Abstract
A male mutation bias is observed across vertebrates, and, where data are available, this bias is accompanied by increased per-generation mutation rates with parental age. While continuing mitotic cell division in the male germline post puberty has been proposed as the major cellular mechanism underlying both patterns, little direct evidence for this role has been found. Understanding the evolution of the per-generation mutation rate among species requires that we identify the molecular mechanisms that change between species. Here, we study the per-generation mutation rate in an extended pedigree of the brown (grizzly) bear, Ursus arctos horribilis. Brown bears hibernate for one-third of the year, a period during which spermatogenesis slows or stops altogether. The reduction of spermatogenesis is predicted to lessen the male mutation bias and to lower the per-generation mutation rate in this species. However, using whole-genome sequencing, we find that both male bias and per-generation mutation rates are highly similar to that expected for a non-hibernating species. We also carry out a phylogenetic comparison of substitution rates along the lineage leading to brown bear and panda (a non-hibernating species) and find no slowing of the substitution rate in the hibernator. Our results contribute to accumulating evidence that suggests that male germline cell division is not the major determinant of mutation rates and mutation biases. The results also provide a quantitative basis for improved estimates of the timing of carnivore evolution.
Keywords: male mutation bias, life history, spermatogenesis, genomics
Significance.
Males often transmit more mutations to their offspring than females, which is thought to be due to the higher number of cell divisions that males undergo before having children. Here, we have examined the number of mutations passed on to children by male and female grizzly bears, who hibernate for one-third of the year. We find that male grizzly bears pass five times as many mutations to their offspring as female grizzly bears, even though cell division is thought to greatly slow down during hibernation. These results cast further doubt on cell division as the major cause of the difference between sexes.
Introduction
The per-generation mutation rate evolves between species. Whole-genome sequencing has revealed that the mutation rate per generation varies by several orders of magnitude across eukaryotes (Lynch 2010) and by at least two-fold among mammals (Chintalapati and Moorjani 2020; Wang et al. 2022). Prior to the advent of large-scale DNA sequencing, early studies of disease mutations in humans uncovered two general patterns in the accumulation of mutations. First, parental age was found to be positively correlated with the probability of observing a de novo mutation: older parents were more likely to have children with inherited diseases (Weinberg 1912; Risch et al. 1987). Second, advanced paternal age better predicted the appearance of disease mutations than advanced maternal age—implying that mutation was male biased (Haldane 1946; Penrose 1955). Both of these patterns have been confirmed by studies that have sequenced large numbers of human pedigrees (Kong et al. 2012; Goldmann et al. 2016; Rahbari et al. 2016; Jónsson et al. 2017), as well as by studies in other mammals (Venn et al. 2014; Thomas et al. 2018; Besenbacher et al. 2019; Lindsay et al. 2019; Wang et al. 2020, 2022; Wu et al. 2020; Bergeron et al. 2021).
The main mechanism proposed to explain both the parental age effect and male-biased mutation is the continuing replication of the male germline in mammals. After a relatively fixed number of mitotic divisions during germline development before puberty in both sexes, the male germline continues to undergo mitotic cell division post puberty (Drost and Lee 1995). Although the polymerases responsible for genome replication have very low error rates (McElhinny et al. 2010a, 2010b), the cell lineage leading to any given spermatozoon can go through hundreds of spermatogenic cell divisions. This difference in the contribution of mutations between males and females informs evolutionary models of the mutation rate, which usually combine a period of constant pre-puberty mutation accumulation in both sexes with a period of increasing mutation post puberty only in males (e.g., Thomas and Hahn 2014; Amster and Sella 2016; Gao et al. 2016; Thomas et al. 2018). Female age has a small effect on the accumulation of mutations (e.g., Goldmann et al. 2016; Jónsson et al. 2017), but very large sample sizes have been needed to detect it; therefore, it is often ignored in these models.
Despite the general acceptance of the male germline replication model of mutation accumulation (but see Hurst and Ellegren 1998), several observations from whole-genome sequencing studies have emerged that do not fit easily within this paradigm. Here we describe four of these patterns (see de Manuel et al. 2022 for further discussion). (1) Spermatogenic cycle length is not predictive of mutation rates: non-human primates with shorter seminiferous epithelial cycle lengths do not show a faster rate of mutation accumulation per year (Wang et al. 2020; Wu et al. 2020). Although the number of these cell cycles may not exactly match the number of replications (Scally 2016; Thomas et al. 2018), shorter cycle lengths should result in more mutations per unit time. (2) In humans, the male bias in mutations exists even in the youngest fathers studied (Gao et al. 2019). If male bias is largely driven by the continued replications of spermatogenesis post puberty, there should be little difference between the sexes immediately post puberty. (3) C→T mutations at CpG sites show a similar degree of male bias as other mutations (Jónsson et al. 2017), even though they are not thought to be associated with polymerase errors during replication. While both sexes can incur exogenous damage at these sites, it is not clear how or why such damage would cause more mutations in males. (4) Studies of somatic mutagenesis have not found a higher mutation rate in tissues that are mitotically active (Abascal et al. 2021). While variation in the somatic mutation rate across tissue types was observed, the mutation rate was not associated with the rate of cellular division in each tissue.
With the growing number of questions surrounding the role of male germline replication in the evolution of the mutation rate, species that experience a slowdown or cessation of this replication represent a potentially illuminating study system. Many mammals (and non-mammals) undergo a period of quiescence or torpor during the winter—what is generally referred to as hibernation (Geiser 2013). Many physiological functions are altered during hibernation; in particular, reproduction and spermatogenesis, with a complete cessation of spermatogenesis in some species (e.g., in black bears [Ursus americanus]; Tsubota et al. 1997). Male germline activity restarts in late winter through a process known as testicular recrudescence.
The brown (grizzly) bear, Ursus arctos horribilis, is a model system for studies of the genetics and physiology of mammalian hibernation (Hershey et al. 2008; McGee et al. 2008; Buffenstein et al. 2014; Rigano et al. 2017; Jansen et al. 2019; Mugahid et al. 2019). During hibernation, bears do not eat, produce minimal-to-no urine, reduce heart rates to 10–15 beats per minute, do not lose bone mass, and have minimal muscle mass loss despite having almost no weight-bearing activity. Brown bears are seasonal breeders, with the peak breeding season occurring in June. After breeding season, the testis becomes reduced in size, and—at least in closely related black bears—spermatogenesis and reproductive steroidogenesis are greatly reduced as the male enters hibernation (Howell-Skalla et al. 2000). Histological studies in Hokkaido brown bears (Ursus arctos yesoensis) found no evidence of spermatogenesis between October and January (Tsubota and Kanagawa 1989), further supporting the idea that spermatogenesis is reduced during hibernation.
