&E staining revealed obvious inflammatory cell infiltration and patchy hemorrhage in the alveolar cavity and the pulmonary interstitium at 24 h post LPS treatment (Fig. 1A). These signs were accompanied by sharp increases in serum IL-6 and interferon (IFN)-γ levels and remarkable changes in body weight and wet-to-dry lung weight ratio (W/D) (Figs. 1B, S2A and S2B). Elevated IL-6 was detected in both the LPS-infected lung tissue and the RAW246.7 cells (Figs. 1C and D). Both DXM and the 300 mg/kg inosine treatment inhibited LPS-induced increases in serum IL-6 and IFN-γ (Fig. 1B), lowered the IL-6 content in LPS-infected mouse lung tissue (Fig. 1C) and mononuclear macrophages (Fig. 1D), and ameliorated LPS-induced lung tissue damage (Fig. 1A). Inosine significantly upregulated serum IL-10 content in the mouse model (Fig. 1B). The 900 mg/kg inosine treatment attenuated H1N1-induced serum IL-6 upregulation and acute lung injury in mice within the first 24 h post-infection (Figs. 1E and F). Inosine had a weaker antagonistic effect than DXM against H1N1-induced histopathology but exerted no adverse effect on mouse body weight (Fig. S2C). The preceding results suggest that inosine has broad-spectrum anti-inflammatory efficacy in COVID-19 therapy by controlling the cytokine storm.
Fig. 1.
Inosine alleviated lipopolysaccharide (LPS)- and H1N1 virus-induced acute inflammatory lung injury. (A) C57BL/6 mice were challenged with 5 mg/kg LPS and administered 300 mg/kg inosine for 24 h. Lung tissue sections were stained with hematoxylin-eosin and injury was examined (n = 6). (B) Mice were treated as described in (A). Serum cytokine and chemokine levels were measured (n = 6). (C) Mice were treated with LPS and/or inosine for 24 h. Interleukin (IL)-6 expression levels in lung tissue were detected by Western blotting. (D) RAW264.7 cells were treated with LPS and inosine for 24 h. IL-6 expression in RAW264.7 cells was measured by Western blotting (n = 3). (E) Mice were infected with Beijing/501/2009 strain (BJ501) and treated with 900 mg/kg inosine for 24 h. Quantitative enzyme-linked immunosorbent assay (ELISA) of serum IL-6 (n = 6). (F) Mice were treated as described in (E). Pathological damage to lung tissue was examined by hematoxylin-eosin (H&E) staining (n = 6). Ctrl: control group; LPS: LPS-induced acute lung injury and no other treatment; LPS + Inosine: LPS-induced acute lung injury followed by inosine treatment; LPS + DXM: LPS-induced acute lung injury followed by dexamethasone treatment as positive control; BJ501: H1N1-induced acute lung injury and no other treatment; BJ501 + Inosine: BJ501-induced acute lung injury followed by inosine treatment; BJ501 + DXM: BJ501-induced acute lung injury followed by dexamethasone treatment as positive control. Data are means ± standard error of the mean (SEM). ∗P < 0.05, ∗∗∗∗P < 0.0001 vs. control. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. LPS or BJ501 group.
3.2. Inosine improved survival in SARS-CoV-2-infected mice by alleviating inflammatory injury rather than through antiviral activity
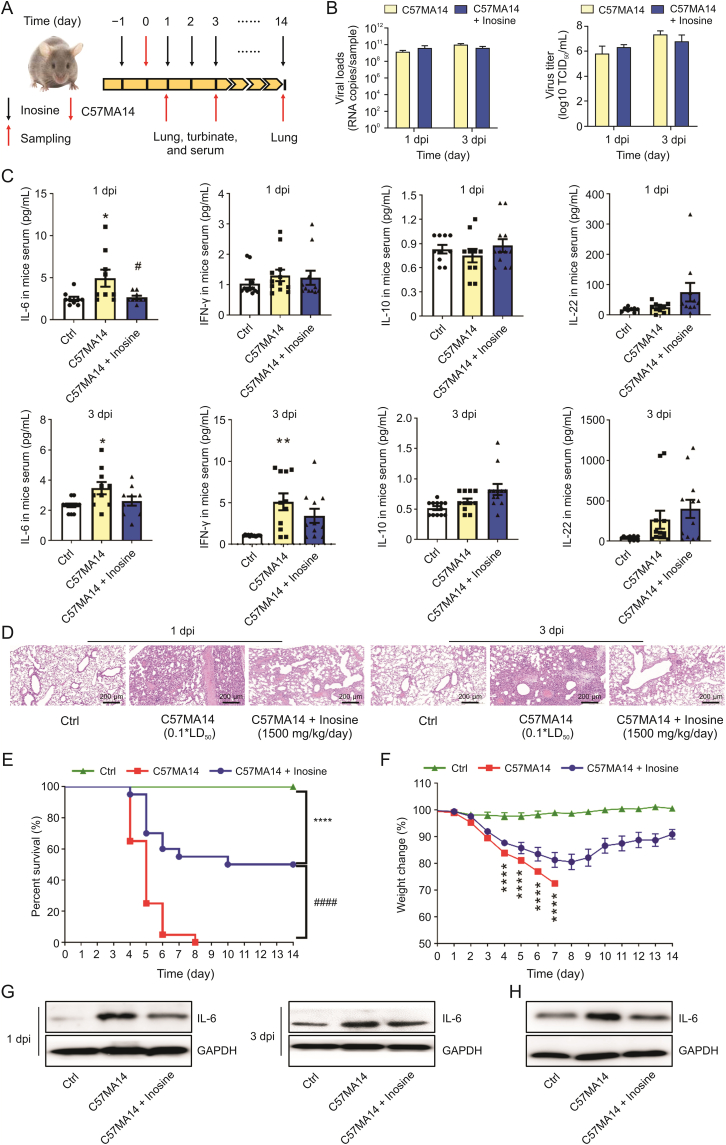
We performed in vivo antiviral assays to determine whether inosine alleviates SARS-CoV-2-induced acute lung injury. The mice were orally administered with single daily inosine doses at 24 h before C57MA14 infection and were sacrificed at 1 day, 3 day or 14 day post infection (dpi) (Fig. 2A). Inosine did not lower SARS-CoV-2 copies and titers in infected mouse lung and nasal turbinate tissues at 1 dpi and at 3 dpi (Figs. 2B and S3A). However, inosine treatment substantially lowered serum IL-6 and IFN-γ and raised serum IL-10 and IL-22 at 1 dpi or 3 dpi (Figs. 2C and S3B). Inosine ameliorated the pathological changes in the lungs and maintained normal pulmonary structure at the early stages of infection (Fig. 2D). After 14 days, all the mice in the SARS-CoV-2 group had died (Fig. 2E). However, daily oral inosine administration impeded weight loss starting at 9 dpi (Fig. 2F) and dramatically improved survival in mice infected with SARS-CoV-2 (Fig. 2E). Inosine also prevented SARS-CoV-2 infection from inducing inflammatory lesions in the lungs of the mice still surviving at 14 dpi (Fig. S3C). Inosine also inhibited pulmonary IL-6 and mononuclear macrophage activation induced by SARS-CoV-2 infection (Figs. 2G and H). Hence, inosine might have improved survival in SARS-CoV-2-infected mice by regulating cytokine release.
