Abstract
RNA therapeutics, including siRNAs, antisense oligonucleotides, and other oligonucleotides, have great potential to selectively treat a multitude of human diseases, from cancer to COVID to Parkinson's disease. RNA therapeutic activity is mechanistically driven by Watson–Crick base pairing to the target gene RNA without the requirement of prior knowledge of the protein structure, function, or cellular location. However, before widespread use of RNA therapeutics becomes a reality, we must overcome a billion years of evolutionary defenses designed to keep invading RNAs from entering cells. Unlike small-molecule therapeutics that are designed to passively diffuse across the cell membrane, macromolecular RNA therapeutics are too large, too charged, and/or too hydrophilic to passively diffuse across the cellular membrane and are instead taken up into cells by endocytosis. However, similar to the cell membrane, endosomes comprise a lipid bilayer that entraps 99% or more of RNA therapeutics, even in semipermissive tissues such as the liver, central nervous system, and muscle. Consequently, before RNA therapeutics can achieve their ultimate clinical potential to treat widespread human disease, the rate-limiting delivery problem of endosomal escape must be solved in a clinically acceptable manner.
Keywords: RNA therapeutics, siRNA, endosomal escape, delivery, ASO
Introduction to the Macromolecular RNA Therapeutic Delivery Problem
Oligonucleotide RNA therapeutics, including siRNAs, charged phosphorothioate (PS) antisense oligonucleotides (ASOs), and neutral phosphorodiamidate morpholino oligomer and peptide nucelic acid ASOs, as well as other oligonucleotides, have great potential to selectively treat a multitude of human diseases, from cancer to COVID to Parkinson's disease. Currently, there are 13 FDA-approved ASOs and siRNA therapeutics targeting cells in the liver, central nervous system (CNS), and muscle [1–3].
Based on the number of late-stage clinical trials, there is an expectation of 15 or more additional approvals over the next 5 years. RNA therapeutics have been shown to knock down genes (siRNAs and PS ASOs) and alter mRNA splicing patterns (PS ASOs and PMO ASOs) in the clinic. There are also preclinical data to support a weaker, but significant, gene knockup mechanism of action of small activating RNAs (saRNAs and ASOs) [1,3].
All of this has been possible due to 40+ years of stellar chemistry resulting in dramatic improvements in metabolic stability, decreased off-target effects, and elimination of innate immune activation, while increasing the delivery potential of RNA therapeutics (Table 1) [4,5]. Indeed, a single dose of inclisiran, an FDA-approved siRNA that targets PCSK9 to treat hypercholesterolemia, results in a 6-month pharmacodynamic (PD) response in patients [6].
Table 1.
Challenges for siRNA and ASO Delivery
| Challenge | siRNA | PS ASO |
|---|---|---|
| Metabolic stability | √ | √ |
| Innate immune activation | √ | √ |
| Off-target effects | √ | √ |
| Toxicity | √ | √ |
| Long duration of response | √ | √ |
| Endosomal escape | X | X |
√ = solved with chemistry; X = not yet solved.
PS, phosphorothioate; ASO, antisense oligonucleotide.
Based largely on the clinical successes of the chemistry, there has been an explosion of RNA therapeutic biotech startups launched in the last several years with ginormous rounds of funding [7].
The mechanism of action of RNA therapeutics relies solely on Watson–Crick base pairing with their cognate target gene RNA, thereby requiring only target gene validation without any prerequisite knowledge of structure or function [4,5]. Impressively, RNA therapeutics have the ability to pharmaco-evolve their sequence to keep pace with mutations in diseases driven by genetic change, including cancer and COVID , a property that is absent from all other clinical modalities.
Not surprisingly, most RNA therapeutics have very limited to no ability to enter cells [1,4]. Indeed, cells have been evolving defense mechanisms to prevent macromolecules, especially RNAs, from entering them for over a billion years [4,8]. Consequently, starting from the dawn of the RNA therapeutic revolution 40+ years ago [9], The Delivery Problem has remained the 800-pound gorilla in the room that needs to be solved before RNA therapeutics can achieve their ultimate clinical potential to treat widespread human diseases.
Small-molecule therapeutics with an intracellular mechanism of action are designed and selected for their ability to not only hit their cognate target but also to passively diffuse across the cell membrane [10,11]. The majority of small-molecule therapeutics are under 500 Da in size, have no charged atoms, and are moderately lipophilic (log P), and they were succinctly summarized in 2001 as Lipinski's rule of five [10,11].
In contrast to small-molecule therapeutics, macromolecular RNA therapeutics are too large, too charged, and/or too hydrophilic to passively diffuse across the cell membrane [4,5]. Indeed, a typical siRNA is >12 kDa in size, has >40 anionic charged phosphodiester (PO) linkages, and has a highly hydrophilic log P value <1. Likewise, a typical PS ASO is 5–8 kDa in size, has >16 anionic charged PS linkages, and a hydrophilic log P value <1.
