Abstract
Background
The prognostic significance of germline variants in homologous recombination repair genes in advanced prostate cancer (PCa), especially with regard to hormonal therapy, remains controversial.
Methods
Germline DNA from 549 Japanese men with metastatic and/or castration-resistant PCa was sequenced for 27 cancer-predisposing genes. The associations between pathogenic variants and clinical outcomes were examined. Further, for comparison, DNA from prostate biopsy tissue samples from 80 independent patients with metastatic PCa were analysed.
Results
Forty-four (8%) patients carried germline pathogenic variants in one of the analysed genes. BRCA2 was most frequently altered (n = 19), followed by HOXB13 (n = 9), PALB2 (n = 5) and ATM (n = 5). Further, the BRCA1, BRCA2, PALB2 and ATM variants showed significant association with a short time to castration resistance and overall survival (hazard ratio = 1.99 and 2.36; 95% CI, 1.15–3.44 and 1.23–4.51, respectively), independent of other clinical variables. Based on log-rank tests, the time to castration resistance was also significantly short in patients with BRCA1, BRCA2, PALB2 or ATM somatic mutations and TP53 mutations.
Conclusions
Germline variants in BRCA1, BRCA2, PALB2 or ATM are independent prognostic factors of the short duration of response to hormonal therapy in advanced PCa.
Subject terms: Prognostic markers, Prostate cancer, Cancer genetics
Background
Inhibition of the androgen receptor (AR) pathway has been the mainstay of treatment for advanced prostate cancer (PCa). The addition of next-generation androgen pathway inhibitors (ARPIs) such as abiraterone, enzalutamide or apalutamide, to androgen deprivation has been established as the standard therapy for metastatic hormone-sensitive PCa [1–3]. Recently, it has also been reported that intensification of AR pathway inhibition by the addition of ARPIs to radiation therapy prolongs metastasis-free survival of high-risk non-metastatic PCa [4]. However, recent genomic studies revealed that there are multiple biological pathways other than the AR pathway that are also important in metastatic PCa progression [5]. One of the key pathways that has received considerable attention is the DNA repair pathway represented by the homologous recombination repair (HRR) and mismatch repair (MMR) pathways. HRR is the major pathway utilised for repair of DNA double-strand break and BRCA genes are the most frequently mutated HRR genes in PCa. Importantly, susceptibility to poly (ADP-ribose) polymerase (PARP) inhibitors is increased in tumours with BRCA gene alterations by the mechanism of synthetic lethality [6]. There are also non-BRCA alterations in the HRR pathway that lead to “BRCAness”, a molecular phenotype shared between tumours with germline or somatic mutations in BRCA1 or BRCA2 genes; PARP inhibitor are expected to be effective in these cases as well [7]. On the other hand, those with mutations in MLH1, MSH2, MSH6 and PSM2 genes have defects in MMR, which results in microsatellite instability; immune-checkpoint inhibitors are expected to be effective in these cases. Importantly, not only are these genes associated with HRR and MMR druggable but rare germline variants in many of these genes are also reported to be highly penetrant PCa-associated mutations [8].
As up to 15% of patients with metastatic PCa harbour germline pathogenic variants in one of the DNA repair pathway-associated genes that are potentially druggable [8], many guidelines, including the NCCN guideline (Version 2. 2022) and the ESMO guideline [9], recommend germline genetic testing for patients with metastatic PCa. The recommendation to test all metastatic PCa patients is also supported by the Philadelphia Prostate Cancer Consensus Conference 2019 [10]. However, in practice, the implementation of genetic tests is still suboptimal around the world, even in the case of metastatic castration-resistant prostate cancer (CRPC), due to factors such as accessibility and cost [11–13]; therefore, subjecting all patients with metastatic PCa to genetic tests is uncommon. Even though a younger age at diagnosis and the presence of family members with Hereditary Breast and Ovarian Cancer (HBOC) Syndrome or Lynch Syndrome are strong predictors of positive genetic tests, no other clinical parameter can be considered by clinicians when making a shared decision to conduct a genetic test on a particular patient. Thus, investigating the clinical characteristics of PCa in patients harbouring germline pathogenic variants is important.
Conflicting reports exist regarding the prognostic value of germline variants in the HRR-associated genes, especially concerning hormonal therapy for metastatic CRPC [13–18]. However, many of these studies used small sample sizes and lacked sufficient data to show a clinical association between the variants and clinical outcomes in multivariable analysis. In addition, different HRR-associated genes were included in each study, which may have confounded the results. Therefore, to add to the existing literature, we conducted a multiple hospital-based retrospective cohort study using a large cohort and explored the clinical implications of germline genetic variants. Specifically, leukocyte DNA was subjected to target sequencing analysis for 27 known cancer-predisposing genes, including HRR and MMR pathway-associated genes. The association between the identified variants and basic clinical factors as well as the duration of response to hormonal therapy and overall survival (OS) were evaluated.
Methods
Study population
Archived blood samples from 549 patients with metastatic PCa or CRPC treated at Akita University, Kyusyu University, University of Occupational and Environmental Health, Miyazaki University, and Kyoto University were used for germline analysis. The samples were randomly collected and archived at each institution from the patients who consented to research use of their blood samples for genetic studies. No exclusion criterion was defined. Therefore, the samples were mostly collected unbiasedly in a consecutive manner from the patients who presented with either metastatic PCa at diagnosis or those who initially presented with localised PCa that had progressed to CRPC. The median follow-up period was 4.7 years (interquartile range (IQR), 2.6–8.9 years) after diagnosis. In terms of hormonal therapy, all patients received androgen deprivation alone or combined androgen blockade with either bicalutamide or flutamide until castration resistance. None of the patients had been treated with docetaxel or ARPIs before becoming castration-resistant. For somatic mutation analysis, diagnostic prostate needle biopsy tissue samples from 80 patients with metastatic hormone-sensitive PCa, who were diagnosed between September 1, 2006 and September 1, 2016, at Otsu Red Cross Hospital, were used. Patients who presented with bulky local tumours were preferentially selected to ensure adequate tumour content; otherwise, the patients were selected consecutively. Given that the tissue samples were not collected for use in a large-scale genomic analysis, matched germline samples were not available for this cohort.
Sample collection and DNA extraction
Germline DNA was extracted from blood samples using a DNeasy Blood & Tissue kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions, and DNA concentration was determined using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). For mutation analysis using the tissue samples, an additional tissue core was biopsied when performing a standard systemic prostate needle biopsy for the initial diagnosis of PCa. After the confirmation of the presence of cancer cells via rapid cytology, the tissue samples were embedded in the optimal cutting temperature (OCT) compound (Tissue-Tek; Sakura Finetek, Torrance, CA, USA), and thereafter, stored at −80 °C until DNA extraction. The OCT compound was later removed from the samples using phosphate-buffered saline, and DNA was extracted using the DNeasy Blood & Tissue kit.
