Highlights
-
•
Lutein, β-carotene, chlorophyll a, chlorophyll b, and 13 anthocyanins were identified for the first time in fruit peel of a color mutant jujube cultivar-‘Sanbianhong’.
-
•
Major distinctions in the metabolic profiles among the five different colored peel groups.
-
•
Key color metabolites that contribute to the pigmentation in S1, S3, and S5 developmental stages were identified.
-
•
The key color metabolites were quantified using standards as calibrators.
Keywords: Jujube peel, Pigmentation, Metabolome, Quantification standards
Abbreviations: AGC, automatic gain control; BHT, butylated hydroxytoluene; FC, fold change; HCD, higher-energy collision dissociation; HESI, heat electrospray ionization; KEGG, Kyoto Encyclopedia of Genes and Genomes; LOD, limit of detection; LOQ, limit of quantification; NCE, normalized collision energy; OPLS-DA, orthogonal projections to latent structures-discriminant analysis; PCA, principle component analysis; PRM, parallel-reaction monitor; RT, retention time; SBF, ‘Sanbianhong’; SRM, selected reaction monitoring; UHPLC-Q-Orbitrap MS, ultra-high performance liquid chromatography quadrupole Orbitrap mass spectrometry; VIP, variable importance in projection
Abstract
The fruit peel of a color mutant jujube cultivar, ‘Sanbianhong’ (SBF), was investigated using an ultra-high performance liquid chromatography quadrupole Orbitrap mass spectrometry (UHPLC-Q-Orbitrap MS) at five ripening stages (S1, Young fruit stage; S2, swelling stage; S3, white-mature stage; S4, pre-mature stage and S5, mature stage). Lutein, β-carotene, chlorophyll a, chlorophyll b, and 13 anthocyanins were identified. Chlorophyll a and cyanidin 3-O-galactoside were considered key color metabolites in S1 with the content of 1.083 mg/g of fresh weight (FW) and 4.585 mg/g of FW, respectively. Delphinidin (0.488 mg/g FW) and cyanidin (6.259 mg/g FW) were identified as the key pigments in S3. Delphinidin 3-O-glucoside (0.256 mg/g FW) was identified as the key anthocyanin in maturity S5. Herein, the identification and quantitation of pigment-related metabolites of SBF were studied for the first time, and the results provide a theoretical basis for understanding the pigment changes of jujube fruit during ripening.
1. Introduction
Jujube (Ziziphus jujuba Mill.), A member of the Rhamnaceae family is widely used as a centuries-old plant. It is also known as the Chinese date or red date. The red date, which is indigenous to China and has been produced and consumed for over 4000 years, is also commonly available in southern and eastern Asia, Australia, and Europe (Gao et al., 2013). As over 700 Ziziphus jujuba cultivars have been discovered in China to date (Guo et al., 2010), makes it the only nation that exports jujube fruit from ∼1.5 million hectors cultivated area with more than 4.2 million tons of fresh jujube fruit exported annually (Yi et al., 2012). Jujube fruit is popular among customers because of its distinct flavor and excellent nutritional content. Moreover, Jujube is also a rich source of physiologically active substances with nutritional and nutraceutical benefits. According to recent research, Jujube fruits have been shown to contain a variety of beneficial constituents, such as flavonoids, anthocyanins, amino acids, vitamin C, polyphenols, polysaccharides, and triterpene acids. Bioactivity analyses of jujube fruit (Chen et al., 2013, Wang et al., 2011) have revealed antioxidant activity (Choi et al., 2011, Xue et al., 2009), immunological activity (Zhao et al., 2006), a potential sedative effect (Chen et al., 2013, Jiang et al., 2007), jaundice mitigation effects (Ebrahimimd et al., 2011), anti-inflammatory effects (Goyal et al., 2011), analgesic, neuroprotective, and antidiabetic effects.
Anthocyanins (found in jujube fruit) have a strong antioxidant activity that may promote health in various disease conditions significantly (Sarker and Oba, 2018, Zhang et al., 2016). Anthocyanins are primarily linked to the production color intensity of their fruit peels (Chen et al., 2012). They are key water-soluble pigments that give fruits their various hues (Shi et al., 2018) for instance, flowers (Polturak et al., 2018), seeds (Li et al., 2019), and vegetables (Zhang et al., 2016), including pink, red, purple, and blue. The color variations in the fruit at different stages of growth are produced by the interaction of these pigments. Chlorophylls (chlorophyll a and chlorophyll b), carotenoids, and anthocyanins are the main pigments in the immature fruit that impact the color appearance i.e., from white to dark green. Eventually, the fruit color changes from orange to red by the decrease in chlorophyll concentrations while the concentration of carotenoid, anthocyanin, and carotenoids (lutein and β-carotene) increases (Gross, 1991). Kevrešan et al. (2013) reported that the total carotenoid content doubles in the breaker phase in pepper. Perrin et al. (2016) reported that carotenoids are highly accumulated in carrot leaves and roots, and they are impacted by environmental variations. However, studies on pigment compounds in jujube are rare. Furthermore, the diversities of anthocyanins found in jujube, along with chlorophyll and carotenoid concentrations, have not been reported yet. Additionally, the chemical and metabolic mechanisms involved in jujube peel coloring were unknown till recently.
In this study, a color mutant jujube cultivar, ‘Sanbianhong’ (SBF) was selected to investigate the change in jujube peel coloration at different ripening stages. Anthocyanin type, chlorophyll content, and carotenoid content in Ziziphus jujuba Mill. peel at five ripening stages (S1–S5) were qualitatively and quantitatively determined using ultra-high performance liquid chromatography quadrupole Orbitrap mass spectrometry (UHPLC-Q-Orbitrap MS) combined with metabolomic method. The jujube peel undergoes three different color changes during development, hence the name ‘Sanbianhong’. After flowering, the peel color is purple-red in young fruit (S1). As the fruit grows, the color gradually changes to bright yellow in the white-mature stage (S2–S3), and the fruit becomes dark red upon maturity (S4–S5) (Fig. 1). Principle component analysis (PCA) and orthogonal projections to latent structures-discriminant analysis (OPLS-DA) were used for the comparison of jujube peel metabolic profiles at different developmental stages. Additionally, the primary components associated with various color production were determined and visualized through volcano plots and hierarchical cluster analysis. Subsequently, the quantification of targeted pigments in jujube peel was achieved via UHPLC-Q-Orbitrap MS. Up to our knowledge, it is the first study that reports the identification and quantification of color variation in Ziziphus jujuba Mill. peel. Furthermore, it provides the theoretical basis to understand the color variation during the ripening process in jujube.
Fig. 1.
Morphological change during fruit development and ripening in SBF. (A) Photographs of the six ripening stages S1–S5 of SBH; (B) Fruit development color a*; (C) Fruit development color h0. Developmental stages S1–S5 corresponding to days 20, 80, 95, 105, and 120 after anthesis.
