Abstract
Purpose
To depict the lncRNA expression during human oocyte maturation and explore the lncRNAs leading to recurrent oocyte maturation arrest.
Methods
LncRNA sequencing was performed on pooled RNA from 20 oocytes of each group (recurrent oocyte maturation arrest (ROMA), of germinal vesicle (GV), metaphase I (MI), or metaphase II (MII) stages. Bioinformatics software was deployed to compare the lncRNA differential expression between the normal and ROMA oocytes. The co-expression of lncRNA/mRNA was illustrated with the Cytoscape software. The pooled RNA from every 10 oocytes of each group (ROMA, GV, MI, MII) was extracted for further qPCR validation.
Results
There were 17 downregulated and 3 upregulated lncRNAs in the ROMA oocyte. Among them, co-expression analysis indicated that NEAT1 and NORAD were both downregulated. Basing on the KEGG enrichment analysis, PRCKA and JAK3 might be the target genes in the PI3K-Akt pathway and modulated by NEAT1 and NORAD. As validated by qPCR, the expressional levels of lncRNA candidates (NEAT1 and NORAD) and their target genes (PRKCA and JAK3) were confirmed to be extremely lower in the ROMA oocyte than in the normal oocyte.
Conclusion
By targeting the PI3K-Akt pathway genes PRKCA and JAK3, the abnormal expression of NEAT1 and NORAD is suggested to impede oocyte maturation and impair oocyte genome integrity.
Keywords: Recurrent oocyte maturation arrest, Long non-coding RNAs, PI3K-Akt signaling pathway, In vitro fertilization and embryo transfer
Introduction
The oocyte at the germinal vesicle (GV) stage develops to the metaphase I (MI) stage following germinal vesicle breakdown. Subsequently, it proceeds to the metaphase II (MII) stage after the extrusion of the first polar body. The final oocyte maturation is triggered by an LH surge at the late follicular phase. The maternal RNA has been the primary genetic transcript in the oocyte until zygotic genome activation (ZGA). The maternal RNA gradually decays as the oocyte matures after joining paternal RNA to activate the embryonic genome. The molecular mechanism of maternal RNA decay is still under investigation as is the role of epigenetic regulation increasingly recognized to be critical for ZGA [1–3].
In the in vitro fertilization and embryo transfer (IVF-ET) protocol, the gonadotrophins are often used to hyperstimulate the ovaries for harvesting more eggs. Human chorionic gonadotrophin (hCG) is administrated to trigger the oocyte maturation at the final stage, mimicking the LH surge in the natural cycle. Most of the retrieved eggs will mature under superphysiological hCG stimulation. However, it was reported that 0.1–1% of patients who underwent the superovulation cycle were inflicted with recurrent oocyte maturation arrest (ROMA). Moreover, the recurrent maturation arrested oocytes were even resistant to the standard protocol of in vitro maturation [4]. Theoretically, those oocytes can arrest at different stages of GV, MI, and MII [5]. In our clinic, around 0.29% of patients suffered from ROMA, in which the MI stage arrest was the most common phenotype. The de novo mutation of coding genes, such as TUBB8, TRIP13, and PATL2, can cause oocyte maturation arrest, but they can only explain a few ROMA cases but not all of them [6–8]. Nevertheless, the abundant non-coding RNAs in the oocyte could be the epigenetic culprits contributing to ROMA.
Long non-coding RNAs (lncRNAs) refer to the non-coding RNAs longer than 200 nt and play versatile roles in RNA epigenetic regulation. LncRNA can facilitate the formation of ribonucleoprotein by serving as a scaffold and bringing together numerous components such as proteins, RNAs, and DNA [9, 10]. By base-pairing, lncRNA can guide the ribonucleoprotein complex to a specific genomic location [11, 12]. As a molecular decoy, lncRNA can bind to protein complexes and prevent the proteins from interacting with their natural targets [13, 14]. LncRNA can also bind to sequester miRNA (miRNA sponge), which activates target mRNA [15, 16]. In addition, some lncRNAs originating from the enhancer region (enhancer RNA) can regulate neighboring genes via cis or trans [17, 18]. The lncRNAs have been confirmed to express in the oocyte and early embryo, and their expression levels change dynamically once the embryo genome is activated during human early embryonic development [19, 20]. Previous studies reported that lncRNAs get involved in the various biological and developmental processes in early embryonic development, such as cell pluripotency induction and maintenance, X chromosome inactivation, and gene imprinting [21]. The expression levels of some lncRNAs in cumulus cells were correlated with the quality of oocytes and early embryos, so those lncRNAs have the potential to predict oocyte development non-invasively [22].
Despite few studies exploring the expression of lncRNA in oocytes and early embryos, those studies only detected the lncRNA expression from matured oocytes to the blastocyst. Furthermore, rare studies have focused on the lncRNA expression during oocyte maturation. Therefore, the present study aimed to depict the lncRNA expression profile during oocyte maturation and explore the lncRNA candidates associated with ROMA.
Materials and methods
Patient recruitment
This study was scrutinized by the Reproductive Ethics Committee of our hospital, and the IRB approval (code: 200701) was obtained before initiating the study. In our clinic, all patients must decide how to deal with their abandoned oocytes in the informed consent. Therefore, only patients who approved donating their abandoned oocytes to scientific research would be recruited into the study.