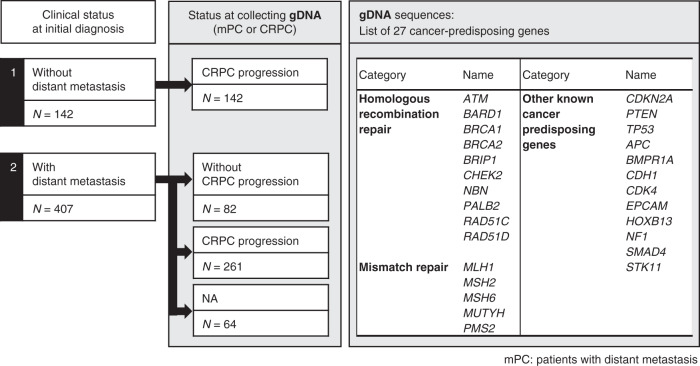
Target sequencing of 27 cancer-predisposing genes
We selected 27 genes based on the 25-gene hereditary cancer panel (Myriad Genetics Laboratories, Salt Lake City, UT, USA) [19]. In addition, NF1 and HOXB13, known to be associated with predisposition to breast cancer and PCa, respectively, were also included (Fig. 1). We analysed the complete coding regions and 2-bp flanking intronic sequences of the genes, except for the exons 10–15 of PMS2 (84,822 bp), using multiplex polymerase chain reaction-based target sequencing, as previously described [20]. Sequencing reads were aligned to the GRCh37 human reference genome assembly. We investigated single nucleotide variants and insertions or deletions using the UnifiedGenotyper and HaplotypeCaller tools of GATK, as previously described [21]. The Best Practice of GATK proposes the use of joint calling for the purpose of discovering germline short variants; however, the method did not work for our sequencing data in previous studies, likely due to very high sequencing depth. Therefore, we developed a custom pipeline in which we individually call all variants from each sample by HaplotypeCaller and UnifiedGenotyper of the GATK software. Next, we calculated alternative allele frequencies for each variant using Samtools to determine the genotype. All custom scripts have been deposited at GitHub (https://github.com/Laboratory-for-Genotyping-Development/TargetSequence.git). The variants with call rates <98%, <20 sequencing reads, and those with strand bias were excluded. After a quality check, 467 genetic variants were identified. Overall, ≥99% of the target region was covered with depth ≥20. The average depth of the samples was 776. The genotype for each individual was determined as described previously [20]. Briefly, when the alternative allele frequency was between 0% and 15%, we assigned “homozygote” to the reference allele. Similarly, when the alternative allele frequency was between 25% and 75%, and between 85% and 100%, we assigned “heterozygote” and “homozygote” to the alternative allele, respectively. If the alternative allele frequency was outside these ranges or a variant position was covered with <20 sequencing reads, “missing genotype” was assigned. The sequencing and variant-calling methods were extensively validated in our previous studies [20–24].
Fig. 1.
Schema describing the patient cohort analysed in the germline study as well as the 27 sequenced genes.
Annotation of germline variants
Variants were assessed for pathogenicity against the ClinVar [25] and SnpEff [26] databases. First, the variants with “pathogenic” or “likely pathogenic” annotations by ClinVar were selected as pathogenic variants. ClinVar version 20210302 was referenced. In our previous study that examined the prevalence of PCa predisposing gene variants in a large cohort of unselected PCa patients and healthy controls, clinical significance was determined using the ACMG/AMP guidelines [21]. In this study, in addition to the annotations using ClinVar, variants were screened using SnpEff and referenced against the previous study. The variants that were determined to be pathogenic in the previous study were also considered to be pathogenic in this study.
Annotation of variants in tissue samples
Given that no paired germline samples were available for this cohort, we applied a conservative mutation call method to identify somatic variants. First, we identified variants with a variant allele frequency ≥10% and a Phred quality score ≥20. Subsequently, these variants were annotated using ANNOVAR [27]. Next, variants registered in the dbSNP or 1000 Genomes Project database were removed [28]. Further, variants suspected to contain sequencing errors were removed by comparing the sequencing data with that obtained for our germline analysis, which were processed similarly after DNA extraction and sequencing. For each variant candidate, we assumed that its allele frequency in the germline mutation study corresponded to the sequence error rate at a specific position. We regarded a variant as a somatic mutation only when its allele frequency in tissues was significantly higher than the sequence error rate at that position (P < 0.05, using a one-sided binomial test). In addition, the variants that were identified in more than three of the 80 patients were also excluded, unless the locus was a mutation hotspot. Furthermore, to ensure the exclusion of potential germline variants, we removed variants with allele frequencies in the range of 40–60% or >99%. Finally, we removed variants in regions with high homology, which could be false-positive calls. The somatic mutation call method was further validated using an independent set of 16 PCa tissue samples in which the presence of at least one somatic mutation in the 27 genes that were studied has been confirmed by whole-exome sequencing with a standard mutation calling method referencing matched germline data (Supplementary Method, Supplementary Table 1).
Acquisition of clinical data
We collected the following clinical data from patient charts: family history of breast, ovary, pancreatic, or prostate cancer; history of breast, colon, or pancreatic cancer; age; prostate-specific antigen (PSA) value; biopsy grade group (GG); Whitmore–Jewett stage; location of metastasis; extent of disease (EOD) score [29], CHAARTED volume [30] at diagnosis for those initially diagnosed with metastatic PCa; duration of ARPI use; CRPC-free time; and OS. The CHAARTED tumour volume categorises patients with metastasis at diagnosis as having high-volume disease and low-volume disease based on metastatic tumour burden and is commonly used to clinically define metastatic PCa with poor outcomes [30]. In this study, CHAARTED volume was determined by the investigators at each institution via retrospective chart review. CRPC was defined according to the criteria established by the Prostate Cancer Working Group 2 [31]. Time to castration resistance was defined as the time from the start of androgen deprivation therapy (ADT) to the date of CRPC. Both OS from initial diagnosis and OS from the commencement of ADT were evaluated. Further, we examined whether the patients showed any signs of neuroendocrine prostate cancer (NEPC) trans-differentiation, and in addition to pathologically confirmed NEPC, we considered the patient as possibly having neuroendocrine changes if the patient had elevated serum NSE or proGRP levels or was diagnosed with NEPC by the investigating physicians based on the discrepancy between PSA and radiographic imaging data.
Ethics statement
The study, which was conducted in accordance with the Declaration of Helsinki, was approved by the ethics committees of RIKEN, Akita University, Kyusyu University, University of Occupational and Environmental Health, Miyazaki University, Kyoto University (approval number G1154), and the Japanese Red Cross Otsu Hospital. All participants at Akita University, Kyusyu University, University of Occupational and Environmental Health, Miyazaki University, and Kyoto University provided written informed consent for the genomic analysis of their blood samples. Regarding the archived biopsy samples obtained from patients at the Japanese Red Cross Otsu Hospital, even though the patients provided informed consent for the use of their material, genomic analysis was not specified in the consent form. Therefore, to prevent patient re-identification, under the guidance of the ethics committee of Kyoto University, all biopsy samples and clinical data were completely anonymized before the study was conducted.
Statistical analyses
Continuous variables were analysed statistically via Student’s t tests, whereas categorical variables were analysed via Fisher’s exact tests or Cochran–Armitage tests. Log-rank tests were performed for survival curve analysis, and the association between clinical and genomic variables and survival outcomes was examined using univariate and multivariable Cox proportional hazards models. The factors that showed significant association with an outcome in the univariate analysis were included in the multivariable analysis. The statistical tests were two-sided, and P values <0.05 were considered statistically significant. All statistical analyses were performed using R package version 3.6.1.