2. Materials and method
2.1. Collection of reagents
Methanol, formic acid, isopropanol, n-hexane, ethanol, methyl tert-butyl ether, butylated hydroxytoluene (BHT), HPLC-grade acetonitrile, and acetone were purchased from Fisher (Pittsburgh, PA, USA), all of which were HPLC-grade. Whereas, an Elix water purification system (Millipore, Billerica, MA, USA) was utilized to prepare in-house deionized water. The reference standards lutein (>96 % purity) and β-carotene (>99 % purity) were acquired from Carote Nature (Lupsingen, Switzerland) and measured by HPLC–PDA. Chlorophyll a (>92 % purity) and chlorophyll b (>92 % purity) were obtained from Sigma-Aldrich (St. Louis, MO, USA), and also measured by HPLC–PDA. The standards of delphinidin-3-O-glucoside (≥95 % purity), cyanidin (≥98 % purity), cyanidian-3,5-O-diglucoside (≥97 % purity), delphinidin (≥98 % purity), and cyanidin 3-O-galactoside (≥98 % purity) were obtained from Extrasynthese (Genay, France). All standards were of HPLC grade.
2.2. Sample collection
The jujube cultivar Ziziphus jujuba Mill. SBF was obtained from the Experimental Station of Jujube at Luoyang Normal University in Luoyang, Henan, China. In order to generate accurate statistical data, six jujube trees with stable phenotypic characteristics, as well as similar health, ages, and heights, were randomly selected. Each jujube tree was taken as a sample, and six-dimensional sampling was performed. The fruits were chosen based on their consistent size, shape, and color without any prior mechanical damage or disease. On days 35, 80, 95, 105, and 120 following anthesis, fruit samples were obtained at five developmental stages. These samples were labeled as S1, S2, S3, S4, and S5 each sample reflecting a distinct developmental stage as shown in Fig. 1. The fruits were divided into pulps and peels with a knife. Pulp with 0.2 cm thickness collected form these five fruits frozen in liquid nitrogen, and stored at −80 °C.
2.3. Pigment metabolite analysis for treated samples
Pigment metabolites were extracted as previously reported (Xue et al., 2020) with some modifications. The freeze-dried samples were crushed at 30 Hz in a mixer mill (MM 400, Retsch) for 1.5 min with a zirconia bead. One hundred milligrams of powder were added to a one-milliliter solution of ethanol, acetone, and n-hexane (1:1:2, v:v:v; 0.01 % BHT) and extracted for 20 min at room temperature in an ultrasonic bath. The supernatant was collected after the centrifugation at 12,000 rpm, 4 °C for 5 min, and the extraction technique was done twice. The dry extract was reconstituted in 0.5 mL of a mixture of acetonitrile, methanol, and methyl tert-butyl ether (51:17:12, v:v:v) after the three supernatants were mixed and dried under the nitrogen gas steam. Later, the supernatant from the solution was centrifuged for 15 min at 12,000 rpm and 4 °C and stored in the dark for UHPLC-MS/MS analysis to prevent the isomerization and photodegradation of carotenoids. Each sample was analyzed six times.
2.4. Color measurement
The peel color of jujube cultivar SBF was determined at each of the five developmental stages. CR-400 Minolta Colorimeter (Osaka, Japan) was used to measure the peel color parameters in the L*, a*, b* and h0 modes, where L* defines lightness (0 = black, 100 = white); a* and b* indicate red-green and blue-yellow; and h0hue = arctan (b*/a*) is the hue angle on the color wheel (0° = red/purple, 90° = yellow, 180° = green, and 270° = blue). The hue of the peel represented by the values of a* and h0 was measured on the equator of fruit.
2.5. Instrumentation condition for qualitative and quantitative analysis of anthocyanins, lutein, chlorophyll and β-carotene
Six biological replicates were evaluated separately for each stage along with the evaluation of 30 random samples for eradicating the analytical biasness. An Orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific, Waltham, MA, USA) equipped with a heat electrospray ionization (HESI) source was used to analyze the sample extracts. A Thermo Hypersil COLD VANQUISH C18 column (1.9 μm, 2.1 mm, 100 mm, Thermo Fisher Scientific, Waltham, MA, USA) was utilized in the HPLC. The mobile phase was made up of 0.1 % formic acid in water (A) isopropanol (B), and the gradient program was set as 60 % B at 0 min, 75 % B at 4 min, 100 % B at 12 min, 98 % B at 15 min, and 60 % B between 16 and 22 min. The injection volume was 5 μL, the temperature was 40 °C, and the 5 μLflow rate was 0.2 mL/min. Full MS scans and a parallel-reaction monitor (PRM) (300–1,800 m/z) were used to perform data-dependent mass spectrometry analysis while adjusting ionization polarity. Data was collected in Q Exactive software using an Orbitrap mass analyzer with a mass resolution of 70,000 at 400 m/z. Each duty cycle, the twenty most energetic precursor ions from a survey scan were picked for MS/MS and identified in an Orbitrap analyzer with a mass resolution of 35,000 at 400 m/z. The HESI probe and transfer capillary temperature were kept at 350 °C, and the automatic gain control (AGC) was set to 1.0 × 106,. The maximum injection time was 100 ms. All tandem mass spectra were generated using the higher-energy collision dissociation (HCD) method with a normalized collision energy (NCE) of 20–80 eV, an AGC target of 2.0 × 105, a maximum injection time of 100 ms, a fill ratio of 1.0 percent, isotope exclusion “on,” and a dynamic exclusion of 5.0 s. The specific ion peaks were examined using MS/MS using Inclusion and Exclusion Indexes, and dynamic exclusion was set for 20 s.
2.6. Qualitative analysis methods
An HPLC-ESI-MS/MS system was used to analyze the parameters were as detailed in section 2.3 metabolites by comparing the accurate precursor ions (Q1), retention duration (RT), product ions (Q3), and fragmentation patterns in a qualitative study of primary and secondary MS data. Data were collected in Xcalibur 4.0 (Thermo-Fisher Scientific, Waltham, MA, USA) with the use of Compound Discover 3.0, Mass Frontier 8 (Thermo-Fisher Scientific), and the publicly available metabolite databases ChemBank (https://chembank.med.harvard.edu/compounds;pubchem), PubChem (https://pubchemblog.ncbi.nlm.nih.gov/) and NIST Chemistry Webbook (https://webbook.nist.gov/). However, the second-order spectrum information was used to identify the metabolites eliminating the repeated signals of NH4+, Na+, K+, and other large molecular weight compounds.
