Abstract
DNA polymorphism of the bp26 gene, coding for a diagnostic protein antigen for brucellosis, was assessed by PCR and restriction fragment length polymorphism analysis using primers to amplify the bp26 gene with its flanking regions. Surprisingly, whereas PCR performed on DNA of the reference strains of the six recognized Brucella species produced a product of the expected size (1,029 bp), PCR performed on DNA of three representative strains from marine mammals (from a seal, a dolphin, and a porpoise) produced a larger product, of about 1,900 bp. Nucleotide sequencing of the 1,900-bp PCR products revealed the presence of an insertion sequence, IS711, downstream of the bp26 gene and adjacent to a Bru-RS1 element previously described as being a hot spot for IS711 insertion. PCR performed on a large number of field strains from different geographic origins and from marine mammal isolates indicated that the occurrence of an IS711 element downstream of the bp26 gene was a feature specific to the marine mammal Brucella strains. Thus, this PCR assay is able to differentiate Brucella terrestrial isolates from marine mammal isolates and could be applied for diagnostic purposes.
Brucellae are gram-negative, facultative, intracellular bacteria that can infect many species of animals, as well as humans. Six species are recognized within the genus Brucella: B. abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae (8). This classification is mainly based on differences in pathogenicity and host preference (8). The main pathogenic species worldwide are B. abortus, which is responsible for bovine brucellosis, B. melitensis, the main etiologic agent of ovine and caprine brucellosis; and B. suis, which is responsible for swine brucellosis. These three Brucella species may cause abortion in their hosts, which results in huge economic losses. B. ovis and B. canis are responsible for ram epididymitis and canine brucellosis, respectively. For B. neotomae, only strains isolated from desert rats have been reported. Distinction between species and biovars is currently performed by differential tests based on phenotypic characterization of lipopolysaccharide antigens, phage typing, dye sensitivity, CO2 requirement, H2S production, and metabolic properties (2).
Brucella strains have also been isolated from a great variety of wildlife species, such as bison, elk, feral swine, wild boars, foxes, hares, African buffalo, reindeer, and caribou (9).
The broad spectrum of Brucella hosts has recently been enlarged to include marine mammals. A number of recent reports have described the isolation and characterization of Brucella strains from a wide variety of marine mammals, such as bottlenose dolphins (Tursiops truncatus), common seals (Phoca vitulina), harbor porpoises (Phocoena phocoena), common dolphins (Delphinus delphis), Atlantic white-sided dolphins (Lagenorhynchus acutus), striped dolphins (Stenella caeruleoalba), hooded seals (Cystophora cristata), grey seals (Halichoerus grypus), a minke whale (Balaenoptera acutorostrata), and an otter (Lutra lutra) (3, 5, 10, 13, 18, 22). These strains were identified as brucellae by their colonial and cell morphology, staining characteristics, biochemical activity, agglutination by monospecific antisera, susceptibility to lysis by a Brucella-specific bacteriophage, and metabolic profiles. However, their overall characteristics were not assimilable to those of any of the six recognized Brucella species. Therefore, it was suggested that they comprise a new species to be called B. maris based on the current classification system (18).
It has been shown, on the basis of DNA-DNA hybridization studies, that the genus Brucella is a highly homogeneous group (>90% DNA homology for all species), and it has been proposed that this genus should comprise only one genomic species (26). Brucella strains isolated from marine mammals also fall into this homogeneous group according to DNA-DNA hybridization (25). Thus, several techniques have been employed to find DNA polymorphisms which would enable the molecular typing of the Brucella species and their different biovars (1, 4, 7, 11, 12, 14, 16, 17, 20, 21, 27).
