Abstract
Background and Aims
Identify novel metabolite associations with blood pressure (BP) salt-sensitivity and hypertension.
Methods and Results
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) Replication study includes 698 Chinese participants who underwent a 3-day baseline examination followed by a 7-day low-sodium feeding and 7-day high-sodium feeding. Latent mixture models identified three trajectories of blood pressure (BP) responses to the sodium interventions. We selected 50 most highly salt-sensitive and 50 most salt-resistant participants for untargeted metabolomics profiling. Multivariable adjusted mixed logistic regression models tested the associations of baseline metabolites with BP salt-sensitivity. Multivariable adjusted mixed linear regression models tested the associations of BP salt-sensitivity with metabolite changes during the sodium interventions. Identified metabolites were tested for associations with hypertension among 1,249 Bogalusa Heart Study (BHS) participants using multiple logistic regression. Fifteen salt-sensitivity metabolites were associated with hypertension in the BHS. Baseline values of serine, 2-methylbutyrylcarnitine and isoleucine directly associated with high salt-sensitivity. Among them, serine indirectly associated with hypertension while 2-methylbutyrylcarnitine and isoleucine directly associated with hypertension. Baseline salt-sensitivity status predicted changes in 14 metabolites when switching to low-sodium or high-sodium interventions. Among them, glutamate, 1-carboxyethylvaline, 2-methylbutyrylcarnitine, 3-methoxytyramine sulfate, glucose, alpha-ketoglutarate, hexanoylcarnitine, gamma-glutamylisoleucine, gamma-glutamylleucine, and gamma-glutamylphenylalanine directly associated with hypertension. Conversely, serine, histidine, threonate and 5-methyluridine indirectly associated with hypertension. Together, these metabolites explained an additional 7% of hypertension susceptibility when added to a model including traditional risk factors.
Conclusions
Our findings contribute to the molecular characterization of BP response to sodium and provide novel biological insights into salt-sensitive hypertension.
Keywords: Untargeted metabolomics, blood pressure salt-sensitivity, hypertension
INTRODUCTION
The number of individuals with hypertension has almost doubled in the past three decades, increasing to over 1.28 billion individuals with high blood pressure (BP) in 2021[1]. As a leading contributor to morbidity and mortality globally, hypertension accounts for 8.7% of total disability-adjusted life-years and 14.0% of total deaths worldwide[2,3]. Randomized controlled trials have established a direct, causal relationship between dietary sodium intake and BP[4–6]. However, BP responses to sodium intake vary considerably between individuals, a phenomenon known as BP salt-sensitivity[4,7,8]. Those with high salt-sensitivity of BP have increased risks of hypertension and related cardiovascular disease[9–12], suggesting that lifestyle interventions targeting this subgroup may be particularly relevant in preventing these conditions. Unfortunately, accurate measurement of BP salt-sensitivity remains costly and time-intensive, requiring a well-controlled feeding study or an acute in-patient protocol[13]. In contrast, molecular technologies are evolving rapidly, reducing costs and allowing for quick and comprehensive biomarker profiling. These scientific advancements provide opportunity to explore molecular phenotyping as an alternative strategy for characterizing sodium-sensitivity of BP in individuals.
The human metabolome is the end-product of complex interactions between genomic, transcriptomic, proteomic, and environmental exposures[14–16]. Given the proximity of metabolome to phenome, metabolomics research has become increasingly utilized to identify novel biomarkers for complex phenotypes. While several reports have identified novel metabolite associations with BP[17–19], only one metabolomics study of salt-sensitivity of BP was conducted previously among a subsample of 103 Dietary Approaches to Stop Hypertension-Sodium (DASH-sodium) participants[20]. This targeted analysis of 57 amino and tricarboxylic acid cycle metabolites identified several metabolites that, together, improved the classification of BP salt-sensitivity beyond race, age, and sex. Although this work was promising, discovery (untargeted) metabolomics approaches that comprehensively identify metabolites across known and unknown biological pathways are still needed. This type of research could identify a broader array of molecular biomarkers for salt-sensitivity of BP, contributing to risk stratification efforts and our general understanding of the biological pathways underlying this complex phenotype.
The current report describes findings from the first untargeted metabolomics study of BP salt-sensitivity, which leveraged stringently collected blood pressure and urinary metabolomics data from 100 Han Chinese participants who underwent a 7-day low-sodium and 7-day high-sodium feeding protocol. Because salt-sensitivity is a recognized hypertension endophenotype[9,10], identified metabolites were further tested for associations with this complex condition in a large, bi-racial sample of 1,249 participants from the Bogalusa Heart study (BHS) who underwent untargeted metabolomics profiling and stringent clinical examination during the 2013 to 2016 study visit.
METHOD
The GenSalt Studies
The GenSalt studies, including the original GenSalt (N=1,906) and subsequent GenSalt Replication (N=698) studies, were separate family-based feeding studies conducted in two independent samples from rural areas of northern China using identical research protocols. The original GenSalt study was conducted from October 2003 to July 2005 while the GenSalt Replication study was conducted from April 2010 to November 2010. Community-based BP screenings were conducted to identify potential probands aged 18–60 years and their sibling, spouses and offspring aged 16 or older. Probands were defined as those with mean systolic BP between 130–160 mmHg, and/or mean diastolic BP between 85–100 mmHg and no use of antihypertension medications. Individuals who had stage 2 hypertension, secondary hypertension, and a history of clinical cardiovascular disease or diabetes, or were pregnant, heavy alcohol drinkers, taking anti-hypertension medications, or currently on a low-sodium diet were excluded from the feeding study. For the current study, we selected the 50 most salt-sensitive and 50 most salt-resistant participants of the GenSalt Replication study to undergo untargeted metabolomics profiling. Selected participants included those with the largest and smallest BP responses to the dietary sodium interventions. Institutional review boards at all participating institutions approved the GenSalt studies. Written informed consents were obtained from all GenSalt participants at baseline prior to the interventions. The consent form was general and included consent for the use of biospecimens for future molecular research.
GenSalt Baseline Data Collection
Standard questionnaires were administered to GenSalt participants by trained staff at baseline to collect information on demographic characteristics (including age and gender), lifestyle risk factors (including physical activity and alcohol drinking), and personal medical history. Physical examinations were conducted during the baseline examination to collect anthropometric and BP measures. Body weight and height were measured twice at baseline with participants in light clothing without shoes. Body mass index (BMI) was calculated as weight in kilograms divided by height in square meters and the mean of the two measures was used for analyses. Obesity was defined as BMI ≥30 kg/m2. Blood pressure (BP) was measured in the morning in triplicate following a standard protocol during each day of the 3-day baseline examination by trained and certified staff using a Hawksley random-zero sphygmomanometer. BP was measured with the participant in a sitting position following 5 minutes of rest. Participants were advised to avoid alcohol drinking, cigarette smoking, having coffee or tea, and exercise for at least 30 minutes prior to their measurements. Hypertension was defined according to the 2017 American College of Cardiology and American Heart Association (ACC/AHA) guidelines as systolic BP greater than or equal to 130 mmHg, diastolic BP greater than or equal to 80 mmHg, or use of antihypertension medication [21], with systolic and diastolic BP estimated as the mean of the nine baseline BP measures for each study participant.