2.7. Quantitative analysis methods
The full MS scans (set from 100 to 1500 m/z) were used for the quantitative study of metabolites. To determine signal intensities, the orbitrap mass spectrometer was used to screen the distinctive ions of each metabolite. Furthermore, Xcalibur 4.0 software was used to accomplish the integration and rectification of chromatographic peaks. Eventually, individual constituent concentrations were calculated using peak areas and calibration curves produced from the corresponding standard compounds. The PRM server of Xcalibur 4.0, was used to gather and process all of the data in the profile based on specific molecular weight. It's a dependable detection method that's similar to QqQ's selected reaction monitoring (SRM) mode, without any instrumental parameter adjustment (Zhang et al., 2018).
For the quantitative analysis of the target color metabolite, each analyte was identified on the basis of its retention time, and their relative abundance. The content of target color metabolite was calculate based on calibration curves according to their relative abundance.
2.8. Calibration curves preparation and determining LOD LOQ and linearity
One milligram of each standard (lutein, chlorophyll a, chlorophyll b, delphinidin, delphinidin-3-O-glucoside, cyanidin 3-O-galactoside, cyanidin, and cyanidian-3,5-O-diglucoside, was dissolved in 1 mL methanol/acetonitrile (3:1) and diluted to generate eleven calibration levels, ranging from 1 × 10−6 to 1 × 10−3 mg/mL. Finally, at each concentration level, the standard solution in triplicates was injected to generate calibration curves. To construct standard curves, the ratios of peak areas were computed and plotted against the corresponding concentrations of standard compounds using the weighted linear regression (least-squares) algorithm (Mari et al., 2015).
The least-squares linear regression model was used to create calibration curves based on the peak area of each authentic standard. Moreover, the linearity of all calibration curves was assessed through correlation coefficients. The limit of detection (LOD) for each authentic standard is defined as the concentration of the authentic standard that gives a signal equal to the background plus three times the blank's standard deviation. The limit of quantification (LOQ) was calculated by infusing successively diluted standard solutions until the signal-to-noise ratio fell below 10 (Mari et al., 2015).
2.9. Statistical analysis
Due to the high dimension dataset characteristics of metabolomics data, combining univariate and multivariate statistical analyses is imperative for accurately identifying differential metabolites. Before analysis, the relative abundances of each metabolite were log-transformed to achieve normality. The amount of each metabolite was compared between time points using Dunnett's test. Consequently, all statistical analyses were performed through Xcalibur 4.0 server.
SIMCA-P version 14.0 (Umetrics, Ume, Sweden, https://www.umetrics.com/simca) was used to evaluate the metabolome data. To maximize the metabolome variations between developmental phases, PCA and OPLS-DA were utilized (Worley & Powers, 2016). To visualize changes in metabolite profiles, the HCA of samples was conducted based on the regions of chromatographic peaks. The squared Euclidean distance was used to determine similarity, and the Ward method was employed for the clustering algorithm (Liang et al., 2013). A retention time window of 0.1 % ± 0.15 min and a mass window of 5.0 ppm ± 2.0 mDa were used to align and analyze metabolites from different files. In the program, the filter condition was selected to “retain entities that occurred in at least 65 % of data in at least one condition.” Significantly, based on variable importance in projection (VIP) and fold change (FC), the volcano graphic depicts the differences in metabolite expression levels between compared samples. The criteria for substantial changes were VIP ≥ 1.0 and FC ≥ 2 or ≤ 0.5 (Cao et al., 2019).
3. Results and discussion
3.1. Changes in phenotype and peel color in SBF
During fruit development, SBF exhibits different patterns of color change (Fig. 1A). The peel color is purple-red in young fruit stage (S1), which is different from the color of most jujube peels in this stage, and then the color gradually fades in swelling stage (S2). As the fruit matures, the peel color changes to a bright yellow with pink spots in the white-mature stage (S3), followed by semi-red in pre-mature stage (S4) and red in mature stage (S5).
Peel colors were visualized in two- and three-dimensional space using an international color measuring system i.e., color spaces and numerical values measured in L*a*b* using the CIELab system (León et al., 2006). In 1999, Segnini applied the method to measure the color of potato chip, the chips occupied a space of 77 < L* < 83, −9 < a* < −4. In the study (Segnini et al., 1999), The two parameters a* and h0 were used to measure color change. The SBF fruit had a hue angle h0 < 30, which appeared as purple-red at the S1 stage (Fig. 1C). The hue angle of SBF decreased from S3 to S5 (white-mature stage to red maturity stage), reaching a constant value of 15. In contrast, the a* values of SBF were higher in S1 (purple-red stage) and S5 (red stage), and lower in S2 and S3 (white-mature stage) (Fig. 1B). Subsequently, the obtained values for the color parameter are comparable to the previously reported study for jujubes. Additionally, they are compatible with the change in fruit peel color from purple-red to red (Almansa et al., 2016).
3.2. PCA revealed differences in peels color metabolite profiles
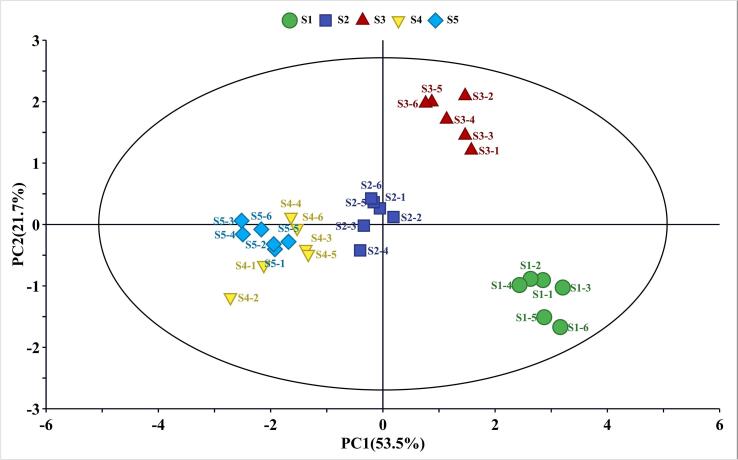
The metabolic alterations in SBF fruit peel during ripening were investigated using the extensively targeted technique. Four major color-forming substances (lutein, β-carotene, anthocyanins, and chlorophyll) were identified from the cultivar. Among them, lutein, β-carotene, chlorophyll a, chlorophyll b, and 13 anthocyanins were annotated (Table S1). A scatter plot of principal components is used to display the underlying structure of multiple variables using PCA, which is a commonly used statistical approach for extracting and rationalizing information from any multivariate description of a biological system. In the PCA plot (Fig. 2), the first component, PC1, represents the main constituent variance of the samples, and it accounts for 53.5 % of the X variance. In contrast, PC2 accounts for a much smaller amount (21.7 %). Together, the first two principal components account for 75.2 % of the metabolic variances for all samples. This indicates that the two main components can explain 75.2 % of the data, and the model is reliable. In Fig. 2, the S1–S3 stage of SBF is clearly separated, suggesting significant differences in color-forming metabolic phenotypes among S1–S3 stages. As a mutant cultivar, SBF color mutation is mainly reflected in the first three stages (purple-red to greenish white), and the PCA results are consistent with the color changes of the fruit. In addition, the plot suggests that S4 and S5 cannot be completely separated. The S4 stage of SBF is a close to S5. One reason may be that the color of S4 is semi-red and S5 is red, and the colors of S4 and S5 partly overlap. The other reason is PCA is an unsupervised method. Subsequently, pairwise comparisons of metabolites were generated using OPLS-DA models (Worley & Powers, 2016), looking for different metabolites in S4 and S5.