Given the annual pause in male germline replication experienced by brown bears, we hypothesized that the per-generation mutation rate and the degree of male bias in mutations from this species would be lower under the male germline replication model of mutation accumulation. That is, we assume that hibernation would reduce the per-generation mutation rate and/or degree of male bias. Here, we test this hypothesis by studying the mutation rate in an extended pedigree of brown bears. Using whole-genome sequencing of the four trios embedded in this pedigree, we find no difference between our estimate of the per-generation mutation rate and its expectation under a model without hibernation. We also find no difference in the degree to which mutations are male-biased compared with other mammals. Further analysis of the per-year mutation rate—estimated via phylogenetic comparison with closely related non-hibernating species—also shows no effect of hibernation. We discuss the implications of these results for our understanding of the cellular basis of mutation rate evolution in mammals.
Results
Testes Size Through Hibernation
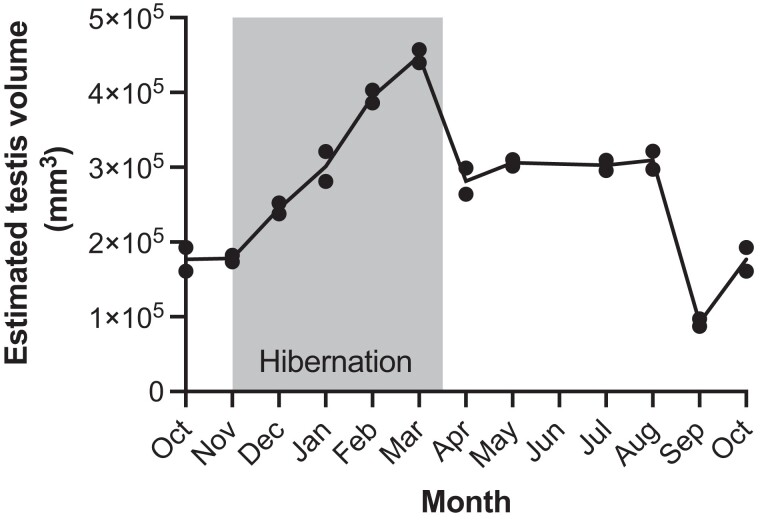
To highlight the physiological and phenotypic changes that the male germline undergoes during hibernation, we measured seasonal variation in testis size from two sexually mature male brown bears across a 4-year period (fig. 1). The results show clear seasonal differences with a testicular recrudescence (regrowth) evident during late hibernation and reduction in size (regression) during the hyperphagia period (August–October). Because sperm production is a lengthy process (taking approximately 2 months), male bears’ peak testis volume occurs before they emerge from the den in the spring. These results confirm other observations of seasonal changes in testis size and function made in both brown and black bears (Tsubota and Kanagawa 1989; Hellgren 1998; White et al. 2005; Spady et al. 2007).
Fig. 1.
Estimated testis volume through hibernation. Two male grizzly bears were sampled so that each bear was measured in each month of the year at least once (measurements were spread across a 4-year period). Dots indicate individual values, and the line is the mean volume. Gray shading indicates the timing of hibernation.
Estimating the Per-generation Mutation Rate
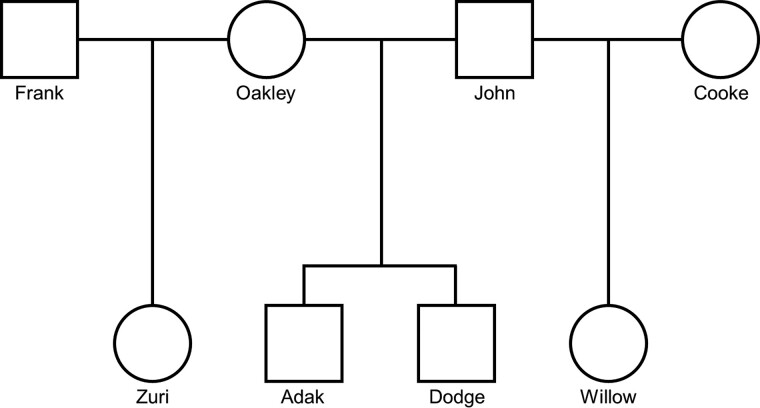
We sequenced the genomes of eight individuals from a large pedigree of captive brown bears kept at the Washington State University Bear Center (fig. 2). Individual samples had an average of 51.1× coverage (min: 46.6×, max: 57.1×), with reads mapped to the brown bear reference genome (NCBI assembly ASM358476v1). The pedigree can be separated into four trios from which independent mutation rate estimates can be made (we observed no candidate mutations shared among siblings). We required that all three individuals in a trio have a minimum (and maximum) depth of high-quality reads for a mutation to be called (Materials and Methods). On average, these filters for “callability” allowed us to examine 1.72 Gb per trio for mutation identification (table 1).
Fig. 2.
Pedigree of bears included in the study. Eight individuals that were part of an extended pedigree were sequenced. The four probands (Zuri, Adak, Dodge, and Willow) each represents the offspring within an independent trio. Males are indicated by squares and females by circles.
Table 1.
Number of Mutations and Mutation Rate Per Trio
| Trio | Proband | Mutations | Callable size (Mb) | Per bp rate (×10−8) | Parental age (y) | |
|---|---|---|---|---|---|---|
| Paternal | Maternal | |||||
| 1 | Zuri | 31 | 1753 | 0.88 | 13 | 12 |
| 2 | Adak | 23 | 1701 | 0.68 | 13 | 12 |
| 3 | Dodge | 31 | 1678 | 0.92 | 13 | 12 |
| 4 | Willow | 30 | 1752 | 0.89 | 13 | 12 |
Note.—Mean mutation rate: 0.84 × 10−8 per bp per generation.
After applying a stringent set of filters, we identified 115 total mutations across the four trios, including one multinucleotide mutation (supplementary table 1, Supplementary Material online). All of the trios have parents that are the same ages (to the nearest year) and consequently we found very little variation in the number of mutations per trio (table 1). To estimate the per base pair mutation rate, we divided the number of mutated bases identified in each trio by twice the number of callable sites (to account for mutations transmitted from both parents; eq. 1). We found the mean per-generation mutation rate in brown bears to be μg = 0.84 × 10−8 per bp (95% confidence interval [CI]: 0.69–1.00) for parents at an average age of 12.5 years across sexes. Table 1 shows the rate estimated for each trio separately. Our approach also produced consistent estimates of the mutation rate, as the stringency of filters was increased (supplementary fig. 1, Supplementary Material online), providing confidence in our results.