Fig. 2.
Inosine ameliorated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced acute lung injury. BALB/c mice were intranasally infected with 0.1∗LD50 C57MA14 and administered with oral inosine for 1 day, 3 days, or 14 days. (A) Schematic of C57MA14 infection and oral inosine administration started at 24 h before infection. Samples were harvested on indicated days after inosine treatment. (B) Viral RNA loads and virus titers in lung tissue were detected by reverse-transcription quantitative real-time polymerase chain reaction (RT-qPCR) and median tissue culture infective dose (TCID50), respectively. (C) Cytokines in mouse sera were quantified by mouse cytokine panel (n = 8−12). (D) Pathological damage to lung tissue was examined by hematoxylin-eosin (H&E) staining (n = 6). (E) Survival and (F) daily weight change in mice infected with C57MA14 and treated with inosine for 14 days (n = 20). (G) IL-6 expression in mouse lung tissue evaluated by Western blotting. (H) C57MA14 (MOI = 0.1) exposure 1 h before 500 μM inosine administration for 24 h. IL-6 expression in RAW264.7 cells was measured by Western blotting (n = 3). dpi: day post infection; Ctrl: control group; C57MA14: SARS-CoV-2-induced acute lung injury and no other treatment; C57MA14 + Inosine: SARS-CoV-2-induced acute lung injury followed by inosine treatment; IL: interleukin; IFN: interferon. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001 vs. control. #P < 0.05, ####P < 0.0001 vs. C57MA14 group.
3.3. STING and GSK3β signaling participated in the acute inflammatory response induced by various infectious agents
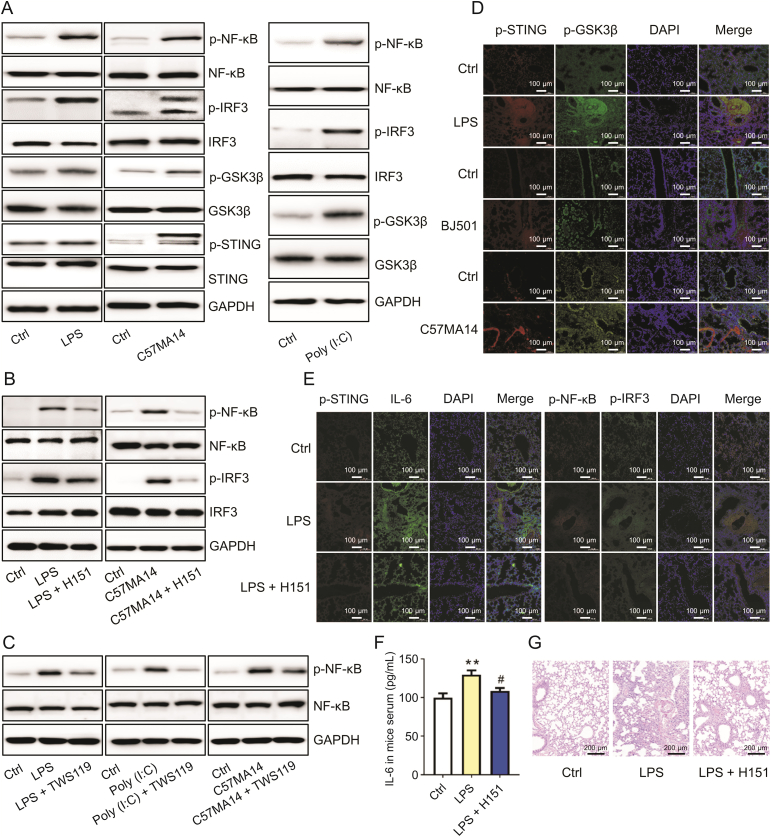
We then sought to identify the innate immune signaling involved in the acute inflammatory response induced various infectious agents. The in vitro experiments showed that LPS and C57MA14 activated STING and GSK3β signaling. Both infectious agents increased STING and GSK3β phosphorylation at Ser365 and Ser9, respectively (Fig. 3A). By contrast, Poly (I:C) only induced GSK3β phosphorylation and had no apparent influence on the STING pathway (Fig. 3A). LPS, Poly (I:C), and SARS-CoV-2 activated the proinflammatory nuclear factors NF-κB and IRF3 (Fig. 3A). STING inhibitor (H151) or GSK3β inhibitor (TWS119) co-administration noticeably reduced NF-κB and IRF3 phosphorylation in RAW246.7 cells infected with LPS, Poly (I:C), or SARS-CoV-2 (Figs. 3B and C). Lung tissues infected with LPS, BJ501, and C57MA14 presented with enhanced STING and GSK3β signaling (Fig. 3D). H151 inhibited STING activation, NF-κB phosphorylation at Ser536, IRF3 phosphorylation at Ser396, and IL-6 expression in LPS-infected lungs (Fig. 3E). It also lowered serum IL-6 and protected the lungs against inflammatory injury at 1 dpi (Figs. 3F and G). Thus, STING and GSK3β might play crucial roles in the acute inflammatory response triggered by LPS, Poly (I:C), and SARS-CoV-2.