Fig. 2.
PCA of the relative differences in metabolites in jujube peels during different ripening stages. Young fruit stage (S1), swelling stage (S2), white-mature stage (S3), pre-mature stage (S4) and red in mature stage (S5). The same color indicates six parallel repeats for each stage. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Differential metabolite analysis upon OPLS–DA
Importantly, when compared to PCA, OPLS–DA improves group separation and makes finding differential metabolites easier. Orthogonal and nonorthogonal variables were evaluated in the OPLS-DA analysis to get undisputed evidence for metabolite differences across inter-group correlations as well as in experimental groups (Trygg & Wold, 2002). Fig. 3A shows the scatter score plot derived from the inter-group comparison in OPLS-DA. The ordinate (t0[1]) reflects the scores of the orthogonal principal components, whereas the abscissa (t[1]) represents the anticipated PC1 values. Moreover, the percentage of predictor (response) variation explained by the model is indicated by R2X. Q2Y denotes the model's cross-validation-estimated prediction performance. The model's prediction ability is represented by Q2. When Q2 > 0.5, the model is regarded as good, while the model is viewed as significant with Q2 > 0.9 (Thévenot et al., 2015). In the comparison between S4 and S5, the OPLS-DA models had high predictability (Q2) and strong goodness of fit (R2X, R2Y) (Q2 = 0.807, R2X = 0.996, R2Y = 0.968). The two phases of SBF formation can be clearly distinguished in the OPLS-DA models, demonstrating significant differences in metabolic profiles of peel hues.
Fig. 3.
OPLS-DA and permutation test results. (A) Score scatter plot of the OPLS-DA model for jujube peels; (B) Permutation test of the OPLS-DA model for jujube peels. The abscissa in (B) indicates the retention degree of permutation (a percentage describing how much of the original sequential order in Y remained unchanged). The point where the percentage equals 1 shows the R2 and Q2 values of the original model. Otherwise, the ordinate represents R2 or Q2. The green dots indicate the R2 values obtained by the permutation test, and the blue square points indicate the Q2 values obtained by the permutation test. The two dotted lines indicate the regression lines for R2 and Q2, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the sequential sequence of the categorical variable Y was randomly altered many times (n = 200) in the permutation test to further assess the predictability of the OPLS-DA model to determine R2 and Q2 values. The findings imply that this step is critical for preventing overfitting in the test model and determining its statistical significance. The original model R2 was close to 1, as shown in Fig. 3B, suggesting that the constructed model well described the real situation in the sample data. Additionally, Q2 was near to 1, indicating that adding more samples to the model might result in a comparable distribution. Furthermore, the degree of permutation retention decreased over time by the increase in Y variables and the decline in the R2 and Q2 model. As a result of these findings, the constructed differential model appears to be reliable and accurate.
3.4. HCA analysis revealed differences in peel color metabolites
HCA is a multivariate statistical analysis approach for classification that may determine homogenous groupings of unlabeled components using a distance metric by evaluating relative similarity or differentially among objects. Therefore, individuals in the same category have the highest homogeneity possible (Rosati et al., 2017). Reliable clustering is achieved when the variability among items belonging to the same cluster is minimal. Consequently, the HCA of the samples was done based on the relative peak areas of differential metabolites generated from the volcano map to determine the similarities and differences of these samples. Significantly, the range approach was used to standardize the metabolite content data to generate a dendrogram of HCA.
The HCA analysis also revealed that the metabolites between the five stages were obviously different (Fig. S1). The concentrations of some metabolites were significantly higher in the S1 stage than other stages, including chlorophyll a, chlorophyll b, lutein, cyanidin 3-O-galactoside, peonidin, cyanidin O-syringic acid, cyanidin 3-O-malonylhexoside, pelargonin, and petunidin 3-O-glucoside. Conversely, some metabolites were significantly higher in the S3 stage, such as delphinidin, cyanidin 3,5-O-diglucoside, β-carotene, pelargonidin, cyanidin, and apigeninidin chloride. Only a few metabolites showed higher levels in the S2, S4, and S5 stages. Thus, the PCA and OPLA-DA analyses together suggest that metabolites in the five stages of jujube fruit development had distinct metabolite profiles. Further analysis of these metabolites, with large differences in content, was then required.
3.5. Identification of differential color metabolites and enrichment analysis during jujube ripening
The differential metabolites were screened by their respective VIP scores and FC values i.e., VIP value of >1 while FC value of ≥2 or ≤0.5 in each sample using the OPLS-DA model. FC indicates whether there is a difference between the experimental and control groups by multiplying the two groups together. Generally, FC ≥ 2 indicates up-regulation, and FC ≤ 0.5 indicates down-regulation. The intensity of the inter-group variations in corresponding metabolites of each group in the model is represented by the VIP value. When the VIP value is more than one, the difference is considered significant (Lourencetti et al., 2018). The differential metabolites were calculated using a volcano plot (Fig. 4A–D) based on the VIP and fold change values. The abscissa represents the fold-change value (logarithm to base 2) of each substance in the group, and each point represents a metabolite in the plot.
Fig. 4.
Differential metabolite screening results for each ripening stage presented using volcano plots. The volcano plot shows the differential metabolite expression levels between S1–S5. Green dots represent downregulated differentially expressed metabolites; orange spots represent upregulated differentially expressed metabolites; and gray spots represent non-differentially expressed metabolites. (A) S1 vs S2; (B) S2 vs S3; (C) S3 vs S4; (D) S4 vs S5. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Two chlorophylls, two carotenoids, and thirteen anthocyanins were detected at each of the five developmental stages (S1–S5). All color production metabolites collected in stage S1 are majorly responsible for the production of the red color before fruit ripening. The findings are substantially in line with those of previous research (Shi et al., 2018). Further analysis of the volcano plot showed that chlorophyll a and a cyanidin 3-O-galactoside were down-regulated and cyanidin was up-regulated in S2 compared to S1 (Fig. 4A). The others showed no significant difference. Comparing S2 and S3 (Fig. 4B), delphinidin and cyanidin were upregulated and cyanidin 3-O-galactoside was downregulated. The others showed no significant difference. From S1 to S3, the peel color of SBF gradually transitioned from purple-red in S1 to yellow in S3. Therefore, the change in color indicates cyanidin 3-O-galactoside and chlorophyll a are one of the key color metabolites involved in the pigmentation in S1, and delphinidin and cyanidin may be considered key pigments in S3. In addition, the change in color from S1 to S3 is not determined by a single variable, but a combination of downregulated chlorophyll A and cyanidin 3-O-galactoside and upregulated delphinidin and cyanidin. By comparing S3 and S4, cyanidin 3,5-O-diglucoside and delphinidin were downregulated, and others showed no significant difference (Fig. 4C). Comparing S4 and S5 (Fig. 4D), delphinidin 3-O-glucoside was upregulated and chlorophyll b and lutein were downregulated. Other metabolites showed no significant difference. From S3 to S5, the color intensity increased gradually from bright yellow in S3 to semi-red stage in S4 and red in S5. Thus, delphinidin 3-O-glucoside may be regarded as the primary anthocyanin implicated in the red pigmentation of jujube peel at S5. These findings are in line with those of Zhang et al., who discovered that delphinidin 3-O-glucoside is one of the key metabolites responsible for the color of ripe jujube fruit's peel (Zhang et al., 2020). Moreover, delphinidin 3-O-glucoside has been reported for red coloration in various plant organs (Stintzing & Carle, 2004).