GenSalt Dietary Intervention
After the 3-day baseline observation, the study participants received a dietary intervention including a 7-day low-sodium diet (3 g of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-sodium diet (18 g of sodium chloride or 307.8 mmol of sodium per day). BP was measured in the morning in triplicate following a standard protocol on days 2, 5, 6 and 7 of each intervention period using a protocol identical to that of the baseline examination. All BP observers were blinded to the dietary intervention phase. During the dietary sodium interventions, dietary potassium intake remained unchanged. Total energy intake was varied according to each participant’s baseline energy intake. All study foods were cooked without salt, and prepackaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure study participants’ compliance to the intervention program, they were required to have their breakfast, lunch, and dinner at the study kitchen under supervision of the study staff during the entire study period. The study participants were instructed to avoid consuming any foods that were not provided by study personnel. One 24-hour and two 8-hour overnight urinary specimens were collected at the baseline examination and on the last 3 days of each intervention period to measure sodium and potassium. Overnight urinary sodium excretion was converted to 24-hour values based on formulas developed from data obtained in a subgroup of study participants[22]. The results from the 24-hour urinary excretions of sodium showed excellent compliance with the study diet[23].
Selection of Highly Salt-Sensitive and Salt-Resistant GenSalt Participants
Latent variable mixture modeling was used to identify subgroups sharing similar underlying trajectories of BP responses to low-sodium and high-sodium interventions, as implemented by the SAS Proc Traj procedure[24]. Systolic BP responses from baseline to low-sodium intervention and responses from low-sodium to high-sodium intervention on days 2, 5, 6, and 7 of each intervention period were used for modeling. We fit models up to the optimal number of trajectories by comparing the Bayesian Information Criterion for each set of trajectories. The model with 3 trajectories fit best, and participants were classified into the highly salt-sensitive, moderately salt-sensitive, or salt-resistant trajectory groups with excellent discrimination[25]. The 50 participants with the highest probability of high salt-sensitivity and the 50 GenSalt Replication study participants with the highest probability of salt-resistance were selected for metabolomics profiling.
GenSalt Metabolomics Profiling
Untargeted, ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) was conducted by Metabolon© using GenSalt 24-hour urine samples collected at three time-points, including the first day of baseline examination and the fifth days of the low-sodium and high-sodium interventions. All urine samples were stored at −80°C since the time of collection. Untargeted metabolomics profiling resulted in the detection and semi-quantification of 1,445 metabolites. These included 972 known biochemical compounds in pathways related to amino acids (n=267), carbohydrates (n=32), cofactors and vitamins (n=39), energy (n=15), lipids (n=170), nucleotides (n=59), peptides (n=40), xenobiotics (n=293), and partially characterized molecules (n=57). An additional 473 unnamed compounds currently lacking chemical standards were also detected. These metabolites were labeled with an “X” followed by numbers (e.g., X-12345) and may be identified upon the eventual acquisition of a matching purified standard (or via classical structural analysis).
Prior to the statistical analysis, additional quality control and manipulation of the metabolite data was undertaken. First, urine metabolomics underwent osmolality-based normalization, which is comparable to creatinine-based normalization23,24 and is used to reduce variability in metabolite values due to differences in total urinary solute concentration25. Next, data filtering removed 73 metabolites that were missing or below the detection threshold in more than 80% of samples. Among the 1,372 metabolites passing quality control, 76 were missing or below the detection threshold in 50% to 80% of the samples. These metabolites were analyzed as ordinal variables after categorization into one of three mutually exclusive groups: 1) missing or below-the-detection-limit; 2) below the median of detectable values; or 3) greater than or equal to the median of detectable values. The remaining 1,296 metabolites were analyzed as continuous variables, where half of the minimum observed value for a particular metabolite was imputed among those with missing or below-the-detection-limit values.
The BHS
The BHS was designed to study the natural history of hypertension and cardiovascular disease (CVD) among a biracial sample (35% African American and 65% white) of residents from Bogalusa, Louisiana. The current BHS core cohort includes 1,298 participants born between 1959 and 1979 who were examined at least twice during childhood and twice during adulthood for CVD risk factors. A total of 1,249 participants underwent untargeted metabolomics profiling and provided measures of BP and other important covariables in the 2013 to 2016 visit cycle.
Data collection methods were described previously in detail[29]. In brief, a standard questionnaire was used to collect information on demographic characteristics, lifestyle, and medical history. Height and weight were measured in duplicate, and BP was measured in triplicate using stringent study protocols. Untargeted metabolomics profiling of serum samples from BHS participants was carried out by Metabolon[19]. Metabolomics data quality control was conducted using procedures identical to those of the GenSalt study. Hypertension was again defined according to the 2017 ACC/AHA guidelines[21]. Written informed consent was obtained when the participants enrolled in the 2013–2016 visit cycle. The consent form included consent for the use of biospecimen for future molecular research. The study was approved by the Tulane University Biomedical Institutional Review.
Statistical Analysis
Characteristics of study participants were presented as means and standard deviations (SDs) for continuous variables and as frequencies (percentages) for categorical variables.
Prior to association analyses, the R package, bestnormalize, was used to normalize continuous metabolite distributions. All metabolites were then standardized to mean=0 and SD=1, so that the scales across metabolites were comparable. Mixed logistic regression models were used to assess whether baseline metabolites predicted salt-sensitivity after adjustment for baseline age, gender, physical activity, alcohol drinking, obesity, and hypertension. To identify metabolites that were altered differently by sodium intervention according to salt-sensitivity status, mixed linear regressions models were employed. Here, we examined the associations of salt-sensitivity status with metabolite changes in response to low-sodium intervention and also in response to high-sodium intervention, again adjusting for baseline age, gender, physical activity, alcohol drinking, obesity, and hypertension. The same mixed linear regression models were also used to estimate adjusted mean changes in metabolite values in response to low-sodium and high-sodium interventions according to salt-sensitivity status. Metabolites that achieved nominal significance (P<0.05) among GenSalt participants were matched to Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolomics pathway database using the Metaboanalyst 5.0[30], a web-based tool, to conduct pathway enrichment and pathway topological analyses.
Nominally significant metabolites were also each tested for association with hypertension in the BHS using multiple logistic regression models that adjusted for age, gender, race, physical activity, alcohol drinking, and obesity. Bonferroni-corrected significance thresholds were employed at this stage of the analysis (0.05/the number of metabolites moved forward from each GenSalt analysis). Metabolites achieving Bonferroni corrected significance in the BHS were categorized into quartiles and their dose response relationships with hypertension were further explored, again employing multivariable logistic regression models with adjustment for age, gender, race, physical activity, alcohol drinking, and obesity. We further assessed the added predictive value and discrimination of the identified salt-sensitivity metabolites together on hypertension status among BHS participants. To minimize the influence of the correlations between the identified metabolites, principal component analysis was done among the identified metabolites and principal components (PCs) with eigenvalue greater than one were selected. We assessed both the incremental R2 (on the liability scale) and change in c-statistics when adding the selected PCs to a model adjusting for traditional risk factors.
Sensitivity analyses to investigate the influence of kidney function on our findings were done by further adjusting for estimated glomerular filtration rate (eGFR) in analyses of BHS participants. Furthermore, to investigate whether imputation of missing and below-the-detection-limit values influenced our findings, sensitivity analyses were carried out excluding participants with missing or below-the-detection-limit values.
