Abstract
Chronic itch is a major symptom of many inflammatory skin diseases. This type of pruritus is thought to be facilitated by cytokines which activate cutaneous nerve fibers; however, the molecular components and mechanisms involved are poorly understood. We found that the cytokine oncostatin M (OSM) is highly upregulated in psoriasis, atopic dermatitis, and cutaneous T-cell lymphoma, diseases associated with chronic itch. OSM receptor (OSMR) is expressed by itch-selective (natriuretic polypeptide B) (Nppb)-neurons and single-cell sequencing showed that OSM is mainly produced by dermal T-cells and monocytes. Unlike canonical pruritogens, OSM does not activate sensory neurons. Instead, it sensitizes neurons by potentiating neural responses to pruritogens and by enhancing neural excitability. Knockout of OSMR in sensory neurons attenuated OSM-sensitized itch and inflammatory itch in mice and pharmacological antagonism of the OSM receptor complex effectively alleviated pruritus in experimental inflammatory dermatitis in a rodent model. Together our results uncover OSM as an itch neuromodulator and reveal OSM signal transduction as a potential target for antipruritic therapy.
One sentence summary:
Itch sensory neurons can be sensitized by OSM released from dermal immune cells.
Itch associated with cutaneous diseases is thought to arise from the activation of sensory neurons which innervate the skin (1). This activation often takes place in the context of skin diseases where the immune system is also activated frequently as a result of barrier disruption (2). For many persistent skin diseases, chronic itch is not only a minor side-effect but severely reduces quality of life of patients (3–6). Since most chronic itch is difficult to treat effectively, it is, globally, a major unmet health concern (7–9).
Pruritic agents, both produced endogenously and those encountered via contact with exogenous sources, stimulate itch-neurons which in turn, via connections to the CNS, produce the percept of itch and evoke the urge to scratch (6, 10–12). In mice, three classes of peripheral itch-neurons have been characterized, those which express the itch receptor Mrgpra3, Mrgprd-neurons, and those which express natriuretic polypeptide B, (Nppb) (13–16). Itch-inducing agents like monoamines (17), lipids (18, 19), proteases (20–22), peptides, and some cytokines including IL-31, IL-4, and IL-13 (4, 23–25) can directly depolarize sensory neurons which evokes pruriception (26, 27). However, the data for cytokines serving as direct pruritogens is limited and remains uncertain. Emphasizing the importance of cytokine reception in itch, agents targeting cytokine receptors have shown clinical value in treating atopic dermatitis (24, 28). During chronic itch the processes that drives the continuous urge to scratch are not well understood but may not be the same as those which elicit acute itch. An example of this is histamine which is known to be a key mediator of acute itch but except in urticaria, is not thought to be a major mediator for chronic itch. The pruritogen(s) involved in chronic itch are still poorly understood and may include underappreciated substances like leukotrienes (29).
In this study, we examined the transcriptomic profile as well as single-cell sequencing data of skin from human subjects with skin diseases associated with chronic itch and uncovered that, in inflammatory skin biopsies, oncostatin M (OSM) is overwhelmingly expressed by several dermal immune cells including T-cells, monocytes, dendritic cells and mast cells. Although, OSM has previously been implicated as a general agent found during inflammation (30–32), a role for this cytokine in itch has not been established. Physiological studies showed OSM directly modulated Nppb-neurons by enhancing neural activity instead of depolarizing neurons, a mechanism very different from canonical pruritogens. This OSM-mediated sensitization resulted in exaggerated itch in inflammatory skin. Genetic studies and single-cell sequencing data suggested OSM also indirectly evoked itch by stimulating stromal cells in skin. Finally, we show that inhibiting OSM receptor complex, in a mouse model, can alleviate itch and skin inflammation in dermatitis. Together, we showed that OSM is the predominant cytokine in skin inflammation which multidimensionally modulates sensory neurons to induce skin pruritus.
Results
Nppb-neurons express multiple cytokine receptors
Given the reported involvement of cytokines in chronic itch we were interested in profiling the expression of their receptors in sensory neurons to more fully describe these interactions (23, 27). Initially, we examined the cytokine receptors expressed by Nppb-neurons since these neurons have been shown to be involved in itch, the expression of the neuropeptide Nppb (released by these cells) is increased in chronic itch (33), and interleukin 31 receptor (IL31ra), a receptor associated with chronic itch (34, 35), is expressed by these cells. Whereas, IL31ra is expressed only in Nppb-neurons (36–39), the expression of other cytokine receptors has not been defined in vivo (40). To determine the receptor transcriptome of Nppb-neurons, we collected dissociated Nppb-neurons (genetically labeled) (Figure 1A) and performed pooled-cell RNA-seq. Sequencing data from pooled Trpm8 and Trpv1-cells (Figure 1B) was used for comparison to identify signaling molecules enriched in Nppb-neurons. As anticipated, this analysis revealed the selective enrichment of the previously reported receptor for Il31ra, as well as the oncostatin M receptor (OSMR) and an important downstream signaling molecule, Janus kinase 1 (Jak1) (Figure 1C). OSMR is thought to form heterodimers with IL31ra to transduce IL-31 signals (31, 41). Also, along with being a co-receptor for IL31 signaling, OSMR acts as a receptor for OSM together with the co-receptor, gp130 (30–32). Analysis of sequencing data showed that Nppb-neurons also express high amounts of gp130 (Il6st) (Figure 1D), indicating that Nppb-neurons express components necessary for the detection of OSM. In addition, we found that Nppb-neurons express Il13ra1 and Il4ra which can form receptor complexes to transduce IL-13 and IL-4 signals respectively (42). The expression amounts of Il13ra1 and Il4ra were about 50-fold lower than that of OSMR, gp130, and Il31ra (Figure 1D and Table 1). To validate these RNA-seq results and to get a better insight into which cells express cytokine receptors, we performed multi-label in situ hybridization (ISH) on both mouse and human dorsal root ganglion (DRG). In agreement with our sequencing results, in mouse DRG, about 90% OSMR-expressing neurons also co-expressed Nppb (Figure 1E, Osmr+Nppb+/Osmr+ = 548/613 = 89%, Osmr+Nppb+/Nppb+ = 548/567 = 92%, n=5 mice), although there is a fraction of neurons which express low amounts of OSMR that are Nppb-negative and Trpv1-positive (Figure S1). The OSMR-neurons also co-expressed the co-receptor gp130 that is broadly expressed in many DRG neurons (Figure 1F, Osmr+gp130+/Osmr+ = 164/166, n=3 mice). In human sensory neurons, similar to what we found in mouse DRG, OSMR and NPPB are highly co-expressed (Figure 1G, 219/223 cells, n=5 donors and S1A). For other cytokine receptor subunits, we found that IL31ra, similar to OSMR, is selectively expressed in Nppb-neurons (Figure 1H). Although Nppb-neurons express Il13ra1 and Il4ra, both receptors are found in many other cell-types and their expression are much lower than that of OSMR or IL31ra (Figure 1I–J). Together these results show that Nppb-neurons, in both mouse and human DRG express receptor complexes for transducing multiple cytokine signals including those for OSM.
Figure 1. Nppb-neurons express multiple cytokine receptor complex.
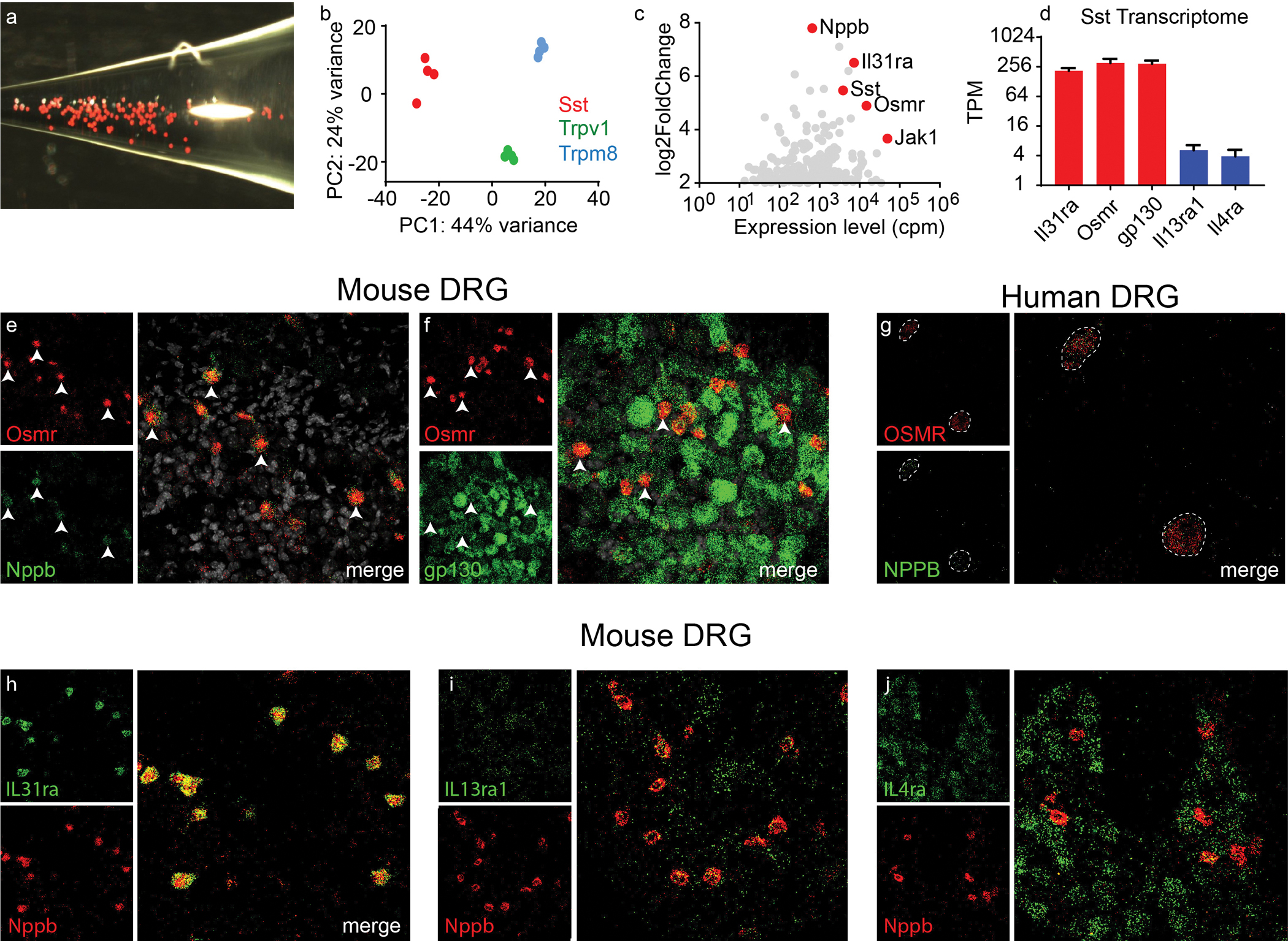
(A) Representative image of dissociated Nppb-neurons from Sst-cre:: AAV9-CAG-Flex-tdTomato mice (B) Principal component analysis of clustered RNAseq data from Sst-, Trpm8-, and Trpv1-neurons (n=4 RNA libraries for each class of neuron). (C) Analysis of differential gene expression between Sst- and Trpm8-neurons with estimated gene abundance, counts per million (cpm). Genes enriched in Sst-neurons are indicated in red. (D) Quantification of transcript abundances, transcript per million, (TPM) of indicated genes; data presented as mean ± SEM. (E) Representative images of multilabel in situ hybridization (ISH) of mouse DRG with indicated genes (F and G). Images of ISH of human DRG hybridized with probes for OSMR and NPPB. (H-J) Images of sections of mouse DRG hybridized with probes to the indicated genes.