3.6. Quantitative analysis of significant pigments causing the color change of jujube peels
Qualitative analysis of the identified compounds was performed via a preliminary fragmentation study based on MS/MS experiments on individual precursor ions. The accurate reaction times (RTs), fragmentation patterns, precursor ion (Q1), and product ion (Q3), were compared to the obtained results by injecting standards under the similar conditions.
Consequently, eight compounds were identified as a primary source involved in the variation of jujube peel color i.e., lutein, chlorophyll a, chlorophyll b, delphinidin, cyanidin, delphinidin-3-O-glucoside, cyanidian-3,5-O-diglucoside, cyanidin 3-O-galactoside. All compounds were tentatively identified using the Orbitrap mass spectra of standards, m/z spectral data, elution order, and retention time.
All of these eight authentic standards are described in Table 1, as well as Figs. 5, S2, and S3. Lutein, represent 569.43115 (M−H)+ ion at m/z, peaked at 4.45 min whereas, the elemental composition was C40H56O2. In the positive ion mode, multiple chemical bonds were broken, and fragments at 119.08582 m/z and 145.10135 m/z, which are characteristic of lutein, were detected in their MS/MS spectra (Fu et al., 2012).
Table 1.
The performance characteristics of eight authentic standards.
| Compound | RT(min) | Molecular formula | Ion | Measured (m/z) | Theoretical (m/z) | MS/MS | Error (ppm) |
|---|---|---|---|---|---|---|---|
|
8.22 | C55H72MgN4O5 | [M−H]+ | 893.54059 | 892.53476 | 615.24286/555.22345 | 0.05 |
|
6.68 | C55H70MgN4O6 | [M−H]+ | 907.52136 | 906.51402 | 629.20935/569.18823 | −0.03 |
|
4.45 | C40H56O2 | [M−H]+ | 569.43115 | 568.42748 | 119.08582/145.10135 | 0.1 |
|
5.08 | C15H11CLO7 | [M−H]+ | 339.34445 | 338.01878 | 304.30768/322.31674 | 0.07 |
|
2.51 | C21H21O12CL | [M−H]+ | 501.37750 | 500.07160 | 256.26258/228.23164 | 0.04 |
|
10.29 | C27H31O16CL | [M−H]+ | 647.45728 | 646.12951 | 448.39413/91.61111 | −0.06 |
|
3.09 | C15H11CIO6 | [M−H]+ | 322.18469 | 322.02386 | 163.11166/267.12057 | 0.2 |
|
3.47 | C21H21CIO11 | [M−H]+ | 485.47491 | 484.07669 | 228.23187/200.20177 | −0.08 |
Fig. 5.
The ion chromatograms of eight authentic standards under the optimal UHPLC-Q-Orbitrap MS conditions. (A) Chlorophyll a; (B) Chlorophyll b; (C) Lutein; (D) Delphinidin; (E) Cyanidin; (F) Cyanidin 3,5-O-diglucoside; (G) Delphinidin 3-O-glucoside; (H) Cyanidin 3-O-galactoside.
When chlorophyll a and chlorophyll b were analyzed by ESI/MS/MS, a different fragmentation pattern was observed. For chlorophyll a, the peak appeared at 6.68 min with an elemental composition of C55H72MgN4O5. The parent ion was at 893.54059 m/z, and the principal daughter ions were at 615.24268 m/z and 555.22339 m/z, together with a series of low molecular weight ions in the positive ion mode. For chlorophyll b, the peak was at 8.20 min and the elemental composition was C55H70MgN4O6. In the positive ion mode, the parent ion was at m/z 907.52014, while the principal daughter ions were at 629.20935 m/z and 569.18823 m/z (Fu et al., 2012).
Five anthocyanins were analyzed by ESI/MS/MS. For delphinidin, the peak appeared at 5.07 min, the (M−H)+ ion was at m/z 339.34442, and the elemental composition was C15H11CLO7. In the positive ion mode, fragments at 304.30768 m/z and 322.31674 m/z were detected in their MS/MS spectra. For cyanidine, it peaked at 10.29 min, with the (M−H)+ ion at m/z 647.45728, and the elemental composition was C27H31O16CI. Fragments at 448.39413 m/z and 91.61111 m/z were detected in the MS/MS spectra. In positive ion mode, the peaks of delphinidin-3-O-glucoside, cyanidian-3,5-O-diglucoside and cyanidin 3-O-galactoside appeared at 2.51, 10.29 and 3.47 min, (M−H)+ ions were at m/z 485.47491, 647.45728 and 501.49789, and the elemental compositions were C21H21O12CL, C27H31O16CL and C21H21CIO11, respectively (Janeiro and Brett, 2007, Schütz et al., 2006, Shi et al., 2018).
Nine concentration levels were tested in triplicate to create standard calibration curves. Highly negative intercepts (y-axis) were detected using a linear regression throughout the whole concentration range. Additionally, when the concentration of analyte increased, the sensitivity (slopes) increased. Furthermore, to represent the detector response as a function of concentration, quadratic fitted curves were employed.
Table 2 shows the calibration curves for all eight analytes as well as their performance characteristics. The correlation coefficients within the calibrating range of all standards were>0.99, indicating that all calibration curves were linear. These results suggest that a standard calibration can be applied for quantitative purposes. The signal-to-noise ratio (S/N) of each peak in the spike injection was used to compute the LOQ and LOD for each compound in each sample. These calculations are based on S/N = 10 for LOQ and S/N = 3 for LOD. The observed values are acceptable for analysis of these compounds in the predicted ranges, although these values differed widely across the eight compounds due to the much higher sensitivity. Table 2 shows that the selected approach is sensitive enough to identify trace components in jujube fruit peel, with LOD and LOQ ranging from 0.01 to 1 ng/mL and 0.5 to 3 ng/mL, respectively.