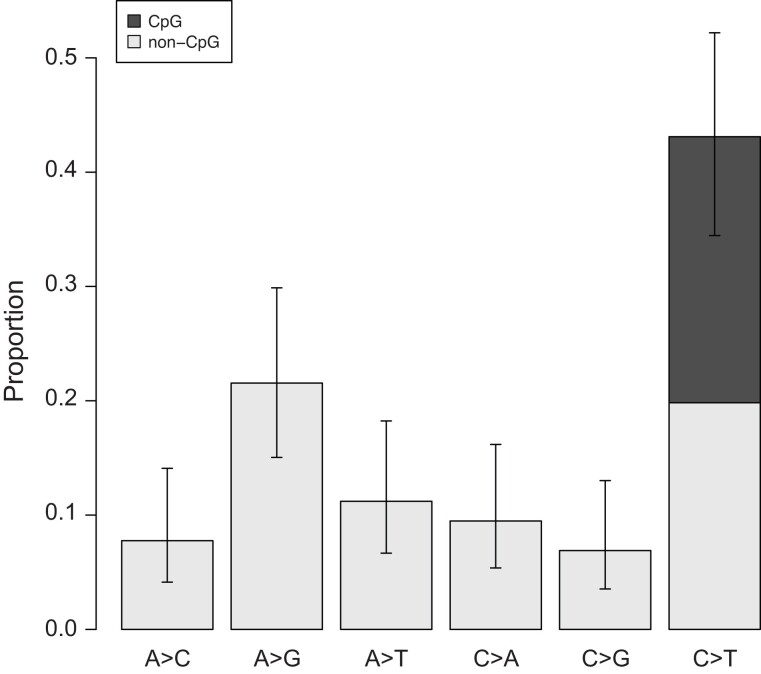
We did not find the mutation spectrum in the bear to be significantly different from the spectrum found in humans (χ2-test, P = 0.30; fig. 3). The transition-to-transversion ratio among mutations was 1.8 (1.3–2.8), comparable with the expectation for SNPs in humans and other mammals. Similarly, we found that a substantial fraction of all mutations were C→T transitions at CpG sites (23%). We estimate the mutation rate at CpG sites in the brown bear to be 2.0 (1.3–2.7) × 10−7 per bp per generation for parents at an average age of 12.5 years across sexes. This roughly order-of-magnitude higher mutation rate at CpG sites is consistent with previous estimates in other species (e.g., Kong et al. 2012).
Fig. 3.
Bear mutation spectrum. The proportion of each mutation class among bear trios, including their reverse complements. Dark gray region indicates the proportion of mutations occurring at CpG sites. Error bars show binomial 95% CI (Wilson score interval).
Testing for an Effect of Hibernation on the Per-generation Mutation Rate
The per-generation mutation rate for brown bears estimated here (μg= 0.84 × 10−8 per bp) is lower than that observed in humans: 1.29 × 10−8 per bp for parents with an average age of 30.1 years (Jónsson et al. 2017). However, parents in the bear trios from this study are less than half the average human age, and the age at puberty in brown bears is also less than half of what it is in humans (4 years for female brown bears, 4.5 for male brown bears; Schwartz et al. 2003; White et al. 2005). A direct comparison of these rates therefore captures differences in reproductive life history between species rather than the potential effects of hibernation on mutation rates.
In order to test for an effect of hibernation, we calculated the expected per-generation mutation rate under two models that consider mutation accumulation up to the point of conception, but with no hibernation. First, we used a “total longevity” model (cf., Wang et al. 2022), where the mutation rate is dependent solely on the age at conception (Materials and Methods). That is, mutation accumulation is constant from birth to conception, with a rate of accumulation in males and females independently estimated from human data (eqs 5 and 6). Using the ages of conception from the bears in our study (Am=13 and Af = 12), we predict a per-nucleotide per-generation mutation rate of E(μg) = 0.62 × 10−8 per bp under the total longevity model. This expected rate is significantly lower than the observed rate (based on a comparison with the 95% CI of the estimated rate), meaning that the mutation rate in brown bears appears to be higher than predicted under this model.
Second, we used an updated parameterization of the “reproductive longevity” model (Thomas et al. 2018). This model separates parental mutation accumulation into two stages: pre and post puberty. Modeling pre-puberty separately allows us to account for the higher rate of mutation in this stage (e.g., Jónsson et al. 2018; Sasani et al. 2019), as well as for the fact that pre-puberty mutation accumulation seems to be relatively constant across species with very different ages at puberty (Thomas et al. 2018; Wang et al. 2020, 2022). Post puberty, we assume that bears follow the same process of mutation accumulation as humans (Materials and Methods). Using the reproductive longevity model, we calculate an expected per-generation mutation rate of E(μg) = 0.88 × 10−8 per bp. This predicted rate is not significantly different from the observed rate. We therefore tentatively conclude that hibernation in brown bears does not appear to lower the per-generation mutation rate, at least relative to the expectations of two models parameterized by human data that include no hibernation.
In addition to an effect on the overall per-generation rate, hibernation could reduce the proportion of paternally derived mutations. Such a reduction may be expected if spermatogenesis experiences a seasonal pause, as suggested by the absence of expressible sperm during hibernation and the presence of testicular regression after the breeding season. We investigated this potential effect of hibernation by examining the parent-of-origin across individual mutations. We were able to phase 26 of the 115 total mutations using read-pair tracing (supplementary table 1, Supplementary Material online). Of the phased mutations, 22 were transmitted by a male parent and 4 were transmitted by a female parent. This proportion of male-biased mutations (84.6%) is highly consistent with the proportion found in humans (80.4%; Jónsson et al. 2017), and not significantly different (χ2 test, P = 0.8). The results again show no detectable effect of hibernation on the male mutation process, with a degree of male bias as expected from uninterrupted germline replication post puberty.
Comparisons Using the Per-year Mutation Rate Estimated from Phylogenies
In order to investigate possible effects of hibernation over a longer time period, we compared the number of substitutions in brown bears to the number in a sister lineage without hibernation (pandas). If hibernation has slowed the rate of mutation accumulation (and has been a trait associated with the brown bear lineage for a long period of time), we expect to observe fewer substitutions in the brown bear genome compared with the panda. Under the standard assumption that for neutral mutations the substitution rate equals the mutation rate (Kimura 1968), this comparison allows us to compare the mutation rate per year between hibernating and non-hibernating sister lineages.
To study substitution rates, we used 4,886 genic alignments among brown bear, panda, ferret, and dog (Materials and Methods). These two outgroups allowed us to compare the tip branch lengths specific to the brown bear and panda. We compared synonymous substitutions per site to minimize the effect of selection, finding the brown bear substitution rate to be 87.6% of that in the panda. The average length of the tip branch leading to brown bear (since its common ancestor with panda) was dS = 0.0212 substitutions/site (S.E. 2.2 × 10−4) and the average length of the panda branch was dS = 0.0242 (S.E. 2.2 × 10−4).