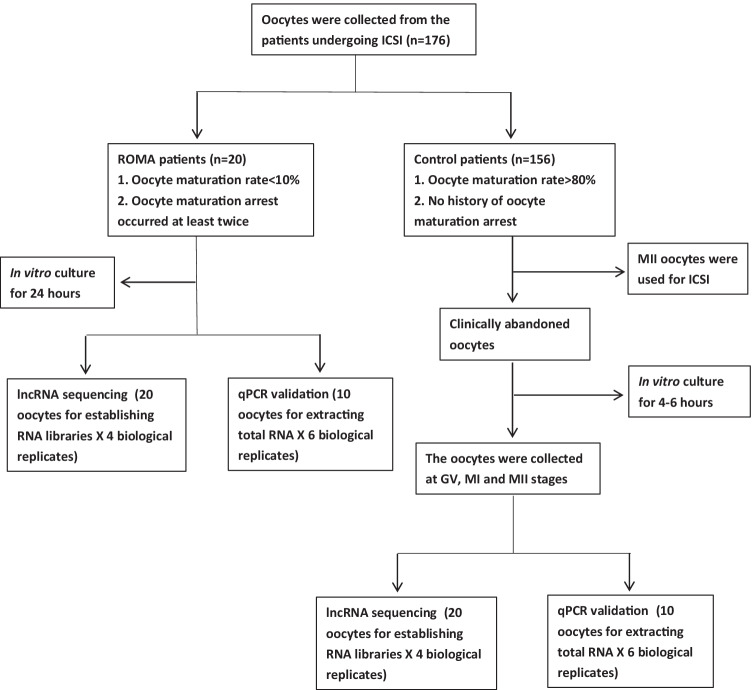
In total, 176 patients undergoing intracytoplasmic injection (ICSI) were recruited into the study (20 ROMA and 156 control patients). The selection criteria for the ROMA patients were as follows (n = 20): (1) more than 90% of oocytes (maturation rate < 10%) were immature on the oocyte retrieval day and stayed immature after in vitro culture for more than 24 h and (2) the patients suffered at least two failures of oocyte maturation. The control group (n = 156) was defined as patients with more than 80% mature oocytes and no history of oocyte maturation arrest. The exclusion criteria of the control group included (1) genetic diseases related to oocyte maturation; (2) endocrine diseases such as diabetes and hyperthyroidism; and (3) immune diseases such as lupus erythematosus, and rheumatoid arthritis. The patient selection flow chart is presented in Fig. 1.
Fig. 1.
The patient selection flow chart
Ovarian stimulation protocol
Regarding the ovarian stimulation protocol, a long-acting GnRH agonist was administrated during the middle luteal phase. Two weeks later, gonadotrophins were injected daily for ovarian stimulation. The injecting dose of gonadotrophins was determined by the ovarian response. The ultrasound examinations and hormone measurements were conducted every 3–4 days to monitor follicular growth. When at least two follicles reached 18 mm, hCG was administrated to trigger oocyte maturation. Oocyte retrieval surgery was performed 34–38 h after hCG administration.
Oocyte collecting criteria
The collected oocytes were denuded before checking the maturation state. For rescuing ROMA oocytes, the immature oocytes were in vitro cultured with Sage IVM media (CooperSurgical, Trumbull, CT, USA) for 24 h at 37 °C in 6% CO2 and then collected for research when they stayed immature. As for the “control oocytes,” they originated from the immature oocytes (GV or MI) abandoned after ICSI manipulation. They were regarded as control oocytes because the oocyte maturation rate of the patients reached 80% (the average maturation rate in our lab). Those oocytes were cultured with IVF media (Vitrolife, Kungsbacka, Sweden) in a 6% CO2 incubator at 37 °C for 4–6 h and collected at GV, MI, and MII stages, relatively.
Those oocytes were vitrified following the manufacturer’s protocol. Briefly, embryos were placed into the Vitri 1™ Cleave Solution for 5 min, then into the Vitri 2™ Cleave Solution for 2 min, and finally into the Vitri 3™ Cleave Solution for 30 s. The embryos were loaded with high-security straws in a maximum of 1 μL, and then placed into the liquid nitrogen for long-term storage. As for the thawing procedure, straws were removed from the cryotube and dipped into the Warm1™ solution for 10 to 30 s, then into the Warm2™ solution for 1 min, and then into the Warm3™ solution for 2 min, and finally into the Warm4™ solution for 5 min. Finally, eggs were transferred to a pre-equilibrated culture dish.
RNA library construction and sequencing
According to the preliminary experiment, the pooled RNA from every 20 oocytes was the premium quantity for establishing RNA libraries. In total, 80 oocytes (20 oocytes for each sequencing sample × 4 biological replicates) in each group (GV, MI, MII, ROMA) were collected for constructing RNA libraries. Additionally, the pooled RNA from every ten oocytes was the premium quantity for qRT-PCR amplification based on our pilot study. Thus, 60 oocytes (10 oocytes for each qPCR reaction × 6 biological replicates) in each group (GV, MI, MII, ROMA) were harvested for qPCR validation, as each reaction was technically repeated triple times.
TRIzol (Invitrogen, USA) was used for extracting total RNA from the thawed oocytes. Fragmented RNAs (approximately 200 bp) were subjected to first-strand and second-strand cDNA synthesis followed by adaptor ligation and amplified by 18 cycles according to instructions of NEBNext® Ultra™ RNA Library Prep Kit for Illumina (NEB, USA). The purified library products were evaluated using Agilent 2200 TapeStation and Qubit® 2.0 (Life Technologies, USA). An eligible RNA library should meet the following prerequisites: (1) the size of the principal peak scattered between 300 and 350 bp; (2) the mass concentration of the RNA library was above 20 ng/μL; (3) the molar concentration of RNA library was above 100 nmol/L. The libraries were paired-end sequenced (sequencing reads were 150 bp) with the Illumina HiSeq 3000 platform.
Pre-processing of sequencing reads
Raw Fastq sequences were treated with Trimmomatic tools (v0.36) to remove trailing sequences below a Phred quality score of 20 and to achieve uniform sequence lengths for downstream clustering processes. Sequencing read quality was inspected using the FastQC software. Adapter removal and read trimming were performed using Trimmomatic. Sequencing reads were trimmed from the end (base quality less than Q20) and filtered by length (less than 25 bp). HISAT2 was used to align the clean reads to the human reference genome hg19 with default parameters. HTSeq v0.6.0 was used to count the reads numbers mapped to each gene.
Identification of lncRNAs and mRNAs
The raw data were firstly filtered to remove low-quality reads, and then the clean data that passed repeated testing was assembled using StringTie based on the reads mapped to the reference genome. The assembled transcripts were annotated using the GffCompare program. The unknown transcripts were used to screen for putative lncRNAs referring to the public databases (http://asia.ensembl.org/info/genome/genebuild/biotypes.html;www.ncbi.nlm.nih.gov/nuccore). They were further screened using CPC/CNCI/Pfamto to distinguish the protein-coding genes from the non-coding genes. Besides, target gene mRNAs were identified using a minimum length and exon number threshold. Generally, transcripts with lengths above 200 nt with predicted ORF were selected as the candidate genes.