RESULTS
The participants’ flowchart was presented as Supplementary Figure 1 and baseline characteristics were shown in Table 1. Highly salt-sensitive GenSalt participants were, on average, older and more likely to be current drinkers and have hypertension compared to their salt-resistant counterparts. BHS participants were predominantly female, and more than half were white, current drinkers, obese, and hypertensive.
Table 1.
Baseline characteristics of GenSalt and BHS participants.
| GenSalt Study | BHS (N=1,249) | ||
|---|---|---|---|
|
| |||
| Salt-Sensitive (N=50) | Salt-Resistant (N=50) | ||
|
| |||
| Age, years, mean (SD) | 51.8 (6.45) | 42.2 (10.6) | 48.2 (5.28) |
| Male, n (%) | 23 (46%) | 26 (52%) | 515 (41%) |
| Race, n (%) | |||
| Asian | 50 (100%) | 50 (100%) | 0 (0%) |
| White | 0 (0%) | 0 (0 %) | 822 (66%) |
| Black | 0 (0%) | 0 (0%) | 427 (34%) |
| Current Drinking, n (%) | 15 (30%) | 6 (12%) | 696 (56%) |
| Physical Activity, MET-hours per day, mean (SD) | 20.6 (12.0) | 18.4 (11.7) | 12.0 (10.9) |
| Obesity, n (%) | 8 (16%) | 9 (18%) | 644 (52%) |
| Hypertension, n (%) | 45 (90%) | 11 (22%) | 775 (62%) |
| Anti-hypertension medication, n (%) | 0 (0%) | 0 (0%) | 433 (35%) |
BHS: Bogalusa Heart Study; SD: standard deviation; MET: metabolic equivalent
Analyses of baseline metabolite values
Although no metabolites achieved Bonferroni corrected significance (P<3.64×10−5), a total of 161 baseline metabolites were nominally associated with BP salt-sensitivity status among GenSalt participants (P<0.05). Twenty-one were inversely associated with high salt-sensitivity (associated with salt-resistance) and 140 were positively associated with high salt-sensitivity (Supplementary Figure 2A and Supplementary Table 1).
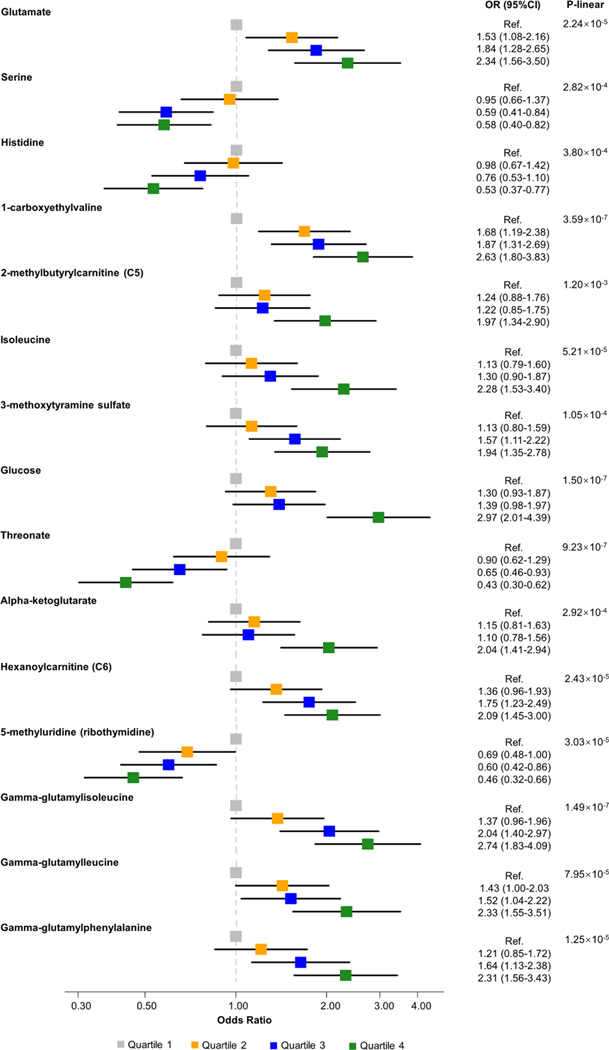
Among the 161 baseline metabolites that predicted salt-sensitivity of BP in GenSalt, 71 were detected and 4 were further associated with hypertension in BHS participants, including 3 named metabolites and 1 unnamed metabolite (Table 2 and Supplementary Table 1). All of the named metabolites came from amino acid sub-pathways, including serine from the glycine, serine and threonine metabolism sub-pathway; and 2-methylbutyrylcarnitine and isoleucine from the leucine, isoleucine and valine metabolism sub-pathway. All three named metabolites conferred increased odds of high salt-sensitivity among GenSalt participants. They also conferred increased odds of hypertension in the BHS except serine, which had a negative association with hypertension (Table 2). The three known metabolites demonstrated significant graded dose response associations with hypertension when categorized into quartiles (Figure 1).
Table 2.
Associations of baseline metabolites, metabolite change in response to low-sodium and high-sodium interventions with high salt-sensitivity among GenSalt participants and corresponding metabolite associations with hypertension among BHS participants.