Consequently, in contrast to small-molecule therapeutics, RNA therapeutics are unable to passively diffuse across the cell membrane and are instead taken up into cells by endocytosis [1–5]. However, similar to the cell membrane, endosomes comprise a lipid bilayer. Therefore, even in semipermissive tissues, such as liver hepatocytes, neurons of the CNS, and skeletal muscle, endosomes entrap 99+% of RNA therapeutics [4,12–14].
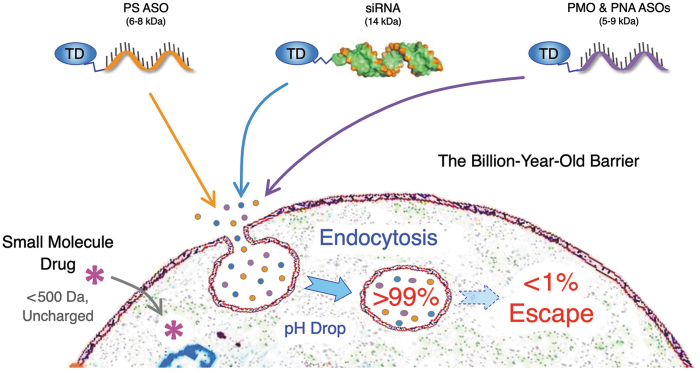
Thus, endosomal escape is The Delivery Problem (Fig. 1).
FIG. 1.
RNA therapeutics, including PS ASOs, siRNAs, PMO ASOs, PNA ASOs, and other oligonucleotides, are too large, too charged, and/or too hydrophilic to passively diffuse across the billion-year-old cell membrane lipid bilayer, but instead require a TD to drive uptake by endocytosis. However, endosomes also comprise a lipid bilayer, and 1% or less of endocytosed RNA therapeutics escape the endosome. Thus, endosomal escape is the rate-limiting delivery step. TD, targeting domain; PS, phosphorothioate; ASO, antisense oligonucleotide.
The Magnitude of the Endosomal Escape Problem
Naked (unconjugated) RNA therapeutics are inefficiently taken up into cells by a variety of endocytotic mechanisms [1,4,14,15]. However, conjugation of a targeting domain such as N-acetylgalactosamine (GalNAc), which binds to the highly expressed liver hepatocyte, asialoglycoprotein receptor (ASGPR) [16], enhances the amount of endocytosed PS ASO 10-fold [17]. Due to their nonprotein-binding anionic charged PO backbone, the vast majority (>95%) of naked siRNAs are cleared by first-pass renal filtration [1,4]. In contrast, 50%–70% of the administered dose of GalNAc-conjugated siRNAs is endocytosed by liver hepatocytes [18].
However, while targeting domains, such as GalNAc, dramatically increases the amount of RNA therapeutics inside endosomes, they do not directly impact the rate of endosomal escape. Recent work from Jiang's group using quantitative NanoSIMS microscopy found that only 1%–2% of GalNAc-PS ASO conjugates escape endosomes in vivo [13]. Likewise, using a highly quantitative approach, Brown et al. found that only <0.3% of the endocytosed siRNAs escape from endosomes in vivo [12].
While endosomally trapped siRNAs serve as a “depot effect” that results in a single siRNA dose achieving very long PD responses due to a constant low level of siRNA leakage into the cytoplasm over months [12], this notion also belies the fact that 99.7% of endocytosed siRNAs never escape into the cytoplasm and fail to engage their target gene RNA.
Thus, the lack of endosomal escape is the singular biggest problem preventing the widespread use of RNA therapeutics for treating cancer, COVID, and a multitude of other diseases.
Mechanisms of Endosomal Escape
Sooner or later, a very small amount of macromolecular RNA therapeutics will cross the endosomal lipid bilayer to enter the cytoplasm of permissive cells, such as liver hepatocytes, neurons of the CNS, and skeletal muscle. That much is certain. However, irrespective of how RNA therapeutics enter endosomes, there is a paucity of rigorous data available on the mechanism(s) that siRNAs or ASOs use to productively escape into the cytoplasm. Likewise, there is no understanding as to why these seemingly permissive cells are in fact permissive.
Unlike insects, mammals do not have functional RNA transporters [4]. Previous studies have estimated that ∼2,000 cytoplasmic siRNAs are required for a maximal RNAi response [19], whereas PS ASOs require ∼50,000 for maximal activity [20]. In the clinics, single-dose GalNAc-siRNA conjugates require 2–3 weeks before achieving a maximal RNAi response [6], giving us a back-of-the-envelope calculation of, on average, ∼5 siRNAs escaping per hour. This is the reason why it takes 2–3 weeks to achieve a maximal RNAi response, which is how the depot effect works to constantly, but ever so slowly, reload new Ago2 complexes over 6 or more months.
As for the putative endosomal escape mechanism(s), there are no rigorous data determining how RNA therapeutics enter the cytoplasm. However, like oil on water, when perturbed, lipid bilayers spontaneously “heal” to close the hole. We speculate that evolution may not have completely selected against rare, very short duration (ns scale), and small-diameter (<10 nm) lipid bilayer perturbations in endosomes, perhaps because there was no selection against small RNAs capable of rapidly traversing such a small hole and they did not carry enough genetic information or catalytic activity to be harmful.