Differential expression analysis
Differential expression was assessed by DESeq using read counts as input. The Benjamini–Hochberg multiple test correction method was enabled. Differentially expressed genes were chosen according to the criteria of fold change > 2 and adjusted p-value < 0.05. Finally, a hierarchical clustering analysis was performed using the R language package Gplots according to the read counts of differential genes in different groups. Moreover, colors represent different clustering information, such as the similar expression pattern in the same group, including similar functions or participating in the same biological process.
Prediction of lncRNA target genes
The interactive network was established by calculating the Pearson correlation coefficient and p value between lncRNAs (NEAT1 and NORAD) and their target genes. The transcripts were filtered using a correlation coefficient (COR) of > 0.85 and a p-value < 0.05 before the Cytoscape software was applied to illustrate the interactive network.
GO terms and KEGG ontology enrichment analysis
The predicted genes of lncRNAs were selected for GO and KEGG ontology enrichment analyses. GO was performed with the KOBAS 3.0 software. GO provides label classification of gene function and gene product attributes (http://www.geneontology.org). GO analysis covers three domains: cellular component (CC), molecular function (MF), and biological process (BP). The differentially expressed genes and the enrichment of different pathways were mapped using the KEGG ontology enrichment with the KOBAS 3.0 software (http://www.genome.jp/kegg). For KEGG enrichment analysis, a p-value < 0.05 was used as the threshold to determine the significant enrichment of the gene sets.
Quantitative PCR
The total RNA from the oocytes was extracted with TRIzol (Invitrogen, USA) following the manufacturer’s instructions. The SuperScript III First-Strand Synthesis System (Invitrogen, USA) was deployed for cDNA synthesis, followed by the qPCR amplification using SYBR® Green qPCR supermixes (Bio-Rad, USA). The Bio-Rad CTX96 real-time PCR detection system was applied for a real-time PCR reaction. The PCR primers of NEAT1, PRKCA, and JAK3 (Table 1) were synthesized referring to the previous studies [23–25]. The expression level relative to GAPDH was calculated using the cyclic threshold (CT) values logarithmically transformed using the 2 − ΔCt function. All the relative expression levels were normalized according to the average value of 6 biological samples in the control group. The PCR reaction of each biological sample was technically repeated triple times.
Table 1.
The primers for qPCR validations
| Primers | Primer sequence |
|---|---|
| NORAD forward | 5′-CTCTGCTGTGGCTGCCC-3′ |
| NORAD reverse | 5′-GGGTGGGAAAGAGAGGTTCG-3′ |
| NEAT1 forward | 5′-AGGCAGGGAGAGGTAGAAGG-3′ |
| NEAT1 reverse | 5′-TGGCATGGACAAGTTGAAGA-3′ |
| PRKCA forward | 5′-CCTCATGTACCACATTCAGCA-3′ |
| PRKCA reverse | 5′-TCTGGGGCGATATAATCTGG-3′ |
| JAK3 forward | 5′-TCGTGACCTCAATAGCCTCATCTC-3′ |
| JAK3 reverse | 5′-CCACTGACACATATGCCCATCTGT-3′ |
| GAPDH forward | 5′-AGCCACATCGCTCAGACAC-3′ |
| GAPDH reverse | 5′-GCCCAATACGACCAAATCC-3′ |
Statistics analysis
The online tool VENNY 2.1 was applied to analyze the intersectant genes. The patients’ demographic data between the ROMA and control groups were compared with Student’s t-test. One-way ANOVA was used for the multiple comparisons of gene expression levels between groups. The statistical analysis was performed using the Prism software (GraphPad version 8.0), with a p-value < 0.05 regarded as statistical significance.
Results
Patient demographics
The patient baseline data were comparable between the ROMA and the control groups (Table 2).
Table 2.
Patient demographic data
| Patient variables | Control (n = 156) | ROMA (n = 20) | p value |
|---|---|---|---|
| Age (years) | 34.13 ± 5.32 | 34.15 ± 5.49 | 0.99 |
| Infertile years | 4.80 ± 3.65 | 5.00 ± 3.01 | 0.79 |
| BMI (kg/m2) | 21.81 ± 3.01 | 22.39 ± 3.09 | 0.28 |
| FSH (IU/L) | 6.61 ± 2.01 | 6.55 ± 1.31 | 0.85 |
| LH (IU/L) | 6.10 ± 3.02 | 5.01 ± 3.02 | 0.20 |
| E2 (ng/L) | 38.25 ± 15.20 | 42.66 ± 19.99 | 0.35 |
| PRL (μg/L) | 20.38 ± 5.76 | 20.82 ± 5.97 | 0.81 |
| T (nmol/L) | 0.26 ± 0.10 | 0.25 ± 0.15 | 0.77 |
| AMH (ng/mL) | 3.17 ± 1.58 | 3.64 ± 2.04 | 0.33 |
| Go dosage (IU) | 1849 ± 684 | 1945 ± 687 | 0.56 |
| Oocyte number | 12.62 ± 7.54 | 11.70 ± 7.27 | 0.60 |
General variables of RNA libraries
In total, 12 RNA libraries were established for data analysis, and each library was sequenced in duplicate (Table 3). The effective read rates were 100%, total mapped rates fluctuated between 87.73 and 93.45%, and unique mapped rates ranged from 84.41 to 90.59%. In total, the number of lncRNAs expressed in the GV, MI, MII, and ROMA oocytes was 3814, 3586, 3047, and 2814, relatively.
Table 3.