As was the case for our trio-based estimates of the per-generation mutation rate, a direct comparison of substitution rates does not provide evidence for an effect of hibernation. The panda has a younger average age at conception in the wild than the brown bear (Wei and Hu 1994; Peng et al. 2001; Aitken-Palmer 2010; Kersey et al. 2010), which can increase the rate of substitution per year relative to the brown bear (Laird et al. 1969; Wu and Li 1985; Thomas and Hahn 2014; Gao et al. 2016). To account for this difference, we calculate the expected difference in per-year mutation rates under a reproductive longevity model (given that it was a much closer fit to the per-generation mutation rate in the previous section). Such a comparison allows us to test whether an effect of hibernation must be invoked to explain the lower substitution rate in the branch leading to brown bears, at least relative to a model that separates rates before and after puberty to account for differences in generation times.
To predict the per-year mutation rate in both brown bear and panda, we used equation (9), applying appropriate ages for each species (Materials and Methods) and using common mutational parameters estimated from humans. Using the ages of puberty and conception estimated from current populations necessarily means that we are assuming these ages have been constant since the split of the two species. We are also assuming that the relationship between age and number of mutations has been constant within each species since their split, and that these mutation predictions are relevant to the synonymous substitution rates estimated from phylogenetic data. We estimated a mean expected per-year mutation rate for brown bears of 8.02 × 10−10 per bp under a reproductive longevity model. We compared expected per-year mutation rates along these two lineages using a range of values for the average age at conception in the wild for brown bears (table 2; supplementary table 2, Supplementary Material online). From this range of life history estimates, we predict the brown bear per-year mutation rate should be 81.2–90.1% of that observed in the panda, assuming the absence of any effect of hibernation. Our results from the phylogenetic analyses (87.6% relative to panda) fall squarely within this range. We therefore conclude that hibernation has not led to a measurable difference in per-year mutation rates between the brown bear and the panda.
Table 2.
Ratio of Expected Yearly Substitution Rates in Brown Bear Relative to Panda at Different Parental Ages in the Brown
| Maternal age at conception | Paternal age at conception | ||
|---|---|---|---|
| 13 | 14 | 15 | |
| 7 | 0.901 | 0.886 | 0.873 |
| 8 | 0.865 | 0.852 | 0.841 |
| 9 | 0.832 | 0.822 | 0.812 |
Note.—Supplementary table 2, Supplementary Material online shows the underlying per-year values for each cell.
Discussion
By sequencing multiple independent trios (fig. 2), we identified 115 de novo nucleotide mutations in brown bears. These mutations allow us to estimate the per-generation mutation rate, the mutation spectrum (fig. 3), the degree of male-biased mutation, as well as the presence of multinucleotide mutations (multiple closely spaced mutations that occur in a single generation; Schrider et al. 2011). Our estimate of the per-generation rate, 0.84 × 10−8 per bp for parents at an average age of 12.5 years, joins a large and growing list of species for which this important evolutionary parameter has been measured (Chintalapati and Moorjani 2020; Yoder and Tiley 2021).
Our most striking result is the absence of an obvious effect of hibernation on the mutation rate, despite the seasonal testicular regression associated with hibernation (fig. 1) and the apparent reduction of spermatogenesis. Comparisons with non-hibernating species of the per-generation mutation rate, the per-year mutation rate, and the degree of male-bias reveal no significant reductions in brown bears under the set of parameters and model assumptions used here. After accounting for the fact that a 12-year-old bear should transmit fewer mutations than the average human included in previous studies, we do not find a lower per-generation mutation rate in brown bears due to hibernation. Similar comparisons of the per-year rate in brown bear with the panda (a non-hibernating species) that take into account differences in life history between these two species also revealed no differences. Our predictions of the per-generation mutation rate with no hibernation were made under two distinct models. Although both models assume that many of the underlying mutation parameters are the same among species, we previously found them to be conserved among multiple mammals (Thomas et al. 2018; Wang et al. 2020, 2022).
There are several non-exclusive mechanisms that can explain the absence of a clear difference in mutation accumulation between hibernating and non-hibernating species. One obvious explanation is that an increasing mutation rate with age and a male bias in mutation are not driven by continuing mitosis during male spermatogenesis. As mentioned in the Introduction (also see de Manuel et al. 2022), there are several patterns from whole-genome sequencing studies that do not appear consistent with the classical role attributed to male germline replication. Although continued male germline replication is an obvious correlate of many of the coarse patterns of mutation accumulation, data from whole-genome sequencing have also uncovered multiple fine-grained patterns that do not fit with this hypothesis. A simple model in which some aspect of mutation repair differs between males and females across most of their lifespan would fit the general trends equally well, and would do much to explain several seemingly paradoxical patterns (Gao et al. 2019; de Manuel et al. 2022). Further investigation of underlying mutational mechanisms may help to add additional detail to this newer model.
Despite the allure of new possible biological models, there are several ways to explain our data in brown bears that are consistent with classical hypotheses for the role of spermatogenesis in mutation accumulation. First, although there is a huge reduction in testis size and an absence of sperm during early hibernation (Tsubota and Kanagawa 1989), spermatogonial cells may be replicating throughout the year. Only a subset of spermatogonial cells is actively dividing even in full-sized testes (Plant 2010), and the absence of sperm may be due to a halt in spermiogenesis rather than spermatogenesis. Under this model, despite all outward appearances, hibernation would have no appreciable effect on male germline replication. A second possibility is that spermatogenesis fully halts during the first part of hibernation, but then accelerates every spring during testicular recrudescence. Alternatively, animals coming out of hibernation often experience an increase in oxidative stress (Orr et al. 2009), though it is not clear whether this would lead to an increase in mutations (Rajaei et al. 2021). An explanation involving increased spermatogenesis would require a puberty-like process that occurs every year, ensuring that the male germline maintains the same total number of cell divisions per year, regardless of hibernation status. A hypothetical mechanism of accelerated replication would also explain the appearance of male bias in mutation number just after puberty in humans (Gao et al. 2019), and the marked similarity in the number of mutations just after puberty across a number of species (Thomas et al. 2018; Wang et al. 2022). Third, it is possible that in addition to a slowdown in spermatogenesis, female bears experience a slowdown in mutation rate during hibernation, one that mirrors a male slowdown. Such a slowdown could be due to metabolic or physiological changes to female bears during hibernation, especially changes in endocrine signaling in reproductive tissues (Hellgren 1998). Such an explanation would help to explain the lack of an effect of hibernation on male mutation bias, but would also seem to imply even lower mutation rates in hibernating species. Finally, these explanations for major patterns of mutation accumulation do not preclude an important, but smaller, role for male germline replication than is currently believed.