Table 2.
The calibration curves, LODs, and LOQs of the eight components.
| Compound | Linearity equation | R2 | Linearity range (ng/mL) | LOD (ng/mL) | LOQ (ng/mL) |
|---|---|---|---|---|---|
|
Y = 1.744 × 104X − 1.419 × 103 | 0.9951 | 15–80,000 | 0.4 | 1.5 |
|
Y = 8.067 × 103X − 5.196 × 103 | 0.9930 | 10–40,000 | 0.03 | 1 |
|
Y = 5.429 × 104X + 6.694 × 104 | 0.9917 | 30–20,000 | 0.02 | 3 |
|
Y = 2.743 × 105X + 4.478 × 107 | 0.9951 | 10–50,000 | 0.05 | 1 |
|
Y = 1.033 × 103X − 2.507 × 104 | 0.9957 | 30–70,000 | 0.5 | 3 |
|
Y = 9.510 × 105X + 6.768 × 107 | 0.9909 | 5–90,000 | 0.01 | 0.5 |
|
Y = 3.015 × 102X + 3.523 × 104 | 0.9934 | 30–85,000 | 1 | 3 |
|
Y = 1.659 × 103X + 1.715 × 105 | 0.9964 | 30–90,000 | 1 | 3 |
The quantification results are displayed in Fig. 6A–H, and each compound's content was determined using the appropriate calibration curve. In a 22-minute run, the data was effectively acquired. The total ion chromatograms (Fig. S4) serve as an analytical fingerprint for pigment-related compound identification and authentication.
Fig. 6.
Quantitative analytical results of eight color metabolites using the standard curve. (A) Chlorophyll a; (B) Chlorophyll b; (C) Lutein; (D) Delphinidin; (E) Cyanidin; (F) Cyanidin 3,5-O-diglucoside; (G) Delphinidin 3-O-glucoside; (H) Cyanidin 3-O-galactoside.
The Q-Orbitrap MS is a powerful high-resolution mass spectrometer that can perform both qualitative and quantitative evaluations simultaneously. Because the Q-Orbitrap MS has such high sensitivity even for some low-content compounds (e.g., lutein [0.001–0.073 mg/g FW]) and high-content compounds (e.g., cyanidin [3.166–3.259 mg/g FW]) investigated at the same time, indicating that low- and high-content constituents could be quantified simultaneously.
The content of chlorophyll a largely varies in jujube fruit peel (Fig. 6A), depending on the developmental stage. As one of the main color metabolites in the early stage of jujube fruit development (S1), the content of chlorophyll a is at a maximum at S1 (1.083 mg/g FW). With the continuous maturation of jujube fruit, chlorophyll a gradually decreased, with the exception of a slight increase at S3. The lowest value, 0.023 mg/g FW, was detected in S5. This result is in line with prior research, which found that chlorophylls play a significant role in the early stages of the ripening process Shi et al. (2018). Cyanidin 3-O-galactoside concentration peaked at 4.585 mg/g FW in S1 subsequently reduced (Fig. 6H). The findings are comparable to those Bastos et al. (2016), who discovered that anthocyanin concentration increased during the start of the Indian jujube fruit pigmentation stage and then reduced until complete maturity. The results are consistent with those of Section 3.1 shown in Fig. 3B and 3C, which statistically indicate that S1 is purple-red. Chlorophyll a (Fig. 6A) and Cyanidin 3-O-galactoside (Fig. 6H) are key color metabolites involved in pigmentation in S1. The content of Cyanidin 3-O-galactoside is 4.2 times greater than chlorophyll a, So the color of the jujube peel is purple-red in S1. Cyanidin and delphinidin are the main anthocyanins found in S3. The concentrations of cyanidin began to increase before S3 and then decreased to S5. The highest value was 6.259 mg/g FW in S3 (Fig. 6G). The concentrations of delphinidin began to increase before S3 and then decreased to S4, with a slight rise in S5. The maximum value was 0.488 mg/g FW in S3 (Fig. 6D). Similar change patterns were found for the contents of cyanidin 3,5-O-diglucoside. The content of cyanidin 3,5-O-diglucoside was highest in S3, but not significantly higher than that in S2. However, there was a significant decline from S3 to S4 (Fig. 6F). Fig. 3B and 3C showed that the red gradually fades from S1 to S3, corresponding to the results revealed in Fig. 6, which indicated that most of the color metabolites decreased in S3 (such as chlorophylls, lutein, delphinidin 3-O-glucoside, and cyanidin 3-O-galactoside), except for the increase in cyanidin, delphinidin and cyanidin 3,5-O-diglucoside. The color presented in S3 results from a variety of color metabolites. delphinidin 3-O-glucoside was considered the primary anthocyanin involved in the red pigmentation of the jujube peel in S5. The 3-O-glucoside content of delphinidin increased first (S1–S2), then decreased (S2–S3), and then increased (S3–S4–S5). The content was significantly higher in S5 (0.256 mg/g FW) than in S4 (Fig. 6E). The change trend of delphinidin 3-O-glucoside corresponded to that in Fig. 3B and 3C, which showed that the red gradually increases from S3 to S4 and S5. The results of the volcano map showed that the content of delphinidin 3-O-glucoside increased significantly, and the other color metabolites had no significant change from S3 to S4 and S5. Meanwhile, the color of the jujube peel gradually turned red, indicating that the content of delphinidin 3-O-glucoside corresponded to the color change of the jujube peel. The results were consistent with an earlier report, which showed that glycosylated anthocyanins contribute significantly to the formation of red color (Nobuhiro et al., 2013). The content of chlorophyll b (Fig. 6B) and lutein (Fig. 6C) decreased sharply from S1 to S2 and then slowly declined until fruit maturity. Compared to S4, chlorophyll b and lutein decreased sharply in S5.
4. Conclusion
Briefly, UHPLC-QqQ-MS was applied for the investigation of color change in fruit peels at different developmental stages in SBF. The findings reveal that there are significant differences in metabolic profiles across the five various colored peel groups. The differentially colored metabolites during each jujube ripening stage were identified through analysis of PCA, OPLS-DA, HCA, and volcano plots. The results suggest that the key pigmentations are distinctive in the early, middle, and late stages of jujube fruit development. Based on standards and calibration curves, the major pigmentation compounds were quantified. The identification and quantitation of color metabolites of SBF are reported for the first time in this paper. Importantly, the findings give a theoretical foundation for understanding jujube fruit pigment changes during ripening, allowing for a better knowledge of the key pigments responsible for ripening fruit color.