The read variables of RNA libraries
| Oocyte samples | Effective reads (%) | Total mapped (%) | Multiple mapped (%) | Uniquely mapped (%) |
|---|---|---|---|---|
| GV-1 | 33,932,434 (100%) | 30,555,997 (90.05%) | 984,131 (2.90%) | 29,571,866 (87.15%) |
| GV-2 | 36,638,364 (100%) | 32,856,839 (89.68%) | 1,014,903 (2.77%) | 31,841,936 (86.91%) |
| GV-3 | 32,629,768 (100%) | 29,522,251 (90.48%) | 888,004 (2.72%) | 28,634,247 (87.75%) |
| GV-4 | 25,925,760 (100%) | 24,228,123 (93.45%) | 743,254 (2.87%) | 23,484,869 (90.59%) |
| MI-1 | 35,920,126 (100%) | 32,427,318 (90.28%) | 1,024,168 (2.85%) | 31,403,150 (87.42%) |
| MI-2 | 32,936,256 (100%) | 29,447,176 (89.41%) | 980,844 (2.98%) | 28,466,332 (86.43%) |
| MI-3 | 32,497,504 (100%) | 29,655,197 (91.25%) | 990,528 (3.05%) | 28,664,669 (88.21%) |
| MI-4 | 34,142,352 (100%) | 30,981,358 (90.74%) | 1,055,072 (3.09%) | 29,926,286 (87.65%) |
| MII-1 | 34,275,096 (100%) | 30,556,956 (89.15%) | 1,122,785 (3.28%) | 29,434,171 (85.88%) |
| MII-2 | 24,449,952 (100%) | 21,740,489 (88.92%) | 803,235 (3.29%) | 20,937,254 (85.63%) |
| MII-3 | 38,783,590 (100%) | 34,023,241 (87.73%) | 1,287,335 (3.32%) | 32,735,906 (84.41%) |
| MII-4 | 29,087,318 (100%) | 26,757,178 (91.99%) | 776,998 (2.67%) | 25,980,180 (89.32%) |
| ROMA-1 | 38,427,514 (100%) | 35,695,813 (92.89%) | 1,180,687 (3.07%) | 34,515,126 (89.82%) |
| ROMA-2 | 39,906,932 (100%) | 37,132,703 (93.05%) | 1,157,668 (2.90%) | 35,975,035 (90.15%) |
| ROMA-3 | 29,965,726 (100%) | 27,654,693 (92.29%) | 954,987 (3.19%) | 26,699,706 (89.10%) |
| ROMA-4 | 31,189,304 (100%) | 29,038,612 (93.10%) | 985,247 (3.16%) | 28,053,365 (89.95%) |
The differential expression of lncRNAs from the GV and MII stages of control oocytes
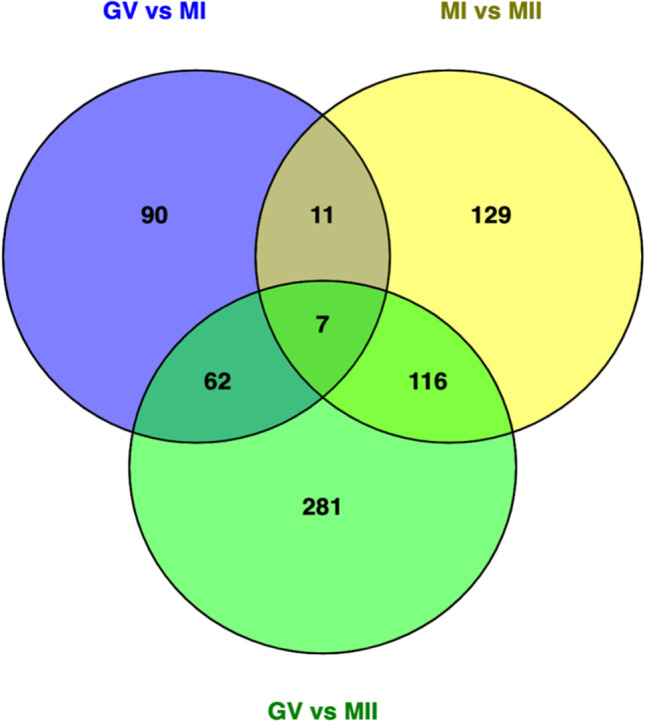
During oocyte maturation, 170 lncRNAs were differentially expressed between GV and MI oocytes, whereas 263 lncRNAs significantly changed from MI to MII oocytes. Additionally, 466 lncRNAs significantly altered from GV to MII (Fig. 2). There were seven lncRNAs constantly altered from the GV to MII stage, in which three lncRNAs were downregulated and four lncRNAs were upregulated (Table 4).
Fig. 2.
The constantly altered lncRNAs in the control oocytes from GV to MII stage
Table 4.
The constantly differential lncRNAs in all stages of control oocytes
| Accession no | Gene name | From GV to MII |
|---|---|---|
| ENST00000441217.1 | ENSG00000235499.1 | Down |
| ENST00000523336.1 | ENSG00000253642.1 | Down |
| ENST00000538335.1 | ENSG00000212694.4 | Up |
| ENST00000555282.1 | ENSG00000258913.1 | Up |
| ENST00000574616.1 | ENSG00000263244.1 | Down |
| NR_033932.1 | RGMB-AS1 | Up |
| NR_109931.1 | LOC101928796 | Up |
Differential expression of lncRNAs between the control and ROMA oocytes
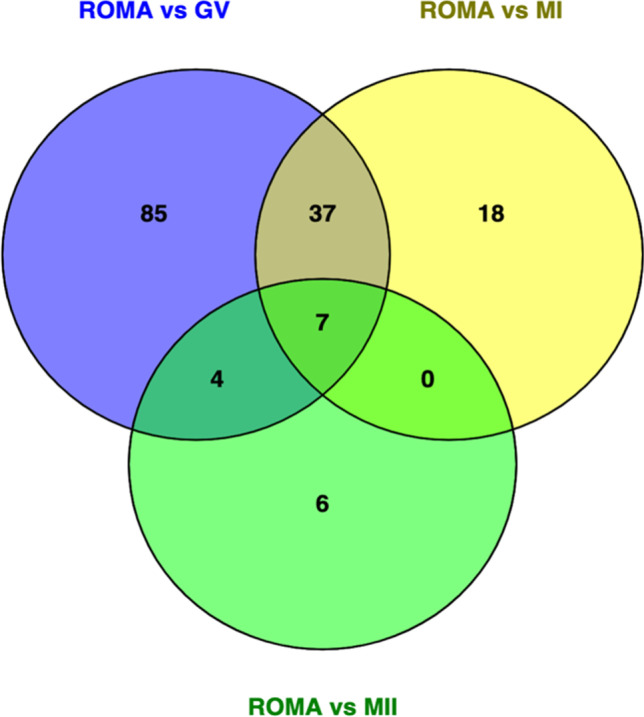
Generally, 135 lncRNAs were differentially expressed between the ROMA and control oocytes at the GV stage. In addition, the abundance of 63 lncRNAs was differential between the ROMA and control oocytes at the MI stage. Besides, the expressional levels of 17 lncRNAs were significantly different between the ROMA and control oocytes at the MII stage. After the intersection, seven lncRNAs (four downregulated and three upregulated) constantly changed throughout the three stages (Fig. 3 and Table 5).