Results
Genetic profile of germline variants and their association with clinical parameters
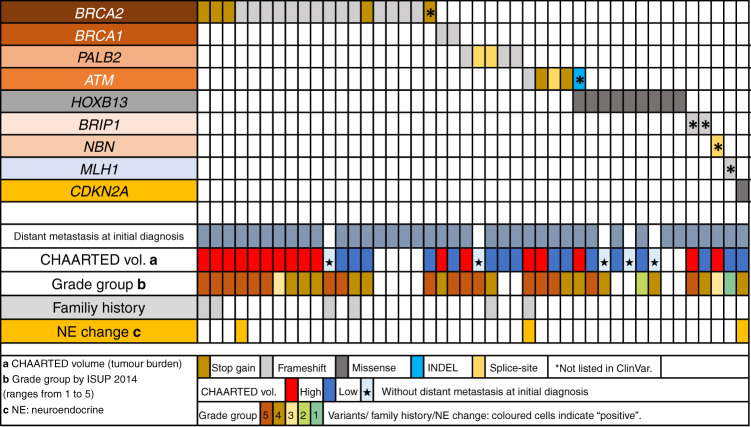
A summarised description of the patient cohort included in this study and the genes examined is shown in Fig. 1; patient and tumour characteristics of the germline variant study are summarised in Table 1. Of the 486 patients for whom data on castration resistance acquisition were available, 404 (83.1%) had become castration-resistant at the time of clinical data collection (Supplementary Table 2). Among the 549 patients, 45 germline pathogenic variants were identified in 44 (8.0%) patients (Supplementary Table 2), and BRCA2 was found to be most frequently mutated (n = 19, 42.2%), followed by HOXB13 (n = 9, 20.0%), PALB2 (n = 5, 11.1%) and ATM (n = 5, 11.1%) (Supplementary Fig. 1). Beside HBOC syndrome-associated genes and HOXB13, one case each of MLH1 and CDKN2A mutations was observed. The genetic and clinical profiles of patients with pathogenic variants are summarised in Fig. 2. Among those diagnosed with metastatic disease, a greater proportion of patients with the BRCA1 or BRCA2 variants had CHAARTED high-volume disease as compared with the patients with the HOXB13 variant; however, this difference was not statistically significant (P = 0.07, two-sided Fischer’s exact test).
Table 1.
Patient and tumour characteristics at initial diagnosis in the germline variant study (n = 549).
| Factor | Group | All patients | Pathogenic germline BRCA1/2, PALB2 or ATM variants | |
|---|---|---|---|---|
| Yes | No | |||
| n = 549 | n = 29 | n = 520 | ||
| Age (median [IQR]) (year old) | 69.7 [64.0, 75.5] | 67.1 [58.5, 72.0] | 69.9 [64.1, 75.6] | |
| Initial PSA (median [IQR]) (ng/mL) | 111 [29.8, 469.3] | 255.5 [60.1, 867.0] | 108.5 [29.1, 432.8] | |
| Albumin (median [IQR]) (g/dL) at diagnosis | 4.2 [3.9, 4.4] | 4.0 [3.9, 4.3] | 4.2 [3.9, 4.4] | |
| LDH (median [IQR]) (U/L) at diagnosis | 192 [169, 237] | 212 [172, 251] | 191 [169, 237] | |
| Family history of cancer | ||||
| Prostate cancer (%) | No | 382 (96.0) | 14 (82.4) | 368 (96.6) |
| Yes | 16 (4.0) | 3 (17.6) | 13 (3.4) | |
| NA | 151 | 12 | 139 | |
| Breast cancer (%) | No | 386 (97.0) | 15 (88.2) | 371 (97.4) |
| Yes | 12 (3.0) | 2 (11.8) | 10 (2.6) | |
| NA | 151 | 12 | 139 | |
| Ovarian cancer (%) | No | 396 (99.5) | 17 (100.0) | 379 (99.5) |
| Yes | 2 (0.5) | 0 (0.0) | 2 (0.5) | |
| NA | 151 | 12 | 139 | |
| Pancreatic cancer (%) | No | 387 (97.2) | 15 (88.2) | 372 (97.6) |
| Yes | 11 (2.8) | 2 (11.8) | 9 (2.4) | |
| NA | 151 | 12 | 139 | |
| Any of the above (%) | No | 360 (90.1) | 11 (64.7) | 349 (91.6) |
| Yes | 38 (9.5) | 6 (35.3) | 32 (8.4) | |
| NA | 151 | 12 | 139 | |
| Past history of cancer | ||||
| Breast cancer (%) | No | 485 (99.8) | 25 (100.0) | 460 (100.0) |
| Yes | 1 (0.2) | 0 (0.0) | 1 (0.2) | |
| NA | 63 | 4 | 59 | |
| Colon cancer (%) | No | 467 (96.1) | 24 (96.0) | 443 (96.1) |
| Yes | 19 (3.9) | 1 (4.0) | 18 (3.9) | |
| NA | 63 | 4 | 59 | |
| Pancreatic cancer (%) | No | 482 (99.2) | 25 (100.0) | 457 (99.1) |
| Yes | 4 (0.8) | 0 (0.0) | 4 (0.9) | |
| NA | 63 | 4 | 59 | |
| Any of the above (%) | No | 463 (95.3) | 24 (96.0) | 439 (95.2) |
| Yes | 23 (4.7) | 1 (4.0) | 22 (4.8) | |
| NA | 63 | 4 | 59 | |
| Biopsy Grade group (%) | 1 | 8 (1.8) | 0 (0.0) | 8 (1.9) |
| 2 | 23 (5.1) | 0 (0.0) | 23 (5.3) | |
| 3 | 33 (7.3) | 1 (4.3) | 32 (7.4) | |
| 4 | 133 (293) | 8 (34.8) | 125 (29.0) | |
| 5 | 257 (56.7) | 14 (60.9) | 243 (56.3) | |
| NA | 95 | 6 | 89 | |
| Jewett stage (%) | A | 4 (0.7) | 0 (0.0) | 4 (0.8) |
| B | 38 (7.0) | 1 (3.4) | 37 (7.2) | |
| C | 56 (10.3) | 1 (3.4) | 55 (10.7) | |
| D1 | 39 (7.2) | 4 (13.8) | 35 (6.8) | |
| D2 | 406 (74.8) | 23 (79.3) | 383 (74.5) | |
| NA | 6 | 0 | 6 | |
| *CHAARTED volume (%) | Low | 153 (43.6) | 9 (39.1) | 144 (43.9) |
| High | 198 (56.4) | 14 (60.9) | 184 (56.1) | |
| NA | 100 | 4 | 96 | |
| Bone metastasis (%) | No | 165 (34.6) | 7 (28.0) | 158 (34.6) |
| Yes | 317 (65.8) | 18 (72.0) | 299 (65.4) | |
| NA | 67 | 4 | 63 | |
| Extent of disease (EOD) (%) | 0 | 165 (36.8) | 7 (29.2) | 158 (37.2) |
| 1 | 117 (26.1) | 5 (20.8) | 112 (26.4) | |
| 2 | 83 (18.5) | 6 (25.0) | 77 (18.1) | |
| 3 | 59 (13.1) | 4 (16.7) | 55 (12.9) | |
| 4 | 25 (5.6) | 2 (8.3) | 23 (5.4) | |
| NA | 100 | 5 | 95 | |
| Lung metastasis (%) | No | 439 (91.7) | 23 (92.0) | 416 (91.6) |
| Yes | 40 (8.4) | 2 (8.0) | 38 (8.4) | |
| NA | 70 | 4 | 66 | |
| Liver metastasis (%) | No | 471 (98.3) | 25 (100.0) | 446 (98.2) |
| Yes | 8 (1.7) | 0 (0.0) | 8 (1.8) | |
| NA | 70 | 4 | 66 | |
PS performance status by Common Toxicity Criteria, Version 2.0; Grade group by ISUP 2014, NA not available.
*CHAARTED volume was assigned only in the metastatic cases.
Fig. 2.
Summary of the genetic and clinical profiles of patients with germline variants in the 27 analysed genes.