Funding sources
This work was supported by the National Natural Science Foundation of China (Grant Nos.: 32001669, 32101565) and Key Scientific Research Projects in Colleges and Universities of Henan Province (Grant No.: 20B180005). We thank LetPub (https://www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
CRediT authorship contribution statement
Hongxia Liu: Conceptualization. Lefei Wang: Writing – review & editing. Hui Liu: Validation. Benliang Deng: Investigation, Project administration. Shipeng Li: Supervision. Xusheng Zhao: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2022.100470.
Contributor Information
Shipeng Li, Email: lsp423000@163.com.
Xusheng Zhao, Email: xshzhao2022@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
HCA analyses of metabolites identified in five stages of jujube fruit development. The colors of the heat map indicate the relative content of each metabolite, from low (green) to high (red).
Supplementary Fig. S2.
The mother ion of eight color metabolites under the optimal UHPLC-Q-Orbitrap MS conditions. (A) Chlorophyll a; (B) Chlorophyll b; (C) Lutein; (D) Delphinidin; (E) Cyanidin; (F) Cyanidin 3,5-O-diglucoside; (G) Delphinidin 3-O-glucoside; (H) Cyanidin 3-O-galactoside.
Supplementary Fig. S3.
The daughter ion of eight color metabolites under the optimal UHPLC-Q-Orbitrap MS conditions. (A) Chlorophyll a; (B) Chlorophyll b; (C) Lutein; (D) Delphinidin; (E) Cyanidin; (F) Cyanidin 3,5-O-diglucoside; (G) Delphinidin 3-O-glucoside; (H) Cyanidin 3-O-galactoside.
Supplementary Fig. S4.
Total ion chromatogram (TIC) of jujube fruit at different ripening stages (S1–S5) in negative ion mode.
Characteristics of the pigmentation identified and quantified in the jujube peels.
References
- Almansa S., Hernández F., Legua P., Nicolás-Almansa M., Amorós A. Physico-chemical and physiological changes during fruit development and on-tree ripening of two Spanish jujube cultivars (Ziziphus jujuba Mill.) Journal of the Science of Food and Agriculture. 2016;96:4098–4105. doi: 10.1002/jsfa.7610. [DOI] [PubMed] [Google Scholar]
- Bastos V.J., Neves L.C., da Silva P.M.C., Shahab M., Colombo R.C., Roberto S.R. Harvest point determination of indian jujube fruit (Ziziphus mauritiana L.) based on physicochemical and functional parameters. Scientia Horticulturae. 2016;213:392–402. doi: 10.1016/j.scienta.2016.10.030. [DOI] [Google Scholar]
- Cao H., Ji Y., Li S., Lu L., Tian M., Yang W., et al. Extensive metabolic profiles of leaves and stems from the medicinal plant Dendrobium officinale Kimura et Migo. Metabolites. 2019;9:215. doi: 10.3390/metabo9100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.S., Zhang D., Wang Y.Q., Li P.M., Ma F.W. Effects of fruit bagging on the contents of phenolic compounds in the peel and flesh of ‘Golden Delicious’, ‘Red Delicious’, and ‘Royal Gala’ apples. Scientia Horticulturae. 2012;142:68–73. doi: 10.1016/j.scienta.2012.05.001. [DOI] [Google Scholar]
- Chen J., Li Z., Maiwulanjiang M., Zhang W.L., Zhan J.Y.X., Lam C.T.W., et al. Chemical and biological assessment of Ziziphus jujuba fruits from China: Different geographical sources and developmental stages. Journal of Agricultural and Food Chemistry. 2013;61:7315–7324. doi: 10.1021/jf402379u. [DOI] [PubMed] [Google Scholar]
- Choi S.H., Ahn J.B., Kozukue N., Levin C.E., Friedman M. Distribution of free amino acids, flavonoids, total phenolics, and antioxidative activities of jujube (Ziziphus jujuba) fruits and seeds harvested from plants grown in Korea. Journal of Agricultural and Food Chemistry. 2011;59:6594–6604. doi: 10.1021/jf200371r. [DOI] [PubMed] [Google Scholar]
- Ebrahimimd S., Ashkani-Esfahani S., Poormahmudibs A. Investigating the efficacy of zizyphus jujuba on neonatal jaundice. Iranian Journal of Pediatrics. 2011;21:320–324. [PMC free article] [PubMed] [Google Scholar]
- Fu W., Magnúsdóttir M., Brynjólfson S., Palsson B.Ø., Paglia G. UPLC-UV-MSE analysis for quantification and identification of major carotenoid and chlorophyll species in algae. Analytical and Bioanalytical Chemistry. 2012;404:3145–3154. doi: 10.1007/s00216-012-6434-4. [DOI] [PubMed] [Google Scholar]
- Gao Q.H., Wu C.S., Wang M. The jujube (Ziziphus Jujuba Mill.) fruit: A review of current knowledge of fruit composition and health benefits. Journal of Agricultural and Food Chemistry. 2013;61:3351–3363. doi: 10.1021/jf4007032. [DOI] [PubMed] [Google Scholar]
- Goyal R., Sharma P.L., Singh M. Possible attenuation of nitric oxide expression in anti-inflammatory effect of Ziziphus jujuba in rat. Journal of Natural Medicines. 2011;65:514–518. doi: 10.1007/s11418-011-0531-0. [DOI] [PubMed] [Google Scholar]
- Gross J. Van Nostrand Reinhold; 1991. Pigments in vegetables: Chlorophylls and carotenoids. [Google Scholar]
- Guo S., Duan J.A., Tang Y.P., Zhu Z.H., Qian Y.F., Yang N.Y., et al. Characterization of nucleosides and nucleobases in fruits of Ziziphus jujuba by UPLC-DAD-MS. Journal of Agricultural and Food Chemistry. 2010;58:10774–10780. doi: 10.1021/jf102648q. [DOI] [PubMed] [Google Scholar]
- Janeiro P., Brett A.M.O. Redox behavior of anthocyanins present in Vitis vinifera L. Electroanalysis. 2007;19:1779–1786. doi: 10.1002/elan.200703941. [DOI] [Google Scholar]
- Jiang J.G., Huang X.J., Chen J., Lin Q.S. Comparison of the sedative and hypnotic effects of flavonoids, saponins, and polysaccharides extracted from Semen Ziziphus jujube. Natural Product Research. 2007;21:310–320. doi: 10.1080/14786410701192827. [DOI] [PubMed] [Google Scholar]
- Kevrešan Ž.S., Mastilović J.S., Mandić A.I., Torbica A.M. Effect of different ripening conditions on pigments of pepper for paprika production at green stage of maturity. Journal of Agricultural and Food Chemistry. 2013;61:9125–9130. doi: 10.1021/jf400424a. [DOI] [PubMed] [Google Scholar]
- León K., Mery D., Pedreschi F., León J. Color measurement in L∗a∗b∗ units from RGB digital images. Food Research International. 2006;39:1084–1091. doi: 10.1016/j.foodres.2006.03.006. [DOI] [Google Scholar]