Table 1:
Transcript abundance of cytokine receptors in Nppb neurons
| Interleukin receptors | gp130 (IL6ST)-coreceptors | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Il31ra* | Il13ra1 | gp130* | Osmr * | Il4ra | Il6ra | Cntfr | Il11ra1 | Il11ra2 | Il27ra |
| 213±29 | 5.1±1.4 | 295±48 | 306±65 | 3.9±1.4 | 0.52±0.28 | 31±9.3 | 9.3±5.7 | 0 | 0.14±0.06 |
The transcript abundance (transcripts per million, TPM ± SEM) of cytokine receptors in Nppb-neurons (averaged from 4 samples).
Expression of OSM is prominently increased in skin disorders associated with chronic itch
Elevated expression of IL-31, IL-4, and IL-13 have been proposed to be causative for some types of itch, but it is unclear if these are the only cytokines involved in modulating pruritic responses (23, 24, 43). To better define cytokine expression in diseased skin associated with inflammatory itch and to use physiologically relevant results for comparison, we examined several RNA-seq datasets from human subjects with different pruritic skin diseases. We analyzed RNA-seq data from skin biopsies of patients with psoriasis (GSE54456, GSE121212) (44), atopic dermatitis (GSE121212) (45), and cutaneous T-cell lymphoma (CTCL) (GSE113113) (46) all conditions which in most cases result in considerable pruritus (43, 47, 48). We found that amounts of OSM transcripts were significantly upregulated in the skin from human patients with psoriasis, p<0.0001, atopic dermatitis, p=0.0002, and were higher compared to controls in CTCL (Figure 2A–C and see Table 2). For the human samples, not all subjects had elevated OSM (Figures 2D), suggesting that there is heterogeneity in these disease groups and there may be several factors which produce itch. In addition, we analyzed RNA-seq datasets from mouse models of atopic-dermatitis, MC903 (GSE90883) (27) and an imiquimod-induced psoriasiform mouse model (GSE86315) (49). In the imiquimod mouse model, OSM was the only significantly upregulated cytokine, p=0.023, and in the MC903 mouse model, OSM expression was significantly increased together with Il-4, (p=0.0016 and p<0.0001, respectively) (Figure 2E–F and see Table 2). Differential gene expression analyses also revealed that IL-6 is upregulated in human psoriasis, atopic dermatitis, and imiquimod mouse model (log2 fold changes are 2.2, 2.1, 1.4, respectively, Table 2), however, the expression of the IL-6 receptor, IL6Ra, is extremely low in Nppb-neurons and is not significantly higher than zero (p=0.16, Table 1). OSM is the cytokine that is commonly upregulated in these inflammatory skin conditions (Figure 2G). To validate our results, we performed ISH on skin from imiquimod or MC903 treated mice. Corroborating our analysis of sequencing results, we found a marked increase in OSM expression in lesioned skin from both models of dermatitis (Figure 2I–J). We also examined OSM expression in skin biopsy samples from psoriasis patients and found high OSM-immunoreactivity in four out of nine specimens we examined (Figure 2L). Furthermore, the OSM-positive staining in these samples often accumulated around peripheral nerve fibers, consistent with potential neuroimmune crosstalk (Figure 2M).
Figure 2. Oncostatin M is upregulated in various inflammatory skin diseases.
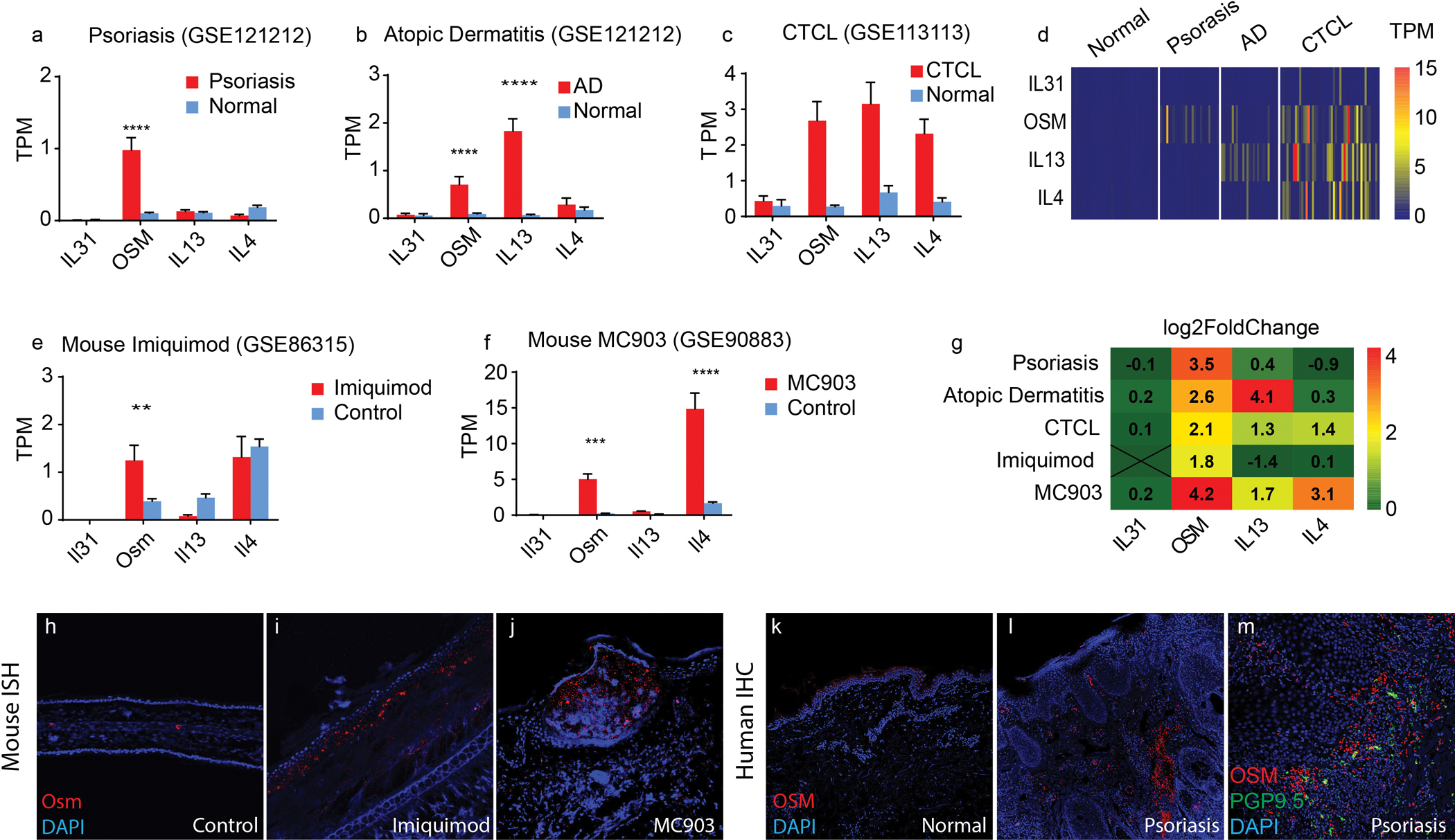
(A-D) Quantification of transcript abundance (transcript per million, TPM) of IL-31, OSM, IL-13 and IL-4 in healthy, psoriasis (A), atopic dermatitis (B), CTCL (C) skin. Accession numbers of data are indicated. (A) 28 patients and 38 healthy controls, (for OSM****p<0.0001). (B) 27 patients and 38 healthy controls (for OSM****p=0.0002 and for IL-13****p<0.0001). (C) 49 patients and 3 healthy controls. (D) The heatmaps display the estimated transcript abundances (TPM) of IL-31, OSM, IL-13, and IL-4 (rows) in each patient (columns) from these datasets. (E-F) Quantification of transcript abundance in the imiquimod-induced psoriasiform dermatitis mouse model (E) and the MC903-induced atopic dermatitis-like mouse model (F). (E) 4 imiquimod-treated and 4 control mice (**p=0.023) (F) n=4 MC903-treated and n=4 control mice, (for OSM***p=0.0016 and for IL4****p<0.0001). Data presented in A-C and E-F are means ± SEM (two-way ANOVA with Sidak post-hoc test). (G) Using DESeq2, the differential gene expression of IL-31, OSM, IL-13 and IL-4 was determined between lesioned and healthy skin samples. Fold-changes are presented on a log2 scale. (H-J) Representative images of ISH of mouse skin hybridized with probes to OSM (red) counterstained with DAPI (blue), (H) normal mouse ear, (I) two weeks after daily imiquimod treatment, and (J) 12 days after daily MC903 treatment. (K-M) Immunohistochemistry of OSM (red) in formalin-fixed parafilm-embedded skin biopsies from a healthy donor (K), and from a psoriasis patient (L). (M) Representative image of OSM immunoreactivity (red) and anti-PGP9.5 positive peripheral nerve fibers (green).
Table 2:
Differential gene expression analyses of interleukins in skin diseases
| Disease | Interleukins | Interleukins for gp130 (IL6ST) complex receptors | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IL-31 | IL-4 | IL-13 | OSM | IL-6 | LIF | CNTF | CTF1 | IL-11 | IL-27 | |
| Psoriasis | −0.1 | −0.9 | 0.4 | 3.5 | 2.2 | −1.6 | −0.9 | −0.6 | 0.6 | 0.4 |
| AD | 0.2 | 0.2 | 3.6 | 2.3 | 2.1 | −1.7 | 0.3 | 0.04 | 0.4 | −0.4 |
| CTCL | 0.09 | 1.4 | 1.3 | 2.1 | −2.2 | −0.8 | 0.4 | 0.4 | 0.5 | 1.3 |
| Imiquimod | N.d. | 0.05 | −1.4 | 1.8 | 1.4 | 0.8 | −0.3 | −0.5 | 0.4 | −0.06 |
Differential gene expression analyses of interleukins in inflammatory skin conditions (inflamed skin vs normal skin). The numbers indicate the expression of genes that are up- or down-regulated for inflamed skin samples versus normal skin (log 2 scale). N.d. indicates not detected. The analyses were performed with R package DESeq2.