Alternatively, as endosomes mature, they fuse to form multivesicular bodies (MVBs) as well as fuse with lysosomes [14]. During the fusion events, there is an inherent breach of the lipid bilayer, which could allow localized RNA therapeutics to escape. Moreover, retro-transport to the Golgi may also place RNA therapeutics in an environment where breaching across the lipid bilayer may be more permissible [14]. Retro-transport has not been thoroughly investigated.
Determining the mechanism of action that allows macromolecular RNA therapeutics to escape across the endosomal lipid bilayer and enter the cytoplasm could open up a significant avenue for therapeutic manipulation. However, when only 1% or less of a given RNA therapeutic escapes over the course of two to three weeks, it is extremely difficult to biochemically ascertain where and when the escape actually occurred.
Indeed, experimental manipulation by inhibitors or knockdown of endolysosomal system genes often results in significant cell biological perturbations that may or may not actually identify genes and pathways involved in endosomal escape in unperturbed cells in vivo. Likewise, RNA therapeutics conjugate to hydrophobic, aromatic fluorescent dyes and dramatically alter the RNA's overall biophysical properties.
Moreover, microscopically following a 0.3% endosomal escape of an siRNA-dye conjugate is no easy task. In our opinion, the most significant methodological advancement in dissecting endosomal escape mechanisms has been the recent utilization of quantitative NanoSIMS microscopy that merely requires substituting Br atoms on the RNA (no dyes) [13]. However, it will take multiple nonperturbing methodologies to identify the endosomal escape mechanism(s).
Current Approaches to Enhance the Endosomal Escape
Given the magnitude of the endosomal escape problem, it is not surprising that there have been a wide variety of approaches over the last four decades to enhance delivery, and hence endosomal escape, of RNA therapeutics (Fig. 2). However, significant enhancements of endosomal escape more often than not come at the expense of cytotoxicity. Indeed, it has been extremely difficult to uncouple the two.
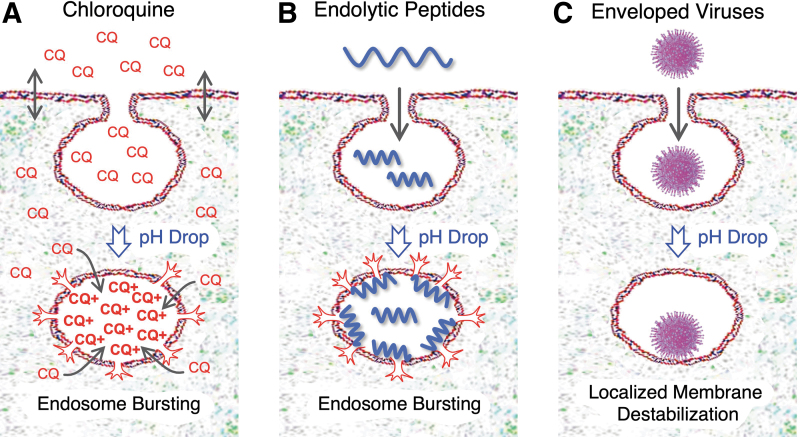
FIG. 2.
Endosomal escape approaches. (A) Small-molecule endolytic agents, such as chloroquine (CQ), are membrane permeable, but become protonated, positively charged (CQ+), and trapped inside the low pH environment of an endosome, resulting in a 1,000-fold or more concentration increase that leads to endosomal bursting. (B) Endolytic peptides and polymers are often codelivered with RNA therapeutics in 10- to 30-fold molar excess and ultimately disrupt the endosomal lipid bilayer membrane, leading to endosomal bursting. (C) Enveloped viruses contain fusogenic peptide domains that are selectively activated in the low pH of endosomes, causing localized endosomal membrane disruption (not endosomal bursting). Adapted from the study by Dowdy [4].
PS backbones
The PS linkage is the mainstay of PS ASOs and the termini of siRNA strands. PS linkages add significant metabolic stability and resistance to RNases [5,15]. While the lone pair of electrons in a PO linkage are in resonance equally between the two nonbridging oxygen atoms, in a PS linkage, the electrons reside 97% of the time on the sulfur atom [5,15]. Because the sulfur atom has a larger outer electron shell than oxygen, the lone pair of electrons has to travel greater distances and this is thought to translate biochemically into a more hydrophobic atomic surface that is capable of binding proteins, whereas the nonbridging oxygen atoms of a PO linkage do not bind proteins [4,5,15].
The enhanced protein binding of PS linkages not only increases the PS ASO's pharmacokinetics by binding to serum proteins, such as albumin, but it also allows PS ASOs to bind to cell surface proteins and thereby either stimulate endocytosis or be passively taken up into endosomes [15]. While a full PS backbone ASO is a critical design component and facilitates endosomal escape, unfortunately only 1%–2% of PS ASOs escape the endosome [13].