Fig. 3.
The intersection of differential genes between the ROMA and control oocytes
Table 5.
The constantly differential lncRNAs between the ROMA and control oocytes
| Accession no | Gene name | Expression in ROMA |
|---|---|---|
| ENST00000554458.1 | ENSG00000258784.1 | Down |
| NR_027451.1 | NORAD | Down |
| ENST00000511993.1 | ENSG00000234828.3 | Up |
| ENST00000440744.2 | ENSG00000230649.2 | Up |
| NR_131012.1 | NEAT1 | Down |
| ENST00000559621.1 | ENSG00000259383.1 | Up |
| ENST00000457290.2 | ENSG00000238273.3 | Down |
The target genes of differential lncRNAs between the control and ROMA oocytes
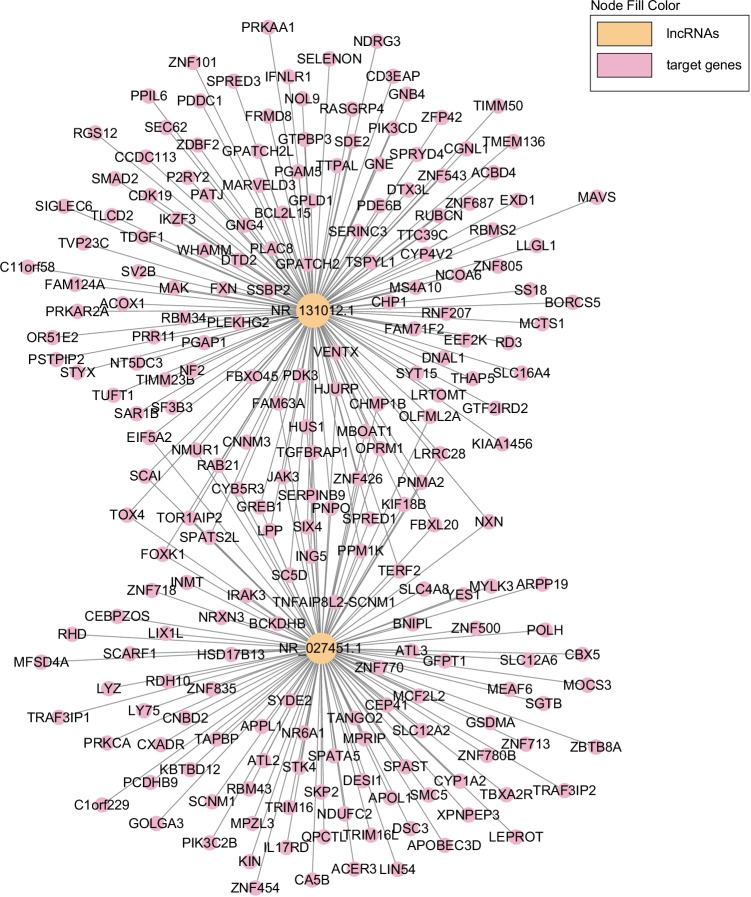
As the well-studied lncRNAs, NEAT1 and NORAD had the most target genes among the constantly differential lncRNAs. Specifically, NEAT1 targeted 136 protein-coding genes, while NORAD was associated with 115 protein-coding genes, and 34 genes were both targeted by NEAT1 and NORAD (Fig. 4). Moreover, the expressional heatmap of target genes is presented in Fig. 5A and B.
Fig. 4.
The target genes of NEAT1 and NORAD
Fig. 5.
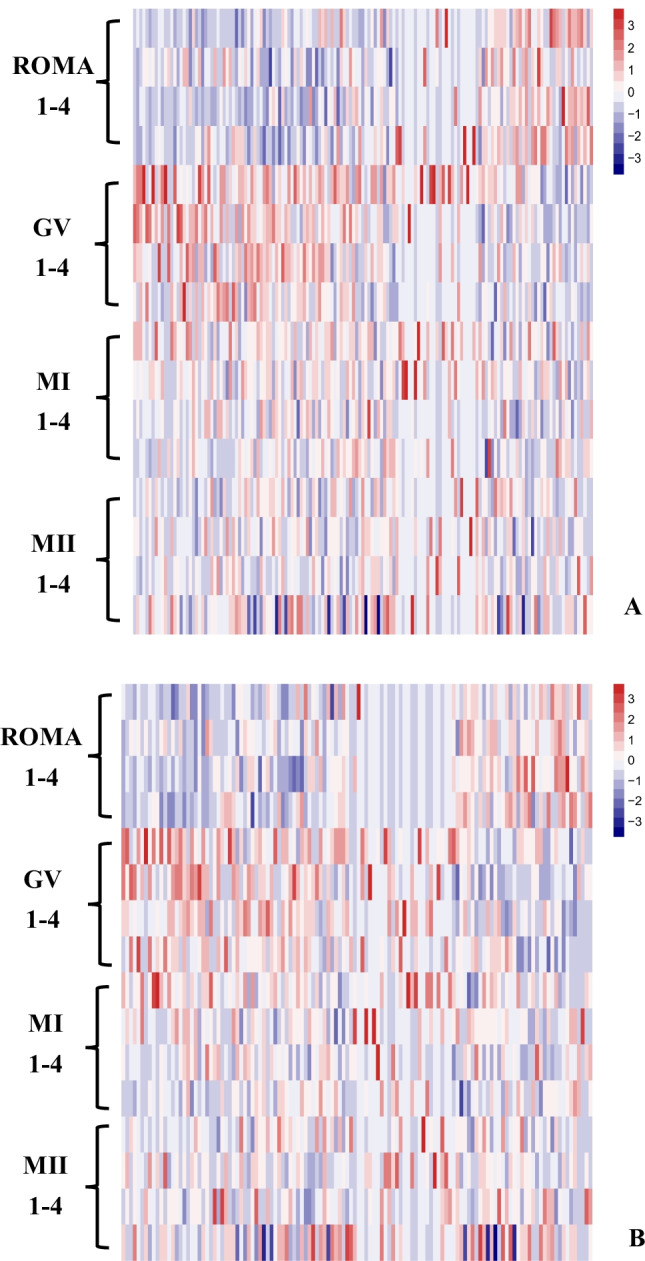
The expressional heatmap of the target genes of (A) NEAT1 and (B) NORAD
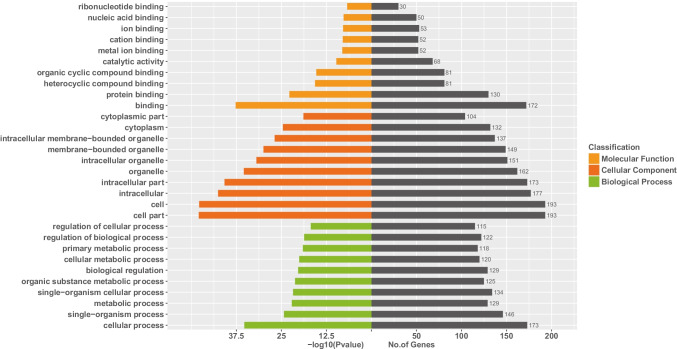
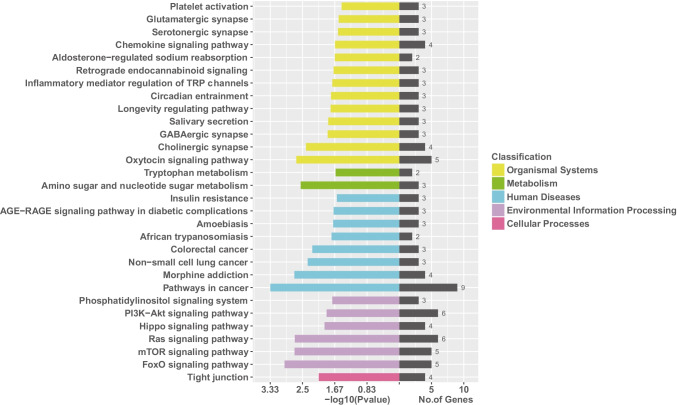