OSM selectively sensitizes Nppb-neurons.
Our findings that OSM is elevated in skin samples of diseased skin associated with chronic itch and OSMR is expressed in itch-sensory neurons, suggest that OSM may contribute to skin pruritus. Therefore, we hypothesized that OSM might act, as a classic pruritogen which directly activates sensory neurons. In order to test this hypothesis, we first examined in vitro if OSM can cause an increase in intracellular calcium by imaging dissociated DRG neurons expressing the calcium reporter GCaMP6. We expected that if OSM activates Nppb-neurons, intracellular calcium would rise in a subset of the GCaMP6-expressing neurons. We imaged 236 capsaicin-responsive DRG neurons, and observed 68 neurons that were activated by leukotriene C4 (LTC4, 100nM), histamine (100μM) and capsaicin (10μM), cells we previously showed express Nppb (19). However, none of these cells, nor any of the other neurons labeled with GCaMP6 responded to OSM (1μg/mL, ~100 fold above ED50) (Figure 3A–B). To further confirm this observation, we performed whole-cell electrophysiological recording on Sst-cre:: AAV9-CAG-Flex-tdTomato neurons (Nppb-neurons) (19, 50). Nppb-neurons are small-diameter neurons with an averaged diameter of 14.2μm ± 2.2μm (Figure 3C). Applying OSM did not evoke noticeable currents on these neurons (Figure 3D). By contrast, histamine (100 μM) evoked sizable transient inward currents (Figure 3D–E). Therefore, OSM does not act like a classic pruritogen as it cannot directly depolarize itch-neurons. Although, OSM does not elicit a rapid depolarization of Nppb-neurons, it could nonetheless stimulate other cellular signaling pathways, such as activation of kinases, which might modulate neuronal activity in other ways. Therefore next, to examine if OSM can modulate neuronal responses to pruritogens, we recorded histamine-evoked whole-cell currents before and after short-term exposure to OSM. Although antihistamines are not useful for treatment of psoriasis, atopic dermatitis and CTCL, the pruritogens that underlie these chronic itch conditions are not well characterized. Therefore, for these mechanistic experiments, we used histamine in lieu of the itch-inducing agents active in chronic itch. In naïve Nppb-neurons, repetitive histamine challenge rapidly desensitizes histamine-evoked currents by 62.5±4.6% (Figure 3F, 3H) Pretreatment with OSM (1μg/mL) for 2 minutes increased histamine-induced currents by 31.8±17.1% (Figure 3G–H). Since Nppb-neurons also express receptor complexes for IL-31 and IL-13, we next tested whether these two cytokines have similar effects on the histamine elicited responses. We found that pretreatment with IL-31, similar to OSM treatment, potentiated histamine-induced currents (Figure 3H, S2A). However, IL-13 pretreatment failed to produce potentiation of histamine responses (Figure 3H, S2B). The OSM-dependent potentiation of histamine induced activation was selective to Nppb-neurons as administration of OSM on Mrgpra3-cre-GFP-neurons failed to potentiate histamine-currents (Figure S2C–D). Given that OSM treatment potentiates histamine responses, we wondered whether OSM might also enhance the baseline excitability of Nppb-neurons. For these studies, to probe for changes in excitability, we examined neuronal activity before and after OSM treatment using a current injection protocol. Naïve Nppb-neurons, in the conditions we employed in our studies, are almost resistant to current injection induced action potential firing, on average firing only a single action potential with an injection of 300-pA (Figure 3I). Short-term OSM treatments (1–2 minutes) did not change the detectable excitability of Nppb-neurons (Figure S2E–G). However, even though short-term exposure did not alter activity of Nppb-neurons, we wondered if longer exposure might. When we incubated neurons with OSM for longer times, the majority of Nppb-neurons displayed tonic firing in response to small injections of current (Figure 3J–K). Similar to OSM, IL-31, exerted the same effect on neuronal excitability, whereas treatment with IL-13 had no detectable effects (Figure 3K, S2H–I).
Figure 3. Oncostatin M sensitizes Nppb-neurons.
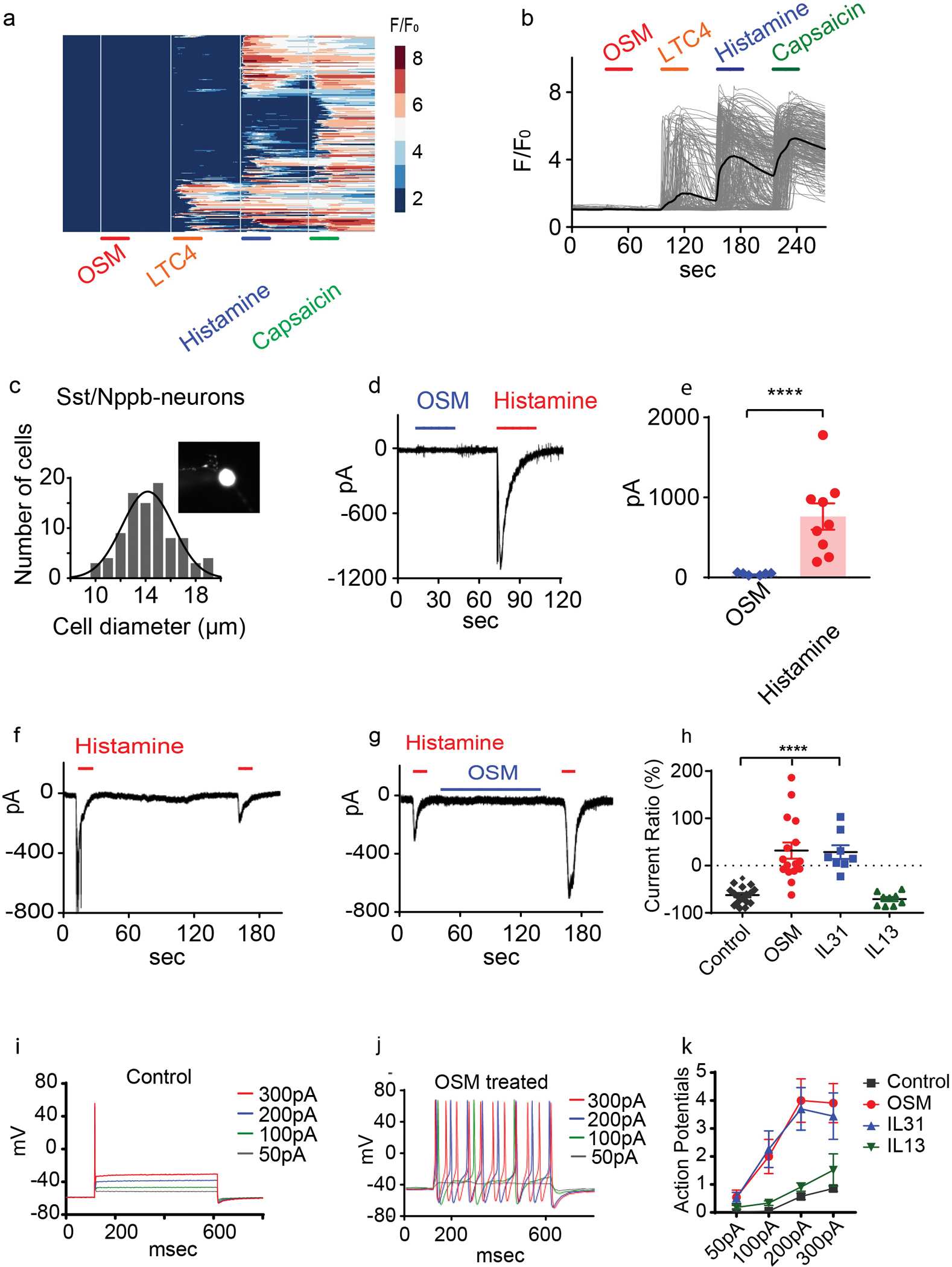
(A-B) Calcium imaging of dissociated Trpv1-GCaMP6 DRG neurons challenged (30 second; indicated with bars) with OSM (1μg/mL), LTC4 (100nM), histamine (10μM), and capsaicin (10μM) (326 neurons n= 3 mice). The colored scale bar represents normalized changes of fluorescence signals. The solid line in B indicates the averaged response of all the capsaicin-responsive neurons. (C) Quantification of cell diameter of Nppb-neurons (insert panel) (n=90). (D-E) Whole-cell voltage-clamp recordings (held at −60mV) of dissociated Nppb-tdTomato neurons exposed to OSM alone (blue line, 1μg/mL), and histamine (red line, 100μM). (E) n=9 neurons from 2 mice, ****p<0.0001, two-tailed Student’s T-test. (F-G). (F-H) Electrophysiological traces of Nppb-neuron responses. (F) Responses to histamine challenge without OSM pretreatment and (G) after pretreatment with OSM (1μg/mL) for 2 minutes. (H) Summary of the cytokine-mediated effects on histamine-evoked currents. Repetitive histamine (n=17), pretreatment with OSM (n=16) or IL-31 (n=8) (for OSM****p<0.0001 and for IL-31 p<0.0001), one-way ANOVA with Dunnett’s post-hoc test. Pretreatment with IL-13 (n=9) (p=0.95). (I-K) Electrophysiological traces of Nppb-neurons under current clamp conditions. (I) Control conditions, and (J) after incubation with OSM (100ng/mL) for 2–3 days. (K) Quantification of effects of cytokine. Prolonged incubation with OSM (n=22) or IL-31 (n=27), IL-13 (n=33), and controls (n=21); ***p=0.0003, ***p=0.0003, and p=0.7950 respectively, two-way ANOVA with Dunnett’s post-hoc test. Data presented as mean ± SEM.