Clinical features of cases with germline HBOC-associated gene variants
Thirty-four cases with germline variants in the HBOC-associated genes (BRCA1, BRCA2, PALB2, ATM, BRIP1 and NBN) were detected. To precisely examine the features of PCa with germline variants in the HBOC-associated genes, we exclusively focused on cases with variants that have a clear pathogenicity annotation (pathogenic or likely pathogenic) based on the ClinVar database (BRCA2, 18 cases; BRCA1, two cases; PALB2, five cases; and ATM, four cases). All the variants were a frameshift deletion, a stop-gain variant, or a splice-site variant, and no missense variant was found. The patient and tumour characteristics of patients with and without known pathogenic variants in the above four HBOC-associated genes are summarised in Table 1. The age at diagnosis was significantly lower in the variant-positive group (median = 67.1 years; IQR, 58.5–72.0 years) than in the variant-negative group (median = 69.9 years; IQR, 64.4–75.6 years; P = 0.04). The PSA level at diagnosis was also significantly higher in the variant-positive group (median = 255. 5 ng/mL; IQR, 60.1–867.0 ng/mL) than in the variant-negative group (median = 108.5 ng/mL; IQR, 29.1–432.8 ng/mL; P = 0.04). Furthermore, the variant-positive group also had a significantly higher number of patients with a family history of HBOC-associated cancers and PCa (P = 0.001 and 0.03, respectively). In all, 92% and 82.6% of the variant-positive and negative groups, respectively, were castration-resistant at the time of the study (Supplementary Table 2). A higher proportion of patients in the variant-positive group were treated with the following agents than that in the variant-negative group: ARPI (72.0% vs 56.9%), docetaxel (64.0% vs 42.9%), or platinum chemotherapy (32.0% vs 14.3%), reflecting the aggressive nature of the cases with HBOC-associated gene mutations.
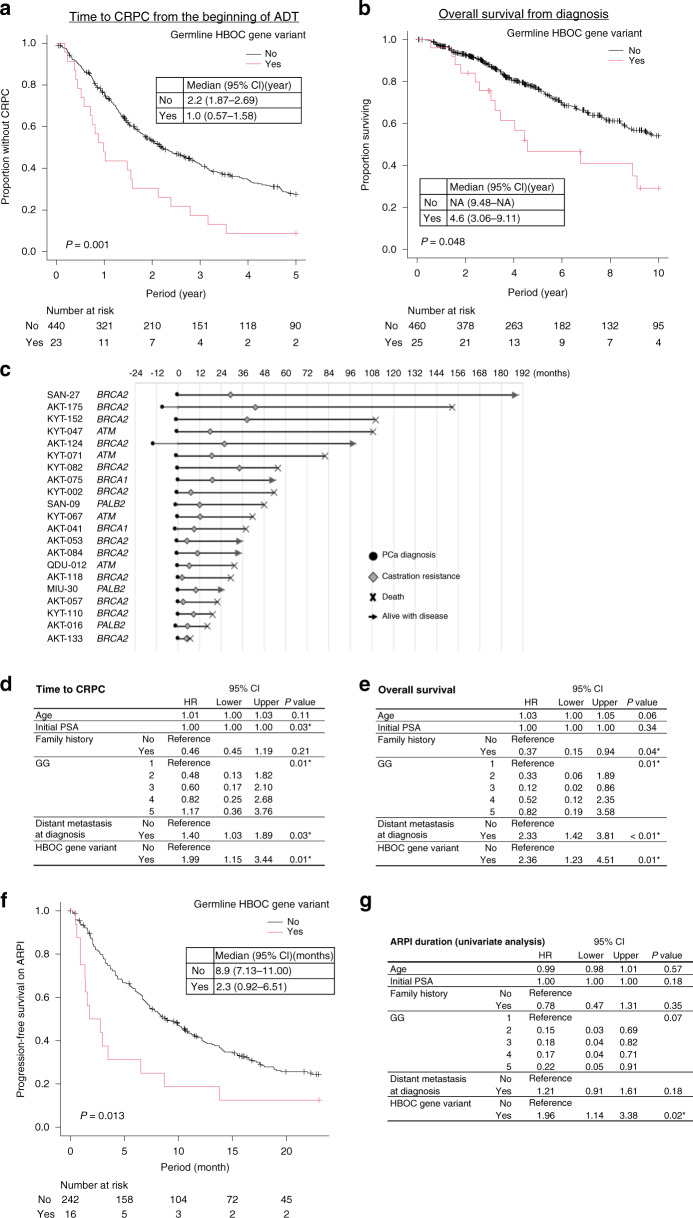
Germline HBOC-associated gene variants and duration of response to hormonal therapy
Next, we examined the existence of any association between germline HBOC-associated gene variants and the duration of response to hormonal therapy. At the time of analysis, 16 (55.2%) and 172 (33.1%) patients with and without germline HBOC-associated gene variants were deceased, respectively. We identified that patients with germline HBOC-associated gene variants had a significantly shorter time to CRPC and a shorter OS (Fig. 3a, b and Supplementary Fig. 2). The time to CRPC and the OS for each case with germline HBOC-associated gene variants are shown as a swimmer plot in Fig. 3c. Importantly, HBOC-associated gene variants were found to be independently associated with a shorter time to CRPC and a shorter OS from diagnosis based on multivariable analysis after adjusting for major clinical variables (Fig. 3d, e). Conversely, no difference in time to CRPC or OS in the presence of the HOXB13 variant was detected (Supplementary Fig. 3A, B). Further, among the patients treated with ARPIs, the time to progression was significantly shorter for cases with germline HBOC-associated gene variants (Fig. 3f, g). Considering that BRCA1 may have a weaker association with the aggressive phenotype of PCa [32], we also conducted a sensitivity analysis excluding BRCA1 from HBOC-associated gene variants; however, due to the small number of BRCA1 carriers, the results were unchanged (Supplementary Fig. 4). Taken together, these findings indicated that the presence of HBOC-associated gene variants is a significant prognostic factor of the duration of response to hormonal therapy.
Fig. 3. Prognostic value of pathogenic germline variants in HBOC-associated genes in patients treated with hormonal therapy.
a Kaplan–Meier curve showing time to castration resistance from the initiation of androgen deprivation therapy (ADT) for patients with and without known pathogenic germline variants in HBOC-associated genes (BRCA1, BRCA2, ATM and PALB2). Group differences were tested by performing log-rank tests. b Kaplan–Meier curve showing overall survival (OS) from the time of diagnosis for patients with and without pathogenic germline variants in HBOC-associated genes. c Swimmer plot of the cases with known pathogenic germline variants in HBOC-associated genes. Cases with insufficient data on time to castration resistance and OS were excluded. d Multivariable Cox regression analysis for the evaluation of the association between clinical and genetic variables and time to castration resistance from the initiation of ADT. Clinical factors that showed significant association with time to castration resistance based on the univariate analysis were included in the multivariable analysis. e Multivariable Cox regression analysis for the evaluation of the association between clinical and genetic variables and OS from initial diagnosis. Clinical factors that showed significant association with OS in the univariate analysis were included in the multivariable analysis. f Kaplan–Meier curve showing progression-free survival (PFS) for androgen receptor pathway inhibitors (ARPIs, abiraterone, or enzalutamide) for patients with and without known pathogenic germline variants in HBOC-associated genes. g Univariate Cox regression analysis for the evaluation of the association between clinical and genetic variables and PFS based on ARPIs. Multivariable analysis was not performed due to the limited number of events.