The addition of single-stranded PS tails on the guide strand of dimer siRNAs (di-siRNAs) has also enhanced delivery for CNS disorders [21], although it remains unclear how much of this effect is due to increased residency in the cerebrospinal fluid of the larger di-siRNA, effectively giving it more shots on goal, versus monomeric siRNAs and/or due to increased endosomal escape from the PS tails [21].
Cholesterol, lipid tails, and saponins
Conjugation of hydrophobic cholesterol or alkyl lipid tails to siRNAs and PS ASOs has been a pillar for enhancing PK, endocytosis, and endosomal escape of RNA therapeutics for several decades [1–5,22]. Indeed, lipidation of PS ASOs can result in a fivefold enhanced endosomal escape [23]. Cholesterol and lipid tails bind to serum lipid particles and proteins, resulting in an increased PK and delivery, especially to the liver and CNS [21,24].
The proposed mechanism of action is that the hydrophobic lipid tail buries itself into the endosomal lipid bilayer. This concentrates the RNA conjugate on the luminal surface of the endosomal membrane, followed by unknown rare membrane-disruptive events, resulting in the conjugate flipping into the cytoplasm. Saponins, such as digitonin, are natural product nonionic detergents that avidly bind cholesterol and have been used for decades to permeabilize mammalian cell membranes to make lysates [25]. Saponins have been used to enhance endosomal escape of coadministered PS ASOs [26] and they are undergoing further optimization for targeted delivery and endosomal selective activation [27].
However, currently, the mechanism of endosomal escape remains unclear. One of the downsides of hydrophobic conjugates is that their biophysical properties tend to dominant the overall PK of the conjugate and negate the ability to incorporate highly selective targeting domains, such as monoclonal antibodies, nanobodies, DARPins, centyrins, and others, for systemic delivery.
All things considered, lipid conjugates are currently at the top of the list as a usable endosomal escape approach for local delivery into the CNS and eye as well as the liver.
Small-molecule endolytics
Some of the earliest work showing the ability to enhance endosomal escape was performed by addition of the small-molecule endolytic, chloroquine, an antimalarial drug [28,29]. Due to two ionizable nitrogen atoms, chloroquine preferentially concentrates 1,000-fold or more in the low pH of endosomes (charged small molecules cannot passively diffuse across lipid bilayers), followed by insertion of its hydrophobic bicyclic aromatic rings into the lipid bilayer, resulting in endosomal lysis [29,30].
While coadministration of chloroquine with RNA therapeutics enhances their endosomal escape, this comes with a high price of cytotoxicity as at the effective concentration, many endosomes/lysosomes undergo lysis [4,30]. Likewise, newer small-molecule endolytics such as Retro-1 [31] and guanabenz [32], which enhance endosomal escape of RNA therapeutics, also have been associated with cytotoxicity concerns, whereas UNC7938 (with two ionizable nitrogen atoms and four hydrophobic aromatic rings) has recently shown significantly enhanced endosomal escape of a PMO in the lungs of mouse models [33].
While targeted endosomal delivery of siRNA conjugates with nigericin, a hydrophobic ionophore that catalyzes the exchange of K+ for H+, can enhance endosomal escape, it does so by bursting the entire endosome [34,35], which is not a clinically acceptable path forward. Overall, due to the uncontrollable problem of hitting many endosomes in many cell types and tissues, regardless of whether or not they contain RNA therapeutic cargo, we do not foresee a clinical development path going forward for using small-molecule endolytic agents, especially those whose mechanism of action results in endosomal bursting and release of the entire endosomal contents into the cytoplasm.
Endolytic peptides
Multiple approaches to addressing the endosomal escape problem have been built off of viral peptides and insect toxins [14]. At its critical concentration, melittin, the active peptide in bee venom, which comprises N-terminal hydrophobic residues and C-terminal cationic residues, forms a pore through the cell membrane and has been heavily investigated for use as an RNA therapeutic endosomal enhancer [36]. While melittin is extremely effective at forming pores, it is far too toxic, too antigenic, and too uncontrollable for use in clinics.
Moreover, due to its cationic charge, melittin aggregates anionic RNA therapeutics, as do all cationic peptides. Endoporter, an amphipathic peptide comprising ionizable His and Leu residues, has also been extensively used to enhance delivery of RNA therapeutics in cell culture [37]. Although it points the direction forward, due to toxicity (although dramatically less than melittin) and requirement for >10-fold molar excess, endoporter is not an acceptable solution for clinical applications.
The most impressive endolytic peptide for enhancing endosomal escape of RNA therapeutics has been a derivative of the influenza fusogenic hemagglutinin 2 (HA2) peptide [38], called INF7 [39]. Brown et al. showed that the GalNAc-INF7 endolytic peptide could enhance endosomal escape of a coadministered GalNAc-siRNA 20-fold in vivo, but this still resulted in less than 0.2% total escape [12].
However, this required a 30-fold molar excess of GalNAc-INF7 endolytic peptide and came at the expense of unacceptable cytotoxicity. Moreover, there remains concern for an adaptive immune response to all endolytic peptides, thereby restricting the number of times the molecule could be used in a specific patient. Consequently, we do not foresee a clinical path forward for the use of this generation of endosome-bursting endolytic peptides to enhance delivery of RNA therapeutics.