OSM is secreted by various dermal immune cells
OSM is a pleiotropic cytokine that is expressed in activated T-cells, macrophages, dendritic cells and neutrophils (51–54). To identify the cellular source of OSM in chronic skin pruritus, we examined published single-cell RNA-seq data of whole skin biopsies collected from cutaneous T-cell Lymphoma patients (GSE128531) (55). In healthy skin biopsies, we detected very few cells expressing OSM and these cells co-expressed pan-leukocyte marker CD45 (Figure 4A–C). The fraction of OSM-expressing cells profoundly increased in CTCL skin biopsies (from 0.37±0.07 % to 6.08±2.6 %, p=0.016) (Figure 4D–F). Since OSM is mainly expressed by CD45 cells, we further characterized these cells and found that most T-cells (CD4+ or CD3+) as well as monocytes (CD14+) co-expressed OSM (Figure 4G–I). A small fraction of OSM-expressing immune cells express CD1C, a marker for dermal dendritic cells and minor number of OSM-expressing cells are marked by tryptase, a marker of mast-cells (Figure 4J–K). When comparing OSM expression with other cytokines including IL-31, IL-13, and IL-4, we found that OSM-expressing cells outnumbered the cells expressing other cytokines (p=0.016, 0.04, 0.02, respectively), suggesting a predominant role of OSM in skin inflammation (Figure 4L–N). To further support the involvement of OSM in inflammatory itch, we analyzed single-cell sequencing data from psoriatic skin biopsies (EGAS00001002927) (56). In agreement with our analysis of CTCL data, OSM was co-expressed with CD4, CD3, CD14 and CD45 cells (Figure 4O–Q). To validate this sequencing data, we performed immunohistochemistry on psoriatic skin biopsies. We found that, in healthy human skin biopsies, the infiltration of CD4+ and OSM+ expressing cells was very low (Figure 4R). However, in 3 out of 5 psoriatic skin biopsies, we observed massive CD4+ cells infiltration as well as strong OSM immunoreactivity in skin samples from psoriasis patients (Figure 4S). Together, these analyses uncovered a pivotal role of OSM in skin inflammation. Furthermore, OSM could be the major cytokine in neuroimmune interactions between sensory neurons and multiple dermal immune cells.
Figure 4. OSM is predominantly expressed in skin T-cells.
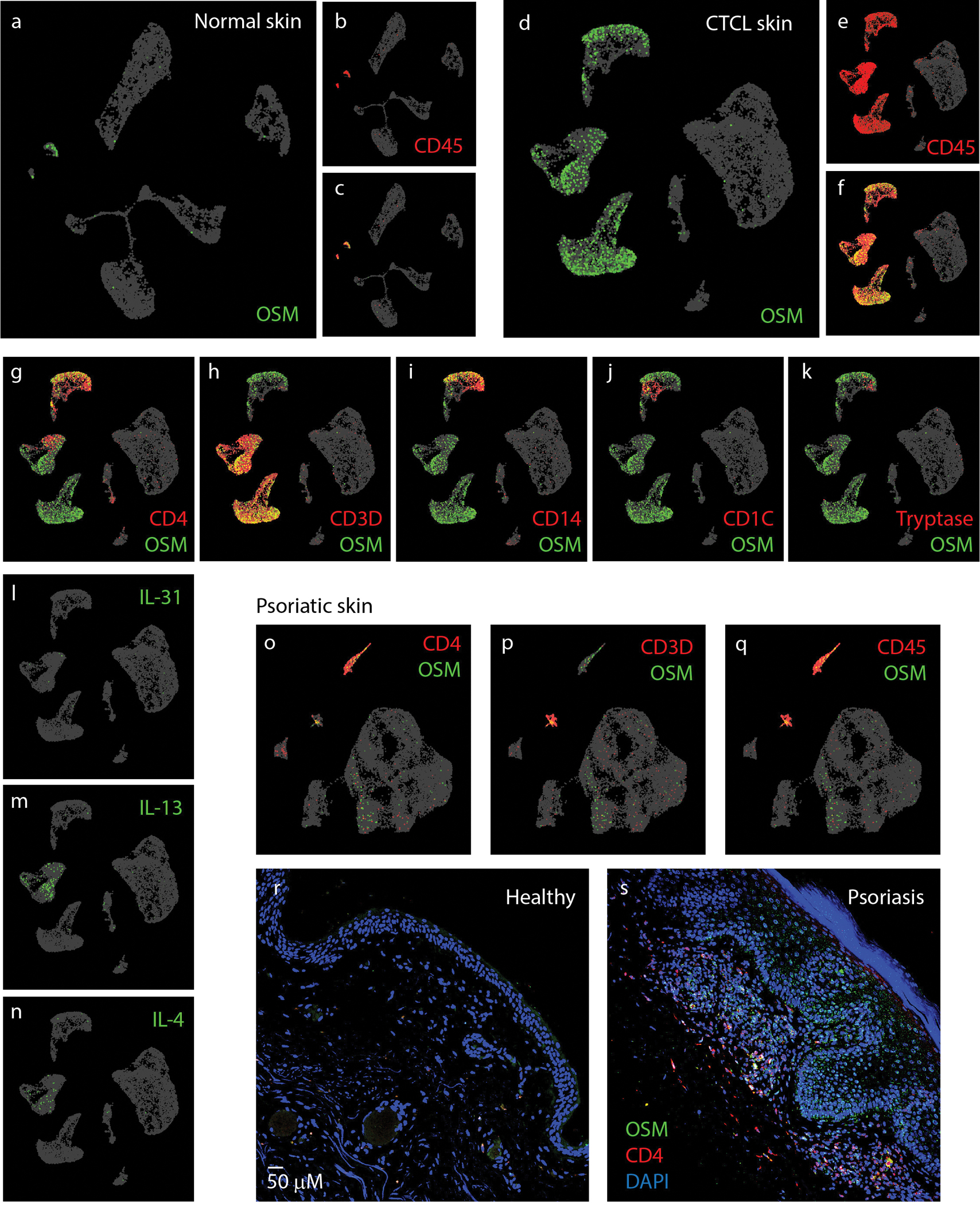
(A-C) UMAP plot of single-cell RNA-sequencing data (GSE128531) pooled from 4 normal skin biopsies (14,179 cells) showing expression pattern of OSM (green) and pan-leukocyte marker CD45 (PTPRC) (red). (D-F) UMAP plot of GSE128531 dataset which included 5 CTCL skin biopsies (30,663 cells) showing expression of OSM (green) and CD45 (red). (G-K) Co-expression of OSM (green) with markers for immune cell genes CD4+ and CD3+ (T-cells) (G-H), CD14+ (monocytes) (I), CD1C (dendritic cells) (J), and tryptase (mast cell) (K). (L-N) Expression of IL-31, IL-13, and IL-4 as indicated. (O-T) UMAP plots of single-cell sequencing data (EGAS00001002927) of psoriatic skin biopsies (21,025 cells pooled from 3 patients). Co-expression of OSM with CD4 (O), CD3 (P) and CD45 (Q). (R-S) Representative images of parafilm-embedded human skin sections immuostained against OSM (green) and CD4 (red) in healthy controls(R) and in psoriatic skin samples (S)
OSM can induce and exaggerate itch.
Our physiological studies suggest that OSM can sensitize itch-selective sensory neurons by increasing neuronal activity and excitability. To examine the functional consequences of this neuromodulation in vivo, we assayed the effects of OSM on behavioral responses in mice. For these studies we injected recombinant mouse OSM (1μg/μL, 10μL) alone or together with histamine intradermally into the nape of the neck and recorded scratching responses. As expected, from our calcium imaging and electrophysiological experiments, OSM evoked only minimal itch-responses shortly after administration (Figure 5A), which was not statistically different from saline injection (in the first 30 minutes after administration). In contrast to its lack of effect on its own, OSM profoundly potentiated histamine and leukotriene-evoked scratching bouts (Figure 5B, C). Although OSM alone did not evoke increases in scratching (over saline) in the first 30 minutes after injection, when we extended our observation time and recorded scratch responses up to 90 minutes after treatment, we found that mice slowly developed intense itch-behavior about 30–40 minutes after OSM injection (Figure 5D). During the 30–60 period, OSM challenge significantly increased scratching bouts compared to saline injected controls (51.6±15.5, p=0.033) (Figure 5E). It is known OSMR, like Il31Ra,(35) is expressed by many stromal cells such as keratinocytes and analysis of single-cell sequencing of skin samples showed that OSMR is expressed by keratinocytes, fibroblasts, and endothelial cells (Figure S3E) and potentially activation of these cells could be responsible for the late-onset scratching induced by OSM administration. Therefore, OSM might be acting on cells in the skin as well as sensory neurons. To investigate whether OSM-potentiates scratching and whether OSM-evoked slow-onset scratching responses caused by peripheral sensitization of sensory neurons, we generated DRG specific Osmr conditional knockout mice (Osmr cKO). These cKO mice were made by crossing Osmrfl/fl mice with Trpv1-Cre animals to permit all Osmr-expression to be eliminated including Nppb-negative Trpv1-positive neurons (Figure S1). Congruent with OSM causing itch by sensitization of sensory neurons, the loss of Osmr in DRG-neurons significantly reduced the scratching bouts evoked by co-administration of OSM and histamine (34.3±5.6 scratching bouts in cKO mice, versus 60.6±6.5 bouts in littermate controls, p=0.007) (Figure 5F). OSM-evoked delayed scratching responses in Osmr cKO mice were also significantly reduced (27.8±8.9 scratching bouts in cKO mice, versus 55.7±10 bouts in control littermates, p=0.049 (Figure 5G). Analysis of the phenotype of knockout mice in the imiquimod model of chronic itch also showed that Osmr in sensory neurons is important for the development of increased scratching and skin hypertrophy (Figure 5H–K). Together, these results suggest that OSM can induce and exaggerate itch by sensitizing sensory neurons.
Figure 5. OSM can induce and exaggerate itch.