Regarding the GO enrichment analysis (Fig. 6), it indicated that the target genes were primarily scattered at the binding (molecular function) part, cell or cell part (cellular components), and the cellular process (biological process). Based on the KEGG enrichment analysis (Fig. 7), PI3K-Akt, Ras, mTOR, and FoxO signaling pathways were profoundly affected by NEAT1 and NORAD.
Fig. 6.
The GO enrichment analysis of the target genes of NEAT1 and NORAD
Fig. 7.
The KEGG enrichment analysis of the target genes of NEAT1 and NORAD
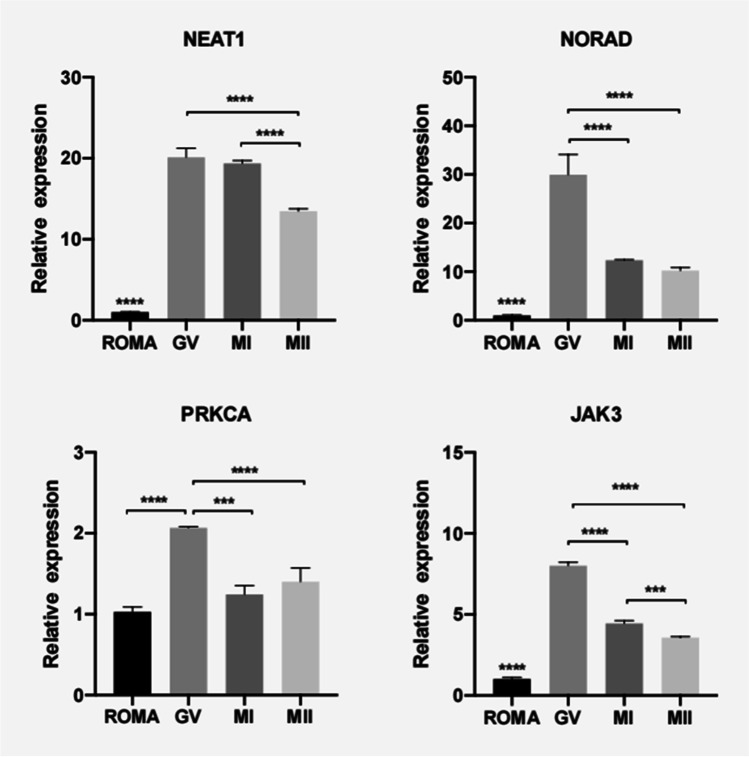
Quantitative RT-PCR validation of lncRNA candidates and their target genes
In the PI3K-Akt signaling pathway, six genes (PRKCA, GNB4, PIK3CD, GNG4, JAK3, PRKAA1) were targeted by NEAT1 or NORAD, of which only PRKCA and JAK3 were abundantly expressed in human oocytes as indicated by mRNA sequencing data. Therefore, lncRNA candidates (NEAT1 and NORAD) and their target genes (PRKCA and JAK3) were selected for further qPCR validation.
As validated by qRT-PCR, the expressional levels of NEAT1, NORAD, and their target gene JAK3 were significantly lower in the ROMA oocytes than in the control oocytes (Fig. 8). In addition, PRKCA, the NORAD target gene, was significantly lower in the ROMA oocytes than in the GV stage control oocytes. In the control oocytes, the expression of NEAT1, NORAD, PRKCA, and JAK3 declined significantly as the oocytes developed from GV to MII stages.
Fig. 8.