The BP26 protein, also named Omp28, has been previously identified as an immunodominant antigen in Brucella infections of cattle, sheep, and humans (6, 19, 23). In the present study, DNA polymorphisms of the bp26 gene, coding for this protein, were assessed by PCR-restriction fragment length polymorphism analysis. Primers were designed to amplify the entire bp26 gene, with its flanking regions, based on the bp26 nucleotide sequence of B. melitensis 16M (GenBank accession no. U45996) (6). The primers used were 26A (forward primer; 5′ GCCCCTGACATAACCCGCTT 3′) and 26B (reverse primer; 5′ GAGCGTGACATTTGCCGATA 3′). PCR was performed on extracted DNAs as described previously (7, 27). Briefly, amplification reaction mixtures were prepared in volumes of 100 μl containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100 (1× PCR buffer; Promega, Charbonnieres, France), a 200 μM each concentration of deoxynucleoside triphosphate, a 1 μM concentration of each primer, 100 ng of genomic DNA, and 5 U of Taq DNA polymerase (Promega). The temperature cycling for the amplification was performed in a GeneAmp PCR system 9600 thermocycler (Perkin-Elmer) as follows: cycle 1 was 94°C for 5 min (denaturation); the next 30 cycles were 58°C for 1 min (annealing), 70°C for 1 min 30 s (extension), and 94°C for 1 min (denaturation); and the last cycle was 58°C for 1 min (annealing) and 70°C for 10 min (extension). The PCR products were run on 1% (wt/vol) agarose gels containing 0.5 μg of ethidium bromide per ml.
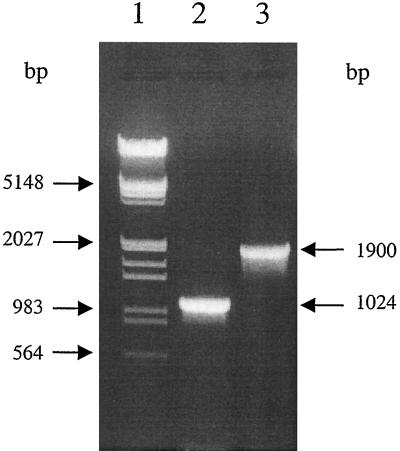
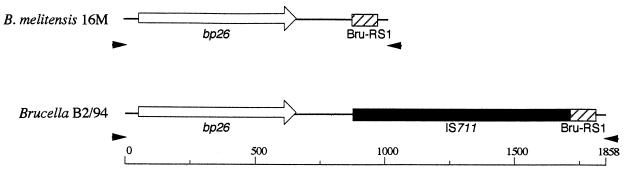
Surprisingly, whereas PCR performed on DNA of the reference strains of the six recognized Brucella species produced a product of the expected size (1,029 bp), PCR performed on DNA of three representative strains from marine mammals (a seal, a dolphin, and a porpoise) produced a larger product, of about 1,900 bp (Fig. 1). The nucleotide sequences of the 1,900-bp PCR products of the three marine Brucella strains (B2/94, B1/94, and B14/94) were determined, and they revealed the presence of an insertion sequence, IS711, downstream of the bp26 gene (Fig. 2). Interestingly, the IS711 element was found adjacent to a Bru-RS1 element described as being a hot spot for IS711 insertion (15). Bru-RS1 is a repeated palindromic DNA element of 103 bp which is highly conserved among brucellae and found more than 35 times in the Brucella genome (15). Such a Bru-RS1 element was previously described to occur downstream of the bp26 gene of B. melitensis 16M (19). Insertion of the IS711 element resulted in duplication of the nucleotides TA (data not shown) at the target site, as previously described for B. ovis (16). To assess whether the occurrence of an IS711 element downstream of the bp26 gene was specific to Brucella strains isolated from marine mammals, PCR with primers 26A and 26B was performed on a large number of field strains of Brucella from different geographic origins and a large number of the recent isolates from different marine mammals (Tables 1 and 2). All terrestrial isolates, including B. ovis strains for which a higher number of IS711 copies have been described (16, 17, 21), showed a PCR profile in an agarose gel with a band of size of 1,029 bp, whereas PCR on all marine mammal isolates showed the typical 1.9-kb band, implying the presence of the IS711 element (data not shown). The bp26 gene by itself, as shown by restriction fragment length polymorphism analysis with different restriction enzymes (AluI, ClaI, EcoRII, EcoRV, HaeII, HaeIII, HinfI, PstI, Sau3A, StyI, and TaqI) and nucleotide sequencing, did not appear to be useful for molecular typing purposes and thus must be rather conserved among brucellae (data not shown). Only a few differences were observed in the bp26 gene, and these were in B. abortus strains (data not shown).
FIG. 1.
PCR-amplified bp26 gene using primers 26A and 26B of B. melitensis 16M (lane 2) and seal isolate B2/94 (lane 3) run on a 1% agarose gel. Lane 1, λ DNA EcoRI/HindIII ladder (Appligene, Illkirch, France).