| GenSalt Baseline | GenSalt Low-Sodium Intervention | GenSalt High-Sodium Intervention | BHS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Metabolite | OR*‡ | 95% CI | P value | Beta*§ | SE | P value | Beta*|| | SE | P value | OR† | 95% CI | P value |
|
| ||||||||||||
| Glutamate | 0.52 | (0.24, 1.13) | 9.78E-02 | −0.48 | 0.21 | 2.34E-02 | 0.44 | 0.33 | 1.85E-01 | 1.42 | (1.22, 1.65) | 3.90E-06 |
| Serine | 2.13 | (1.20, 3.78) | 1.00E-02 | −1.06 | 0.27 | 8.84E-05 | 0.72 | 0.29 | 1.17E-02 | 0.76 | (0.67, 0.86) | 2.49E-05 |
| Histidine | 0.68 | (0.30, 1.54) | 3.60E-01 | −0.50 | 0.28 | 7.29E-02 | 0.64 | 0.23 | 4.70E-03 | 0.74 | (0.64, 0.85) | 1.88E-05 |
| 1 -carboxyethylvaline | 0.97 | (0.47, 2.00) | 9.38E-01 | 0.11 | 0.32 | 7.25E-01 | −0.61 | 0.30 | 4.60E-02 | 1.44 | (1.25, 1.66) | 3.83E-07 |
| 2-methylbutyrylcarnitine (C5) | 2.02 | (1.14, 3.60) | 1.65E-02 | −0.25 | 0.25 | 3.12E-01 | 0.58 | 0.30 | 4.86E-02 | 1.32 | (1.14, 1.51) | 1.25E-04 |
| Isoleucine | 2.45 | (1.07, 5.60) | 3.36E-02 | −0.58 | 0.30 | 5.43E-02 | 0.29 | 0.35 | 4.08E-01 | 1.30 | (1.13, 1.50) | 2.76E-04 |
| 3-methoxytyramine sulfate | 1.88 | (0.87, 4.06) | 1.10E-01 | −0.28 | 0.27 | 3.03E-01 | −0.65 | 0.31 | 3.39E-02 | 1.31 | (1.15, 1.50) | 7.16E-05 |
| Glucose | 2.01 | (0.89, 4.49) | 9.10E-02 | −0.11 | 0.31 | 7.12E-01 | 0.77 | 0.29 | 8.61E-03 | 1.49 | (1.30, 1.71) | 9.63E-09 |
| Threonate | 1.29 | (0.61, 2.74) | 5.03E-01 | −0.71 | 0.27 | 8.86E-03 | 0.39 | 0.34 | 2.46E-01 | 0.73 | (0.65, 0.83) | 2.28E-06 |
| Alpha-ketoglutarate | 1.37 | (0.55, 3.42) | 5.00E-01 | 0.47 | 0.27 | 7.38E-02 | −0.61 | 0.27 | 2.29E-02 | 1.33 | (1.17, 1.51) | 1.94E-05 |
| Hexanoylcarnitine (C6) | 0.89 | (0.38, 2.05) | 7.76E-01 | −0.39 | 0.28 | 1.52E-01 | 0.84 | 0.26 | 1.48E-03 | 1.31 | (1.15, 1.49) | 6.18E-05 |
| 5-methyluridine (ribothymidine) | 2.28 | (0.82, 6.34) 1.14E | 1.14E-01 | −0.87 | 0.28 | 1.64E-03 | 0.43 | 0.38 | 2.61E-01 | 0.71 | (0.62, 0.81) | 7.06E-07 |
| Gamma-glutamylisoleucine¶ | 1.14 | (0.55, 2.38) | 7.22E-01 | −0.68 | 0.29 | 1.82E-02 | 0.46 | 0.25 | 6.96E-02 | 1.50 | (1.29, 1.74) | 5.99E-08 |
| Gamma-glutamylleucine | 0.95 | (0.37, 2.41) | 9.11E-01 | −0.61 | 0.29 | 3.43E-02 | 0.62 | 0.26 | 1.60E-02 | 1.39 | (1.20, 1.62) | 1.55E-05 |
| Gamma-glutamylphenylalanine | 0.96 | (0.43, 2.13) | 9.16E-01 | −0.63 | 0.30 | 3.41E-02 | 0.74 | 0.30 | 1.45E-02 | 1.42 | (1.23, 1.64) | 1.91E-06 |
Adjusted for age, gender, physical activity, drinking, obesity and hypertension.
Adjusted for age, gender, race, physical activity, drinking, and obesity.
Beta estimate corresponds to each standard deviation larger metabolite value at baseline
Beta estimate corresponds to each standard deviation larger metabolite change during low-sodium intervention.
Beta estimate corresponds to each standard deviation larger metabolite change during high-sodium intervention.
Indicates compounds that have not been officially confirmed based on a standard.
Figure 1.
Plots depicting associations of hypertension among BHS participants with identified metabolites
Analyses of metabolite responses to sodium intervention
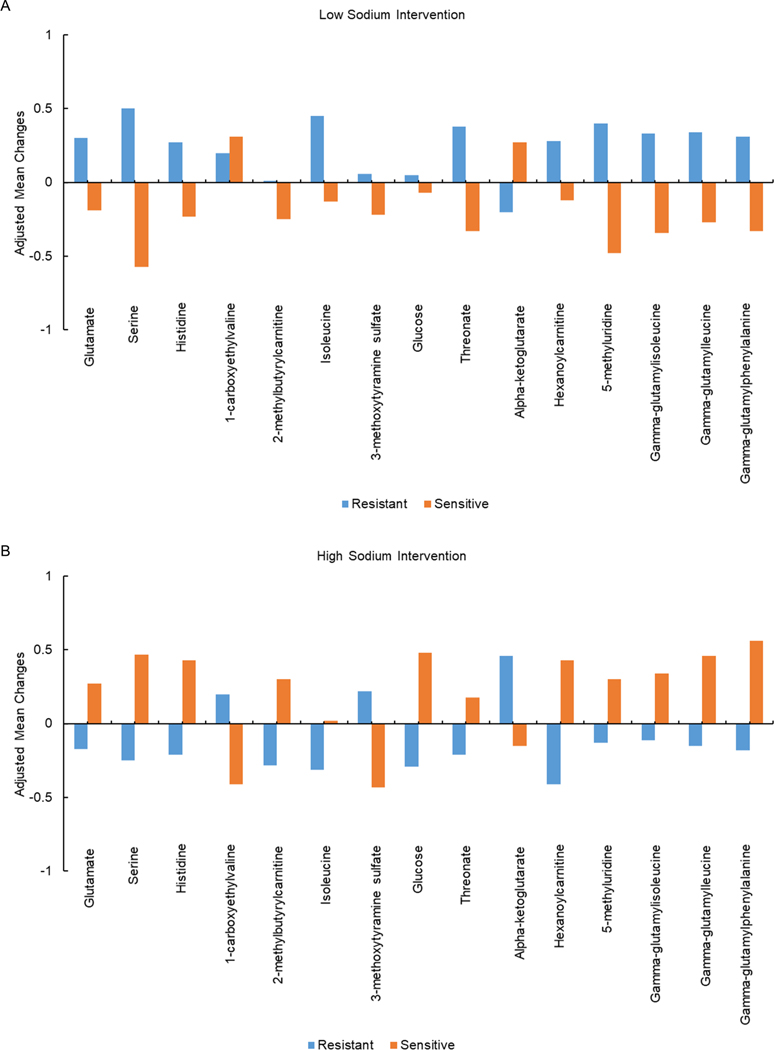
Among GenSalt participants, salt-sensitivity status nominally associated with changes in the values of 97 metabolites when switching from usual diet to the low-sodium intervention (Supplementary Figure 2B and Supplementary Table 2), as well as changes in the values of 153 metabolites when switching from low-sodium to high-sodium intervention (Supplementary Figure 2C and Supplementary Table 3). None achieved Bonferroni corrected significance. Among the 215 metabolites differentially altered by low-sodium and/or high-sodium interventions according to salt-sensitivity status, 112 metabolites were detected in the BHS. Of those, 16 metabolites, including 14 named metabolites, also associated with hypertension in the BHS. Adjusted mean changes of these 14 metabolites along with isoleucine during low-sodium and high-sodium intervention according to salt-sensitivity status are presented in Figure 2 and include: glutamate, serine, histidine, 1-carboxyethylvaline, 2-methylbutyrylcarnitine, isoleucine and 3-methoxytyramine sulfate from amino acid sub-pathways (glutamate metabolism; glycine, serine and threonine metabolism; histidine metabolism; leucine, isoleucine and valine metabolism; and tyrosine metabolism); glucose from a carbohydrate sub-pathway (glycolysis, gluconeogenesis, and pyruvate metabolism); threonate from a cofactors and vitamins sub-pathway (ascorbate and aldarate metabolism); alpha-ketoglutarate from an energy sub-pathway (TCA cycle); hexanoylcarnitine from a lipid sub-pathway (fatty acid metabolism); 5-methyluridine from a nucleotide sub-pathway (pyrimidine metabolism); along with gamma-glutamylisoleucine, gamma-glutamylleucine, and gamma-glutamylphenylalanine from a peptide sub-pathway (gamma-glutamyl amino acid).