Our own work on endosomal escape peptides has produced both insights and mixed results. The highly cationic guanidinium group on Arg residues of the transActivator of Transcription (TAT) protein transduction domain peptide, also called a cell-penetrating peptide, stimulates endocytosis and facilitates endosomal escape [40,41]. However, all guanidinium-containing escape domains, including TAT, are extremely sticky and adhere to cell surfaces, anionic plasma components, including glycosides, proteins, lipids, and nucleic acids, including anionic RNA therapeutics [42]. Therefore, cationic peptides have predominantly been successfully used in vivo only with neutral backbone PMO or PNA oligonucleotides [43].
While investigating the endosomal delivery mechanism of TAT, we showed a ∼10-fold improved delivery of a tester TAT-Cre recombinase protein (37 kDa) into cells when it was codelivered with the TAT-HA2 peptide that directs the influenza HA2 endosomal escape peptide into the same endosomal compartments [43]. To avoid potential concerns for HA2 immunogenicity, we synthesized and screened a library of short, hydrophobic, endosomal escape domain (EED) peptides <6 amino acids, which is below the length required for an adaptive immune response [30].
We performed a GFP complementation screen with EED peptides by linking them to a TAT peptide that was conjugated through a disulfide bond to the beta-11 GFP peptide fragment [30]. None of the alkyl residue EED peptides showed increased activity, whereas EED peptides containing two or three hydrophobic aromatic Trp and/or Phe residue combinations showed an approximately eightfold enhanced endosomal escape [30].
Unfortunately, due to too much solution-exposed hydrophobicity, Trp/Phe-containing EEDs conjugated to siRNAs underwent a hydrophobic collapse, whereby the aromatic residues aggregated and precipitated the conjugate molecule, resulting in significant cell surface toxicity. While these results identified a potential direction to enhance endosomal escape, significant additional chemistry improvements to mask the hydrophobicity are needed to address hydrophobic collapse and hence cytotoxicity.
Dynamic polyconjugate and ionizable lipid nanoparticles
Building on earlier work with the melittin peptide, Rozema et al. developed a ∼50-kDa endolytic dynamic polyconjugate (DPC) polymer containing cationic primary amines, hydrophobic butyl groups, GalNAc-targeting domains, and siRNAs [44]. The primary amines are masked (neutralized) with PEG through an acid-labile maleic anhydride bond that is stable in plasma (pH ∼7.4), but unstable in the low pH (<6) of endosomes.
The result is an endosomal selective conversion of neutral DPC into a cationic, active endolytic polymer inside endosomes. DPC was further refined to replace the somewhat unstable pH bond for a more stable cathepsin B protease cleavable bond [45]. Unfortunately, similar to small-molecule endolytic agents, the DPC mechanism of action results in lysis of the entire endosome, releasing all of its contents into the cytoplasm of the cell. Guidry et al. also determined that after DPC escapes or bursts the endosome, it can then retro-escape from the cytoplasm across the cell membrane lipid bilayer back out of the cell, resulting in significant cytotoxicity [46].
The pH-sensitive DPC-siRNA was tested in several clinical trials against HBV (ARC-520); however, it was ultimately dropped due to an FDA clinical hold based on deaths in a primate toxicology study [47]. Overall, DPC is an interesting endosome-activated escape approach, but requires further refinement into a form that initiates only localized endosomal membrane disruption and not endosomal bursting.
Patisiran, the first FDA-approved siRNA, targets the transthyretin (TTR) gene and is delivered in an ionizable (MC3) lipid nanoparticle (LNP) [2]. Mechanistically similar to the selective unmasking approach of DPCs, the surfaces of ionizable lipid LNPs are effectively neutral in plasma, but due to an ideal pKa of 6.4, they become selectively cationic inside endosomes [2]. There are no cationic lipids in nature.
Therefore, ionized cationic lipids are thought to ion-pair with endosomal anionic lipids to form a cone structure that destabilizes lipid bilayers by driving transition into an inverted hexagonal HII phase, resulting in endosomal escape of the LNP into the cytoplasm [48]. However, due to the advent of extreme metabolic stabilizing chemistry combined with their small size (<14 kDa) and conjugation of targeting domains, it is doubtful that there will be another LNP-delivered siRNA (or ASO) taken into the clinics. Indeed, vutrisiran, a GalNAc-siRNA-targeting TTR that has finished phase III clinical trials and is being evaluated for FDA approval, is due to replace patisiran [49].
In contrast to relatively small siRNAs and ASOs, much larger mRNA vaccines (∼1 megaDa) and nonviral DNA vectors (∼3 megaDa) require LNPs (∼100 megaDa) for protection from degradative enzymes, avoidance of the innate immune system, and to facilitate endosomal escape [1,2]. Similar to DPC, the key development of ionizable LNPs is the masking in plasma and their selective activation inside endosomes (due to their ∼6.4 pKa), which drives localized endosomal membrane disruption, not endosomal bursting [50].