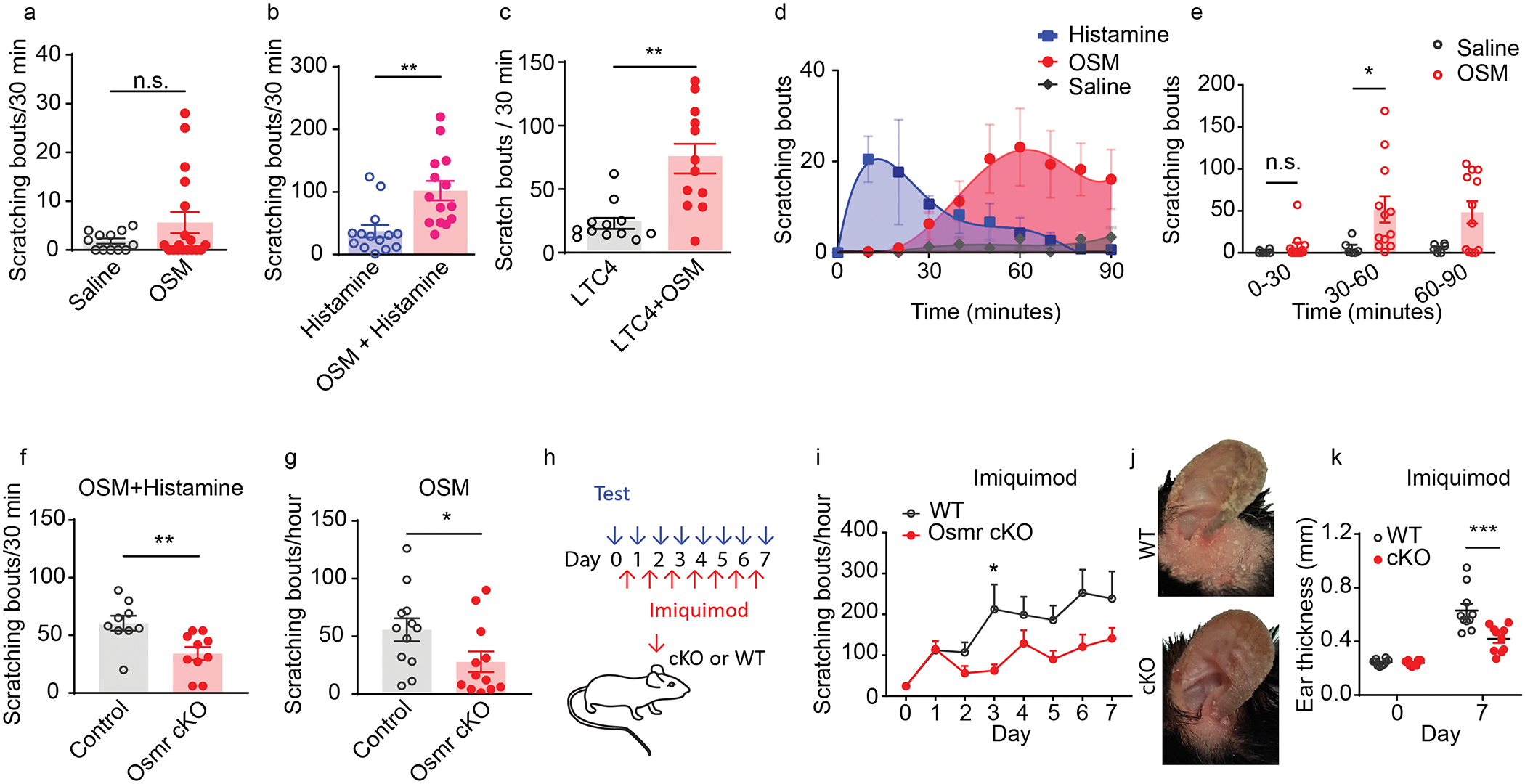
(A) Scratching bouts elicited by intradermal injection of recombinant mouse OSM (1μg/μL, 10μL, n=18) in the first 30 minutes after injection, compared to saline injection (n=12, ns p=0.1) and (B) by injection of histamine alone (10μg, n=18), and by co-injection of histamine with OSM (10μg, n=14), **p=0.002 and (C) by LTC4 alone (n=12) and co-injection LTC4 with OSM (n=12), **p=0.0005. A-C, data presented as mean ± SEM, two-tailed Student’s T-test. (D) Examination of time-course of OSM (n=12) and histamine (n=8) induced scratching responses and (E) quantification of scratching bouts, *p=0.033 ANOVA Sidak’s multiple comparison test. (F-G) Examination of the effects of sensory-neurons specific knockout of Osmr on OSM-induced itch behavior. (F) Co-administration of OSM and histamine (10μg in 10μL) into cKO mice (n=10) during 30-minute trials (using automated recording of scratch bouts compared to littermate controls (n=9), **p=0.007. (G) OSM injection in cKO mice (n=12) compared to control littermates (n=12), *p=0.049. F-G, data presented as mean ± SEM, two-tailed Student’s T-test. (H-K) Examination of the effect of sensory-neurons specific elimination of Osmr expression in the imiquimod model of chronic itch. (H) Schematic depicting the time course for the development of imiquimod-induced chronic itch. Imiquimod was topically applied to mouse ears for 7 consecutive days. Scratching bouts were measured daily prior to imiquimod treatment and the severity of skin inflammation was assessed by measurements of ear thickness. (I) Osmr Trpv1-cre cKO mice (n=10) compared to wild-type control mice (n=10) at day 3, *p=0.023 (ANOVA Sidak’s multiple comparison test). (J) Representative images of ears of control and Osmr cKO mice treated with imiquimod at day 7. (K) Quantification at Day 7 of ear thickness in cKO mice (n=10) compared to control littermates (n=10) (ANOVA Sidak’s multiple comparison test, ***p=0.0001), data presented as mean ± SEM.
Pharmacologically inhibition of OSM signaling reduces inflammatory itch
Findings from our bioinformatic analyses, and physiological and behavioral studies suggest that OSM evokes inflammatory skin pruritus through multiple pathways. We next tested whether systemic disruption of the OSM signaling cascade alleviates itch-behavior in a model of chronic skin inflammation. Since a selective OSM receptor inhibitor is not commercially available, we used a gp130 inhibitor, SC144 (57), to inhibit the OSMR/gp130 receptor complex. Initially, we tested whether SC144 would inhibit potentiation of histamine-responses in vitro. We performed whole-cell recordings on Nppb-neurons and examined the effects of SC-144 (10μM) treatment upon OSM-mediated sensitization. Indeed, inhibition of OSM signaling effectively prevented sensitization of Nppb-neurons (Figure 6A–B). At the behavioral level, SC-144 treatment (10mg/kg) almost completely inhibited OSM-evoked delayed scratching responses (Figure 6C). Showing the selectivity of this reagent, histamine-evoked immediate scratching was not affected by SC-144 administration (Figure 6D). Although the SC-144 could inhibit activation of gp130-receptor complexes other than gp-130-OSMR, other potential co-receptors for gp130 are not expressed at amounts expected to be functional in Nppb-neurons (Table 1). In addition, apart from IL-6, the expression of cytokines, that can activate other gp130-receptor complexes, are not upregulated in the imiquimod model of itch and in patients’ samples of skin diseases associated with chronic itch (Table 2). These results indicate, at a behavioral level, that disrupting OSMR/gp130 receptor signaling can inhibit OSM evoked itch through Nppb-neurons. To test if this strategy is also effective for inflammatory skin pruritus, we employed a mouse model of psoriasis (58, 59) and tested whether SC-144 can reduce severity of dermatitis and its associated itch phenotypes. In this model mice received daily imiquimod treatment (TARO, 0.05g/mouse/day) to induce psoriasiform dermatitis (Figure 6E) and at day 7 these animals display intense scratching behavior directed to the site of imiquimod administration. We found that SC-144 treatment greatly reduced spontaneous scratching behavior (Figure 6F–G). In addition, this treatment also mitigated skin inflammation, with reduced ear thickness in SC-144 treated mice (Figure 6H). Together these results are consistent with OSM being a mediator promoting persistent itch during chronic skin inflammation (Figure 6i).
Figure 6. Disruption of OSM signaling mitigates cutaneous inflammatory scratching.
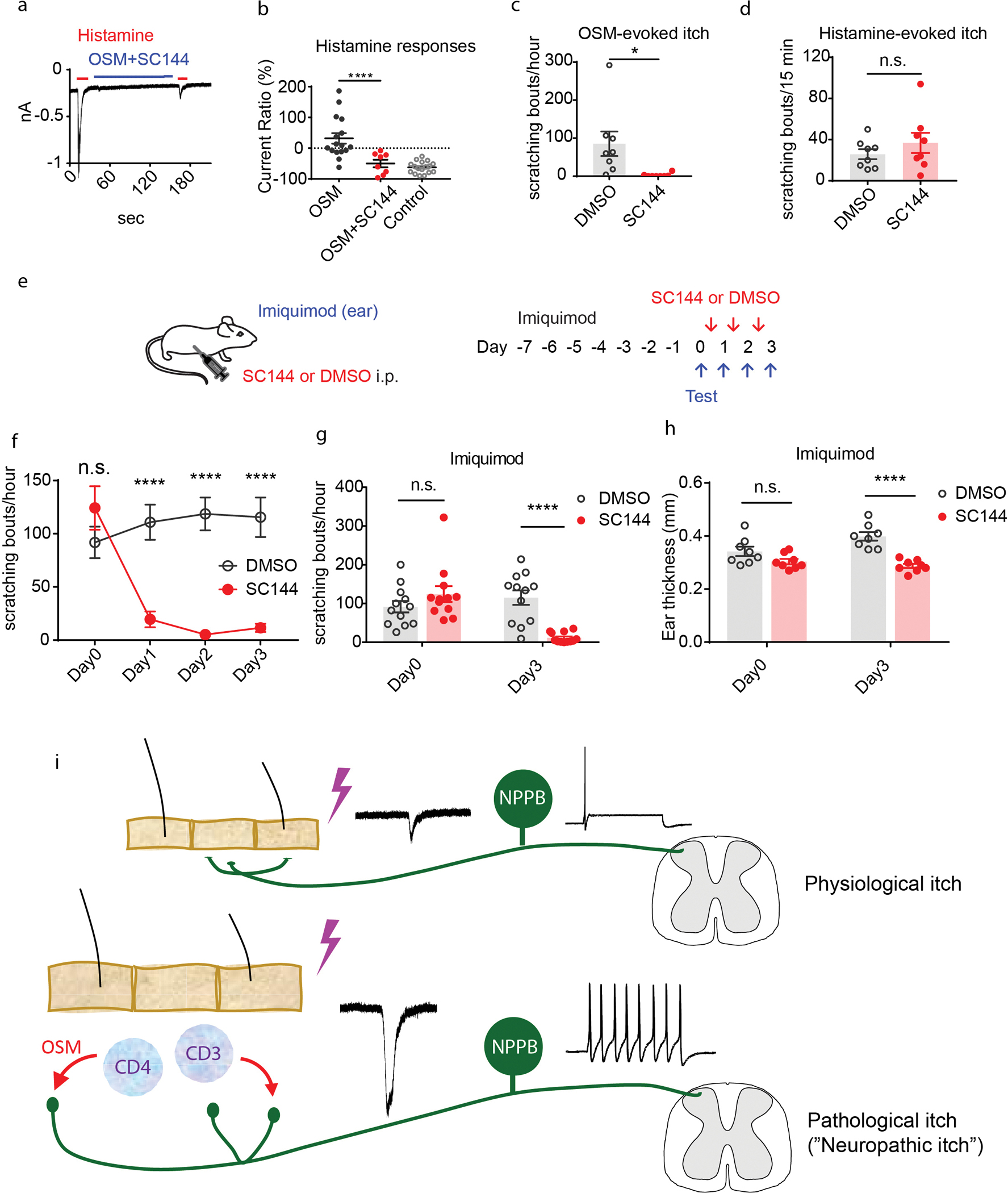
(A-B) Representative traces of voltage-clamp recording of Nppb-neurons. (A) Administration of histamine (red line) or histamine after treatment with SC144 (10μM) and OSM (1μg/mL) (blue line). (B) Quantification of repetitive histamine-evoked currents after OSM-treatment (n=16 neurons) compared with OSM + SC-144 treatment (n=8), ****p=0.0008. (C) Effects of SC144 on OSM-induced delayed scratching. 30–60 minutes post treatment mice administered SC144 (n=8) compared with control mice (n=8) injected with vehicle (DMSO), *p=0.02, data presented as mean ± SEM, two-tailed Student’s T-test. (D) Effects of SC144 on histamine scratching. During the first 15 minutes after histamine injection, scratching bouts in the presence of SC144 (n=8) compared with control DMSO (n=8), ns p=0.3, two-tailed Student T-test, data presented as means ± SEM. (E) Experimental design of tests on the efficacy of SC144 for itch in the imiquimod-induced psoriasiform-dermatitis model. Mouse ears were topically treated with imiquimod for 7 consecutive days to establish dermatitis. Immediately before SC144 or vehicle treatment, numbers of scratching bouts were measured (Day 0). At Day 1, 2, and 3, SC144 or vehicle was intraperitoneally injected into mice 30 minutes before behavioral tests. (F-H) Examination of the effects of SC144 on imiquimod-induced itch and skin inflammation. (F) At Day 0, before treatment, scratching bouts in SC144 compared to DMSO treatment group, ns p=0.36. In DMSO group, scratching bouts compared with SC144 treatment, at Day 1, 2, and 3, ****p<0.001 (ANOVA Sidak’s multiple comparison test). (G) Quantification of scratching bouts for control DMSO group (n=12) compared to the SC144-treated group (n=12), at Day 0 ns p=0.15 and at day 3, ****p<0.0001 (ANOVA Sidak’s multiple comparison test). (H) Quantification of ear thickness, in the SC-144 treatment group compared to the control DMSO group at Day 3, p<0.0001 (ANOVA Sidak’s multiple comparison test), data presented as mean ± SEM. (I) Schematic of possible mechanism for itch sensitization during chronic skin inflammation. In healthy skin, pruritogens transiently activate peripheral Nppb-nerve fibers and elicit transient itch sensation. During chronic skin inflammation, immune cells infiltrate inflamed skin and release OSM. The Nppb-nerve fibers are sensitized by OSM and exhibit hyperactivity and hyperexcitability which can lead to exaggerated persistent itch. In addition, the loss or alteration of epidermal nerve fibers may contribute to altered itch responses (89, 90).