Fig. 3.
Infection activated stimulator of interferon genes (STING) and glycogen synthase kinase-3-beta (GSK3β) signaling. (A) After 10 ng/mL lipopolysaccharide (LPS), C57MA14 (MOI = 0.1), or 50 μg/mL Poly (I:C) infection for 24 h, RAW264.7 cells were collected and lysed with specified antibodies for Western blot (n = 3). (B, C) RAW264.7 cells were subjected to LPS or Poly (I:C) and treated with 10 μM H151 (B) or 10 μM TWS119 (C) for 24 h. NF-κB and IRF3 phosphorylation levels were measured by immunoblot (n = 3). (D) Mice were challenged with 5 mg/kg LPS, 0.1∗LD50 Beijing/501/2009 strain (BJ501), or 0.1∗LD50 C57MA14. Lung tissue sections were stained with p-STING (red) and p-GSK3β (green) antibodies. Nuclei were stained with 40,6-diamidino-2-phenylindole (DAPI) (blue) (n = 5). (E) Mice were stimulated with LPS and treated with 7 mg/kg H151 for 24 h. Lung tissue sections were subjected to immunofluorescence assay with p-STING (red), IL-6 (green), p-IRF3 (green), and p-NF-κB (red) antibodies. Nuclei were stained with DAPI (blue) (n = 5). (F) Mice were treated as described in (D). Serum IL-6 in serum was measured by enzyme-linked immunosorbent assay (ELISA) (n = 5). (G) Mice were treated as described in (D). Lung tissue sections were subjected to hematoxylin-eosin (H&E) staining (n = 5). p-GSK3β: anti-phospho-GSK3β; p-STING: anti-phospho-STING; p-IRF3: anti-phospho-IRF3, p-NF-κB: anti-phospho-NF-κB; DAPDH: glyceraldehyde-3-phosphate dehydrogenase. ∗∗P < 0.01 vs. control. #P < 0.05 vs. LPS group.
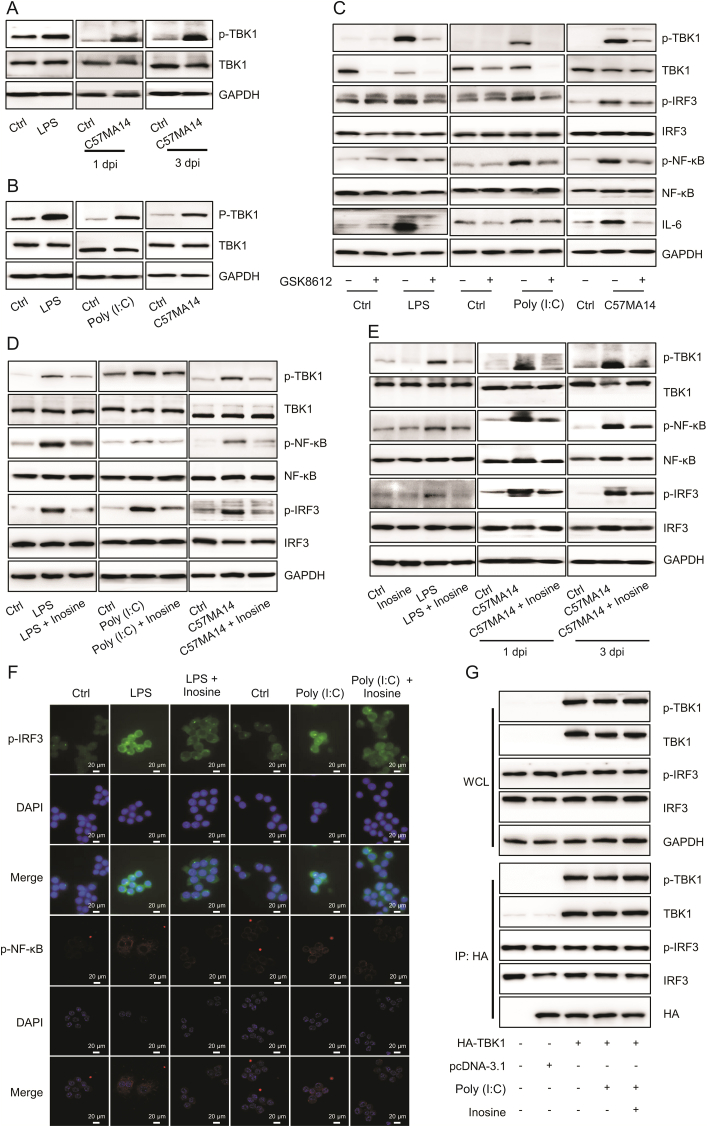
3.4. The inhibition of TBK1 phosphorylation also attenuated IL-6 upregulation induced by LPS, Poly (I:C), and SARS-CoV-2
TBK1 is essential for activating NF-κB and releasing IL-6 downstream [22]. TBK1 phosphorylation at Ser172 was detected in lung tissues and cells infected with LPS, Poly (I:C), or SARS-CoV-2 (Figs. 4A and B). After GSK8612 (TBK1 inhibitor) suppressed TBK1 phosphorylation, IRF3 and NF-κB phosphorylation and IL-6 expression also decreased in RAW246.7 cells infected with LPS, Poly (I:C), or SARS-CoV-2 (Fig. 4C). In vivo experiments disclosed that GSK8612 lowered serum IL-6 levels (Fig. S4A) and attenuated acute inflammatory lung injury in LPS- and H1N1-infected mice (Fig. S4B). GSK8612 significantly inhibited LPS- and H1N10-induced TBK1, IRF3, and NF-κB phosphorylation (Fig. S4C). Similarly, inosine suppressed TBK1, IRF3, and NF-κB phosphorylation promoted by LPS, SARS-CoV-2, or Poly (I:C) infection (Figs. 4D and E and S4D). The inhibition of TBK1 phosphorylation by inosine drastically reduced the levels and nuclear translocation of phosphorylated IRF3 and NF-κB (Figs. 4F and S4E). However, inosine did not hinder binding between TBK1 and IRF3 in HEK293 cells overexpressing the former (Fig. 4G). Therefore, TBK1 phosphorylation is strongly associated with the proinflammatory response and IL-6 upregulation promoted by the foregoing infectious agents and is a potential inosine target.