Figure 2.
Adjusted mean changes in metabolite value during a.) Low-sodium intervention; and b.) High-sodium intervention.
Compared to salt-resistance of BP, high salt-sensitivity of BP associated with relative decreases in 1-carboxyethylvaline and alpha-ketoglutarate during high-sodium intervention. For the remaining 12 metabolites, high salt-sensitivity of BP associated with relative increases metabolite values in response to high-sodium, or conversely, relative decreases in metabolite values in response to low-sodium. Each standard deviation or quartile increase in glutamate, 1-carboxyethylvaline, 2-methylbutyrylcarnitine, 3-methoxytyramine sulfate, glucose, alpha-ketoglutarate, hexanoylcarnitine, gamma-glutamylisoleucine, gamma-glutamylleucine, and gamma-glutamylphenylalanine metabolites associated with higher odds of hypertension in the BHS. In contrast, each standard deviation or quartile increase in serine, histidine, threonate, and 5-methyluridine associated with lower odds of hypertension in the BHS (Table 2 and Figure 2).
Results of sensitivity analyses further adjusting for eGFR were consistent with those of the main analysis (Supplementary Table 4). Among four identified metabolites with missing values imputed to half of the detection limit, sensitivity analyses excluding those participants with imputed values were also similar to those of the main analysis (Supplementary Table 5).
Pathway analysis
Supplementary Table 6 shows the forty KEGG biochemical pathways that were identified among salt-sensitive metabolites. Two biochemical pathways, aminoacyl-tRNA biosynthesis (P=1.24×10−5, FDR=1.05×10−3, pathway impact=0.17) and arginine biosynthesis (P=9.08×10−4, FDR=3.81×10−2, pathway impact=0.19), demonstrated significant enrichment for identified metabolites (Supplementary Table 6 and Supplementary Figure 3).
Added value of salt-sensitivity metabolites in the prediction of hypertension
Supplementary Figure 4 depicts the pairwise Pearson correlations of the 15 salt-sensitivity metabolites that associated with hypertension in the BHS. Aside from the strong correlations of gamma-glutamylisoleucine and gamma-glutamylphenylalanine (r=0.81), the pairwise correlations of metabolites were generally modest to moderate (r<0.7), with 5 PCs explaining 66.8% of the variance in the 15 metabolites (Supplementary Figure 5). Addition of these 5 PCs to the age, gender, race, physical activity, drinking, and obesity adjusted model captured an additional 7.1% of the variance in hypertension risk (R2 on the liability scale) and improved the C-statistic from 0.73 to 0.78 (P<0.0001; Table 3 and Supplementary Figure 6).
Table 3.
Comparison of R2 and C-statistics before and after addition of identified salt-sensitivity metabolites.
| R2 | C-statistics | |
|---|---|---|
|
| ||
| Model 1* | 0.1427 | 0.7297 |
| Model 1* + 5 PCAs | 0.2140 | 0.7782 |
| Difference | 0.0713 | 0.0485 |
| P value | <.0001 | <.0001 |
Model 1 was adjusted for age, gender, race, physical activity, drinking, and obesity.
DISCUSSION
In the first untargeted metabolomics study of salt-sensitivity of BP, we identified 161 baseline metabolites that were associated with BP salt-sensitivity. In addition, salt-sensitivity status was associated with changes in the values of 215 metabolites under low and/or high sodium conditions. Among the 336 identified salt-sensitivity metabolites, 15 were robustly associated with hypertension in a large, biracial sample of BHS participants and included 7 metabolites, nearly half, from amino acid pathways, along with 3 peptide metabolites and 1 metabolite from each of the carbohydrate, cofactor and vitamin, energy, lipid, and nucleotide pathways. An important role for amino acids in BP salt-sensitivity was further supported by pathway analyses which demonstrated significant enrichment of identified metabolites with two amino acid-related KEGG pathways, aminoacyl-tRNA and arginine biosynthesis. Together, the 15 salt-sensitivity metabolites provided additional predictive value for hypertension, significantly improving discrimination and explaining a further 7.1% of hypertension risk beyond traditional risk factors. In aggregate, our findings implicate new molecular mechanisms underlying BP response to dietary sodium intake and identify novel biomarkers of individual sodium-sensitivity and hypertension status.
Among the seven identified salt-sensitivity metabolites from the amino acid pathway, serine and histidine showed inverse associations with hypertension. Baseline serine values positively associated with high salt-sensitivity, while salt-sensitivity status predicted changes in both metabolites during sodium interventions. Given previous reports of high salt-sensitivity as an important risk factor for hypertension[9,10], the positive baseline association of serine with high salt-sensitivity and inverse relation with hypertension status may seem counterintuitive. However, there is increasing evidence that both high salt-sensitivity and salt-resistance of BP increase hypertension risk compared to moderate salt-sensitivity status[25,31] Since we compared highly salt-sensitive to salt-resistant participants, these findings may reflect beneficial effects of increased serine on salt-resistance of BP. Our findings of protective associations of both serine and histidine with hypertension are consistent with previous reports. For example, serine plays an important role in the production of nitric oxide and antioxidants[18,19,32], which have been shown to have BP lowering effects. In addition, histidine has important anti-oxidant and anti-inflammatory properties[33]. Higher values of this metabolite have been inversely related to blood pressure[34] and associated with lower incidence of coronary heart disease[33]. Although mechanistic studies are needed, we provide early evidence that serine and histidine may lower hypertension risk through sodium-related pathways.
Among the remaining five salt-sensitivity metabolites from amino acid pathways, glutamate, 1-carboxyethylvaline, 2-methylbutyrylcarnitine, and 3-methoxytramine sulfate were differentially altered by sodium intervention according to salt-sensitivity status. Furthermore, higher baseline values of 2-methylbutyrylcarnitine and isoleucine were associated with higher odds of salt-sensitivity. All five metabolites were positively associated with hypertension in the current study. Glutamate is an excitatory neurotransmitter and its metabolism is regulated by the glutathione cycle[35]. Excess oxidative stress induced by glutamate is a plausible biological pathway underlying the observed association[36,37]. Although 3-methoxytyramine sulfate is poorly characterized, its parent metabolite, 3-methoxytyramine, is a dopamine metabolite. Dopamine metabolism can cause dysregulated water and salt reabsorption, which is thought to be a key feature of salt-sensitive hypertension[38,39]. Isoleucine, 1-carboxyethylvaline, and 2-methylbutyrylcarnitine are all involved in branched chain amino acid (BCAA) metabolism[40,41]. Although this is the first human study to implicate BCAA metabolites in human salt-sensitivity of BP, isoleucine were previously shown to be elevated in Dahl salt-sensitive rats[42], and 2-methylbutyrylcarnitine was previously shown to change significantly during sodium restriction in humans[43]. Furthermore, the 1-carboxyethylvaline metabolite was previously associated with decreased Shannon diversity index, a marker of gut microbial health that has previously been linked to numerous cardiometabolic conditions[44]. These promising findings provide novel biological insights into high salt-sensitivity in humans and are among the first to identify associations of BCAA metabolites underlying this condition. In total, our findings provide compelling evidence that these metabolites could be part of the biological pathway linking dietary sodium intake to BP.