Clinical features of cases with somatic mutations in HBOC-associated genes
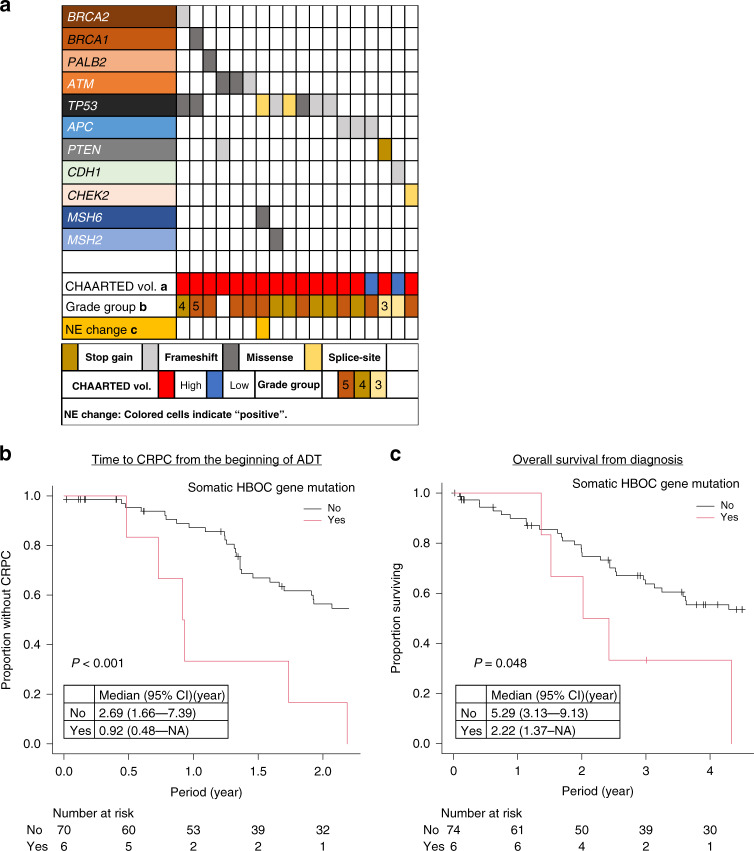
Reportedly, in PCa, somatic and germline variants in HBOC-associated genes exert a similar clinical impact in terms of response to PARP inhibitors [6]. Therefore, to further confirm the association between HBOC-associated gene variants and the duration of response to hormonal therapy, we also sequenced PCa tissues from an independent cohort of patients with metastatic prostate cancer and examined the somatic aberrations of these genes. The patient and tumour characteristics in the somatic mutation study are summarised in Table 2. The proportion of patients with pathogenic somatic mutations was 23% (18 patients) (Supplementary Fig. 5 and Supplementary Table 4). The most frequently mutated gene was TP53 (n = 8), followed by APC (n = 3), and ATM (n = 3). The genetic and clinical profiles of patients with somatic mutations are summarised in Fig. 4a. Six patients had somatic mutations in BRCA1, BRCA2, ATM, or PALB2. No difference in age and PSA levels at diagnosis between the groups with and without was noted. The two groups also showed a similar percentage of patients with GG4 or higher-grade cancer (83.3% vs. 89.0%). However, all cases with mutations had CHAARTED high-volume disease, whereas a quarter of the patients in the non-mutated group had CHAARTED low-volume disease. In addition, the time to CRPC in the mutated group was significantly shorter than that in the non-mutated group (Fig. 4b). The mutated group also tended to show shorter OS than the non-mutated group (2.2 years vs. 5.3 years, P = 0.048) (Fig. 4c). Reportedly, TP53 aberrations are associated with poor outcomes in metastatic PCa [33, 34]. Similar to previous studies, cases with TP53 mutations had a significantly shorter time to CRPC; however, the difference in OS did not show statistical significance, possibly owing to the small number of cases involved in the analysis (Supplementary Fig. 6A, B).
Table 2.
Patient and tumour characteristics in the somatic mutation study (n = 80).
| Factor | Group | All patients | Somatic BRCA1/2, PALB2 or ATM mutations | |
|---|---|---|---|---|
| Yes | No | |||
| n = 80 | n = 6 | n = 74 | ||
| Age (median [IQR]) (year old) | 73.5 [66.0, 80.0] | 73.5 [66.0, 81.8] | 73.5 [66.3, 79.8] | |
| Initial PSA (median [IQR]) (ng/mL) | 505.1 [133.3, 1912.0] | 309.0 [222.5, 620.5] | 507.6 [129.5, 1944.0] | |
| Albumin (median [IQR]) (g/dL) | 4.1 [3.7, 4.4] | 4.2 [4.1, 4.2] | 4.1 [3.7, 4.4] | |
| LDH (median [IQR]) (U/L) | 207 [174, 262] | 258 [224, 301] | 207 [172, 261] | |
| Biopsy Grade Group (%) | 1 | 1 (1.3) | 0 (0.0) | 1 (1.4) |
| 2 | 3 (3.8) | 0 (0.0) | 3 (4.1) | |
| 3 | 5 (6.3) | 1 (16.7) | 4 (5.5) | |
| 4 | 21 (26.6) | 2 (33.3) | 19 (26.0) | |
| 5 | 49 (62.0) | 3 (50.0) | 46 (63.0) | |
| NA | 1 | 0 | 1 | |
| Jewett stage (%) | A-C | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| D1 | 8 (10.1) | 0 (0.0) | 8 (11.0) | |
| D2 | 71 (88.8) | 6 (100.0) | 65 (89.0) | |
| NA | 1 | 0 | 1 | |
| Metastasis at initial diagnosis (%) | No | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Yes | 80 (100.0) | 6 (100.0) | 74 (100.0) | |
| CHAARTED volume (%) | Low | 18 (22.8) | 0 (0.0) | 18 (24.7) |
| High | 61 (77.2) | 6 (100.0) | 55 (75.3) | |
| NA | 1 | 0 | 1 | |
| Bone metastasis (%) | No | 24 (30.0) | 2 (33.3) | 22 (29.7) |
| Yes | 56 (70.0) | 4 (66.7) | 52 (70.3) | |
| Extent of disease (EOD) (%) | 0 | 18 (22.5) | 2 (33.3) | 16 (21.6) |
| 1 | 14 (17.5) | 0 (0.0) | 14 (18.9) | |
| 2 | 20 (25.0) | 3 (50.0) | 17 (23.0) | |
| 3 | 15 (18.8) | 0 (0.0) | 15 (20.3) | |
| 4 | 13 (16.3) | 1 (16.7) | 12 (16.2) | |
| Lung metastasis (%) | No | 72 (90.0) | 4 (66.7) | 68 (91.9) |
| Yes | 8 (10.0) | 2 (33.3) | 6 (8.1) | |
| Liver metastasis (%) | No | 77 (96.3) | 5 (83.3) | 72 (97.3) |
| Yes | 3 (3.8) | 1 (16.7) | 2 (2.7) | |
| Acquisition of castration resistance (%) | No | 32 (42.1) | 0 (0.0) | 32 (45.7) |
| Yes | 44 (57.9) | 6 (100.0) | 38 (54.3) | |
| NA | 4 | 0 | 4 | |
| Possible NE change (%) | No | 77 (96.3) | 6 (100.0) | 71 (95.9) |
| Yes | 3 (3.8) | 0 (0.0) | 3 (4.1) | |
NE neuroendocrine, NA not available.
Fig. 4. Summary of the genetic and clinical profiles of patients with somatic variants in the 27 analysed genes.
a Summary of the genetic and clinical profiles of patients with somatic mutations in the 27 analysed genes. b Kaplan–Meier curve showing time to castration resistance from initiation of androgen deprivation therapy (ADT) for patients with or without somatic mutations in HBOC-associated genes (BRCA1, BRCA2, PALB2 or ATM). Group differences were tested by performing log-rank tests. c Kaplan–Meier curve showing overall survival from the time of diagnosis for patients with and without somatic mutations in HBOC-associated genes.