Fig. 4.
Inosine inhibited TANK-binding kinase 1 (TBK1)-mediated interleukin (IL)-6 production. (A) Western blot was used to detect p-TBK1 expression in mouse lung tissues (n = 3). (B) RAW264.7 cells were stimulated with 10 ng/mL lipopolysaccharide (LPS), C57MA14 (MOI = 0.1), or 50 μg/mL Poly (I:C) for 24 h. p-TBK1 expression was measured by Western blotting (n = 3). (C) RAW264.7 cells were exposed to LPS, Poly (I:C), or C57MA14, co-treating with or without GSK8612 for 24 h. TBK1, nuclear factor kappa B (NF-κB), and interferon regulatory factor (IRF3) phosphorylation levels and IL-6 expression were determined by immunoblot (n = 3). (D) RAW264.7 cells were challenged with LPS, C57MA14, or Poly (I:C) and treated with 500 μM inosine for 24 h. Immunoblots of p-TBK1, p-NF-κB, and p-IRF3 in RAW264.7 cells (n = 3). (E) Western blotting was used to measure p-TBK1, p-IRF3, and p-NF-κB expression in mouse lung tissues (n = 3). (F) Confocal fluorescence images of RAW264.7 cells infected with LPS or Poly (I:C) and treated with 500 μM of inosine for 24 h (n = 3). (G) HEK293 cells were transfected with plasmids encoding HA-tagged TBK1. Cell lysates were precipitated with anti-HA antibody. Immunoprecipitate was detected by Western blotting using specified antibodies (n = 3). p-TBK1: anti-phospho-TBK1; p-IRF3: anti-phospho-IRF3; p-NF-κB: anti-phospho-NF-κB; WCL: whole cell lysate; IP: immunoprecipitation; HA: hemagglutinin antigen; dpi: day post infection.
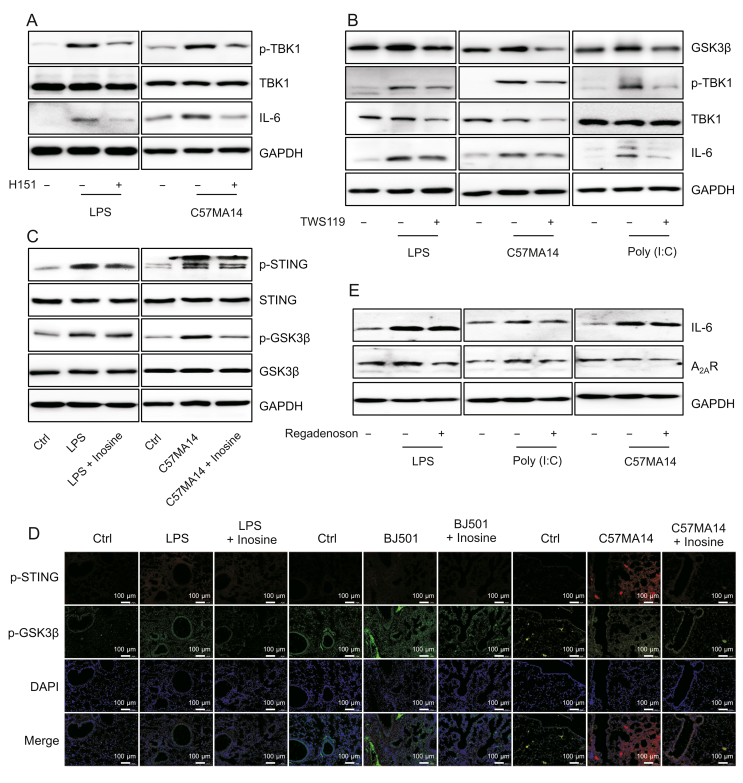
3.5. STING and GSK3β promoted TBK1 phosphorylation and IL-6 expression
To determine whether innate immune signaling is implicated in TBK1 phosphorylation and IL-6 upregulation, we inhibited STING or GSK3β in mononuclear macrophages infected with LPS, Poly (I:C), or SARS-CoV-2. H151 and TWS119 significantly inhibited LPS- and SARS-CoV-2-induced TBK1 phosphorylation as well as IL-6 expression in infected cells (Figs. 5A and B). TWS119 negated Poly (I:C)-induced increases in TBK1 phosphorylation and cytoplasmic IL-6 content (Fig. 5B). In vitro and in vivo experiments revealed that inosine inhibited STING and GSK3β phosphorylation induced by various infectious agents (Figs. 5C and D). A2AR mediated the anti-inflammatory action of inosine by downregulating IL-6. Nevertheless, the A2AR agonist regadenoson could not oppose IL-6 upregulation in infected RAW246.7 cells (Fig. 5E). STING, GSK3β, and TBK1 evoke acute inflammatory responses to LPS, Poly (I:C), or SARS-CoV-2 infection. The preceding results indicate that inosine may target STING and GSK3β. As both of these are upstream TBK1 regulators, inosine indirectly suppresses TBK1 phosphorylation and IL-6 upregulation induced by multiple stimuli.