Models of mutation accumulation are an essential part of understanding the evolution of mutation and the mutation rate. Per-generation and per-year mutation rates are keys to many evolutionary inferences, from estimates of divergence times to explanations for the maintenance of sexual reproduction. Understanding the factors underlying changes in these rates is therefore necessary for researchers to form a comprehensive picture of many aspects of evolution. Interestingly, the results presented here are easily accommodated by current models of mutation. Even for models that were explicitly constructed with male replication in mind (e.g., Thomas and Hahn 2014; Amster and Sella 2016; Gao et al. 2016; Thomas et al. 2018), a simple re-parameterization that is agnostic to the causes of male bias will almost always result in the same outcome. For example, the amount of mutation accumulation in males post puberty, μgM1, need not depend on cell-division rates for the predictions used here to hold (eq. 8). Despite the success of such models, one outstanding question is how much our analyses will suffer if models remain phenomenological, and how much our science will improve if we fully incorporate molecular mechanism. There are clearly different processes of accumulation for different types of mutation—for instance, there is no parental age effect for structural mutations (Brandler et al. 2016; Belyeu et al. 2021; Thomas et al. 2021)—and often rates are not correlated among types of mutation (Ho and Schaack 2021). One overall goal for the field will therefore be a general model of the cellular mechanisms that drive mutation rate evolution, a goal that will benefit from mutation rate studies in a wide variety of organisms and for a wide variety of different mutation types.
Materials and Methods
Animals
Brown bears (U. a. horribilis Linnaeus 1758) were housed at the Washington State University Bear Research, Education and Conservation Center (WSU Bear Center, Pullman, WA, USA) in accordance with the Bear Care and Colony Health Standard Operating Procedures approved by the Washington State Institutional Animal Care and Use Committee protocol #6546. The bears at WSU Bear Center hibernate from November to mid-to-late March.
Testes Measurements
Two adult males were periodically anesthetized as previously described (Ware et al. 2012) over a 4-year period. Both males were 13 years old at the time of first measurement. The final data set includes measurements at roughly monthly intervals between January and December, though each bear was only measured approximately three times in any particular year. Once each bear was anesthetized, each testis was manually palpated and externalized with gentle pressure. Paired testes measurements, including skin, were then made using a caliper micrometer (Mitutoyo, model 505-681) to the nearest 0.1 mm. The length (L) and width (W) of the testes were measured three times, and the average values for each testis were recorded. An estimated testis volume was then derived for each testis using the formula, W2 × L (as described by Gorman and Zucker 1995), and the two testis values were added together to generate a total estimated testis volume per individual.
DNA Extraction and Quantification
Samples from an extended pedigree with four embedded trios (n = 8 individuals; fig. 2) were used for per-generation mutation rate estimates. Blood was collected (∼5 ml) from the jugular vein into PAXgene Blood DNA Tubes. DNA was extracted using the PAXgene Blood DNA Kit following the standard protocol for whole blood with no modifications. DNA was quantified with the high sensitivity double-stranded (ds) DNA Assay Kit (Qubit dsDNA HS Assay Kit; Thermo Fisher Scientific, #Q32854) on the Qubit 2.0 fluorometer.
DNA Sequencing
Extracted DNA was sequenced at the Baylor College of Medicine Human Genome Sequencing Center (Houston, TX, USA). Standard polymerase chain reaction–free libraries were prepared using KAPA Hyper PCR-free library reagents (KK8505; KAPA Biosystems). Total genomic DNA was sheared into fragments of approximately 200–600 bp and purified using AMPure XP beads. Sheared DNA molecules were subjected to double size selection with different ratios of AMPure XP beads to select a narrow size band of sheared DNA molecules for library preparation. This was followed by DNA end-repair and 3′-adenylation before the ligation of barcoded adapters. Library quality was evaluated by fragment analysis and qPCR assay. The resulting libraries were sequenced on an Illumina NovaSeq 6000, producing 150 bp paired-end reads.
Mutation Identification
Sequenced reads were aligned with BWA-MEM version 0.7.12-r1039 (Li 2013) to the domestic brown bear reference genome, ASM358476v1 (Taylor et al. 2018). Picard MarkDuplicates v. 1.105 (Broad Institute 2019) was used to identify and mark duplicate reads from the BAM files. We used GATK v. 4.1.2.0 (Van der Auwera et al. 2013) to call variants using best practices. HaplotypeCaller was used to generate gVCF files for each sample and joint genotype calling across samples was performed with GenotypeGVCFs. We applied GATK hard filters: (SNPs: “QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0”) and removed calls that failed.
We used the same pipeline for identifying autosomal de novo mutations from called variants as in our previous work (Wang et al. 2020, 2022), which we summarize here: An initial set of candidate mutations was identified as “Mendelian violations” in each trio. Specifically, we looked for violations where both parents were reference homozygous and the offspring was heterozygous for an alternate allele. As this is the most common type of genotyping error (Wang et al. 2021), we then apply the following filters to the initial set of candidates to get a set of high-confidence candidates:
Read depth at the candidate site must be between 20 and 80 for every individual in the trio. Sites with too few reads are likely to be sampling errors, whereas sites with too many reads are likely to be from repetitive regions.
High genotype quality (GQ) in all individuals (GQ > 60).
Candidate mutations must be present on reads from both the forward and reverse strand in the offspring.
Candidate mutations must not be present in any reads from either parent.
Candidate mutations must not be present in any other samples (except siblings).
Candidate mutation must not have low allelic depth in the offspring (allelic balance > 0.30).
We assessed the sensitivity of our mutation rate estimates across a range of stringency criteria and found them to be in good agreement across reasonable filter limits (supplementary fig. 1, Supplementary Material online). The distribution of allelic balances was also centered at 0.5 (supplementary fig. 2, Supplementary Material online).
Per-generation Mutation Rate Estimate
In order to transform the identified number of de novo mutations into a rate per-base per-generation, we need an accurate count of the number of bases at which mutations could have been identified in each trio. As in previous work, we applied existing strategies that considered differences in coverage and filtering among sites (Besenbacher et al. 2019; Wang et al. 2020, 2022), and that estimate false-negative rates from this filtering. Briefly, the number of identified mutations was divided by the total number of “callable sites.” Callable sites are a product of the number of sites covered by the appropriate sequencing depth and the estimated probability that such a site would be called correctly given that it was a true de novo mutation. The mutation rate is then calculated as:
| (1) |
where μs,i is the per-base mutation rate for trio i, Nmut,i is the number of mutated bases in trio i, and Ci(x) is the callability of site x in that trio. This strategy assumes that the ability to call each individual in the trio correctly is independent, allowing us to estimate Ci(x) as:
| (2) |
where Cc, Cp, and Cm are the probability of calling the child, father, and mother correctly for trio i. These values are estimated by applying the same set of stringent filters to high-confidence calls from each trio. For heterozygous variants in the child,
| (3) |
where Nhet,all is the number of variants in the offspring where one parent is homozygous reference and the other parent is homozygous alternate, leading to high confidence in the child heterozygote call, and Nhet,filtered is the set of such calls that pass our child-specific candidate mutation filters. The parental callability, Cp(x) and Cm(x), was estimated in a similar manner, by calculating the proportion of remaining sites in each after the application of the stringent mutation filters. Based on our previous results and the results of comparisons of our pipeline to those from other research groups when applied to common data sets (Bergeron et al. 2022), we assume our pipeline produces no (or very few) false positives.