FIG. 2.
Schematic view deduced from nucleotide sequencing of the bp26 gene and flanking regions of B. melitensis 16M and seal isolate B2/94. Arrowheads indicate the locations of the primers used for PCR.
TABLE 1.
Brucella reference, vaccine, and field strains from terrestrial mammals used in this study
| Species | Biovar | Straina | Host or source | Geographic origin |
|---|---|---|---|---|
| B. abortus | 1 | 544 (ATCC 23448; BCCN R4)b | Cattle | England |
| 1 | B19 (BCCN V1)c | Cattle | United States | |
| 1 | 99S (BCCN R20) | Cattle | United States | |
| 1 | 2308 (BCCN R23) | Cattle | United States | |
| 1 | BCCN 92-73 | Cattle | France | |
| 1 | BCCN 94-44 | Human | Algeria | |
| 1 | BCCN 95-19 | Chamois | France | |
| 1 | BCCN 95-50 | Cattle | Argentina | |
| 1 | BCCN 95-51 | Cattle | Argentina | |
| 1 | BCCN 95-55 | Cattle | Costa Rica | |
| 1 | BCCN 96-62 | Cattle | Italy | |
| 2 | 86/8/59 (ATCC 23449; BCCN R5)b | Cattle | England | |
| 3 | Tulya (ATCC 23450; BCCN R6)b | Human | Uganda | |
| 3 | BCCN 91-90 | Cattle | Greece | |
| 3 | BCCN 92-25 | Cattle | France | |
| 3 | BCCN 92-104 | Cattle | Guinea | |
| 3 | BCCN 93-15 | Cattle | Spain | |
| 3 | BCCN 93-26 | Dromedary | Soudan | |
| 3 | BCCN 94-18 | Cattle | France | |
| 3 | BCCN 94-19 | Cattle | France | |
| 3 | BCCN 94-63 | Cattle | France | |
| 3 | BCCN 95-7 | Cattle | France | |
| 3 | BCCN 95-12 | Cattle | France | |
| 4 | 292 (ATCC 23451; BCCN R7)b | Cattle | England | |
| 5 | B3196 (ATCC 23452; BCCN R8)b | Cattle | England | |
| 6 | 870 (ATCC 23453; BCCN R9)b | Cattle | Africa | |
| 9 | C68 (ATCC 23455; BCCN R11)b | Cattle | England | |
| R | 45/20 (BCCN V2)c | Cattle | England | |
| R | RB51 (BCCN V5)c | Cattle | United States | |
| B. melitensis | 1 | 16M (ATCC 23456; BCCN R1)b | Goat | United States |
| 1 | Rev.1 (BCCN V4a)c | Goat | Mexico | |
| 1 | BCCN 75-478 | Sheep | Israel | |
| 1 | BCCN 87-92 | Human | United States | |
| 1 | BCCN 88-42 | Sheep | Israel | |
| 1 | BCCN 90-61 | Sheep | South Africa | |
| 1 | BCCN 92-70 | Human | France | |
| 1 | BCCN 92-106c | Unknown | Algeria | |
| 1 | BCCN 93-2 | Human | France | |
| 1 | BCCN 93-4 | Human | France | |
| 1 | BCCN 94-37 | Human | France | |
| 1 | BCCN 96-28 | Sheep | Israel | |
| 2 | 63/9 (ATCC 23457; BCCN R2)b | Goat | Turkey | |
| 3 | Ether (ATCC 23458; BCCN R3)b | Goat | Italy | |
| 3 | BCCN 83-198 | Human | Spain | |
| 3 | BCCN 90-112 | Cattle | Greece | |
| 3 | BCCN 92-80 | Sheep | Spain | |
| 3 | BCCN 92-118 | Human | Tunisia | |
| 3 | BCCN 94-16 | Cattle | France | |
| 3 | BCCN 95-30 | Sheep | Italy | |
| 3 | BCCN 95-36 | Goat | Italy | |
| 3 | BCCN 96-32 | Sheep | Israel | |
| 3 | BCCN 96-142 | Human | France | |
| 3 | BCCN 96-146 | Human | Algeria | |
| B. suis | 1 | 1330 (ATCC 23444; BCCN R12)b | Swine | United States |
| 1 | BCCN 95-13 | Human | New Caledonia | |
| 1 | BCCN 96-138a | Human | Argentina | |
| 1 | BCCN 98-21 | Human | France | |
| 1 | BCCN 98-43 | Unknown | Argentina | |
| 2 | Thomsen (ATCC 23445; BCCN R13)b | Swine | Denmark | |
| 2 | BCCN 93-75 | Swine | Spain | |
| 2 | BCCN 93-80 | Swine | Spain | |
| 2 | BCCN 94-2 | Boar | Belgium | |
| 2 | BCCN 94-9 | Hare | France | |
| 2 | BCCN 94-11 | Boar | France | |
| 2 | BCCN 97-59 | Swine | France | |
| 2 | BCCN 97-100 | Swine | France | |
| 2 | BCCN 97-107 | Swine | France | |
| 2 | BCCN 98-9 | Boar | France | |
| 3 | 686 (ATCC 23446; BCCN R14)b | Swine | United States | |