Three correlated peptide metabolites (all r>0.60), gamma-glutamylisoleucine, gamma-glutamylleucine, and gamma-glutamylphenylalanine, were also differentially altered in response to sodium intervention according to salt-sensitivity status. Each of these metabolites displayed a direct association with hypertension. In general, gamma glutamyl amino acids are mainly biosynthesized during glutathione degradation[45] and regulated by gamma-glutamyl-transferase, an enzyme previously linked to hypertension and purported to exert its influence through oxidative stress and inflammation pathways[46]. Previously, Derkach and colleagues reported significant changes in gamma-glutamyl metabolites after sodium restriction[43]. We add to these results by further implicating gamma-glutamyl amino acid metabolites in salt-sensitive hypertension.
The five remaining salt-sensitivity metabolites identified by the current study were all differentially altered by dietary sodium intervention according to salt-sensitivity status. Among them, three directly associated with hypertension, including glucose from the carbohydrate pathway, alpha-ketoglutarate from the energy pathway and hexanoylcarnitine from the lipid pathway, and two indirectly associated with hypertension, including threonate from the cofactors and vitamins pathway and 5-methyluridine from the nucleotide pathway. Our findings are in line with previous reports implicating elevated glucose as a risk factor for BP salt-sensitivity and hypertension[13,43,47]. In contrast, alpha-ketoglutarate and hexanoylcarnitine have not been related to BP phenotypes previously. Alpha-ketoglutarate is a rate-determining metabolite in the tricarboxylic acid (TCA) and is a precursor for glutamate and glutamine[48,49]. Hexanoylcarnitine has been associated with adiposity phenotypes[50] and cardiovascular disease symptoms[51,52]. Furthermore, its function as an oxidation intermediate may provide a likely biological explanation for the observed association with high salt-sensitivity and BP[51]. Among the two metabolites inversely associated with hypertension, threonate is an ascorbate metabolite synthesized from vitamin C degradation[53], and higher levels have been related to increased intake of green leafy vegetables[54]. Very limited research on the role of 5-methyluridine, a common modification during the maturation of transfer RNA[55], in human disease has been conducted, and we are the first to report its association with a BP related trait. In aggregate, these findings support a role of glucose in salt-sensitive hypertension and propose novel biological pathways underlying this condition.
The current study has several strengths. To our knowledge, this is the first untargeted metabolomics study of BP salt-sensitivity conducted to date, allowing us to comprehensively assess the relation between metabolites across multiple biological pathways and salt-sensitivity of BP. Furthermore, GenSalt uses a rigorous protocol for measuring sodium-sensitivity, including a stringent dietary sodium feeding and multiple measures of BP on multiple days of the baseline examination and each sodium intervention period. The interventional GenSalt design and repeated metabolite measures collected at baseline and the end of each intervention phase further provided a unique opportunity to directly examine changes in metabolites in response to sodium, providing novel mechanistic insights into the relation between dietary sodium intake and BP. In addition, by utilizing data from the BHS, we were able to further examine the relevance of BP salt-sensitivity metabolites in hypertension in a racially diverse sample, more consistent with the demographics of the US population. Certain limitations should also be mentioned. The small sample size of GenSalt metabolomics study subsample may have limited our power to uncover metabolites with relatively small effect sizes. Furthermore, despite robust sensitivity analyses in the BHS, we cannot rule out confounding due to estimated glomerular filtration rate and other variables that were not measured in the GenSalt study. In addition, metabolomics profiling in GenSalt was conducted using urinary biospecimens but only serum metabolomics data was available in in the BHS. Although some studies have reported correlations of metabolite values across urine and serum [54,56], lack of association with hypertension could have resulted from differences in biospecimens. Moreover, if certain metabolite findings were ancestry specific, their relevance to hypertension could not be adequately assessed in the black and white BHS participants. These differences might have led to false negative results. On the other hand, the metabolites robustly identified across studies may be very generalizable to diverse populations. Finally, our findings differed from those of the previous targeted metabolomics study of salt-sensitivity conducted among DASH-sodium participants[20]. Among the 5 metabolites identified in DASH-sodium (including β-aminoisobutyric acid, homocysteine, citrulline, lysine and cystine), only citrulline, lysine and cystine were detected and tested in GenSalt. These three metabolites did not attain nominal significance in our study. Most DASH-sodium participants (>95%) were white or Black, with typical American diets, while our study participants were all of Chinese Han ancestry and followed dietary patterns common to northern, rural China. Metabolites levels are known to be influenced by ancestries, diet and other environmental factors which could explained the inconsistent findings observed [57–59]. However, we cannot rule out the possibility of false positive or false negative findings, given the small sample sizes in both studies.
In total, the current study identified 15 salt-sensitivity related metabolites that may play a role in the development of hypertension. Together, these metabolites enhanced the prediction of hypertension over models that included traditional risk factors alone. These data suggest that metabolomics information could represent a valuable tool in efforts to molecularly characterize sodium-sensitivity of BP. Furthermore, our findings provide novel insights into the biological mechanisms underlying salt-sensitive hypertension, supporting a role for BCAAs, gamma glutamyl amino acids, and both fatty acid and pyrimidine metabolism underlying this complex condition.
Supplementary Material
Highlights.
One hundred and sixty one baseline metabolites were associated with BP salt-sensitivity.
BP salt-sensitivity status was associated with changes in the values of 215 metabolites under low and/or high sodium conditions.
Among the 336 identified salt-sensitivity metabolites, 15 were robustly associated with hypertension.
Together, the 15 salt-sensitivity metabolites explained an additional 7% of hypertension susceptibility when added to a model including traditional risk factors.
ACKNOWLEDGMENTS
We are grateful for the contribution of all staff members who were involved in conducting the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) Replication study and the Bogalusa Heart Study. We extend our gratitude to the participants of GenSalt Replication study and Bogalusa Heart Study.
The Genetic Epidemiology Network of Salt Sensitivity study (JH) was supported by the National Heart, Lung, and Blood Institute of the NIH under award numbers U01HL072507, R01HL087263, and R01HL090682, and the National Institute of General Medical Sciences of the NIH under award number P20GM109036. The Bogalusa Heart Study (LAB) was supported by the National Institute on Aging of the NIH under award numbers RF1AG041200 R21AG051914, and the National Institute of General Medical Sciences of the NIH under award number P20GM109036.
Funding
The Genetic Epidemiology Network of Salt Sensitivity study (JH) was supported by the National Heart, Lung, and Blood Institute of the NIH under award numbers U01HL072507, R01HL087263, and R01HL090682, and the National Institute of General Medical Sciences of the NIH under award number P20GM109036. The Bogalusa Heart Study (LAB) was supported by the National Institute on Aging of the NIH under award number RF1AG041200, and the National Institute of General Medical Sciences of the NIH under award number P20GM109036.