Phasing Mutations
We used read-pair tracing to determine the parent of origin (“phase”) for mutations across all of our trios. We did this by applying WhatsHap 1.0 (Patterson et al. 2015) in read-based phasing mode for each individual separately, and then matched informative blocks bearing the mutation to their parent of origin according to the rules of Mendelian inheritance. Ambiguous blocks, including any that showed genotype inconsistencies between parent and offspring, were left unphased.
Per-year Mutation Rate Estimate
To estimate long-term rates of molecular evolution, we identified orthologs from brown bear (ASM358476v1), panda (Ailuropoda melanoleuca, ASM200744v2), dog (Canis lupus familiaris, Cfam_1.0), and ferret (Mustela furo, MusPutFur1.0) using OrthoFinder v. 2.5.2 (Emms and Kelly 2019) with DIAMOND v. 0.9.27 (Buchfink et al. 2021) as the sequence search program. Only orthogroups with single-copy orthologs were considered in the analysis. We were not able to confidently place genes on the bear X chromosome, but excluded the set of genes with human orthologs on the X from all further comparisons.
Protein-coding sequences for each orthogroup containing all four species were aligned by codon using GUIDANCE2 (Sela et al. 2015) with MAFFT v. 7.471 (Katoh and Standley 2013). GUIDANCE2 provides quality scores for each residue and column of the alignment. Scores were used to remove unreliable sequence: low-confidence residues with scores <0.93 were converted into gaps. Columns with gaps and N's were removed from the alignments using trimAl v. 1.4.rev22 (Capella-Gutiérrez et al. 2009). Additionally, alignments with sequences that were shorter than 200 bp were filtered. This process resulted in a total of 4,886 gene alignments that were considered for further analysis.
Synonymous substitutions per site (dS) for each branch were estimated using HyPhy (Pond et al. 2005) with the FitMG94.bf model (https://github.com/veg/hyphy-analyses). We assumed that every gene had the same topology: (((U. a. horribilis, A. melanoleuca), M. furo), C. l. familiaris). The average tip branch lengths leading to brown bear and panda were obtained by taking the mean of dS values across all genes, after removing genes where either of the two tip branches was longer than 0.2 (which we took as an indication of poor alignment). Despite the absence of an absolute time estimate for the split between brown bear and panda, their comparison as sister lineages provides an equal amount of time for substitutions to have accumulated in each species, and we therefore refer to the estimated distances as substitution rates.
Expected Per-generation Mutation Rate in the Absence of Hibernation
To compare the estimated per-generation mutation rate obtained in our bear pedigrees to that expected in a non-hibernating species that is otherwise equivalent, we used two models. The first model is what we refer to as the “total longevity” model of mutation accumulation (cf., Wang et al. 2022). This model can be used to predict the expected per-generation mutation rate taking into account the calendar age of parents at conception. Given a known relationship between parental age and the number of mutations inherited by offspring—and assuming that mutations accumulate at a constant rate across the lifespan of the parents—the total longevity model estimates the expected per-generation mutation rate, E(μg), from contributions made by the male (μgM) and female (μgF) parents. Since autosomes spend half their time in males and half their time in females, the expectation in the total longevity model becomes:
| (4) |
In order to obtain μgM and μgF, we use the linear model presented in Jónsson et al. (2017); this assumes that the rate of accumulation in humans is a good proxy for that expected in bears. This model estimates the number of mutations from each parent as a function of only age at conception, denoted here as AM and AF for males and females, respectively:
| (5) |
| (6) |
To arrive at a per-nucleotide mutation rate, we divide both terms by 2.72 × 109 bp, the average callable genome size in Jónsson et al. (2017). Using the ages of conception from the bears in our study (AM=13 and AF = 12), we predict a per-nucleotide per-generation mutation rate of E(μg) = 0.62 × 10−8 under the total longevity model.
The second model we use is the “reproductive longevity” model first described in Thomas et al. (2018), but with updated parameters. There is strong evidence that the germline experiences different rates of mutation accumulation across different life stages (e.g., Rahbari et al. 2016; Jónsson et al. 2018; Sasani et al 2019). This model divides the expected per-generation mutation rate into contributions from two different life stages with different rates of mutation accumulation: before and after puberty. There are therefore four different mutation rates that contribute to the overall mutation rate: females before puberty (μgF0), females after puberty (μgF1), males before puberty (μgM0), and males after puberty (μgM1). Similar to the total longevity model above, we can estimate the expected per-generation mutation rate from the reproductive longevity model as:
| (7) |
To calculate the number of mutations expected at puberty (μgM0 and μgF0), we again use rates estimated from humans. Given a human age of puberty of 12 years for both males and females, we can use equations (5) and (6) to get these values by substituting human puberty ages for AM and AF.
We estimate μgF1 and μgM1 as a function of the rate of mutation accumulation post puberty, and the reproductive longevity (RL) of the parents:
| (8) |
| (9) |
where RLM is the difference between the age of puberty in males (PM) and the age of the male parent at conception of his offspring (AM):
| (10) |
RLF is similarly calculated as the difference between the age of puberty and the age of offspring conception in females (i.e., RLF = AF − PF). RL therefore accounts for the amount of time mutations have had to accumulate post puberty, whereas μgM1 and μgF1 describe the number of such post-puberty mutations given the model of mutation estimated from humans (Jónsson et al. 2017). These per-year estimates are again divided by the callable genome size, 2.72 × 109 bp, to yield per-nucleotide rates.
The ages of conception of the bears in our study are AM = 13 and AF = 12, whereas the ages of puberty in brown bears are PM = 4.5 and PF = 4 (Schwartz et al. 2003; White et al. 2005). Using these values, we predict a per-nucleotide per-generation mutation rate of E(μg) = 0.88 × 10−8 under the reproductive longevity model. Although all of these calculations assume that many parameter values are the same between humans and bears, we have found them to be remarkably well-conserved across species (Thomas et al. 2018; Wang et al. 2020, 2022).
Expected Per-year Mutation Rate in the Absence of Hibernation
The model described above can be extended to calculate the expected per-year mutation rate as a function of differing life histories (Ségurel et al. 2014; Thomas and Hahn 2014; Amster and Sella 2016; Gao et al. 2016). We used the above estimates along with parameters from the life histories of brown bears and pandas to calculate the expected per-year mutation rate, without regard for hibernation status.