Fig. 5.
Stimulator of interferon genes (STING) and glycogen synthase kinase-3-beta (GSK3β) promoted TANK-binding kinase 1 (TBK1) phosphorylation and interleukin (IL)-6 expression. (A) RAW264.7 cells were infected with 10 ng/mL lipopolysaccharide (LPS) or C57MA14 (MOI = 0.1) and treated with 1 μM H151 for ≤24 h. Western blotting was used to measure p-TBK1 and IL-6 expression in cell lysates (n = 3). (B) RAW264.7 cells were stimulated with LPS, C57MA14, or Poly (I:C) and treated with 10 μM of TWS119 for 24 h. Representative Western blot of p-TBK1 and IL-6 in cells (n = 3). (C) RAW264.7 cells were subjected to LPS or C57MA14 and treated with 500 μM of inosine for 24 h. Total proteins were isolated and p-STING and p-GSK3β levels were probed (n = 3). (D) Mice were subjected to 5 mg/kg LPS or 0.1∗LD50 C57MA14 infection and administered inosine for 24 h. Lung tissue sections were analyzed by immunofluorescent p-STING (red) and p-GSK3β (green) co-staining. Nuclei were stained with DAPI (blue) (n = 5). (E) RAW264.7 cells were infected with LPS, C57MA14, or Poly (I:C), accompanying with regadenoson co-treatment for 24 h, and then subjected to Western blotting using indicated antibodies (n = 3). p-NF-κB: anti-phospho-NF-κB; p-STING: anti-phospho-STING; p-GSK3β: anti-phospho-GSK3β; DAPI: 40,6-diamidino-2-phenylindole; Ctrl: control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
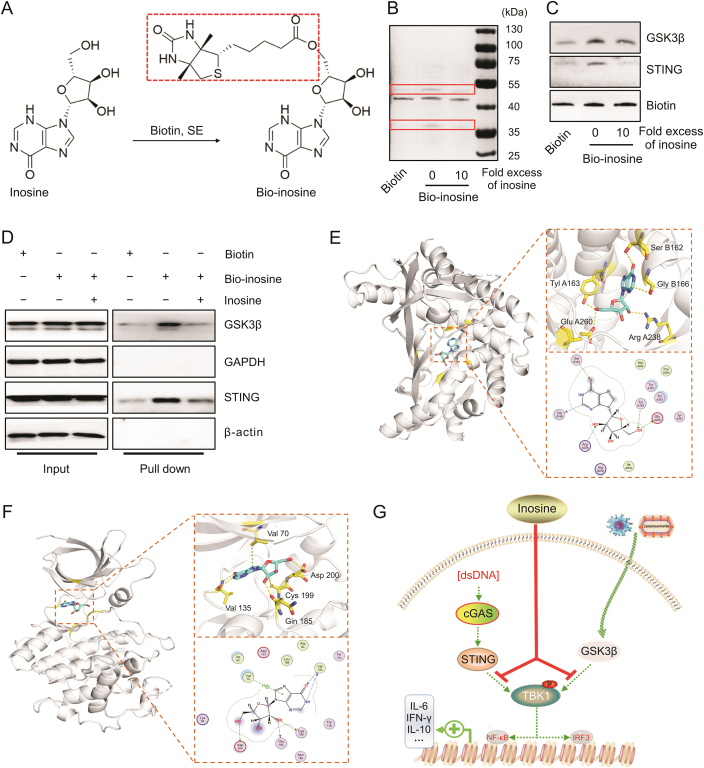
3.6. Inosine inhibited TBK1 phosphorylation by interfering with STING and GSK3β activation
We performed a biotin pulldown assay and molecular docking to clarify the mechanism by which inosine inhibits TBK1 phosphorylation. First, we introduced long-chain biotin via esterification of the hydroxyl group to construct a biotinylated inosine (Fig. 6A). It was confirmed that the latter also had anti-inflammatory efficacy as it suppressed proinflammatory signals (STING or GSK3β) and TBK1-mediated IL-6 expression in LPS- and Poly (I:C)-infected cells (Figs. S5A and B). Next, our pulldown analysis with silver staining disclosed that, relative to biotin, bio-inosine significantly enhanced protein precipitation at 33–35 kDa and 40–55 kDa. However, unlabeled inosine at tenfold the bio-inosine concentration competitively reduced the precipitation of proteins targeted by bio-inosine (Fig. 6B). Western blotting demonstrated that the precipitated protein bands included STING and GSK3β (Fig. 6C). Hence, binding occurred directly between inosine and these proteins. Flag-tagged STING and GSK3β proteins were expressed in the HEK293 cells (Fig. S5C). Cell lysates were also tested in the biotin pulldown assays with and without bio-inosine. The latter precipitated flag-tagged STING and GSK3β. Tenfold excess unlabeled free inosine blocked binding between bio-inosine and STING or GSK3β (Fig. 6D). We performed a docking analysis to clarify the mode of inosine binding to STING or GSK3β. Figs. 6E and F show that inosine binds human STING in the pocket between the two chains along with the kinase binding domain of human GSK3β. Van der Waals and π–π stacking interactions formed between inosine and STING or GSK3β (Figs. 6E and F). The docking scores of inosine with human STING and GSK3β were −6.77 kcal/mol and −8.16 kcal/mol, respectively. The foregoing discoveries suggest that inosine directly targets STING and GSK3β, inhibits their phosphorylation, and suppresses the TBK1-mediated proinflammatory response.
Fig. 6.