QRT-PCR validation of the expression levels of differential lncRNAs and their target genes in the ROMA and control oocytes. Asterisks indicate statistically significant differences between two groups
Discussion
As the primary genetic transcript, the maternal RNA dramatically changes amid oocyte maturation. Maternal RNA decays gradually, followed by embryonic genome activation. It has been reported that lncRNAs may play versatile roles in early embryonic development [19–21]. However, reports on the association of lncRNAs with oocyte maturation are still lacking. For the first time, the present study reveals the expression profile of lncRNAs during oocyte maturation and further explores lncRNA candidates associated with arrested oocyte maturation.
lncRNAs can regulate maternal RNA in epigenetic modification, transcription, post-transcription, translation, and post-translation [26]. By directly or indirectly interacting with chromatin, lncRNAs can exert a trans or cis regulation in chromatin transcription. Besides that, lncRNA can modulate RNA transcription by forming chromatin loops or targeting regulatory elements. Regarding epigenetic modification, lncRNAs can play a trans-acting role by binding proteins to sequence motifs and the RNA structure or base-pairing with target RNAs [27, 28].
As oocytes mature, the lncRNA dynamic could get involved in erasing maternal RNA and activating the embryonic genome. The altering lncRNAs from the GV to MII stage were more than the ones from the GV to MI stage and from the MI to the MII stage. It implied that lncRNAs dramatically changed as the oocyte developed from the GV to MII stage since MI was just an intermediate stage between GV and MII.
When the target gene analysis was performed on the differentially expressed lncRNAs, both NEAT1 (nuclear paraspeckle assembly transcript 1, NR_131012.1) and NORAD (non-coding RNA activated by DNA damage, NR_027451.1) were the well-studied lncRNAs and manifested broad biological functions. NEAT1 is a vital component of nuclear paraspeckles and can be classified into two isoforms: NEAT1_1 (3.7 kb) and NEAT1_2 (23 kb). As an oncogene, NEAT1 could suppress some miRNAs such as miR-449-5p, miR-377-3p, and miR-204 to promote the proliferation, migration, and invasion of oncogenic cells [29, 30]. Interestingly, the previous study suggested that the suppression of NEAT1 will inhibit cell growth by enhancing the apoptosis level and inflammatory cytokines in osteoarthritis chondrocytes [31]. Similarly, the abnormal downregulation of NEAT1 in the ROMA oocyte might facilitate the oocyte growth and induce apoptosis [32]. Moreover, the NEAT1 knockout mice failed to produce corpus luteum and manifested infertility [33]. Non-coding RNA activated by DNA damage (NORAD) plays a pivotal role in maintaining genome integrity [34]. Once the DNA damage occurs, the increasingly expressed NORAD can sequester the pumilio (PUM) proteins, stabilize PUM targets, and maintain genome integrity [23]. Therefore, NORAD modulates a wide range of cell biological processes such as proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT). The dysregulation of NORAD is closely related to cell growth abnormality or tumorigenesis [35, 36]. In addition, the NORAD-deficient mice manifested premature aging due to the augmented activity of PUMILIO proteins [37]. In brief, the oocyte maturation arrest might be caused by the suppressive expression of NORAD via the following mechanisms: (1) The decreasing NORAD was insufficient to sequester the PUM proteins and maintain the genome integrity. (2) The suppressed expression of NORAD may disrupt oocyte maturation due to its essential role in modulating cell growth.
Among the most affected signaling pathways, the PI3K-Akt signaling pathway has been indispensable for oogenesis and folliculogenesis [38–40]. MYT1 can inactivate CDK1 (also called CDC2 or P34CDC2) and maintain the oocyte meiotic state by phosphorylating threonine 14 and 15. Insulin or insulin growth factor (IGF) could induce PKB via the mediation of the PI3K-Akt signaling pathway, suppress MYT1 relevant pathway, and initiate the resumption of oocyte meiosis [41]. In the ROMA oocytes, the suppression of PRKCA and JAK3 could impair the mediating function of the PI3K-Akt pathway and block the meiotic event. As validated by qRT-PCR, the target genes PRKCA and JAK3 were suppressed from the GV to the MI stage, which meant the dysregulation of NEAT1 and NORAD started to intervene in oocyte maturation during germinal vesicle breakdown and had a profound impact on later oocyte development.
The expression levels of NEAT1 and NORAD declined as the oocyte matured. The possible explanation could be that the declination of both lncRNAs facilitates erasing maternal RNAs and activating the embryonic genome. However, their suppressive expression may disrupt the normal process of oocyte maturation and lead to oocyte maturation arrest. Besides that, the low expression of NORAD further impairs oocyte genome stability by disrupting the genome monitor system.
There were some limitations existing in the present study. Firstly, the oocytes in the control group were harvested after in vitro incubation, which might cause variations compared to the oocyte that spontaneously matured in vivo. Secondly, the control oocytes were collected from the leftovers after ICSI, so they had unavoidable defects in representing the genuine normal oocytes. The third limitation was that the results obtained from pooling leftover oocytes might cover the heterogeneity of individual patients.
In summary, the present study uncovers expression profiles of lncRNAs during oocyte maturation and explored the lncRNA candidates associated with oocyte arrested maturation. The findings reveal the epigenetic machinery regulating oocyte maturation and the possible pathological mechanism leading to ROMA. As the critical lncRNAs, NEAT1 and NORAD modulate many cellular biological processes. Furthermore, NORAD is indispensable for maintaining genome integrity. Therefore, by targeting PI3K-Akt pathway genes, the extreme suppression of NEAT1 and NORAD could impede oocyte maturation and impair oocyte genome integrity.
Acknowledgements
The authors would like to acknowledge the assistance of Ruibo Biological Co. in conducting lncRNA sequencing and data analysis.
Author contribution
Conceptualization, L.Y.; data curation, Y.H.; formal analysis, L.Z.; methodology, W.L. and X.H.; writing — original draft, L.Y.; writing — review and editing, Y.X. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the grants from the National Natural Science Funding (No. 81671523), the Natural Science Funding of Guangdong Province (No. 2015A030313140 and No. 2017A030313895), and the Funding of Yat-sen Scholarship for Young Scientist.
Declarations
The study has been scrutinized by the reproductive ethics committee of Sun Yat-sen Memorial Hospital. An Institutional Review Board (IRB) approval (code no. 200701) was obtained before conducting the study.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Competing interests
The authors declare no competing interests.