- Li J., Yang P., Yang Q., Gong X., Ma H., Dang K., et al. Analysis of flavonoid metabolites in buckwheat leaves using UPLC-ESI-MS/MS. Molecules. 2019;24:1310. doi: 10.3390/molecules24071310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Ma M., Su W. Fingerprint analysis of Hibiscus mutabilis L. leaves based on ultra performance liquid chromatography with photodiode array detector combined with similarity analysis and hierarchical clustering analysis methods. Pharmacognosy Magazine. 2013;9:238–243. doi: 10.4103/0973-1296.113277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourencetti N.M.S., Wolf I.R., Lacerda M.P.F., Valente G.T., Zanelli C.F., Santoni M.M., et al. Transcriptional profile of a bioethanol production contaminant Candida tropicalis. AMB Express. 2018;8:166. doi: 10.1186/s13568-018-0693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari A., Montoro P., D'Urso G., Macchia M., Pizza C., Piacente S. Metabolic profiling of Vitex agnus castus leaves, fruits and sprouts: Analysis by LC/ESI/(QqQ)MS and (HR) LC/ESI/(Orbitrap)/MSn. Journal of Pharmaceutical and Biomedical Analysis. 2015;102:215–221. doi: 10.1016/j.jpba.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Nobuhiro S., Yuki M., Yutaka A., Masachika O., Masaki M., Naoyuki U., et al. Recent advances in understanding the anthocyanin modification steps in carnation flowers. Scientia Horticulturae. 2013;163:37–45. doi: 10.1016/j.scienta.2013.07.029. [DOI] [Google Scholar]
- Perrin F., Brahem M., Dubois-Laurent C., Huet S., Jourdan M., Geoffriau E., et al. Differential pigment accumulation in carrot leaves and roots during two growing stages. Journal of Agricultural and Food Chemistry. 2016;64:906–912. doi: 10.1021/acs.jafc.5b05308. [DOI] [PubMed] [Google Scholar]
- Polturak G., Heinig U., Grossman N., Battat M., Leshkowitz D., Malitsky S., et al. Transcriptome and metabolic profiling provides insights into betalain biosynthesis and evolution in Mirabilis jalapa. Molecular Plant. 2018;11:189–204. doi: 10.1016/j.molp.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Rosati S., Agostini V., Knaflitz M., Balestra G. Muscle activation patterns during gait: A hierarchical clustering analysis. Biomedical Signal Processing and Control. 2017;31:463–469. doi: 10.1016/j.bspc.2016.09.017. [DOI] [Google Scholar]
- Sarker U., Oba S. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Scientific Reports. 2018;8:12349. doi: 10.1038/s41598-018-30897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz K., Persike M., Carle R., Schieber A. Characterization and quantification of anthocyanins in selected artichoke (Cynara scolymus L.) cultivars by HPLC–DAD–ESI–MS n. Analytical and Bioanalytical Chemistry. 2006;384:1511–1517. doi: 10.1007/s00216-006-0316-6. [DOI] [PubMed] [Google Scholar]
- Segnini S., Dejmek P., Öste R. A low cost video technique for colour measurement of potato chips. LWT – Food Science and Technology. 1999;32:216–222. doi: 10.1006/fstl.1999.0536. [DOI] [Google Scholar]
- Shi Q., Zhang Z., Su J., Zhou J., Li X. Comparative analysis of pigments, phenolics, and antioxidant activity of Chinese jujube (Ziziphus jujuba Mill.) during fruit development. Molecules. 2018;23:1917. doi: 10.3390/molecules23081917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzing F.C., Carle R. Functional properties of anthocyanins and betalains in plants, food, and in human nutrition. Trends in Food Science and Technology. 2004;15:19–38. doi: 10.1016/j.tifs.2003.07.004. [DOI] [Google Scholar]
- Thévenot E.A., Roux A., Xu Y., Ezan E., Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. Journal of Proteome Research. 2015;14:3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- Trygg J., Wold S. Orthogonal projections to latent structures (O-PLS) Journal of Chemometrics. 2002;16:119–128. doi: 10.1002/cem.695. [DOI] [Google Scholar]
- Wang B.N., Liu H.F., Zheng J.B., Fan M.T., Cao W. Distribution of phenolic acids in different tissues of jujube and their antioxidant activity. Journal of Agricultural and Food Chemistry. 2011;59:1288–1292. doi: 10.1021/jf103982q. [DOI] [PubMed] [Google Scholar]
- Worley B., Powers R. PCA as a practical indicator of OPLS-DA model reliability. Current Metabolomics. 2016;4:97–103. doi: 10.2174/2213235X04666160613122429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q., Fan H., Yao F., Cao X., Liu M., Sun J., et al. Transcriptomics and targeted metabolomics profilings for elucidation of pigmentation in Lonicera japonica flowers at different developmental stages. Industrial Crops and Products. 2020;145 doi: 10.1016/j.indcrop.2019.111981. [DOI] [Google Scholar]
- Xue Z., Feng W., Cao J., Cao D., Jiang W. Antioxidant activity and total phenolic contents in peel and pulp of Chinese jujube (Ziziphus jujubamill) fruits. Journal of Food Biochemistry. 2009;33:613–629. doi: 10.1111/j.1745-4514.2009.00241.x. [DOI] [Google Scholar]
- Yi X.K., Wu W.F., Zhang Y.Q., Li J.X., Luo H.P. Thin-layer drying characteristics and modeling of Chinese jujubes. Mathematical Problems in Engineering. 2012;2012:1–18. doi: 10.1155/2012/386214. [DOI] [Google Scholar]
- Zhang G., Chen S., Zhou W., Meng J., Deng K., Zhou H., et al. Rapid qualitative and quantitative analyses of eighteen phenolic compounds from Lycium ruthenicum Murray by UPLC-Q-Orbitrap MS and their antioxidant activity. Food Chemistry. 2018;269:150–156. doi: 10.1016/j.foodchem.2018.06.132. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Wang L., Liu Z., Zhao Z., Zhao J., Wang Z., et al. Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chemistry. 2020;312 doi: 10.1016/j.foodchem.2019.125903. [DOI] [PubMed] [Google Scholar]
- Zhang S.L., Deng P., Xu Y.C., Lü S.W., Wang J.J. Quantification and analysis of anthocyanin and flavonoids compositions, and antioxidant activities in onions with three different colors. Journal of Integrative Agriculture. 2016;15:2175–2181. doi: 10.1016/s2095-3119(16)61385-0. [DOI] [Google Scholar]
- Zhao Z., Li J., Wu X., Dai H., Gao X., Liu M., et al. Structures and immunological activities of two pectic polysaccharides from the fruits of Ziziphus jujuba Mill. cv. jinsixiaozao Hort. Food Research International. 2006;39:917–923. doi: 10.1016/j.foodres.2006.05.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the pigmentation identified and quantified in the jujube peels.