Discussion
With the availability of PARP inhibitors as well as immune-checkpoint inhibitors, growing attention is given to the role of DNA repair–associated genes in PCa. However, even though genetic testing is recommended in several guidelines for patients with metastatic PCa [9, 10], in real-world settings, the test criteria (who, when and what) are still not standardised globally. For example, in the USA, several clinical germline multigene panels specifically designed for PCa are available, and all panels include BRCA1 and BRCA2 genes; however, in Japan, BRACAnalysis (Myriad Genetics, Salt Lake City, UT, USA) is the only approved germline panel for PCa patients, and the test is only available to CRPC patients. Under these circumstances, in addition to known risk factors such as age at diagnosis and family history, information on the prognostic and predictive significance of the variants tested is also important when making a shared decision to conduct genetic testing.
In localised prostate cancer, it has been reported that patients harbouring germline BRCA1 or BRCA2 variants tend to present with higher-grade, higher-stage disease, increased rates of lymph node involvement, and also show shorter cancer-specific survival [35–37]. Conversely, conflicting reports exist regarding the prognostic value of germline variants in HRR-associated genes in metastatic PCa [13, 18]. These contradictory results are primarily due to the sample sizes of these studies and the different genes included. In the PROREPAIR-B study [16], the impact of BRCA1, BRCA2, ATM and PALB2 germline variants on cause-specific survival (CSS) from the diagnosis of CRPC was evaluated. Even though there was no association between genetic variants and CSS when all genes were included, BRCA2 carriers had a significantly shorter CSS (17.4 vs. 33.2 months; P = 0.027). Another retrospective study analysed 319 patients with mCRPC and reported that patients with deleterious germline variants in BRCA1, BRCA2, ATM, PALB2 or CDK12 have a significantly shorter time from ADT initiation to CRPC (11.8 vs. 19.0 months, P = 0.031) and also shorter progression-free survival (PFS) on first-line AR-targeted therapy (3.3 vs. 6.2 months, P = 0.01) by log-rank test [14]. Conversely, pooled data from international studies in which the association between germline DNA repair gene variants and OS after CRPC as well as the duration of response to ARPI were tested showed no difference between patients with and without variants (3.2 vs. 3.0 years, P = 0.37 and 8.3 vs. 8.3 months, P = 0.94, respectively) [17]. However, the study included genes other than the four genes focused on in the present study, such as CHEK2, MSH1, and NBN, for which the clinical implications are less clear. Another study analysed the association between pathogenic BRCA/ATM variants and response to ARPI, revealing superior outcomes in those with BRCA/ATM variants (hazard ratio (HR) 0.52 (95% CI 0.28–0.98) for PFS and HR 0.34 (95% CI 0.12–0.99) for OS, respectively); however, only nine patients with pathogenic variants were included, and therefore, the study may be underpowered [15]. This study focused on cases with HRR-associated gene variants whose pathogenicity was confirmed based on the ClinVar database, and the presence of these variants evidently represented an independent prognostic factor of a shorter time to CRPC after adjusting for major clinical variables and also a shorter duration of response to ARPI based on univariate analysis.
Further, in this study, we investigated the prevalence of a set of known highly penetrant cancer-predisposing genes in lethal PCa among Japanese patients. The prevalence of BRCA2, BRCA1 and ATM mutations were 3.4, 0.4 and 0.9%, respectively. In a previous study that included 7636 unselected Japanese PCa patients and 12,366 healthy males, the observed prevalence of BRCA2, BRCA1 and ATM mutations were 1.1, 0.2 and 0.5% in patients with PCa and 0.2, 0.1 and 0.2% in the healthy males, respectively [21]. Similar to observations in Caucasians [8], our data revealed that the prevalence of mutations in genes mediating DNA repair increases as PCa progresses from a localised to a metastatic state. Additionally, these data also showed that the prevalence of these mutations in Japanese males at all disease states, from healthy controls to cases with lethal PCa, is lower than that in Caucasians [8, 16], except for the PALB2 gene (0.9%), whose prevalence has never been previously reported as higher than 0.5% in Caucasians. Differences in the locations of mutations between different ethnic groups have also been reported. In the present study, nine of the 19 BRCA2 variants were either p.Ile1859fs or p.Arg2318* variants, which reportedly, are frequently mutated in Japanese patients with breast cancer; however, they have not been identified in a large cohort study involving Caucasians [38, 39]. In addition, all HOXB13 variants identified in this study were either p.Gly132Glu or p.Gly17Val variants, which reportedly, are novel subpopulation-specific PCa-associated variants among Japanese [21]. There were no patients with p.Gly84Glu and p.Gly135Glu variants that have been reported from European and Chinese patients, respectively [40, 41].
We also investigated the mutational landscapes of the same genes in tissue samples from patients with metastatic PCa. The most frequently mutated gene was TP53 (n = 8, 10.0%) followed by APC (n = 3, 3.8%), and ATM (n = 3, 3.8%). Large-scale comprehensive profiling of advanced PCa reported that mutations (excluding copy number changes and fusions) of these three genes to be present in 46.7, 8.0 and 7.3% of the cases, respectively [5]. Although the prevalence of TP53 and PTEN mutations recorded in the present study were significantly lower than the previously reported values, a ctDNA analysis involving Japanese patients with CRPC also showed that the prevalence of alterations in the two genes were significantly lower (17.5% and 4.9%, respectively) [42]; this might be one of the characteristics of advanced PCa in Japanese patients. Similar to the results of germline analysis, somatic mutations in BRCA1, BRCA2, ATM and PALB2 were also found to be significantly associated with time to CRPC, and they showed nominal association with OS. A study exploring the genomic correlates of response to ARPI using ctDNA identified BRCA2 and ATM mutations to be significantly associated with a shorter duration of therapy [33]. We have also similarly shown in another study that BRCA2 and ATM mutations in ctDNA constitute an independent prognostic factor of PFS in patients treated with ARPIs for mCRPC even after adjusting for other clinical and genomic factors, including TP53, RB1 and AR aberrations [42]. Conversely, a study that involved the use of tissues showed a longer duration of response to abiraterone in patients with DNA repair gene defects [43]. However, the study included genes other than those investigated in this study, such as FANCA, RAD51B and RAD51C. The technical difficulty associated with detecting low-frequency and sub-clonal mutations in this study without matched germline samples as well as the small sample size possibly affected our results. Therefore, a large-scale somatic variant study to obtain more accurate and reliable results is necessary in the future.
Several potential clinical implications for the findings of the present study exist, depending on the timing of genetic tests. First, if a patient has not undergone a genetic test by the time of castration resistance, the association between short time to CRPC and HRR gene alterations can be important information to prompt a patient to undergo a genetic test because those with a short time to CRPC are more likely to test positive and could be treated by PARP inhibitors. Second, if a genetic test is performed at the time of initial diagnosis, even though HRR alterations are not predictive markers of AR pathway inhibition, those with HRR gene alterations may benefit from a new co-targeting strategy combining AR-targeted agents and PARP inhibitors as the first-line therapy for metastatic hormone-sensitive PCa. Several clinical trials are ongoing to test this hypothesis.
This study had some notable limitations. First, it was a hospital-based retrospective study, and the samples were randomly collected, which may raise selection bias. However, there were no exclusion criteria upon sample collection; primarily, the samples were collected consecutively. Second, samples were collected at different periods at each institution. Therefore, large heterogeneity in the therapeutic management, which possibly affected the clinical outcomes, especially OS, is present. However, given that all the samples were collected during the period when the use of ARPI or docetaxel for castration-sensitive PCa was not reimbursed in Japan, all patients were treated with either combined androgen blockade by bicalutamide/flutamide or by ADT alone. Therefore, until the time of CRPC, all the patients received a similar treatment. Another limitation is the lack of copy number analysis of the tissue samples. Homozygous loss of BRCA2 is an important genomic alteration reported in some cases of PCa. We attempted to examine copy number loss of several PCa-associated tumour suppressors including PTEN, TP53, RB1, and BRCA2 by digital PCR. However, as copy number loss is often focal, we could not reliably detect copy number loss by this approach. Finally, owing to the small number of cases with HBOC-associated gene variants, we could not analyse each gene separately. ATM is a signalling kinase that is activated by DNA double-strand break and activates downstream signalling proteins such as CHEK2. Since ATM is located at the upper stream of the HRR pathway compared to BRCA1 and BRCA2, impact of ATM variants in the HRR pathway may be different compared to BRCA1 or BRCA2 variants [44–47]. In addition, unlike ovarian cancer, the association between BRCA1 variants and PCa or its aggressiveness might be weaker compared to that seen with BRCA2 variants [32, 48]. However, in this study, limited by sample size, we could not detect any apparent difference clinically between the genes. In the future, examination of the association between the pathogenic variants in each gene and the clinical outcomes is necessary.