| 4 | 40 (ATCC 23447; BCCN R15)b | Reindeer | Former USSR | |
| 5 | 513 (BCCN R21)* | Wild rodent | Former USSR | |
| B. ovis | 63/290 (ATCC 25840; BCCN R17)b | Sheep | Africa | |
| Reo 198 (BCCN R22) | Sheep | United States | ||
| BCCN 76-247 | Sheep | France | ||
| BCCN 76-250 | Sheep | France | ||
| BCCN 91-66 | Sheep | Spain | ||
| BCCN 91-70 | Sheep | Spain | ||
| BCCN 91-208 | Sheep | Spain | ||
| BCCN 91-264 | Sheep | Argentina | ||
| BCCN 91-266 | Sheep | Argentina | ||
| BCCN 97-41 | Sheep | Argentina | ||
| BCCN 98-47 | Sheep | Argentina | ||
| B. canis | RM6/66 (ATCC 23365; BCCN R18)b | Dog | United States | |
| D519 (BCCN C1) | Dog | Madagascar | ||
| Hoy 1066 (BCCN C3) | Dog | United States | ||
| BCCN 87-62 | Dog | Canada | ||
| BCCN 87-65 | Dog | Canada | ||
| BCCN 96-104 | Dog | Romania | ||
| BCCN 96-121 | Dog | France | ||
| BCCN 97-60 | Dog | Argentina | ||
| B. neotomae | 5K33 (ATCC 23459; BCCN R16)b | Desert rat | United States |
ATCC, American Type Culture Collection; BCCN, Brucella Culture Collection, Nouzilly, France.
Reference strain.
Vaccine strain.
TABLE 2.
Marine mammal sources of Brucella strains used in this study
| Latin name | Common name | Brucella strain | Geographic origin |
|---|---|---|---|
| Balaenoptera acutorostrata | Minke whale | B202R | Norway |
| Cystophora cristata | Hooded seal | M2006/94/6 | Scotland |
| Delphinus delphis | Common dolphin | B14/94, M644/93/1, M452/97/2 | Scotland |
| Halichoerus grypus | Grey seal | M2375/94/3 | Scotland |
| Lagenorhynchus acutus | White-sided dolphin | M997/94/2, M2438/95/1, M18/96/1, M181/97/1, M2788/97/1 | Scotland |
| Lagenorhynchus albirostris | White-beaked dolphin | M870/97/1 | Scotland |
| Lutra lutra | Otter | M1771/94/1 | Scotland |
| Phoca vitulina | Common seal | B2/94, M2357/93/1, M2466/93/4, M2533/93/1, M292/94/1, M336/94/1, M339/94/2, M972/94/1, M490/95/1, M514/96/4 | Scotland |
| Phocoena phocoena | Porpoise | B1/94, M1068/91/2, M39/94/1, M1570/94/1, M1661/94/2, M515/96/2, M854/98/8, M1747/98/3 | Scotland |
| Stenella caeruleoalba | Striped dolphin | M2194/94/1, M40/95/1 | Scotland |
| Tursiops truncatus | Bottlenose dolphin | 7763/2 | France |
IS711 elements, also known as IS6501 (21), have been described as useful targets for molecular characterization of Brucella species and biovars based on the number and distribution of IS711 copies within the bacterial genomes (4, 16, 17, 21, 24). IS711-based fingerprints were described as stable, species specific, and to some extent biovar specific. B. ovis has been shown to carry a larger number of IS711 copies than the other Brucella species (16, 17, 21). Recently, it has been shown that Brucella strains isolated from marine mammals have more copies of IS711 than all classical species except B. ovis (3). Bricker et al. (3) cloned one of these IS711 elements and its flanking regions to develop a PCR assay which would be specific for strains isolated from marine mammals. However, they obtained an amplification product of the expected size for all marine mammal isolates and not for the classical Brucella species and biovars, except for B. ovis. The PCR assay of our study with primers 26A and 26B, although developed by chance, has the advantage of discriminating between all terrestrial isolates, including B. ovis, and the marine mammal isolates. This simple PCR assay could have several uses in the future, such as possibly tracing these marine mammal strains if they are transmitted to livestock.