Footnotes
Competing Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- [1].Zhou B, Carrillo-Larco RM, Danaei G, Riley LM, Paciorek CJ, Stevens GA, et al. Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet (London, England) 2021;398:957. 10.1016/S0140-6736(21)01330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Forouzanfar MH, Liu P, Roth GA, Ng M, Biryukov S, Marczak L, et al. Global burden of hypertension and systolic blood pressure of at least 110 to 115mmHg, 1990–2015. JAMA - J Am Med Assoc 2017;317:165–82. 10.1001/jama.2016.19043. [DOI] [PubMed] [Google Scholar]
- [3].Stanaway JD, Afshin A, Gakidou E, Lim SS, Abate D, Abate KH, et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018;392:1923–94. 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].He FJ, Li J, MacGregor GA. Effect of longer term modest salt reduction on blood pressure: Cochrane systematic review and meta-analysis of randomised trials. BMJ 2013;346. 10.1136/bmj.f1325. [DOI] [PubMed] [Google Scholar]
- [5].Mozaffarian D, Fahimi S, Singh GM, Micha R, Khatibzadeh S, Engell RE, et al. Global Sodium Consumption and Death from Cardiovascular Causes. N Engl J Med 2014;371:624–34. 10.1056/NEJMoa1304127. [DOI] [PubMed] [Google Scholar]
- [6].Afshin A, Sur PJ, Fay KA, Cornaby L, Ferrara G, Salama JS, et al. Health effects of dietary risks in 195 countries, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019;393:1958–72. 10.1016/S0140-6736(19)30041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen L, He FJ, Dong Y, Huang Y, Harshfield GA, Zhu H. Sodium Reduction, Metabolomic Profiling, and Cardiovascular Disease Risk in Untreated Black Hypertensives: A Randomized, Double-Blind, Placebo-Controlled Trial. Hypertension 2019;74:194–200. 10.1161/HYPERTENSIONAHA.119.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu Y, Shi M, Dolan J, He J. Sodium sensitivity of blood pressure in Chinese populations. J Hum Hypertens 2019. 10.1038/s41371-018-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He J, Kelly TN, Zhao Q, Li H, Huang J, Wang L, et al. Genome-wide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet 2013;6:598–607. 10.1161/CIRCGENETICS.113.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barba G, Galletti F, Cappuccio FP, Siani A, Venezia A, Versiero M, et al. Incidence of hypertension in individuals with different blood pressure salt-sensitivity: Results of a 15-year follow-up study. J Hypertens 2007;25:1465–71. 10.1097/HJH.0b013e3281139ebd. [DOI] [PubMed] [Google Scholar]
- [11].Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension, vol. 37, Lippincott Williams and Wilkins; 2001, p. 429–32. 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- [12].Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, et al. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet 1997;350:1734–7. 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- [13].Elijovich F, Weinberger MH, Anderson CAM, Appel LJ, Bursztyn M, Cook NR, et al. Salt sensitivity of blood pressure : A scientific statement from the American Heart Association. Hypertension 2016;68:e7–46. 10.1161/HYP.0000000000000047. [DOI] [PubMed] [Google Scholar]
- [14].Ussher JR, Elmariah S, Gerszten RE, Dyck JRB. The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease. J Am Coll Cardiol 2016;68:2850–70. 10.1016/j.jacc.2016.09.972. [DOI] [PubMed] [Google Scholar]
- [15].McGarrah RW, Crown SB, Zhang GF, Shah SH, Newgard CB. Cardiovascular metabolomics. Circ Res 2018;122:1238–58. 10.1161/CIRCRESAHA.117.311002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Arnett DK, Claas SA. Omics of blood pressure and hypertension. Circ Res 2018;122:1409–19. 10.1161/CIRCRESAHA.118.311342. [DOI] [PubMed] [Google Scholar]
- [17].Menni C, Graham D, Kastenmüller G, Alharbi NHJ, Alsanosi SM, Mcbride M, et al. Metabolomic Identification of a Novel Pathway of Blood Pressure Regulation Involving Hexadecanedioate. Hypertension 2015;66:422–9. 10.1161/HYPERTENSIONAHA.115.05544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dietrich S, Floegel A, Weikert C, Pischon T, Boeing H, Drogan D. Identification of Serum Metabolites Associated with Incident Hypertension in the European Prospective Investigation into Cancer and Nutrition-Potsdam Study. Hypertension 2016;68:471–7. 10.1161/HYPERTENSIONAHA.116.07292. [DOI] [PubMed] [Google Scholar]
- [19].He WJ, Li C, Mi X, Shi M, Gu X, Bazzano LA, et al. An untargeted metabolomics study of blood pressure: Findings from the Bogalusa Heart Study. J Hypertens 2020;38:1302–11. 10.1097/HJH.0000000000002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cheng Y, Song H, Pan X, Xue H, Wan Y, Wang T, et al. Urinary metabolites associated with blood pressure on a low- or high-sodium diet. Theranostics 2018;8:1468–80. 10.7150/thno.22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Himmelfarb CD, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults a report of the American College of Cardiology/American Heart Association Task Force on Clinical practice guidelines. Hypertension 2018;71:E13–115. 10.1161/HYP.0000000000000065. [DOI] [PubMed] [Google Scholar]
- [22].GenSalt: Rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 2007;21:639–46. 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gu D, Zhao Q, Chen J, Chen JC, Huang J, Bazzano LA, et al. Reproducibility of blood pressure responses to dietary sodium and potassium interventions: The GenSalt study. Hypertension 2013;62:499–505. 10.1161/HYPERTENSIONAHA.113.01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].JONES BL, NAGIN DS, ROEDER K. A SAS Procedure Based on Mixture Models for Estimating Developmental Trajectories. Sociol Methods Res 2001;29:374–93. 10.1177/0049124101029003005. [DOI] [Google Scholar]
- [25].He J, Huang J-F, Li C, Chen J, Lu X, Chen J-C, et al. Sodium Sensitivity, Sodium Resistance, and Incidence of Hypertension. Hypertension 2021:155–64. 10.1161/hypertensionaha.120.16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Khamis MM, Holt T, Awad H, El-Aneed A, Adamko DJ. Comparative analysis of creatinine and osmolality as urine normalization strategies in targeted metabolomics for the differential diagnosis of asthma and COPD. Metabolomics 2018;14. 10.1007/s11306-018-1418-9. [DOI] [PubMed] [Google Scholar]
- [27].Kennedy AD, Miller MJ, Beebe K, Wulff JE, Evans AM, Miller LAD, et al. Metabolomic Profiling of Human Urine as a Screen for Multiple Inborn Errors of Metabolism. Genet Test Mol Biomarkers 2016;20:485–95. 10.1089/gtmb.2015.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nam SL, Paulina de la Mata A, Dias RP, Harynuk JJ. Towards standardization of data normalization strategies to improve urinary metabolomics studies by gc×gc-tofms. Metabolites 2020;10:1–13. 10.3390/metabo10090376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Foster TA, Berenson GS. Measurement error and reliability in four pediatric cross-sectional surveys of cardiovascular disease risk factor variables-The Bogalusa Heart Study. J Chronic Dis 1987;40:13–21. 10.1016/0021-9681(87)90092-0. [DOI] [PubMed] [Google Scholar]