Inosine directly targeted stimulator of interferon genes (STING) and glycogen synthase kinase-3-beta (GSK3β). (A) Synthesis and structure of biotinylated inosine (bio-inosine). (B, C) Bio-inosine or biotin was added to streptavidin-agarose beads and incubated. Lysates prepared from RAW264.7 were added to the streptavidin-agarose beads with bio-inosine or biotin. Precipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). Gel was stained with silver (B) and proteins were detected by Western blotting as indicated (C). (D) STING and GSK3β plasmids were transfected into HEK293 cells. After 24 h transfection, pulldown was performed using streptavidin-agarose beads and subjected to Western blotting. (E) Schematic diagram of binding between inosine and STING protein. Inosine binds STING in pocket between two chains. Protein is shown in gray cartoon. Ligand is represented by cyan stick. (F) Schematic diagram of binding between inosine and GSK3β protein. Inosine combined with GSK3β kinase binding domain. Protein is shown in gray cartoon. Ligand is represented by cyan stick. (G) Schematic diagram of mechanism by which inosine regulates cytokine release. Biotin, SE: Biotin-N-hydroxysuccinimide ester.
4. Discussion
Uncontrolled, hyperactivate inflammatory responses are believed to be the principal triggers of SARS-CoV-2-induced acute lung injury and are closely associated with COVID-19 severity and mortality. The present study identified inosine as a broad-spectrum anti-inflammatory that might effectively alleviate acute inflammatory lung injury induced by multiple infectious agents. Inosine substantially improved survival in SARS-CoV-2-infected mice mainly by suppressing the release of IL-6 and other proinflammatory cytokines. Inosine interacted with STING and GSK3β, inhibited their phosphorylation, significantly decreased TBK1 phosphorylation, suppressed the proinflammatory nuclear factors NF-κB and IRF3, and downregulated proinflammatory IL-6 in response to pathogen attack and invasion. This study demonstrated the critical role of TBK1 in acute inflammatory lung injury evoked by various infectious agents and indicated that inosine and other drugs targeting TBK1 are potential therapeutic strategies against COVID-19.
Megakaryocytes and monocytes synthesize and secrete the proinflammatory cytokines, which are major clinical features of COVID-19. These cells and substances promote disease progression via acute inflammatory lung injury [30,31]. SARS-CoV-2 and other infectious agents indeed trigger severe pulmonary inflammatory responses including the pathological features of pneumonia and cytokine storms [32,33]. Effective cytokine storm management can improve outcomes and survival in patients with COVID-19. Standard clinical anti-cytokine therapies include glucocorticoid and cytokine antagonist administration. In contrast, inosine suppresses but does not abolish the release of the proinflammatory cytokine IL-6 induced by SARS-CoV-2. Nevertheless, it also upregulates the anti-inflammatory cytokine IL-10. Hence, it can attenuate the cytokine response. For these reasons, inosine significantly improved survival in SARS-CoV-2-infected mice even without directly inhibiting viral replication. Inosine indistinguishably alleviated the lung tissue damage and lowered the serum levels of IL-6. This factor is thought to be a major component of the cytokine storm and is correlated with COVID-19 severity and mortality. In fact, inosine administration might prevent pulmonary injuries resulting from acute inflammation caused by multiple stimuli. Hence, inosine might deploy the same anti-cytokine storm mechanisms including IL-6 inhibition against SARS-CoV-2, other viruses, and bacterial pathogens.
Our study demonstrated that SARS-CoV-2 activated the NF-κB transcription factor (TF) that induces cytokine genes in response to viral or bacterial infection. Early in infection, phosphorylated NF-κB and its cascade co-activators induce IL-6 and TNF-α. Phosphorylation of the TF IRF3 was also detected in SARS-CoV-2-infected macrophages and lungs. After IRF3 senses viral RNA and cytoplastic DNA, it induces type І IFN secretion [34,35]. Nevertheless, neither the present nor previous studies detected any significant elevation in the type І IFN response in severe SARS-CoV-2 infection (Fig. S2B). By contrast, SARS-CoV-2 infection remarkably upregulated serum IFN-γ at 3 dpi. Therefore, IRF3 activation might contribute to IFN-γ induction. A recent study reported that IL-6 is a non-canonical interferon-stimulated gene (ISG) responding to IFN signaling [36]. For this reason, IRF3 activation might be implicated in SARS-CoV-2-induced IL-6 expression. SARS-CoV-2-elicited cytokine release may be correlated with multiple cellular pathways or networks such as NF-κB and IRF3 signaling. LPS, Poly (I:C), and H1N1 induced NF-κB and IRF3 phosphorylation which, in turn, first upregulated IL-6 and TNF-α and then IFN-γ. In response to all infectious agents, inosine had excellent anti-cytokine efficacy as it suppressed the release of IL-6 mainly by inhibiting NF-κB and IRF3 phosphorylation.
Purinergic receptors are expressed by a wide range of immune cells and might have various anti-inflammatory effects [37]. A2AR mediates the anti-cytokine effects of inosine in LPS-induced acute lung injury [38,39]. However, selective adenosine A2AR agonists did not abrogate elevated inflammation associated with transcription signaling or IL-6 upregulation in response to multiple stimuli. Thus, the anti-inflammatory effects of inosine are not A2AR-dependent. TBK1 is the gatekeeper of bacterial and viral-induced inflammatory signaling crossroads [22,40] and is strongly phosphorylated in macrophages and lung tissues infected with LPS, Poly (I:C), H1N1, or SARS-CoV-2. TBK1 phosphorylation is essential for the recruitment and phosphorylation of the downstream TFs NF-κB and IFR3. TBK1 also promotes the nuclear translocation of phosphor-NF-κB and phosphor-IFR3 which, in turn, regulate proinflammatory cytokine expression [22]. The inhibition of TBK1 phosphorylation via inosine or the administration of a selective TBK1 inhibitor significantly attenuated the NF-κB and IRF3 responses and acute inflammatory pulmonary injury but allowed IL-6 upregulation. In a TBK-1-dependent manner, inosine downregulated IL-6 and improved the morphology and histology of lungs infected with SARS-CoV-2 and other agents.