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes from bp26 to IS711 have been deposited in GenBank under accession numbers AF242532, AF242533, and AF242534.
Acknowledgments
We thank G. Foster, B. Garin-Bastuji, and J. Godfroid for the gift of the Brucella strains isolated from marine mammals. We are grateful to J. M. Verger for helpful discussions.
REFERENCES
- 1.Allardet-Servent A, Bourg G, Ramuz M, Pages M, Bellis M, Roizes G. DNA polymorphism in strains of the genus Brucella. J Bacteriol. 1988;170:4603–4607. doi: 10.1128/jb.170.10.4603-4607.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alton G G, Jones L M, Angus R D, Verger J M. Techniques for the brucellosis laboratory. Paris, France: Institut National de la Recherche Agronomique; 1988. [Google Scholar]
- 3.Bricker B J, Ewalt D R, MacMillan A P, Foster G, Brew S. Molecular characterization of Brucella strains isolated from marine mammals. J Clin Microbiol. 2000;38:1258–1262. doi: 10.1128/jcm.38.3.1258-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker B J, Halling S M. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. 1994;32:2660–2666. doi: 10.1128/jcm.32.11.2660-2666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clavareau C, Wellemans V, Walravens K, Tryland M, Verger J M, Grayon M, Cloeckaert A, Letesson J J, Godfroid J. Phenotypic and molecular characterization of a Brucella strain isolated from a minke whale (Balaenoptera acutorostrata) Microbiology. 1998;144:3267–3273. doi: 10.1099/00221287-144-12-3267. [DOI] [PubMed] [Google Scholar]
- 6.Cloeckaert A, Salih-Alj Debbarh H, Vizcaino N, Saman E, Dubray G, Zygmunt M S. Cloning, nucleotide sequence, and expression of the Brucella melitensis bp26 gene coding for a protein immunogenic in infected sheep. FEMS Microbiol Lett. 1996;140:139–144. doi: 10.1016/0378-1097(96)00169-3. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert A, Verger J M, Grayon M, Grépinet O. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology. 1995;141:2111–2121. doi: 10.1099/13500872-141-9-2111. [DOI] [PubMed] [Google Scholar]
- 8.Corbel M J, Brinley-Morgan W J. Genus Brucella Meyer and Shaw 1920, 173AL. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 377–388. [Google Scholar]
- 9.Davis D S. Brucellosis in wildlife. 1990. pp. 321–334. . K. Nielsen and J. R. Duncan (ed.), Animal brucellosis. CRC Press, Boca Raton, Fla. [Google Scholar]
- 10.Ewalt D R, Payeur J B, Martin B M, Cummins D R, Miller W G. Characteristics of a Brucella species from a bottlenose dolphin (Tursiops truncatus) J Vet Diagn Investig. 1994;6:448–452. doi: 10.1177/104063879400600408. [DOI] [PubMed] [Google Scholar]
- 11.Fekete A, Bantle J A, Halling S M, Stich R W. Amplification fragment length polymorphism in Brucella strains by use of polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7778–7783. doi: 10.1128/jb.174.23.7778-7783.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ficht T A, Bearden S W, Sowa B A, Marquis H. Genetic variation at the omp2 porin locus of the brucellae: species-specific markers. Mol Microbiol. 1990;4:1135–1142. doi: 10.1111/j.1365-2958.1990.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 13.Foster G, Jahans K L, Reid R J, Ross H M. Isolation of Brucella species from cetaceans, seals and an otter. Vet Rec. 1996;138:583–586. doi: 10.1136/vr.138.24.583. [DOI] [PubMed] [Google Scholar]