To calculate expected mutation rates per year, E(μy), we sum the mutational contribution from each life stage per generation, and weight these contributions by the amount of time spent in each:
| (11) |
Here, because we are interested in the long-term evolution of mutation rates, we use the average age of conception of bears in the wild. We used a mean of AF = 8 (with a range between 7 and 9; Schwartz et al. 2003) and a mean AM = 14 (with a range between 13 and 15; F. van Manen, personal communication) in all calculations for brown bears. Using these estimates for the average ages of conception and estimates for the average age of puberty given in the previous section, we calculate a mean expected per-year mutation rate for grizzly bears of E(μy) = 8.02 × 10−10 per bp (supplementary table 2, Supplementary Material online). For panda, we used average ages of puberty of PM = PF = 4.5 (Janssen et al. 2006; Steinman et al. 2006) and average ages of conception in the wild of AF = 7 and AM = 8 (Wei and Hu 1994). These parameters yield an expected per-year mutation rate for pandas of E(μy) = 9.41 × 10−10 per bp.
Supplementary Material
Acknowledgments
The authors thank Julia Lowe and Teann Manser for assistance with analyses, and veterinarian Ahmed Tibary and the students and staff of the WSU Bear Center for bear handling and care. Frank van Manen (US Geological Survey) kindly answered our questions about breeding ages in the wild. This work was supported by the Indiana University Precision Health Initiative, internal funds from Baylor College of Medicine, Interagency Grizzly Bear Committee, USDA National Institute of Food and Agriculture (McIntire–Stennis project 1018967), International Association for Bear Research and Management, T.N. Tollefson and Mazuri Exotic Animal Nutrition, the Raili Korkka Brown Bear Endowment, Nutritional Ecology Endowment, and the Bear Research and Conservation endowment at Washington State University. Laurence Hurst, Aylwyn Scally, and an anonymous reviewer all provided helpful feedback.
Contributor Information
Richard J Wang, Department of Biology, Indiana University, Bloomington, Indiana 47405.
Yadira Peña-Garcia, Department of Biology, Indiana University, Bloomington, Indiana 47405.
Madeleine G Bibby, School of Biological Sciences, Washington State University, Pullman, Washington 99164.
Muthuswamy Raveendran, Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas 77030; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030.
R Alan Harris, Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas 77030; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030.
Heiko T Jansen, Department of Integrative Physiology and Neuroscience, Washington State University, Pullman, Washington 99164.
Charles T Robbins, School of Biological Sciences, Washington State University, Pullman, Washington 99164; School of the Environment, Washington State University, Pullman, Washington 99164.
Jeffrey Rogers, Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas 77030; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030.
Joanna L Kelley, School of Biological Sciences, Washington State University, Pullman, Washington 99164.
Matthew W Hahn, Department of Biology, Indiana University, Bloomington, Indiana 47405; Department of Computer Science, Indiana University, Bloomington, Indiana 47405.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
NCBI SRA for short reads SRR11336675-78 and SRR11336682-85.
Literature Cited
- Abascal F, et al. 2021. Somatic mutation landscapes at single-molecule resolution. Nature 593:405–410. [DOI] [PubMed] [Google Scholar]
- Aitken-Palmer C. 2010. Assessment of male giant panda seasonal reproduction, sexual maturity and comparative sperm cryotolerance. University of Maryland, College Park.
- Amster G, Sella G. 2016. Life history effects on the molecular clock of autosomes and sex chromosomes. Proc Natl Acad Sci U S A. 113:1588–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyeu JR, et al. 2021. De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families. Am J Hum Genet. 108:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron LA, et al. 2021. The germline mutational process in rhesus macaque and its implications for phylogenetic dating. GigaScience 10:giab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron LA, et al. 2022. Mutationathon: towards standardization in estimates of pedigree-based germline mutation rates. eLife 11:e73577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besenbacher S, Hvilsom C, Marques-Bonet T, Mailund T, Schierup MH. 2019. Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat Ecol Evol. 3:286–292. [DOI] [PubMed] [Google Scholar]
- Brandler WM, et al. 2016. Frequency and complexity of de novo structural mutation in autism. Am J Hum Genet. 98:667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 18:366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R, Nelson OL, Corbit KC. 2014. Questioning the preclinical paradigm: natural, extreme biology as an alternative discovery platform. Aging 6:913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapati M, Moorjani P. 2020. Evolution of the mutation rate across primates. Curr Opin Genet Dev. 62:58–64. [DOI] [PubMed] [Google Scholar]
- de Manuel M, Wu FL, Przeworski M. 2022. A paternal bias in germline mutation is widespread in amniotes and can arise independently of cell division numbers. eLife 11:e80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost JB, Lee WR. 1995. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 25:48–64. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, et al. 2019. Overlooked roles of DNA damage and maternal age in generating human germline mutations. Proc Natl Acad Sci U S A. 116:9491–9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wyman MJ, Sella G, Przeworski M. 2016. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 14:e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser F. 2013. Hibernation. Curr Biol. 23:R188–R193. [DOI] [PubMed] [Google Scholar]
- Goldmann JM, et al. 2016. Parent-of-origin-specific signatures of de novo mutations. Nat Genet. 48:935–939. [DOI] [PubMed] [Google Scholar]
- Gorman MR, Zucker I. 1995. Testicular regression and recrudescence without subsequent photorefractoriness in Siberian hamsters. Am J Physiol. 269:R800–R806. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. 1946. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen. 13:262–271. [DOI] [PubMed] [Google Scholar]
- Hellgren EC. 1998. Physiology of hibernation in bears. Ursus 10:467–477. [Google Scholar]
- Hershey JD, Robbins CT, Nelson OL, Lin DC. 2008. Minimal seasonal alterations in the skeletal muscle of captive brown bears. Physiol Biochem Zool. 81:138–147. [DOI] [PubMed] [Google Scholar]
- Ho EKH, Schaack S. 2021. Intraspecific variation in the rates of mutations causing structural variation in Daphnia magna. Genome Biol Evol. 13:evab241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell-Skalla L, Bunick D, Nelson R, Bahr J. 2000. Testicular recrudescence in the male black bear (Ursus americanus): changes in testicular luteinizing hormone-, follicle-stimulating hormone-, and prolactin-receptor ribonucleic acid abundance and dependency on prolactin. Biol Reprod. 63:440–447. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Ellegren H. 1998. Sex biases in the mutation rate. Trends Genet. 14:446–452. [DOI] [PubMed] [Google Scholar]
- Jansen HT, et al. 2019. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun Biol. 2:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen DL, et al. 2006. Significant medical issues and biological reference values for giant pandas from the Biomedical Survey. In: Zhang A, Wildt DE, Janssen DL, Zhang H, Ellis S, editors. Giant pandas: biology, veterinary medicine and management. Cambridge: Cambridge University Press. p. 59–86. [Google Scholar]