- [30].Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 2021;49:388–96. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nierenberg JL, Li C, He J, Gu D, Chen J, Lu X, et al. Blood pressure genetic risk score predicts blood pressure responses to dietary sodium and potassium the GenSalt Study (Genetic epidemiology network of salt sensitivity). Hypertension 2017;70:1106–12. 10.1161/HYPERTENSIONAHA.117.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Natarajan P, Peloso GM, Zekavat SM, Montasser M, Ganna A, Chaffin M, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun 2018;9:1–12. 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yu B, Li AH, Muzny D, Veeraraghavan N, De Vries PS, Bis JC, et al. Association of Rare Loss-Of-Function Alleles in HAL, Serum Histidine: Levels and Incident Coronary Heart Disease. Circ Cardiovasc Genet 2015;8:351–5. 10.1161/CIRCGENETICS.114.000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mels CM, Delles C, Louw R, Schutte AE. Central systolic pressure and a nonessential amino acidmetabolomics profile: TheAfrican Prospective study on the Early Detection and Identification of Cardiovascular disease andHypertension. J Hypertens 2019;37:1157–66. 10.1097/HJH.0000000000002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sedlak TW, Paul BD, Parker GM, Hester LD, Snowman AM, Taniguchi Y, et al. The glutathione cycle shapes synaptic glutamate activity. Proc Natl Acad Sci U S A 2019;116:2701–6. 10.1073/pnas.1817885116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coyle JT, Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science (80- ) 1993;262:689–95. 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- [37].Parfenova H, Basuroy S, Bhattacharya S, Tcheranova D, Qu Y, Regan RF, et al. Glutamate induces oxidative stress and apoptosis in cerebral vascular endothelial cells: Contributions of HO-1 and HO-2 to cytoprotection. Am J Physiol - Cell Physiol 2006;290. 10.1152/ajpcell.00386.2005. [DOI] [PubMed] [Google Scholar]
- [38].Harris RC, Zhang MZ. Dopamine, the kidney, and hypertension. Curr Hypertens Rep 2012;14:138–43. 10.1007/s11906-012-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Contreras F, Fouillioux C, Bolívar A, Simonovis N, Hernández-Hernández R, Armas-Hernandez MJ, et al. Dopamine, hypertension and obesity. J Hum Hypertens 2002;16:S13–7. 10.1038/sj.jhh.1001334. [DOI] [PubMed] [Google Scholar]
- [40].Sass JO, Ensenauer R, Röschinger W, Reich H, Steuerwald U, Schirrmacher O, et al. 2-Methylbutyryl-coenzyme A dehydrogenase deficiency: Functional and molecular studies on a defect in isoleucine catabolism. Mol Genet Metab 2008;93:30–5. 10.1016/j.ymgme.2007.09.002. [DOI] [PubMed] [Google Scholar]
- [41].Neinast M, Murashige D, Arany Z. Branched Chain Amino Acids. Annu Rev Physiol 2019;81:139–64. 10.1146/annurev-physiol-020518-114455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang L, Hou E, Wang Z, Sun N, He L, Chen L, et al. Analysis of metabolites in plasma reveals distinct metabolic features between Dahl salt-sensitive rats and consomic SS.13BN rats. Biochem Biophys Res Commun 2014;450:863–9. 10.1016/j.bbrc.2014.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Derkach A, Sampson J, Joseph J, Playdon MC, Stolzenberg-Solomon RZ. Effects of dietary sodium on metabolites: The Dietary Approaches to Stop Hypertension (DASH)–Sodium Feeding Study. Am J Clin Nutr 2017;106:1131–41. 10.3945/ajcn.116.150136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol 2019;37:1217–28. 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- [45].Bachhawat AK, Yadav S. The glutathione cycle: Glutathione metabolism beyond the γ-glutamyl cycle. IUBMB Life 2018;70:585–92. 10.1002/iub.1756. [DOI] [PubMed] [Google Scholar]
- [46].Kunutsor SK, Apekey TA, Cheung BMY. Gamma-glutamyltransferase and risk of hypertension: A systematic review and dose-responsemeta-analysis of prospective evidence. J Hypertens 2015;33:2373–81. 10.1097/HJH.0000000000000763. [DOI] [PubMed] [Google Scholar]
- [47].Ferrannini E, Cushman WC. Diabetes and hypertension: The bad companions. Lancet 2012;380:601–10. 10.1016/S0140-6736(12)60987-8. [DOI] [PubMed] [Google Scholar]
- [48].Wu N, Yang M, Gaur U, Xu H, Yao Y, Li D. Alpha-ketoglutarate: Physiological functions and applications. Biomol Ther 2016;24:1–8. 10.4062/biomolther.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zdzisińska B, Żurek A, Kandefer-Szerszeń M. Alpha-Ketoglutarate as a Molecule with Pleiotropic Activity: Well-Known and Novel Possibilities of Therapeutic Use. Arch Immunol Ther Exp (Warsz) 2017;65:21–36. 10.1007/s00005-016-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Muniandy M, Velagapudi V, Hakkarainen A, Lundbom J, Lundbom N, Rissanen A, et al. Plasma metabolites reveal distinct profiles associating with different metabolic risk factors in monozygotic twin pairs. Int J Obes 2019;43:487–502. 10.1038/s41366018-0132-z. [DOI] [PubMed] [Google Scholar]
- [51].Vorkas PA, Shalhoub J, Lewis MR, Spagou K, Want EJ, Nicholson JK, et al. Metabolic Phenotypes of Carotid Atherosclerotic Plaques Relate to Stroke Risk: An Exploratory Study. Eur J Vasc Endovasc Surg 2016;52:5–10. 10.1016/j.ejvs.2016.01.022. [DOI] [PubMed] [Google Scholar]
- [52].Relationship between the plasma acylcarnitine profile and cardiometabolic risk factors in adults diagnosed with cardiovascular diseases - ScienceDirect n.d. https://www-sciencedirect-com.libproxy.tulane.edu/science/article/pii/S000989812030190X (accessed March 14, 2021). [DOI] [PubMed]
- [53].Wang H, Jiang J, Hu P. Determination of l-threonate in human plasma and urine by high performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci 2006;834:155–62. 10.1016/j.jchromb.2006.02.057. [DOI] [PubMed] [Google Scholar]
- [54].Playdon MC, Sampson JN, Cross AJ, Sinha R, Guertin KA, Moy KA, et al. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr 2016;104:776–89. 10.3945/ajcn.116.135301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chang YH, Nishimura S, Oishi H, Kelly VP, Kuno A, Takahashi S. TRMT2A is a novel cell cycle regulator that suppresses cell proliferation. Biochem Biophys Res Commun 2019;508:410–5. 10.1016/j.bbrc.2018.11.104. [DOI] [PubMed] [Google Scholar]
- [56].Lau CHE, Siskos AP, Maitre L, Robinson O, Athersuch TJ, Want EJ, et al. Determinants of the urinary and serum metabolome in children from six European populations. BMC Med 2018;16. 10.1186/s12916-018-1190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Patel MJ, Batch BC, Svetkey LP, Bain JR, Turer CB, Haynes C, et al. Race and Sex Differences in Small-Molecule Metabolites and Metabolic Hormones in Overweight and Obese Adults. OMICS 2013;17:627. 10.1089/OMI.2013.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Menni C, Zhai G, MacGregor A, Prehn C, Römisch-Margl W, Suhre K, et al. Targeted metabolomics profiles are strongly correlated with nutritional patterns in women. Metabolomics 2013;9:506–14. 10.1007/S11306-012-0469-6/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nicholson G, Rantalainen M, Maher AD, Li JV., Malmodin D, Ahmadi KR, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol 2011;7:525. 10.1038/MSB.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.