Discussion
OSM is a pleiotropic cytokine which belongs to the IL-6 family of cytokines (31, 60). Its receptor, OSMR, forms heterodimers with either IL31RA or gp130 (IL6ST) to transduce IL-31 or OSM, respectively (30, 31, 61). It is well known that OSM contributes to many physiological processes, including hematopoiesis, bone remodeling, and liver development. However, OSM can also cause many deleterious conditions including Th1 and Th2 mediated skin and lung inflammation, arthritis, colitis, and some cancers (30, 32, 61). Although the role of OSM or OSMR in itch was not explored, it was reported missense mutations in OSMR in Familiar Primary Localized Cutaneous Amyloidosis are associated with severe pruritus (62–64), suggesting that OSM/OSMR axis may play roles in skin pruritus which may include perturbations of sensory neurons function and/or an imbalance of cytokine receptor signaling in keratinocytes. Here we showed that OSMR is preferentially expressed by itch-selective sensory neurons. OSM expression amounts are elevated in various skin diseases linked with itch. These data strongly suggested OSM can directly modulate itch-selective sensory neurons during chronic skin inflammation. Our functional studies showed that the mode of action of OSM is different from classic pruritogens and cytokines IL-4, IL-13, or IL31 (26, 27, 33); OSM acts to augment pruritogen induced activity. Firstly, OSM causes potentiation of neural responses to pruritogens. The latter studies were performed with histamine as a pruritogen since the active pruritogens in these types of itch is not well characterized. Therefore, although we used histamine in our mechanistic studies, we cannot exclude that other pruritogens might produce different effects on responses in itch-neurons. Secondly, OSM has the longer-term effect of converting action potential firing of sensory neurons from phasic to tonic. The process of cytokine mediated sensitization of sensory neurons caused a relatively fast increase in activity as well as a much slower modification of neural activity. The latter effect might be due to changes in gene expression (65). The rapid effect of OSM was similar to that reported to be elicited by inflammatory mediators to heat sensitization (66, 67). The long-term modulation of OSM on neuronal excitability also has precedence in that interleukin-1 was reported to enhance excitability causing allodynia (68, 69). Our observation that OSM also evoked late-onset scratching responses parallels results from human subjects where it was reported that cutaneous challenge with IL-31 induces late-onset itch (70).
Our bulk and single-cell sequencing analyses revealed the overwhelming expression of OSM compared to other well-known cytokines, including IL-31, IL-13, or IL-4. Therefore, it is tempting to speculate that OSM might play a major role on neuronal activity in many cutaneous diseases where barrier disruption leads to inflammation (2, 4) and chronic itch. However, there was also heterogeneity in increased cytokines expression with elevated expression of OSM, IL-31, IL-13, and IL-4 in different samples. Since the quantification of these cytokines were made at fixed times, the differences in expression of cytokines may reflect difference in stages of disease, or they could reflect differences in types of inflammation. In addition, we have no information on the itch phenotype of these subjects.
Single cell sequencing data also revealed that OSM is expressed by many dermal immune cells including T-cells, monocytes, dendritic cells and mast cells. This suggests OSM might be a universal cytokine for neuroimmune communications between sensory neurons and dermal immune cells, which might explain its prevalence in many types of pruritus. In a mouse model of psoriasis chronic itch, we showed that either the elimination of OSMR from sensory neurons or the interference of OSMR signaling (with SC-144) reduced scratching. Although these results are predictive of OSMR signaling in psoriasis, future studies will be needed to determine whether it is similarly effective for other types of inflammatory itch. We also note that OSMR is expressed in keratinocytes and that the expression of IL-6 is increased and at least some of the effects of SC-144 treatment may be on non-sensory neurons and non Nppb-neurons (that express IL6Ra). Indeed, this broader effect may account for the larger reduction of scratch elicited by SC-144 compared to elimination of OSMR from sensory neurons and may also be a benefit for treatment of inflammatory skin conditions that may have a component of pain. In the future, it will be important to use Osmr specific antagonist to determine whether the effects of SC-144 are selective for Osmr or occur via inhibition of other gp130-receptor mediated processes. In addition, further preclinical and clinical studies are required to determine the effectiveness of OSMR antagonism and to examine whether adverse effects and toxicity are associated with these agents. Regardless of the exact mechanisms, SC-144 shows potential for treatment of inflammatory skin pruritus.
Chronic itch is a common symptom and a major complaint in dermatology clinics (71, 72). From a clinical perspectives, itch is classified into several categories including dermatologic, systemic, neurologic, psychogenic, mixed, and others (73). Recently it has been suggested that neurologic itch can be further divided into neuropathic itch and neurogenic itch (3, 5, 72, 74). Neuropathic itch, a term borrowed from neuropathic pain (75, 76) conceptually involves induction of neuronal hyperexcitability. However, the molecular features of neuropathic itch are not well-defined. Pruritogens, such as histamine, serotonin, chloroquine, and leukotriene activate G-protein coupled-receptors which subsequently depolarize sensory neurons evoking relatively short-term onset scratching responses (19, 77). Our electrophysiological recordings showed histamine quickly desensitized Nppb-neurons and therefore this type of pruritogen probably does not on their own cause long-term hyperexcitability. Here we found that OSM modulates sensory neurons and evokes itch by slowly increasing neuronal sensitivity and excitability. Therefore, this cytokine-mediated neuronal modulation seems to fit the mechanistic definition of neuropathic itch suggested by others (3, 5, 72, 74). Furthermore, and more importantly, since OSMR is a common subunit for transducing OSM and IL-31 signaling, antagonizing OSMR may be a promising strategy to control chronic inflammatory itch for conditions where expression of cytokines are elevated.
Materials and Methods
Study Design
The primary research objective was to determine the cytokines involved in production of inflammatory pruritus and whether itch could be reduced by antagonism of OSMR. All other hypotheses were related to these objectives. The research subjects and units of investigation were primary-cultured DRG cells, DRG tissue from mice and human donors, and mice in controlled laboratory experiments. Sample sizes for in vitro and cell-based electrophysiological assays were those used by other labs in the field. For animal experiments, sample sizes were based on experience and were of a size generally used in the itch field. Data was not excluded in any studies.
Statistical Analysis
Prism 7.0 (GraphPad Software) was used for statistical analyses. Differences between mean values were analyzed using unpaired two-tailed Student’s t test or ANOVA for multiple comparisons as indicated in figure legends. Differences were considered significant for ∗p < 0.05. Exact p values, definition and number of replicates as well as definitions of center and dispersion are given in the respective figure legend. No statistical method was employed to predetermine sample sizes.
Materials and methods
Animals
All experiments using mice followed NIH guidelines and were approved by the National Institute of Dental and Craniofacial Research ACUC. Mice were housed in small social groups (4–5 animals) in individually ventilated cages under 12-hour light/dark cycles and fed ad libitum. 6–10-week old animals of both genders were used in all experiments. C57BL/6N wild-type mice were purchased from Envigo and all other genetically modified mice were bred in house. Sst-IRES-Cre knock-in (JAX# 013044 (78)) Trpv1-IRES-Cre knock-in (JAX# 017769 (79)) Trpm8-Cre BAC-transgenic mice (80) that drive expression of Cre recombinase in genetically identifiable cell populations have been described previously (19). To avoid non-specific labeling at developmental stages, we intraperitoneally injected with AAV9-CAG-Flex-eGFP or AAV9-CAG-Flex-tdTomato (10 μL, 1×1013 vg/mL) into these Cre mouse lines at postnatal day 2–4. Sensory-neuron specific Osmr KO mice were generated by crossing Osmr fl/fl mouse line (JAX#011081) with Trpv1-Cre mice. Genotyping of offspring from all breeding steps was performed with genomic DNA isolated from tail snips and allele-specific primer pairs.