- Jónsson H, et al. 2017. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549:519–522. [DOI] [PubMed] [Google Scholar]
- Jónsson H, et al. 2018. Multiple transmissions of de novo mutations in families. Nat Genet. 50:1674–1680. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey DC, et al. 2010. Parallel and seasonal changes in gonadal and adrenal hormones in male giant pandas (Ailuropoda melanoleuca). J Mammal. 91:1496–1507. [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217:624–626. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. 2012. Rate of de novo mutations and the importance of father's age to disease risk. Nature 488:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird CD, McConaughy BL, McCarthy BJ. 1969. Rate of fixation of nucleotide substitutions in evolution. Nature 224:149–154. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 1303.3997. [Google Scholar]
- Lindsay SJ, Rahbari R, Kaplanis J, Keane T, Hurles ME. 2019. Similarities and differences in patterns of germline mutation between mice and humans. Nat Commun. 10:4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trends Genet. 26:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny SAN, et al. 2010a. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 6:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny SAN, et al. 2010b. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 107:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee ME, et al. 2008. Decreased bone turnover with balanced resorption and formation prevent cortical bone loss during disuse (hibernation) in grizzly bears (Ursus arctos horribilis). Bone 42:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugahid DA, et al. 2019. Proteomic and transcriptomic changes in hibernating grizzly bears reveal metabolic and signaling pathways that protect against muscle atrophy. Sci Rep. 9:19976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AL, Lohse LA, Drew KL, Hermes-Lima M. 2009. Physiological oxidative stress after arousal from hibernation in Arctic ground squirrel. Comp Biochem Physiol A Mol Integr Physiol. 153:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, et al. 2015. Whatshap: weighted haplotype assembly for future-generation sequencing reads. J Comput Biol. 22:498–509. [DOI] [PubMed] [Google Scholar]
- Peng J, Jiang Z, Hu J. 2001. Status and conservation of giant panda (Ailuropoda melanoleuca): a review. Folia Zoologica Praha. 50:81–88. [Google Scholar]
- Penrose L. 1955. Parental age and mutation. Lancet 266:312–313. [DOI] [PubMed] [Google Scholar]
- Plant TM. 2010. Undifferentiated primate spermatogonia and their endocrine control. Trends Endocrinol Metab. 21:488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. 2005. Hyphy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Rahbari R, et al. 2016. Timing, rates and spectra of human germline mutation. Nat Genet. 48:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaei M, et al. 2021. Mutability of mononucleotide repeats, not oxidative stress, explains the discrepancy between laboratory-accumulated mutations and the natural allele-frequency spectrum in C. elegans. Genome Res. 31:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano K, et al. 2017. Life in the fat lane: seasonal regulation of insulin sensitivity, food intake, and adipose biology in brown bears. J Comp Physiol B. 187:649–676. [DOI] [PubMed] [Google Scholar]
- Risch N, Reich E, Wishnick M, McCarthy J. 1987. Spontaneous mutation and parental age in humans. Am J Hum Genet. 41:218. [PMC free article] [PubMed] [Google Scholar]
- Sasani TA, et al. 2019. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. eLife 8:e46922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A. 2016. Mutation rates and the evolution of germline structure. Phil Trans R Soc B. 371:20150137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hourmozdi JN, Hahn MW. 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr Biol. 21:1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CC, et al. 2003. Reproductive maturation and senescence in the female brown bear. Ursus 14:109–119. [Google Scholar]
- Ségurel L, Wyman MJ, Przeworski M. 2014. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 15:47–70. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43:W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady TJ, Lindburg DG, Durrant BS. 2007. Evolution of reproductive seasonality in bears. Mamm Rev. 37:21–53. [Google Scholar]
- Steinman KJ, et al. 2006. Endocrinology of the giant panda and application of hormone technology to species management. In: Zhang A, Wildt DE, Janssen DL, Zhang H, Ellis S, editors. Giant pandas: biology, veterinary medicine and management. Cambridge: Cambridge University Press. p. 198–230. [Google Scholar]
- Taylor GA, et al. 2018. The genome of the North American brown bear or grizzly: Ursus arctos ssp. horribilis. Genes (Basel). 9:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, Hahn MW. 2014. The human mutation rate is increasing, even as it slows. Mol Biol Evol. 31:253–257. [DOI] [PubMed] [Google Scholar]
- Thomas GWC, et al. 2018. Reproductive longevity predicts mutation rates in primates. Curr Biol. 28:3193–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, et al. 2021. Origins and long-term patterns of copy-number variation in rhesus macaques. Mol Biol Evol. 38:1460–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota T, Kanagawa H. 1989. Annual changes in serum testosterone levels and spermatogenesis in the Hokkaido brown bear Ursus arctos yesoensis. J Mammal Soc Jpn. 14:11–17. [Google Scholar]
- Tsubota T, et al. 1997. Seasonal changes in spermatogenesis and testicular steroidogenesis in the male black bear Ursus americanus. Reproduction 109:21–27. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, et al. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.11–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn O, et al. 2014. Strong male bias drives germline mutation in chimpanzees. Science 344:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, et al. 2020. Paternal age in rhesus macaques is positively associated with germline mutation accumulation but not with measures of offspring sociability. Genome Res. 30:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Radivojac P, Hahn MW. 2021. Distinct error rates for reference and non-reference genotypes estimated by pedigree analysis. Genetics 217:iyaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, et al. 2022. De novo mutations in domestic cat are consistent with an effect of reproductive longevity on both the rate and spectrum of mutations. Mol Biol Evol. 39:msac147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JV, Nelson OL, Robbins CT, Jansen HT. 2012. Temporal organization of activity in the brown bear (Ursus arctos): roles of circadian rhythms, light, and food entrainment. Am J Physiol. 303:R890–R902. [DOI] [PubMed] [Google Scholar]
- Wei F, Hu J. 1994. Studies on the reproduction of giant panda in Wolong Natural Reserve. Acta Theriol Sin. 14:243–248. [Google Scholar]
- Weinberg W. 1912. Zur vererbung des zwergwuchses. Arch Rassen-u Gesel Biolog. 9:710–718. [Google Scholar]
- White D, Berardinelli JG, Aune KE. 2005. Age variation in gross and histological characteristics of the testis and epididymis in grizzly bears. Ursus 16:190–197. [Google Scholar]
- Wu C-I, Li W-H. 1985. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 82:1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FL, et al. 2020. A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS Biol. 18:e3000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Tiley GP. 2021. The challenge and promise of estimating the de novo mutation rate from whole-genome comparisons among closely related individuals. Mol Ecol. 30:6087–6100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NCBI SRA for short reads SRR11336675-78 and SRR11336682-85.