The Enveloped Virus Endosomal Escape Mechanism—Evolutionary Perfection
Certainly, the most impressive and perhaps the most efficient mechanism that nature has evolved to escape the endosome is that of enveloped viruses [51]. Enveloped viruses are similar in size to LNPs, are taken up into cells by endocytosis, and require an endosomal escape mechanism. In contrast to the dismal <1% endosomal escape of RNA therapeutics, enveloped viruses have a whopping 30%–70% endosomal escape efficiency [52,53]. The most well studied of the enveloped viral escape mechanisms is that of the HA protein from influenza virus [53]. HA is a homotrimer comprising two domains: an outer HA1 receptor-binding domain that masks an inner HA2 fusogenic domain [54].
HA1 and HA2 are proteolytically cleaved from each other during viral production, but remain tightly bound to each other. HA1 binding to its target sialic acid glycoprotein receptors on the cell surface stimulates endocytotic uptake of the virus [54]. The low pH (<6) of endosomes causes a gross conformational change that results in HA1 shedding and exposes HA2's fusogenic peptide that buries itself into the endosomal lipid bilayer, thereby driving membrane fusion and escape. SARS-CoV-2's spike (S) protein has evolved a similar endosomal escape mechanism utilizing S1 and S2 domains [55].
Thus, nature's most evolutionarily efficient endosomal escape mechanism drives localized endosomal membrane disruption, not endosomal bursting.
How to Pull off the Great Endosomal Escape!
The endosomal escape of RNA therapeutics remains the problem to solve before we can begin to tackle the myriad of currently undruggable diseases. Depot effects, where >99+% of the RNA therapeutic remains trapped inside an endosome, are preventing us from treating cancer, pandemic virus, and Parkinson's disease patients with RNA therapeutics. We believe that an effective depot of 50% or even 20% or less of a slow-releasing RNA therapeutic would suffice for maintaining long PD responses, thereby freeing up 50% or more of the RNA therapeutic designated for rapid, enhanced endosomal escape.
While it is easy to escape the endosome with endolytic agents that cause the endosome to burst, the hard part is achieving significant levels of escape without killing the cell or causing toxicities in the process. With that critical balance in mind, of all the natural approaches that have evolved to escape endosomes, we believe that the HA and spike mechanism of enveloped viruses causing localized endosomal membrane disruption presents the best potential path forward for dramatically enhancing the delivery of RNA therapeutics in a nontoxic clinically acceptable manner.
Unfortunately, because HA and spike are large proteinaceous machines as well as the immunogens for vaccines, we cannot merely conjugate them onto RNA therapeutics. Likewise, even if it were possible to dramatically reduce their size with peptide mimetics while maintaining a high endosomal selective escape function (which has yet to be done), these peptides would still potentially be recognized by the adaptive immune system.
To master the endosomal escape of RNA therapeutics, we need a novel EED chemical entity with the following properties:
-
(1)
It achieves a 10-fold or greater enhanced endosomal escape that is tunable for different disease indications and RNA therapeutics.
-
(2)
It covalently attaches to the RNA therapeutic, thereby avoiding acting on non-RNA cargo-containing endosomes.
-
(3)
It is masked and/or inert in plasma with a foolproof endosomal activation mechanism.
-
(4)
The escape mechanism is localized endosomal membrane disruption, not endosome bursting.
-
(5)
It is metabolically stable to prevent premature activation and degradation.
-
(6)
It does not biophysically dominate or interfere with the targeting domain.
-
(7)
It is not of a peptide or protein origin, thereby allowing repetitive dosing and avoidance of the adaptive immune system.
-
(8)
By-products are nontoxic at the cellular and systemic levels.
-
(9)
It is capable of undergoing large-scale synthesis for eventual clinical trials.
Fortunately for us, the entire history of success in developing RNA therapeutics was built on the backs of chemists. They are the undisputed heroes of this story. Therefore, to pull off the Great Endosomal Escape, we need them to synthesize new and unique EEDs that incorporate all of these properties.
Acknowledgments
The authors greatly appreciate the input and help of their colleagues: C. Bradshaw, C. Brown, M. Manoharan, M. Maier, L. Sepp-Lorenzino, P. Seth, and M. Stanton.
Author Disclosure Statement
S.F.D. is a member of the Scientific Advisory Boards of Ceptur Therapeutics, Deep Genomics, Generation Bio, Korro Bio, and NeuBase Therapeutics and cofounder of Clear Skies Bio.
Funding Information
This work was supported by Aligning Science Across Parkinson's (The Michael J. Fox Foundation), National Institutes of Health (NCI and NINDS), and Ono Foundation. R.L.S. was supported by an NCI CBIO training grant. The grant numbers are NCI = R21 CA267883, NINDS = R21 NS116663.