RNA-sequencing and analysis
RNA-seq libraries were produced from pooled DRG neurons as described previously (19). Briefly, between 150–250 eGFP- or tdTomato-positive cells per mouse were hand-picked with borosilicate glass pipettes and micromanipulator. Each picking session included one sample per genotype to control for potential experimental bias. A total of 4 independent samples per genotype were generated. Total RNA was extracted with a PicoPure RNA isolation kit (ThermoFisher Scientific) using the manufacturer’s instructions. cDNA was produced and amplified using the Ovation® RNA-seq system V2 (NuGEN) according the manufacturer’s protocol. Sequencing libraries were generated using the Nextera XT method (Illumina, San Diego, CA), pooled and sequenced in 126 bp paired-end mode on a HiSeq 1000 instrument (Illumina). Public-available RNA-seq raw data (fastq files) were downloaded from The Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) with SRA toolkit (81). Sequence reads were mapped to GRCm38v79 reference transcriptome, read counts and transcript abundances were estimated with Kallisto (82) on NIH Biowulf computer clusters. Differential gene expression and single-cell RNA-sequencing analysis was performed with DESeq2 (83) and Seurat (84).
In situ hybridization
DRGs were dissected from mice with various genotypes. Multi-labeling ISH was performed using the RNAscope® technology (ACD) according to the manufacturer’s instructions. Probes against Osmr, Nppb, Il6st (gp130), Jak1, Il31ra, Il13ra1, Il4ra, and Trpv1 in conjunction with the RNAscope® multiplex fluorescent development kit were used. Images were collected on an Eclipse Ti (Nikon) confocal laser-scanning microscope.
Primary cell cultures
Primary cultures of DRG neurons were dissected and dissociated as described previously (85, 86). Briefly, DRG were harvested and incubated in 5 mg/mL Collagenase/Dispase (Millipore-Sigma) for 30 minutes. Cells were mechanically dissociated with a pipette, seeded on poly-D lysine-coated coverslips and cultured with DMEM/F12, supplemented with 10% FBS, penicillin/streptomycin, 100 ng/mL NGF, 50 ng/mL GDNF.
Calcium imaging
For calcium imaging DRG neurons were transfected with AAV9.Syn.GCaMP6f.WPRE.SV40 (2 μL, 1×1013 vg/mL, AV-9-PV2824) (87). After 48 hours of culture, coverslips with DRG neurons expressing calcium indicator GCaMP6f were mounted on a DMi8 microscope (Leica) in HBSS buffer (in mM: 140 NaCl, 2 CaCl2, 10 HEPES, 4 KCl, 1 MgCl2, pH 7.4) and constantly super fused from a gravity-fed six-channel system (VC-6, Warner Instruments). Imaging was performed with an ORCA-Flash 4.0 C1440 digital CMOS camera (Hamamatsu, Bridgewater, NJ) at 1 Hz. Fluorescence intensity in hand-drawn regions of interest was extracted using HCImage (Hamamatsu) and plotted against time. A R package, Pheatmap, was used to generate a heatmap plot to visualize fluorescence signals in all the neurons.
Electrophysiology
For labeling Nppb-neurons, DRG neurons collected from Sst-Cre mice were transfected with AAV9-CAG-Flex-tdTomato (2 μL, 1×1013 vg/mL, from UNC Vector Core) and cultured on cover slips for 48–72 hours. Whole cell current and voltage clamps were performed on Sst-Cre DRG neurons expressing tdTomato. The morphology of selected neurons was examined under bright filed. Only the cells displaying smooth membranes with extended pseudounipoloar axons were selected for recording (Figure 3c). Data were recorded with Axon 700B amplifier and pCLAMP 11 software (Molecular Devices, Sunnyvale). Pipette electrodes were pulled with P-97 microelectrode puller (Sutter Instrument) from borosilicate glass (WPI, Inc) with resistances of 2–4 MΩ. Extracellular solution contained (in mM) 140 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, and 10 Glucose with pH of 7.4 and Osmolality 300 mOsm/kg. Pipette solution contained 140 KCl, 1 MgCl2, 1 EGTA, 10 HEPES, 3 ATP, and 0.5 GTP with pH of 7.4 and osmolality of 300 mOsm/kg.
Mouse behavioral measurements
For all behavioral paradigms, the experimenter was blind to the genotype of mice under study. Ear-tag numbers were read after the experiment and results were unblinded at the end of testing sessions. All behavioral experiments were conducted during the light cycle at ambient temperature (∼23°C). Behavioral assessment of scratching behavior was conducted as described previously (12). Briefly, mice were injected subcutaneously into the nape of the neck with histamine (Sigma-Aldrich) or recombinant murine oncostatin M (LSBio). Compounds were diluted in saline and injection volume was always 10 μl. Scratching behavior was recorded for 30 minutes. In OSM experiments, scratching behavior was monitored for one hour. For pharmacokinetics experiments of histamine and OSM, the observation time was extended to 90 minutes. In SC144 experiment, histamine-induced scratching behavior was recorded for 15 minutes. One bout was defined as scratching behavior toward the injection site between lifting the hind leg from the ground and either putting it back on the ground or guarding the paw with the mouth. In some behavioral tests, scratching bouts were automatically counted with MicroAct system (Neuroscience, Inc). Briefly, a magnet (1 mm in diameter, 3 mm in length) was implanted under the skin of dorsal hind-paw at least 7 days before the behavioral tests (88). About 30 minutes before tests, mice were placed into a special observation chamber with a ring coil. Vertical movements of the implanted magnets will induce currents which were recorded. Scratching bouts were determined using MicroAct software.
Chronic dermatitis model
Imiquimod-induced psoriasiform dermatitis model was established on sex balanced C57BL/6 mice (6–10 weeks old, 20–25g) as previously described (14, 15). Briefly, mice were anesthetized with isoflurane and topical imiquimod cream (TARO) was applied to mouse ear (0.05g/mouse) once a day for consecutive 7–10 days.
Supplementary Material
(A) Semi-quantitative ISH of human DRG visualizing OSMR and NPPB expression; higher magnification of image in figure 1G. DAPI stained glia cells (grey) surround neuron cell bodies. (B) Representative image of ISH of mouse DRG with probes to Osmr and Nppb. Arrows indicate a subpopulation of Trpv1-neurons with low amounts of Osmr transcripts (but not Nppb).
(A-B) Sample voltage clamp electrophysiology recordings of Nppb-neurons showing effects of treatment histamine when combined with IL-31 and IL-13. (A) Treatment with IL31, and (B) with IL13 (blues lines) and administration of histamine (red lines). (C-D) Examination of the responses of MrgprA3-neurons to OSM. (C) Sample traces of the effects of OSM treatment on histamine-evoked currents in Mrgpra3-neurons (n=17). (D) Quantification of the responses of Sst-Ai9 neurons and MrgprA3-neurons to OSM. (E-G) Examination of the effects of short-term OSM treatment on neuronal excitability of Nppb-neurons (n=9). (E-F) Sample electrophysiological traces of Sst-Ai9-neurons to current injection before (E) and 2 minutes after administration of OSM (F). (G) Quantification of the effects of short-term exposure to OSM. (H-I) Sample traces showing effects of current injection on Sst-Ai9 neurons exposed to prolonged IL31 and IL13 treatment. Sample traces of effects of long exposure to IL31 (H) and IL-13 (I).
(A-B) ISH with indicated genes, of trigeminal ganglia from wildtype mice (A) and Osmr cKO mice (B). (C) Quantification of the expression of Osmr relative to the pan-neuronal marker β3-tubulin (Tubb3), Osmr+/Tubbs+ = 54/362 neurons n=3 wildtype mice, Osmr+/Tubbs+ = 5/533 neurons n=3 Osmr cKO mice, ***p=0.0009, Two-tailed Student T-test. (D) Quantification of the percentage of Trpv1 cells relative to Tubb3+ neurons, Trpv1+/Tubb3+ = 92/362 neurons n=3 wildtype mice; Trpv1+/Tubb3+ = 152/535 neurons n=3 Osmr cKO mice; p=0.3, Two-tailed T-test. (E) Analysis of single-cell sequencing analysis of data from CTCL skin samples (GSE128531) highlighting the expression of OSMR (magenta) in indicated cell types.
Acknowledgments
This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, project ZIADE000721-19 (MAH). Human tissue was obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov) and DNA sequencing were carried out by the NIDCR Genomics and Computational Biology Core.
Footnotes
Competing interests
The authors declare no competing interests.
Data and materials availability
All the data for this study is present in the main text or in the Supplementary Materials.