The results of our results demonstrated that inosine did not interfere with TBK1 phosphorylation via direct binding. Rather, it suppressed STING and GSK3β phosphorylation. STING and GSK3β antagonists markedly reduced SARS-CoV-2- and LPS-induced increases in TBK1 phosphorylation and IL-6 expression. STING is a scaffold for the phosphorylation of TBK1 and itself [41]. Residues 1–341 formed dimers or orchestrated oligomers to trigger STING and TNK1 phosphorylation. The STING dimers had α-type loops in the cytosolic ligand-binding domain of STING and participated in the formation of tetramer interfaces for cyclic GMP-AMPP (cGAMP) binding. Cyclic GMP-AMPP is an endogenous secondary messenger of cGAS signaling that participates in STING phosphorylation and IFN production [41,42]. Inosine made no contact with the C-terminal residues of STING which are the TBK1 binding sites. However, inosine bound human STING residues in the pocket between the two chains. One of these (Glu260) is adjacent to a tetramer interface site (Gln273) [41]. Inosine may inhibit the phosphorylation of STING by interfering with its dimerization or its interaction with cGAMP. Our in silico docking and biotin pulldown assays confirmed that inosine directly interacts with GSK3β, Val70, and Val 135. The latter residues are implicated in GSK3β inhibition [43]. When viral infections are perceived, ubiquitinated tripartite motif 9 short isoforms (TRIM9s) bridge activated GSK3β and TBK1 [35] and the latter is then autophosphorylated. STING and GSK3β play critical roles in TBK1 phosphorylation while inosine indirectly suppresses TBK1 activation induced by SARS-CoV-2 and other infectious agents.
5. Conclusions
The present study empirically demonstrated that inosine administration improved survival in severe SARS-CoV-2 infection through its immunomodulatory but not immunosuppressive efficacy against acute inflammatory lung injury. As presented in Fig. 6G, inosine indirectly suppresses the phosphorylation of TBK1 at the crossroad of multiple innate immune responses. In this manner, it hinders overactivation of downstream cascade signaling and excessive proinflammatory cytokine release. Hence, inosine is potentially an excellent broad-spectrum anti-inflammatory agent. Future research should evaluate its clinical efficacy in the treatment of severe and critical COVID-19. Furthermore, TBK1 is a putative therapeutic target in the attenuation of cytokine storms and acute inflammatory lung injury induced by SARS-CoV-2 and other infectious agents.
CRediT author statement
Ningning Wang: Methodology, Investigation, Visualization, Writing - Original draft preparation; Entao Li: Methodology, Investigation, Writing - Original draft preparation; Huifang Deng: Methodology, Writing - Original draft preparation; Lanxin Yue: Investigation; Lei Zhou: Visualization; Rina Su: Investigation; Baokun He: Visualization; Chengcai Lai and Gaofu Li: Methodology; Yuwei Gao: Funding acquisition, Project administration, Supervision, Writing - Reviewing and Editing; Wei Zhou: Conceptualization, Visualization, Supervision, Writing - Original draft preparation, Reviewing and Editing; Yue Gao: Conceptualization, Funding acquisition, Project administration, Supervision, Reviewing and Editing.
Declaration of competing interest
Yue Gao, Wei Zhou, Yuwei Gao, Ningning Wang and Entao Li report a pending China patent application (Patent No.: 202111541575.0). Yue Gao, Wei Zhou, Yuwei Gao and Ningning Wang are the inventors of a pending PTC patent application (Patent No.: PCT/CN2022/078715) entitled “Application of inosine in the treatment for COVID-19”.
Acknowledgments
This work was supported by grants from the Young Elite Scientists Sponsorship Program by CAST (Grant No.: 2021-QNRC1-03) and the National Key Research and Development Program of China (Grant No.: 2020YFC0845400). We thank Prof. Hui P. Chen for providing advice on the experiments.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.10.002.
Contributor Information
Yuwei Gao, Email: yuwei0901@outlook.com.
Wei Zhou, Email: zhouweisyl802@163.com.
Yue Gao, Email: gaoyue@bmi.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Martinez Mesa A., Cabrera César E., Martín-Montañez E., et al. Acute lung injury biomarkers in the prediction of COVID-19 severity: Total thiol, ferritin and lactate dehydrogenase. Antioxidants (Basel) 2021;10 doi: 10.3390/antiox10081221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards A.M., Baric R.S., Saphire E.O., et al. Stopping pandemics before they start: Lessons learned from SARS-CoV-2. Science. 2022;375:1133–1139. doi: 10.1126/science.abn1900. [DOI] [PubMed] [Google Scholar]
- 3.Zhou W., Liu Y., Tian D., et al. Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduct. Target Ther. 2020;5:18. doi: 10.1038/s41392-020-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dal-Ré R., Becker S.L., Bottieau E., et al. Availability of oral antivirals against SARS-CoV-2 infection and the requirement for an ethical prescribing approach. Lancet Infect. Dis. 2022;22 doi: 10.1016/S1473-3099(22)00119-0. e231–e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karki R., Sharma B.R., Tuladhar S., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cron R.Q., Caricchio R., Chatham W.W. Calming the cytokine storm in COVID-19. Nat. Med. 2021;27:1674–1675. doi: 10.1038/s41591-021-01500-9. [DOI] [PubMed] [Google Scholar]
- 7.Dequin P.F., Heming N., Meziani F., et al. Effect of hydrocortisone on 21-day mortality or respiratory support among critically ill patients with COVID-19: A randomized clinical trial. JAMA. 2020;324:1298–1306. doi: 10.1001/jama.2020.16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomazini B.M., Maia I.S., Cavalcanti A.B., et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: The CoDEX randomized clinical trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang R., Zhu C., Wang J., et al. Corticosteroid therapy is associated with the delay of SARS-CoV-2 clearance in COVID-19 patients. Eur. J. Pharmacol. 2020;889 doi: 10.1016/j.ejphar.2020.173556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fajgenbaum D.C., June C.H. Cytokine storm. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W., Liu Y., Xu B., et al. Early identification of patients with severe COVID-19 at increased risk of in-hospital death: A multicenter case-control study in Wuhan. J. Thorac. Dis. 2021;13:1380–1395. doi: 10.21037/jtd-20-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L.Y.C., Hoiland R.L., Stukas S., et al. Confronting the controversy: Interleukin-6 and the COVID-19 cytokine storm syndrome. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.03006-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McElvaney O.J., Curley G.F., Rose-John S., et al. Interleukin-6: Obstacles to targeting a complex cytokine in critical illness. Lancet Respir. Med. 2021;9:643–654. doi: 10.1016/S2213-2600(21)00103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones S.A., Hunter C.A. Is IL-6 a key cytokine target for therapy in COVID-19? Nat. Rev. Immunol. 2021;21:337–339. doi: 10.1038/s41577-021-00553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z., Zhang X., Lei X., et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct. Target Ther. 2021;6 doi: 10.1038/s41392-021-00800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto N., Mitoma H., Kim T., et al. Helicase proteins DHX29 and RIG-I cosense cytosolic nucleic acids in the human airway system. Proc. Natl. Acad. Sci. USA. 2014;111:7747–7752. doi: 10.1073/pnas.1400139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y., Liu Y., An W., et al. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Invest. 2019;129:546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park A., Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan S., Shafiei M.S., Longoria C., et al. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife. 2021;10 doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rana A.K., Rahmatkar S.N., Kumar A., et al. Glycogen synthase kinase-3: A putative target to combat severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Cytokine Growth Factor Rev. 2021;58:92–101. doi: 10.1016/j.cytogfr.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marineau A., Khan K.A., Servant M.J. Roles of GSK-3 and β-catenin in antiviral innate immune sensing of nucleic acids. Cells. 2020;9 doi: 10.3390/cells9040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yum S., Li M., Fang Y., et al. TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2100225118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan S., Torres A.G., Ribas de Pouplana L. Inosine in biology and disease. Genes (Basel) 2021;12 doi: 10.3390/genes12040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liaudet L., Mabley J.G., Pacher P., et al. Inosine exerts a broad range of antiinflammatory effects in a murine model of acute lung injury. Ann. Surg. 2002;235:568–578. doi: 10.1097/00000658-200204000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liaudet L., Mabley J.G., Soriano F.G., et al. Inosine reduces systemic inflammation and improves survival in septic shock induced by cecal ligation and puncture. Am. J. Respir. Crit. Care Med. 2001;164:1213–1220. doi: 10.1164/ajrccm.164.7.2101013. [DOI] [PubMed] [Google Scholar]
- 26.Chen L.P., Cai Y.M., Li J.S. Medication rules of famous veteran traditional Chinese medicine doctor in treatment of chronic bronchitis based on implicit structure model. Zhongguo Zhong Yao Za Zhi. 2017;42:1609–1616. doi: 10.19540/j.cnki.cjcmm.2017.0052. [DOI] [PubMed] [Google Scholar]
- 27.Ding C., Song Z., Shen A., et al. Small molecules targeting the innate immune cGAS‒STING‒TBK1 signaling pathway. Acta Pharm. Sin. B. 2020;10:2272–2298. doi: 10.1016/j.apsb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Liu W., Liang Z. Endophytic bacteria from Pinellia ternata, a new source of purine alkaloids and bacterial manure. Pharm. Biol. 2015;53:1545–1548. doi: 10.3109/13880209.2015.1016580. [DOI] [PubMed] [Google Scholar]
- 29.Ogasawara M., Yoshii K., Jun W., et al. Identification of guanine, guanosine, and inosine for α-amylase inhibitors in the extracts of the earthworm Eisenia fetida and characterization of their inhibitory activities against porcine pancreatic α-amylase. Enzyme Microb. Technol. 2020;142 doi: 10.1016/j.enzmictec.2020.109693. [DOI] [PubMed] [Google Scholar]
- 30.Zhu L., Yang P., Zhao Y., et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696.e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren X., Wen W., Fan X., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184 doi: 10.1016/j.cell.2021.10.023. 1895–1913.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravindra N.G., Alfajaro M.M., Gasque V., et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang R.T., Wu D., Meliton A., et al. Experimental lung injury reduces krüppel-like factor 2 to increase endothelial permeability via regulation of RAPGEF3-Rac1 signaling. Am. J. Respir. Crit. Care Med. 2017;195:639–651. doi: 10.1164/rccm.201604-0668OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S., Cai X., Wu J., et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347 doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 35.Stetson D.B., Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Sang E.R., Tian Y., Miller L.C., et al. Epigenetic evolution of ACE2 and IL-6 genes: Non-canonical interferon-stimulated genes correlate to COVID-19 susceptibility in vertebrates. Genes (Basel) 2021;12 doi: 10.3390/genes12020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.dos Anjos F., Simões J.L.B., Assmann C.E., et al. Potential therapeutic role of purinergic receptors in cardiovascular disease mediated by SARS-CoV-2. J. Immunol. Res. 2020;2020 doi: 10.1155/2020/8632048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovászi M., Németh Z.H., Gause W.C., et al. Inosine monophosphate and inosine differentially regulate endotoxemia and bacterial sepsis. FASEB J. 2021;35 doi: 10.1096/fj.202100862R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milne G.R., Palmer T.M. Anti-inflammatory and immunosuppressive effects of the A2A adenosine receptor. Sci. World J. 2011;11:320–339. doi: 10.1100/tsw.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Y., Liu Q., Tian S., et al. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 2016;26:613–628. doi: 10.1038/cr.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C., Shang G., Gui X., et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567:394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burdette D.L., Monroe K.M., Sotelo-Troha K., et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D., Liu L., Pang L., et al. Biological evaluation and energetic analyses of novel GSK-3β inhibitors. J. Cell. Biochem. 2018;119:3510–3518. doi: 10.1002/jcb.26522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.