References
- 1. Hammond SM, Aartsma-Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, Covello G, Denti MA, Desviat LR, et al. (2021). Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol Med 13:e13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kulkarni JA, Witzigmann D, Thomson SB, Chen S, Leavitt BR, Cullis PR and van der Meel R. The current landscape of nucleic acid therapeutics. (2021). Nat Nanotechnol 16:630–643. [DOI] [PubMed] [Google Scholar]
- 3. Corey DR, Damha MJ and Manoharan M. (2022). Challenges and opportunities for nucleic acid therapeutics. Nucleic Acid Ther 32:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dowdy SF. (2017). Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 35:222–229. [DOI] [PubMed] [Google Scholar]
- 5. Khvorova A and Watts JK. (2017). The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fitzgerald K, White S, Borodovsky A, Bittencourt BR, Strahs A, Clausen V, Wijngaard P, Horton JD, Taubel J, et al. (2017). A Highly Durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med 376:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Philippidis Top 10 RNA-Based Biopharmas A.. Genetic Engin. & Biotech News. www.genengnews.com/topics/omics/top-10-rna-based-bio-pharmas/ Accessed April 21, 2022.
- 8. Dowdy SF and Levy M. (2018). RNA therapeutics (Almost) comes of age: Targeting, delivery and endosomal escape. Nucleic Acid Ther 28:107–108. [DOI] [PubMed] [Google Scholar]
- 9. Zamecnik PC and Stephenson ML. (1978). Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A 75:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipinski CA, Lombardo F, Dominy BW and Feeney PJ. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. [DOI] [PubMed] [Google Scholar]
- 11. Lipinski CA. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341. [DOI] [PubMed] [Google Scholar]
- 12. Brown CR, Gupta S, Qin J, Racie T, He G, Lentini S, Malone R, Yu M, Matsuda S, et al. (2020). Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res 48:11827–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He C, Migawa MT, Chen K, Weston TA, Tanowitz M, Song W, Guagliardo P, Iyer KS, Bennett CF, et al. (2021). High-resolution visualization and quantification of nucleic acid-based therapeutics in cells and tissues using Nanoscale secondary ion mass spectrometry (NanoSIMS). Nucleic Acids Res 49:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Juliano RL. (2018). Intracellular trafficking and endosomal release of oligonucleotides: what we know and what we don't. Nucleic Acid Ther 28:166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crooke ST, Baker BF, Crooke RM, and XH Liang. (2021). Antisense technology: an overview and prospectus. Nat Rev Drug Discov 20:427–453. [DOI] [PubMed] [Google Scholar]
- 16. Morell AG, Gregoriadis G, Scheinberg IH, Hickman J and Ashwell G. (1971). The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem 246:1461–1467. [PubMed] [Google Scholar]
- 17. Prakash TP, Graham MJ, Yu J, Carty R, Low A, Chappell A, Schmidt K, Zhao C, Aghajan M, et al. (2014). Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetylgalactosamine improved potency 10-fold in mice. Nuc Acid Res 42:8696–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel'in AV, et al. (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 19. Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T and Lieberman J. (2015). Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol 33:870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buntz A, Killian T, Schmid D, Seul H, Brinkmann U, Ravn J, Lindholm M, Knoetgen H, Haucke V and Mundigl O. (2019). Quantitative fluorescence imaging determines the absolute number of locked nucleic acid oligonucleotides needed for suppression of target gene expression. Nucleic Acids Res 47:953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alterman JF, BGodinho MDC, Hassler MR, Ferguson CM, Echeverria D, Sapp E, Haraszti RA, Coles AH, Conroy F, et al. (2019). A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat Biotechnol 37:884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soutschek. (2004). Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432:173–178. [DOI] [PubMed] [Google Scholar]
- 23. Wang S, Allen N, Prakash TP, Liang XH and Crooke ST. (2019). Lipid conjugates enhance endosomal release of antisense oligonucleotides into cells. Nucleic Acid Ther 29:245–255. [DOI] [PubMed] [Google Scholar]
- 24. Biscans A, Coles A, Haraszti R, Echeverria D, Hassler M, Osborn M and Khvorova A. (2019). Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res 47:1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fiskum G, Craig SW, Decker GL and Lehninger AL. (1980). The cytoskeleton of digitonin-treated rat hepatocytes. Proc Natl Acad Sci U S A 77:3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang M, Wu B, Shah SN, Lu P and Lu Q. (2018). Saponins enhance exon skipping of 2′-O-methyl phosphorothioate oligonucleotide in vitro and in vivo. Drug Des Devel Ther 12:3705–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sapreme Technologies. https://sapreme-technologies.com Accessed April 21, 2022.