- 14.Grimont F, Verger J M, Cornelis P, Limet J, Lefèvre M, Grayon M, Régnault B, Van Broeck J, Grimont P A D. Molecular typing of Brucella with cloned DNA probes. Res Microbiol. 1992;143:55–65. doi: 10.1016/0923-2508(92)90034-l. [DOI] [PubMed] [Google Scholar]
- 15.Halling S M, Bricker B J. Characterization and occurrence of two repeated palindromic DNA elements of Brucella spp.: Bru-RS1 and Bru-RS2. Mol Microbiol. 1994;14:681–689. doi: 10.1111/j.1365-2958.1994.tb01306.x. [DOI] [PubMed] [Google Scholar]
- 16.Halling S M, Tatum F M, Bricker B J. Sequence and characterization of an insertion sequence, IS711, from Brucella ovis. Gene. 1993;133:123–127. doi: 10.1016/0378-1119(93)90236-v. [DOI] [PubMed] [Google Scholar]
- 17.Halling S M, Zehr E S. Polymorphism in Brucella spp. due to highly repeated DNA. J Bacteriol. 1990;172:6637–6640. doi: 10.1128/jb.172.12.6637-6640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahans K L, Foster G, Broughton E S. The characterisation of Brucella strains isolated from marine mammals. Vet Microbiol. 1997;57:373–382. doi: 10.1016/s0378-1135(97)00118-1. [DOI] [PubMed] [Google Scholar]
- 19.Lindler L E, Hadfield T L, Tall B D, Snellings N J, Rubin F A, Van De Verg L L, Hoover D, Warren R L. Cloning of a Brucella melitensis group 3 antigen gene encoding Omp28, a protein recognized by the humoral immune response during human brucellosis. Infect Immun. 1996;64:2490–2499. doi: 10.1128/iai.64.7.2490-2499.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercier E, Jumas-Bilak E, Allardet-Servent A, O'Callaghan D, Ramuz M. Polymorphism in Brucella strains detected by studying distribution of two short repetitive DNA elements. J Clin Microbiol. 1996;34:1299–1302. doi: 10.1128/jcm.34.5.1299-1302.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouahrani S, Michaux S, Sri Widada J, Bourg G, Tournebize R, Ramuz M, Liautard J P. Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J Gen Microbiol. 1993;139:3265–3273. doi: 10.1099/00221287-139-12-3265. [DOI] [PubMed] [Google Scholar]
- 22.Ross H M, Jahans K L, MacMillan A P, Reid R J, Thompson P M, Foster G. Brucella species infection in North Sea seal and cetacean populations. Vet Rec. 1996;138:647–648. doi: 10.1136/vr.138.26.647. [DOI] [PubMed] [Google Scholar]
- 23.Rossetti O L, Arese A I, Boschiroli M L, Cravero S L. Cloning of Brucella abortus gene and characterization of expressed 26-kilodalton periplasmic protein: potential use for diagnosis. J Clin Microbiol. 1996;34:165–169. doi: 10.1128/jcm.34.1.165-169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemulapalli R, McQuiston J R, Schurig G G, Sriranganathan N, Halling S M, Boyle S M. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin Diagn Lab Immunol. 1999;6:760–764. doi: 10.1128/cdli.6.5.760-764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verger, J. M., M. Grayon, A. Cloeckaert, M. Lefèvre, E. Ageron, and F. Grimont. Classification of Brucella strains isolated from marine mammals using DNA-DNA hybridization and ribotyping. Res. Microbiol., in press. [DOI] [PubMed]
- 26.Verger J-M, Grimont F, Grimont P A D, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Bacteriol. 1985;35:292–295. [Google Scholar]
- 27.Vizcaino N, Verger J M, Grayon M, Zygmunt M S, Cloeckaert A. DNA polymorphism at the omp-31 locus of Brucella spp.: evidence for a large deletion in Brucella abortus, and other species-specific markers. Microbiology. 1997;143:2913–2921. doi: 10.1099/00221287-143-9-2913. [DOI] [PubMed] [Google Scholar]