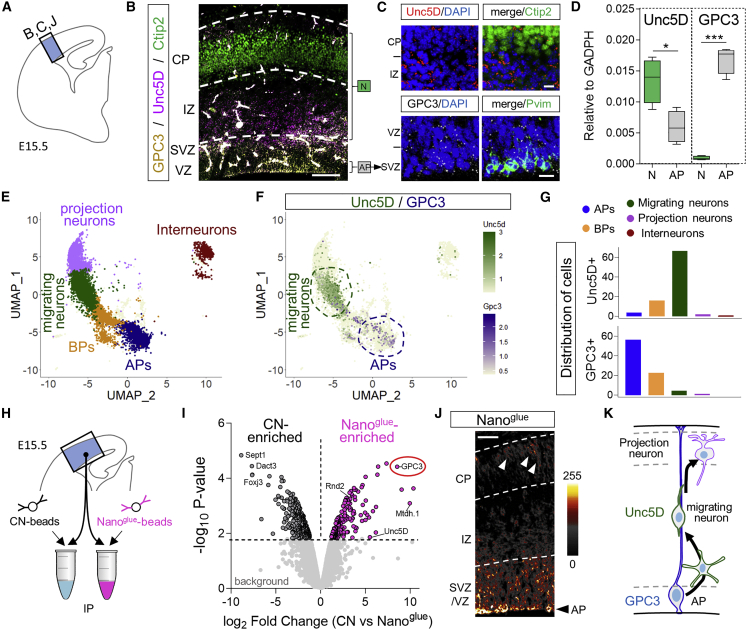
Figure 5.
GPC3 is expressed by cortical apical progenitor cells
(A) Cortical region shown in (B), (J), and magnified in (C).
(B) Double in situ hybridization (ISH) for Unc5D (magenta) and GPC3 (yellow) shows their expression in the cortex at E15.5. ISH is combined with the neuronal marker Ctip2 (green). The layers enriched in neurons (N) and apical progenitors (AP) are indicated.
(C) Upper panels show the ISH for Unc5D (red) combined with the neuronal marker Ctip2 (green). Lower panels show the ISH for GPC3 (white) and the apical progenitor marker Pvim (green). The location of the CP, IZ, SVZ, and VZ layers are indicated. Nuclear staining with DAPI is shown in blue.
(D) Unc5D and GPC3 expression levels normalized to GADPH in neurons and APs, using RNA profiling data published in Florio et al. (2015) (GSE65000). Unc5D mRNA is high in neurons (N), while GPC3 mRNA is enriched in AP cells. ∗p < 0.05, ∗∗∗p < 0.001, two-tailed Student’s t test.
(E) UMAP visualization of single-cell data from E15.5 mouse cortex published in di Bella et al., (2021) (GSE153164). Basal progenitor (BP).
(F) Combined plot of Unc5D (green) and GPC3 (magenta) mRNA expression per cell. Most of Unc5D-expressing cells belong to the migrating neuron cluster (dashed green line), GPC3-expressing cells are enriched in the AP cluster (blue dashed line).
(G) Quantification of the distribution of Unc5D- and GPC3-positive cells.
(H) Cortical region used for pull down with Nanoglue.
(I) Volcano plot showing enriched proteins in control (black) and Nanoglue (pink) pull downs revealed by mass spectrometry. Non-significant proteins are represented in light gray.
(J) Immunostaining for GPC3 using Nanoglue coupled to fluorescent streptavidin on coronal section of E15.5 mouse cortex. The image is colored based on the intensity of the staining. Arrowheads indicate staining that resembles the pattern of AP fibers.
(K) Summary model showing Unc5 and GPC3 expression patterns. Scale bars represent 200 μm (B), (J, top left) and 20 μm (C).
See also Figure S5.