In summary, we have shown that germline and somatic variants in BRCA1, BRCA2, ATM and PALB2 genes have prognostic significance in Japanese individuals. This information could affect the patient’s decision to undergo a genetic test and should be shared with patients when recommending a genetic test to those with metastatic PCa.
Supplementary information
Acknowledgements
We thank Ms. Eriko Komaki at the Department of Urology, Kyoto University for her technical assistance. The super-computing resource was provided by the Human Genome Center, Institute of Medical Science, the University of Tokyo.
Author contributions
SA, MS, SN, NT, NF, HN and YM designed the study. HK, KM, YM, and SA wrote the manuscript. YI, NH and YM performed the sequencing and bioinformatics analyses. HK, KM and SA performed the statistical analyses. HK, KM, MS, SN, NT, NF, KO, SH, SI, TS, TG, T Kobayashi, T Kamoto, ME, TH and OO contributed to sample collection and clinical data acquisition. SA and OO acquired funding.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (grant number 20H03814). SA received research funding from the Takeda Science Foundation to partially support this study.
Data availability
The sequence data of this study can be accessed at the National Bioscience Database Center (NBDC) under accession number JGAS000509. As the current study used archival blood samples, a statement regarding the deposition of genomic data to public repositories was not included in some of the informed consent forms used in this study. Therefore, a public depository of genomic data was not possible considering the Personal Information Protection Law in Japan for some of the samples included in the present study. However, secondary use of genomic and clinical data is allowed under certain conditions. Please contact the corresponding author for details.
Competing interests
SA received research funding from Astellas Pharma, AstraZeneca, and Tosoh, outside of the submitted work and honoraria from Janssen Pharmaceutical, AstraZeneca, Astellas Pharma, Sanofi, Bayer and Takeda Pharmaceutical. MS received research funding from Daiichi Sankyo Company and honoraria from Janssen Pharmaceutical, AstraZeneca and Astellas Pharma. SN received honoraria from Janssen Pharmaceutical, Bayer, AstraZeneca, Takeda Pharmaceutical, Sanofi, Nippon Shinyaku and Astellas Pharma. NF received funding from Takeda Pharmaceutical and Sanofi and honoraria from Janssen Pharmaceutical, Takeda Pharmaceutical, Astellas Pharma and Nippon Shinyaku. T Kobayashi received funding from AstraZeneca and Chugai Pharmaceutical and honoraria from Janssen Pharma, AstraZeneca, Chugai Pharmaceutical, Bayer, MSD, Sanofi, Takeda, Astellas, Nippon Shinyaku, Nihon Kayaku, Merck and Pfizer. ME received research funding from Sanofi, Bayers, Astellas Pharma, Ono Pharmaceutical and Takeda Pharmaceutical and honoraria from Ono Pharmaceutical, Takeda Pharmaceutical, Novartis, Pfizer, Bristol Myers Squibb, Janssen Pharmaceutical, MSD and Merck. TH received research funding from Takeda Pharmaceutical, Astellas Pharma, Daiichi Sankyo Company, Sanofi and Bayer and honoraria from Janssen Pharmaceutical, Takeda Pharmaceutical, Astellas Pharma, Daiichi Sankyo Company, AstraZeneca, Sanofi and Bayer. OO received research funding from Shimazu, Astellas Pharma, and Chugai Pharmaceutical and honoraria from Sanofi, Nihon Kayaku, MSD, Bayer, Daiichi Sankyo Company, Ono Pharmaceutical, Nippon Shinyaku and Takeda Pharmaceutical.
Ethics approval and consent to participate
The study, which was conducted in accordance with the Declaration of Helsinki, was approved by the ethics committees of RIKEN, Akita University, Kyusyu University, University of Occupational and Environmental Health, Miyazaki University, Kyoto University (approval number G1154), and the Japanese Red Cross Otsu Hospital. All participants at Akita University, Kyusyu University, University of Occupational and Environmental Health, Miyazaki University, and Kyoto University provided written informed consent for the genomic analysis of their blood samples. Regarding the archived biopsy samples obtained from patients at the Japanese Red Cross Otsu Hospital, even though the patients provided informed consent for the use of their material, genomic analysis was not specified in the consent form. Therefore, to prevent patient re-identification, under the guidance of the ethics committee of Kyoto University, all biopsy samples and clinical data were completely anonymized before the study was conducted.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hiroko Kimura, Kei Mizuno.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01915-2.
References
- 1.Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med. 2017;377:352–60.. doi: 10.1056/NEJMoa1704174. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong AJ, Azad AA, Iguchi T, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, et al. Improved survival with enzalutamide in patients with metastatic hormone-sensitive prostate cancer. J Clin Oncol. 2022;40:1616–22. doi: 10.1200/JCO.22.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi KN, Agarwal N, Bjartell A, Chung BH, Pereira de Santana Gomes AJ, Given R, et al. Apalutamide for metastatic, castration-sensitive prostate cancer. N Engl J Med. 2019;381:13–24. doi: 10.1056/NEJMoa1903307. [DOI] [PubMed] [Google Scholar]
- 4.Attard G, Murphy L, Clarke NW, Cross W, Jones RJ, Parker CC, et al. Abiraterone acetate and prednisolone with or without enzalutamide for high-risk non-metastatic prostate cancer: a meta-analysis of primary results from two randomised controlled phase 3 trials of the STAMPEDE platform protocol. Lancet. 2022;399:447–60.. doi: 10.1016/S0140-6736(21)02437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med. 2020;383:2345–57.. doi: 10.1056/NEJMoa2022485. [DOI] [PubMed] [Google Scholar]
- 7.Boussios S, Rassy E, Shah S, Ioannidou E, Sheriff M, Pavlidis N. Aberrations of DNA repair pathways in prostate cancer: a cornerstone of precision oncology. Expert Opin Ther Targets. 2021;25:329–33.. doi: 10.1080/14728222.2021.1951226. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker C, Castro E, Fizazi K, Heidenreich A, Ost P, Procopio G, et al. Prostate cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31:1119–34.. doi: 10.1016/j.annonc.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Giri VN, Knudsen KE, Kelly WK, Cheng HH, Cooney KA, Cookson MS, et al. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol. 2020;38:2798–811. doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shore N, Ionescu-Ittu R, Yang L, Laliberté F, Mahendran M, Lejeune D, et al. Real-world genetic testing patterns in metastatic castration-resistant prostate cancer. Future Oncol. 2021;17:2907–21.. doi: 10.2217/fon-2021-0153. [DOI] [PubMed] [Google Scholar]
- 12.Leith A, Ribbands A, Last M, Gayle A, Payne S, McCrea C, et al. Genomic/genetic testing patterns for patients with metastatic castrate-resistant prostate cancer: Results from a real-world study in the United States. J Clin Oncol. 2021;39(6_suppl):49. doi: 10.1200/JCO.2021.39.6_suppl.49. [DOI] [Google Scholar]