- 28. Juliano RL, Wang L, Tavares F, Brown EG, James L, Ariyarathna Y, Ming X, Mao C and Suto M. (2018). Structure-activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides. Nucleic Acids Res 46:1601–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Juliano RL. (2021). Chemical manipulation of the endosome trafficking machinery: implications for oligonucleotide delivery. Biomedicines 9:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lönn P, Kacsinta AD, Cui XS, Hamil AS, Kaulich M, Gogoi K and Dowdy. SF (2016). Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci Rep 6:32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ming X, Carver K, Fisher M, Noel R, Cintrat JC, Gillet D, Barbier J, Cao C, Bauman J and Juliano RL. (2013). The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Res 41:3673–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Osborn MF, Alterman JF, Nikan M, Cao H, Didiot MC, Hassler MR, Coles AH and Khvorova A. (2015). Guanabenz (Wytensin™) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res 43:8664–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dang Y, van Heusden C, Nickerson V, Chung F, Wang Y, Quinney NL, Gentzsch M, Randell SH, Moulton HM, et al. (2021). Enhanced delivery of peptide-morpholino oligonucleotides with a small molecule to correct splicing defects in the lung. Nucleic Acids Res 49:6100–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rangasamy L, Chelvam V, Kanduluru AK, Srinivasarao M, Bandara NA, You F, Orellana EA, Kasinski AL and Low PS. (2018). New mechanism for release of endosomal contents: osmotic lysis via nigericin-mediated K+/H+ exchange. Bioconjug Chem 29:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orellana EA, Abdelaal AM, Rangasamy L, Tenneti S, Myoung S, Low PS and Kasinski AL. (2019). Enhancing MicroRNA activity through increased endosomal release mediated by nigericin. Mol Ther Nucleic Acids 16:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Houa KK, Panb H, Schlesingerc PH and Wickline SA. (2015). A role for peptides in overcoming endosomal entrapment in siRNA delivery–a focus on melittin. Biotechnol Adv 33:931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tesz GJ, Aouadi M, Prot M, Nicoloro SM, Boutet E, Amano SU, Goller A, Wang M, Guo CA, et al. (2011). Glucan particles for selective delivery of siRNA to phagocytic cells in mice. Biochem J 436:351–362. [DOI] [PubMed] [Google Scholar]
- 38. Oliveira S, van Rooy I, Kranenburg O, Storm G and Schiffelers RM. (2007). Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int J Pharm 331:211–214. [DOI] [PubMed] [Google Scholar]
- 39. Mastrobattista E, Koning GA, van Bloois L, Filipe AC, Jiskoot W and Storm G. (2002). Functional characterization of an endosome-disruptive peptide and its application in cytosolic delivery of immunoliposome-entrapped proteins. J Biol Chem 277:27135–27143. [DOI] [PubMed] [Google Scholar]
- 40. Schwarze SR, Ho A, Vocero-Akbani A and Dowdy SF. (1999). In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285:1569–1572. [DOI] [PubMed] [Google Scholar]
- 41. Wadia JS, Stan RV and Dowdy SF. (2004). Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med 10:310–315. [DOI] [PubMed] [Google Scholar]
- 42. Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, et al. (2014). Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol 32:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gait MJ, Arzumanov AA, McClorey G, Godfrey C, Betts C, Hammond S and Wood MJA. (2019). Cell-penetrating peptide conjugates of steric blocking oligonucleotides as therapeutics for neuromuscular diseases from a historical perspective to current prospects of treatment. Nucleic Acid Ther 29:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, et al. (2007). Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA 104:12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rozema DB, Blokhin AV, Wakefield DH, Benson JD, Carlson JC, Klein JJ, Almeida LJ, Nicholas AL, Hamilton HL, et al. (2015). Protease-triggered siRNA delivery vehicles. J Control Release 209:57–66. [DOI] [PubMed] [Google Scholar]
- 46. Guidry EN, Farand J, Soheili A, Parish CA, Kevin NJ, Pipik B, Calati KB, Ikemoto N, Waldman JH, et al. (2014). Improving the in vivo therapeutic index of siRNA polymer conjugates through increasing pH responsiveness. Bioconjug Chem 25:296–307. [DOI] [PubMed] [Google Scholar]
- 47. Arrowhead Pharmaceuticals. Study of ARC-520 in Participants With Hepatitis Virus e Antigen B(HBeAg) Positive Chronic Hepatitis Virus B. https://clinicaltrials.gov/ct2/show/NCT02452528 Accessed April 21, 2022.
- 48. Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, et al. (2010). Rational design of cationic lipids for siRNA delivery. Nat Biotechnol 28:172–176. [DOI] [PubMed] [Google Scholar]
- 49. Alnylam Pharmaceuticals. Vutrisiran. www.alnylam.com/alnylam-rnai-pipeline/ Accessed April 21, 2022.
- 50. Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, et al. (2013). Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol 31:638–646. [DOI] [PubMed] [Google Scholar]
- 51. Hernandez LD, Hoffman LR, Wolfsberg TG and White JM. (1996). Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol 12:627–661. [DOI] [PubMed] [Google Scholar]
- 52. Lagache T, Danos O and Holcman D. (2012). Modeling the step of endosomal escape during cell infection by a nonenveloped virus. Biophys J 102:980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Staring J, Raaben M and Brummelkamp. TR (2018). Viral escape from endosomes and host detection at a glance. J Cell Sci 131:jcs216259. [DOI] [PubMed] [Google Scholar]
- 54. Russell CJ, Hu M and Okda FA. (2018). Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol 26:841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jackson CB, Farzan M, Chen B and Choe H. (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]