Footnotes
Lina Wei and Huayang Xia are considered similar in author order.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cook MS, Blelloch R. Small RNAs in germline development. Curr Top Dev Biol. 2013;102:159–205. doi: 10.1016/B978-0-12-416024-8.00006-4. [DOI] [PubMed] [Google Scholar]
- 2.Banisch TU, Goudarzi M, Raz E. Small RNAs in germ cell development. Curr Top Dev Biol. 2012;99:79–113. doi: 10.1016/B978-0-12-387038-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 3.Tang F. Small RNAs in mammalian germline: tiny for immortal. Differentiation. 2010;79:141–146. doi: 10.1016/j.diff.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Lu Y, Ferrer-Buitrago M, et al. Patients with a high proportion of immature and meiotically resistant oocytes experience defective nuclear oocyte maturation patterns and impaired pregnancy outcomes. Reprod Biomed Online. 2018;36:396–407. doi: 10.1016/j.rbmo.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 5.Mrazek M, Fulka J., Jr Failure of oocyte maturation: possible mechanisms for oocyte maturation arrest. Hum Reprod. 2003;18:2249–2252. doi: 10.1093/humrep/deg434. [DOI] [PubMed] [Google Scholar]
- 6.Wang AC, et al. Mutation analysis of the TUBB8 gene in primary infertile women with arrest in oocyte maturation. Gynecol Endocrinol. 2018;34:900–904. doi: 10.1080/09513590.2018.1464138. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, et al. Bi-allelic missense pathogenic variants in TRIP13 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet. 2020;107:15–23. doi: 10.1016/j.ajhg.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, et al. Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet. 2017;101:609–615. doi: 10.1016/j.ajhg.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Johnsson P, et al. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert MS, et al. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cazalla D, Yario T, Steitz JA, Steitz J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darrow EM, Chadwick BP. Boosting transcription by transcription: enhancer-associated transcripts. Chromosome Res. 2013;21:713–724. doi: 10.1007/s10577-013-9384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 20.Huang K, et al. The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell. 2014;15:410–415. doi: 10.1016/j.stem.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouckenheimer J, et al. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum Reprod Update. 2016;23:19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 22.Burnik Papler T, et al. Transcriptomic analysis and meta-analysis of human granulosa and cumulus cells. PLoS ONE. 2015;10:e0136473. doi: 10.1371/journal.pone.0136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elguindy MM, et al. PUMILIO, but not RBMX, binding is required for regulation of genomic stability by noncoding RNA NORAD. Elife. 2019;8:e48625. doi: 10.7554/eLife.48625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goode B, et al. A recurrent kinase domain mutation in PRKCA defines chordoid glioma of the third ventricle. Nat Commun. 2018;9:810. doi: 10.1038/s41467-018-02826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts JL, et al. Janus kinase 3 (JAK3) deficiency: clinical, immunologic, and molecular analyses of 10 patients and outcomes of stem cell transplantation. Blood. 2004;103:2009–2018. doi: 10.1182/blood-2003-06-2104. [DOI] [PubMed] [Google Scholar]
- 26.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. Mechanisms and functions of long non-coding rnas at multiple regulatory levels. Int J Mol Sci. 2019;20(22):5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife. 2020;9:e60583. doi: 10.7554/eLife.60583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, et al. NEAT1_2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018;9:380. doi: 10.1038/s41419-018-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Long noncoding RNA nuclear enriched abundant transcript 1 (NEAT1) regulates proliferation, apoptosis, and inflammation of chondrocytes via the miR-181a/glycerol-3-phosphate dehydrogenase 1-like (GPD1L) axis. Med Sci Monit. 2019;25:8084–8094. doi: 10.12659/MSM.918416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, et al. LncRNA NEAT1 promotes proliferation of chondrocytes via down-regulation of miR-16-5p in osteoarthritis. J Gene Med. 2020;22:e3203. doi: 10.1002/jgm.3203. [DOI] [PubMed] [Google Scholar]
- 32.Lv Y, et al. LncRNA nuclear-enriched abundant transcript 1 regulates hypoxia-evoked apoptosis and autophagy via mediation of microRNA-181b. Mol Cell Biochem. 2020;464:193–203. doi: 10.1007/s11010-019-03660-2. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, Hirose T. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ventura A. NORAD: defender of the genome. Trends Genet. 2016;32:390–392. doi: 10.1016/j.tig.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki N, et al. Long noncoding RNA NORAD regulates transforming growth factor-β signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci. 2018;109:2211–2220. doi: 10.1111/cas.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, et al. Noncoding RNA activated by DNA damage (NORAD): biologic function and mechanisms in human cancers. Clin Chim Acta. 2019;489:5–9. doi: 10.1016/j.cca.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 37.Kopp F, Elguindy MM, Yalvac ME, Zhang H, Chen B, Gillett FA, Lee S, Sivakumar S, Yu H, Xie Y, Mishra P, Sahenk Z, Mendell JT. PUMILIO hyperactivity drives premature aging of Norad-deficient mice. Elife. 2019;8:e42650. doi: 10.7554/eLife.42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie L, et al. Insulin-like growth factor I promotes oocyte maturation through increasing the expression and phosphorylation of epidermal growth factor receptor in the zebrafish ovary. Mol Cell Endocrinol. 2016;419:198–207. doi: 10.1016/j.mce.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Grøndahl ML, et al. The dormant and the fully competent oocyte: comparing the transcriptome of human oocytes from primordial follicles and in metaphase II. Mol Hum Reprod. 2013;19:600–617. doi: 10.1093/molehr/gat027. [DOI] [PubMed] [Google Scholar]
- 40.Carvalho KF, et al. Mitofusin 1 is required for oocyte growth and communication with follicular somatic cells. FASEB J. 2020;34:7644–7660. doi: 10.1096/fj.201901761R. [DOI] [PubMed] [Google Scholar]
- 41.Filatov M, Khramova Y, Semenova M. Molecular mechanisms of prophase I meiotic arrest maintenance and meiotic resumption in mammalian oocytes. Reprod Sci. 2019;26:1519–1537. doi: 10.1177/1933719118765974. [DOI] [PubMed] [Google Scholar]