- 13.Kolinsky MP, Niederhoffer KY, Kwan EM, Hotte SJ, Hamilou Z, Yip SM, et al. Considerations on the identification and management of metastatic prostate cancer patients with DNA repair gene alterations in the Canadian context. Can Urol Assoc .J. 2022;16:132–43.. doi: 10.5489/cuaj.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annala M, Struss WJ, Warner EW, Beja K, Vandekerkhove G, Wong A, et al. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol. 2017;72:34–42. doi: 10.1016/j.eururo.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Antonarakis ES, Lu C, Luber B, Liang C, Wang H, Chen Y, et al. Germline DNA-repair gene mutations and outcomes in men with metastatic castration-resistant prostate cancer receiving first-line abiraterone and enzalutamide. Eur Urol. 2018;74:218–25.. doi: 10.1016/j.eururo.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro E, Romero-Laorden N, Del Pozo A, Lozano R, Medina A, Puente J, et al. PROREPAIR-B: a prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2019;37:490–503. doi: 10.1200/JCO.18.00358. [DOI] [PubMed] [Google Scholar]
- 17.Mateo J, Cheng HH, Beltran H, Dolling D, Xu W, Pritchard CC, et al. Clinical outcome of prostate cancer patients with germline DNA repair mutations: retrospective analysis from an international study. Eur Urol. 2018;73:687–93.. doi: 10.1016/j.eururo.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messina C, Cattrini C, Soldato D, Vallome G, Caffo O, Castro E, et al. BRCA mutations in prostate cancer: prognostic and predictive implications. J Oncol. 2020;2020:4986365. doi: 10.1155/2020/4986365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenthal ET, Bernhisel R, Brown K, Kidd J, Manley S. Clinical testing with a panel of 25 genes associated with increased cancer risk results in a significant increase in clinically significant findings across a broad range of cancer histories. Cancer Genet. 2017;218–219:58–68. doi: 10.1016/j.cancergen.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Momozawa Y, Akiyama M, Kamatani Y, Arakawa S, Yasuda M, Yoshida S, et al. Low-frequency coding variants in CETP and CFB are associated with susceptibility of exudative age-related macular degeneration in the Japanese population. Hum Mol Genet. 2016;25:5027–34.. doi: 10.1093/hmg/ddw335. [DOI] [PubMed] [Google Scholar]
- 21.Momozawa Y, Iwasaki Y, Hirata M, Liu X, Kamatani Y, Takahashi A, et al. Germline pathogenic variants in 7,636 Japanese patients with prostate cancer and 12,366 controls. J Natl Cancer Inst. 2019;112:369–76. [DOI] [PMC free article] [PubMed]
- 22.Hashimoto M, Saito Y, Nakagawa R, Ogahara I, Takagi S, Takata S, et al. Combined inhibition of XIAP and BCL2 drives maximal therapeutic efficacy in genetically diverse aggressive acute myeloid leukemia. Nat Cancer. 2021;2:340–56.. doi: 10.1038/s43018-021-00177-w. [DOI] [PubMed] [Google Scholar]
- 23.Saiki R, Momozawa Y, Nannya Y, Nakagawa MM, Ochi Y, Yoshizato T, et al. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. Nat Med. 2021;27:1239–49.. doi: 10.1038/s41591-021-01411-9. [DOI] [PubMed] [Google Scholar]
- 24.Momozawa Y, Sasai R, Usui Y, Shiraishi K, Iwasaki Y, Taniyama Y, et al. Expansion of cancer risk profile for BRCA1 and BRCA2 pathogenic variants. JAMA Oncol. 2022;8:871–8. [DOI] [PMC free article] [PubMed]
- 25.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soloway MS, Hardeman SW, Hickey D, Raymond J, Todd B, Soloway S, et al. Stratification of patients with metastatic prostate cancer based on extent of disease on initial bone scan. Cancer. 1988;61:195–202. doi: 10.1002/1097-0142(19880101)61:1<195::AID-CNCR2820610133>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N Engl J Med. 2015;373:737–46. doi: 10.1056/NEJMoa1503747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyberg T, Frost D, Barrowdale D, Evans DG, Bancroft E, Adlard J, et al. Prostate cancer risks for male BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Eur Urol. 2020;77:24–35. doi: 10.1016/j.eururo.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–57.. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 34.De Laere B, Oeyen S, Mayrhofer M, Whitington T, van Dam PJ, Van Oyen P, et al. Outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2019;25:1766–73.. doi: 10.1158/1078-0432.CCR-18-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castro E, Goh C, Leongamornlert D, Saunders E, Tymrakiewicz M, Dadaev T, et al. Effect of BRCA mutations on metastatic relapse and cause-specific survival after radical treatment for localised prostate cancer. Eur Urol. 2015;68:186–93. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–57. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghose A, Moschetta M, Pappas-Gogos G, Sheriff M, Boussios S. Genetic aberrations of DNA repair pathways in prostate cancer: translation to the clinic. Int J Mol Sci. 2021;22:9783. [DOI] [PMC free article] [PubMed]
- 38.Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–11.. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momozawa Y, Iwasaki Y, Parsons MT, Kamatani Y, Takahashi A, Tamura C, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083. doi: 10.1038/s41467-018-06581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–9. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Qu L, Chen Z, Xu C, Ye D, Shao Q, et al. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2013;73:169–75. doi: 10.1002/pros.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno K, Sumiyoshi T, Okegawa T, Terada N, Ishitoya S, Miyazaki Y, et al. Clinical impact of detecting low-frequency variants in cell-free DNA on treatment of castration-resistant prostate cancer. Clin Cancer Res. 2021;27:6164–73.. doi: 10.1158/1078-0432.CCR-21-2328. [DOI] [PubMed] [Google Scholar]
- 43.Hussain M, Daignault-Newton S, Twardowski PW, Albany C, Stein MN, Kunju LP, et al. Targeting androgen receptor and DNA repair in metastatic castration-resistant prostate cancer: results from NCI 9012. J Clin Oncol. 2018;36:991–9. doi: 10.1200/JCO.2017.75.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokolova AO, Marshall CH, Lozano R, Gulati R, Ledet EM, De Sarkar N, et al. Efficacy of systemic therapies in men with metastatic castration resistant prostate cancer harboring germline ATM versus BRCA2 mutations. Prostate. 2021;81:1382–9. doi: 10.1002/pros.24236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall CH, Sokolova AO, McNatty AL, Cheng HH, Eisenberger MA, Bryce AH, et al. Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur Urol. 2019;76:452–8. doi: 10.1016/j.eururo.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carreira S, Porta N, Arce-Gallego S, Seed G, Llop-Guevara A, Bianchini D, et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov. 2021;11:2812–27. doi: 10.1158/2159-8290.CD-21-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah S, Rachmat R, Enyioma S, Ghose A, Revythis A, Boussios S. BRCA mutations in prostate cancer: assessment, implications and treatment considerations. Int J Mol Sci. 2021;22:12628. [DOI] [PMC free article] [PubMed]
- 48.Taza F, Holler AE, Fu W, Wang H, Adra N, Albany C, et al. Differential activity of PARP inhibitors in BRCA1-versus BRCA2-altered metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2021;5:1200–20. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data of this study can be accessed at the National Bioscience Database Center (NBDC) under accession number JGAS000509. As the current study used archival blood samples, a statement regarding the deposition of genomic data to public repositories was not included in some of the informed consent forms used in this study. Therefore, a public depository of genomic data was not possible considering the Personal Information Protection Law in Japan for some of the samples included in the present study. However, secondary use of genomic and clinical data is allowed under certain conditions. Please contact the corresponding author for details.