References
- 1.Yosipovitch G, Greaves MW, Schmelz M, Itch. Lancet 361, 690–694 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Yosipovitch G et al. , Skin Barrier Damage and Itch: Review of Mechanisms, Topical Management and Future Directions. Acta Derm Venereol 99, 1201–1209 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Misery L et al. , Neuropathic pruritus. Nat Rev Neurol 10, 408–416 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lerner EA, Pathophysiology of Itch. Dermatol Clin 36, 175–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hachisuka J, Chiang MC, Ross SE, Itch and neuropathic itch. Pain 159, 603–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikoma A, Steinhoff M, Stander S, Yosipovitch G, Schmelz M, The neurobiology of itch. Nat Rev Neurosci 7, 535–547 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Kini SP et al. , The impact of pruritus on quality of life: the skin equivalent of pain. Arch Dermatol 147, 1153–1156 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Hay RJ et al. , The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol 134, 1527–1534 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Leader B, Carr CW, Chen SC, Pruritus epidemiology and quality of life. Handb Exp Pharmacol 226, 15–38 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bautista DM, Wilson SR, Hoon MA, Why we scratch an itch: the molecules, cells and circuits of itch. Nat Neurosci 17, 175–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoon MA, Molecular dissection of itch. Curr Opin Neurobiol 34, 61–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNeil B, Dong X, Peripheral mechanisms of itch. Neurosci Bull 28, 100–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han L et al. , A subpopulation of nociceptors specifically linked to itch. Nat Neurosci 16, 174–182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra SK, Hoon MA, The cells and circuitry for itch responses in mice. Science 340, 968–971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatto G, Smith KM, Ross SE, Goulding M, Neuronal diversity in the somatosensory system: bridging the gap between cell type and function. Curr Opin Neurobiol 56, 167–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu L et al. , Enhanced excitability of MRGPRA3- and MRGPRD-positive nociceptors in a model of inflammatory itch and pain. Brain 137, 1039–1050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meixiong J et al. , Activation of Mast-Cell-Expressed Mas-Related G-Protein-Coupled Receptors Drives Non-histaminergic Itch. Immunity 50, 1163–1171 e1165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill RZ, Morita T, Brem RB, Bautista DM, S1PR3 Mediates Itch and Pain via Distinct TRP Channel-Dependent Pathways. J Neurosci 38, 7833–7843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solinski HJ et al. , Nppb Neurons Are Sensors of Mast Cell-Induced Itch. Cell Rep 26, 3561–3573 e3564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama T, Lerner EA, Carstens E, Protease-activated receptors and itch. Handb Exp Pharmacol 226, 219–235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy VB, Iuga AO, Shimada SG, LaMotte RH, Lerner EA, Cowhage-evoked itch is mediated by a novel cysteine protease: a ligand of protease-activated receptors. J Neurosci 28, 4331–4335 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy VB et al. , Redefining the concept of protease-activated receptors: cathepsin S evokes itch via activation of Mrgprs. Nat Commun 6, 7864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storan ER, O’Gorman SM, McDonald ID, Steinhoff M, Role of cytokines and chemokines in itch. Handb Exp Pharmacol 226, 163–176 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Trier AM, Kim BS, Cytokine modulation of atopic itch. Curr Opin Immunol 54, 7–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campion M et al. , Interleukin-4 and interleukin-13 evoke scratching behaviour in mice. Exp Dermatol 28, 1501–1504 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Cevikbas F et al. , A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J Allergy Clin Immunol 133, 448–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oetjen LK et al. , Sensory Neurons Co-opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell 171, 217–228 e213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mack MR, Kim BS, The Itch-Scratch Cycle: A Neuroimmune Perspective. Trends Immunol 39, 980–991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F et al. , A basophil-neuronal axis promotes itch. Cell, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards CD, The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm 2013, 512103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.H. M. Hermanns, Oncostatin M and interleukin-31: Cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev 26, 545–558 (2015). [DOI] [PubMed] [Google Scholar]
- 32.West NR, Owens BMJ, Hegazy AN, The oncostatin M-stromal cell axis in health and disease. Scand J Immunol 88, e12694 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Meng J et al. , New mechanism underlying IL-31-induced atopic dermatitis. J Allergy Clin Immunol 141, 1677–1689 e1678 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Bagci IS, Ruzicka T, IL-31: A new key player in dermatology and beyond. J Allergy Clin Immunol 141, 858–866 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Nattkemper LA et al. , Cutaneous T-cell Lymphoma and Pruritus: The Expression of IL-31 and its Receptors in the Skin. Acta Derm Venereol 96, 894–898 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Chiu IM et al. , Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usoskin D et al. , Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Li CL et al. , Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26, 967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen MQ, Wu Y, Bonilla LS, von Buchholtz LJ, Ryba NJP, Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS One 12, e0185543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H et al. , The Neuropeptide Y Y2 Receptor Is Coexpressed with Nppb in Primary Afferent Neurons and Y2 Activation Reduces Histaminergic and IL-31-Induced Itch. J Pharmacol Exp Ther 372, 73–82 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Dillon SR et al. , Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat Immunol 5, 752–760 (2004). [DOI] [PubMed] [Google Scholar]
- 42.LaPorte SL et al. , Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollanazar NK, Smith PK, Yosipovitch G, Mediators of Chronic Pruritus in Atopic Dermatitis: Getting the Itch Out? Clin Rev Allergy Immunol 51, 263–292 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Li B et al. , Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 134, 1828–1838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsoi LC et al. , Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J Invest Dermatol 139, 1480–1489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Querfeld C et al. , Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile. Cancer Immunol Res 6, 900–909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahern K, Gilmore ES, Poligone B, Pruritus in cutaneous T-cell lymphoma: a review. J Am Acad Dermatol 67, 760–768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yosipovitch G, Bernhard JD, Clinical practice. Chronic pruritus. N Engl J Med 368, 1625–1634 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Swindell WR et al. , Imiquimod has strain-dependent effects in mice and does not uniquely model human psoriasis. Genome Med 9, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J et al. , Circuit dissection of the role of somatostatin in itch and pain. Nat Neurosci 21, 707–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Repovic P, Benveniste EN, Prostaglandin E2 is a novel inducer of oncostatin-M expression in macrophages and microglia. J Neurosci 22, 5334–5343 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cross A, Edwards SW, Bucknall RC, Moots RJ, Secretion of oncostatin M by neutrophils in rheumatoid arthritis. Arthritis Rheum 50, 1430–1436 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Tanaka M, Miyajima A, Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol 149, 39–52 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Boniface K et al. , Oncostatin M secreted by skin infiltrating T lymphocytes is a potent keratinocyte activator involved in skin inflammation. J Immunol 178, 4615–4622 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Gaydosik AM et al. , Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors. Clin Cancer Res 25, 4443–4454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng JB et al. , Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep 25, 871–883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu S, Grande F, Garofalo A, Neamati N, Discovery of a novel orally active small-molecule gp130 inhibitor for the treatment of ovarian cancer. Mol Cancer Ther 12, 937–949 (2013). [DOI] [PubMed] [Google Scholar]
- 58.van der Fits L et al. , Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol 182, 5836–5845 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Sakai K et al. , Mouse model of imiquimod-induced psoriatic itch. Pain 157, 2536–2543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rose-John S, Interleukin-6 Family Cytokines. Cold Spring Harb Perspect Biol 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West NR et al. , Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 23, 579–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arita K et al. , Oncostatin M receptor-beta mutations underlie familial primary localized cutaneous amyloidosis. Am J Hum Genet 82, 73–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin MW et al. , Novel IL31RA gene mutation and ancestral OSMR mutant allele in familial primary cutaneous amyloidosis. Eur J Hum Genet 18, 26–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saeedi M et al. , A novel missense mutation in oncostatin M receptor beta causing primary localized cutaneous amyloidosis. Biomed Res Int 2014, 653724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J et al. , The Cytokine TGF-beta Induces Interleukin-31 Expression from Dermal Dendritic Cells to Activate Sensory Neurons and Stimulate Wound Itching. Immunity 53, 371–383 e375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langeslag M et al. , Oncostatin M induces heat hypersensitivity by gp130-dependent sensitization of TRPV1 in sensory neurons. Mol Pain 7, 102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Obreja O et al. , Fast modulation of heat-activated ionic current by proinflammatory interleukin 6 in rat sensory neurons. Brain 128, 1634–1641 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Binshtok AM et al. , Nociceptors are interleukin-1beta sensors. J Neurosci 28, 14062–14073 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stemkowski PL, Noh MC, Chen Y, Smith PA, Increased excitability of medium-sized dorsal root ganglion neurons by prolonged interleukin-1beta exposure is K(+) channel dependent and reversible. J Physiol 593, 3739–3755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hawro T et al. , Interleukin-31 does not induce immediate itch in atopic dermatitis patients and healthy controls after skin challenge. Allergy 69, 113–117 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Wong LS, Wu T, Lee CH, Inflammatory and Noninflammatory Itch: Implications in Pathophysiology-Directed Treatments. Int J Mol Sci 18, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinhoff M, Schmelz M, Szabo IL, Oaklander AL, Clinical presentation, management, and pathophysiology of neuropathic itch. Lancet Neurol 17, 709–720 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Stander S et al. , Clinical classification of itch: a position paper of the International Forum for the Study of Itch. Acta Derm Venereol 87, 291–294 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Steinhoff M, Oaklander AL, Szabo IL, Stander S, Schmelz M, Neuropathic itch. Pain 160 Suppl 1, S11–S16 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Jensen TS et al. , A new definition of neuropathic pain. Pain 152, 2204–2205 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Treede RD et al. , Neuropathic pain: redefinition and a grading system for clinical and research purposes. Neurology 70, 1630–1635 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Liu Q et al. , Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 139, 1353–1365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taniguchi H et al. , A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cavanaugh DJ et al. , Restriction of transient receptor potential vanilloid-1 to the peptidergic subset of primary afferent neurons follows its developmental downregulation in nonpeptidergic neurons. J Neurosci 31, 10119–10127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yarmolinsky DA et al. , Coding and Plasticity in the Mammalian Thermosensory System. Neuron 92, 1079–1092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leinonen R, Sugawara H, Shumway M, International C Nucleotide Sequence Database, The sequence read archive. Nucleic Acids Res 39, D19–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Z et al. , Targeting human Mas-related G protein-coupled receptor X1 to inhibit persistent pain. Proc Natl Acad Sci U S A 114, E1996–E2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tseng P-YZ, Qin; Li Zhea; Dong Xinzhong, MrgprX1 mediates neuronal excitability and itch through tetrodotoxin-resistant sodium channels. Itch 4, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen TW et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Inagaki N et al. , Involvement of unique mechanisms in the induction of scratching behavior in BALB/c mice by compound 48/80. Eur J Pharmacol 448, 175–183 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Nguyen MQ, Le Pichon CE, Ryba N, Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Renthal W et al. , Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron 108, 128–144 e129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Semi-quantitative ISH of human DRG visualizing OSMR and NPPB expression; higher magnification of image in figure 1G. DAPI stained glia cells (grey) surround neuron cell bodies. (B) Representative image of ISH of mouse DRG with probes to Osmr and Nppb. Arrows indicate a subpopulation of Trpv1-neurons with low amounts of Osmr transcripts (but not Nppb).
(A-B) Sample voltage clamp electrophysiology recordings of Nppb-neurons showing effects of treatment histamine when combined with IL-31 and IL-13. (A) Treatment with IL31, and (B) with IL13 (blues lines) and administration of histamine (red lines). (C-D) Examination of the responses of MrgprA3-neurons to OSM. (C) Sample traces of the effects of OSM treatment on histamine-evoked currents in Mrgpra3-neurons (n=17). (D) Quantification of the responses of Sst-Ai9 neurons and MrgprA3-neurons to OSM. (E-G) Examination of the effects of short-term OSM treatment on neuronal excitability of Nppb-neurons (n=9). (E-F) Sample electrophysiological traces of Sst-Ai9-neurons to current injection before (E) and 2 minutes after administration of OSM (F). (G) Quantification of the effects of short-term exposure to OSM. (H-I) Sample traces showing effects of current injection on Sst-Ai9 neurons exposed to prolonged IL31 and IL13 treatment. Sample traces of effects of long exposure to IL31 (H) and IL-13 (I).
(A-B) ISH with indicated genes, of trigeminal ganglia from wildtype mice (A) and Osmr cKO mice (B). (C) Quantification of the expression of Osmr relative to the pan-neuronal marker β3-tubulin (Tubb3), Osmr+/Tubbs+ = 54/362 neurons n=3 wildtype mice, Osmr+/Tubbs+ = 5/533 neurons n=3 Osmr cKO mice, ***p=0.0009, Two-tailed Student T-test. (D) Quantification of the percentage of Trpv1 cells relative to Tubb3+ neurons, Trpv1+/Tubb3+ = 92/362 neurons n=3 wildtype mice; Trpv1+/Tubb3+ = 152/535 neurons n=3 Osmr cKO mice; p=0.3, Two-tailed T-test. (E) Analysis of single-cell sequencing analysis of data from CTCL skin samples (GSE128531) highlighting the expression of OSMR (magenta) in indicated cell types.
Data Availability Statement
All the data for this study is present in the main text or in the Supplementary Materials.


