Abstract
Mesenchymal stem cells (MSCs) have potential role in organ regeneration therapy. Previous work indicating that MSCs confer protection against liver disease. Here, we aimed to determine the potential application in liver regeneration of human placenta-derived MSCs extracellular vesicles (hPMSCs-EVs) via experimental hepatectomy. hPMSCs-EVs were administered intravenously 24 h before 70% partial hepatectomy, the specific composition of hPMSCs-EVs was identified by sequencing and validated by the quantitative polymerase chain reaction, including circ-RBM23. The role of circ-RBM23 in L02 cell was evaluated and it was found that circ-RBM23 knockdown inhibited L02 cell proliferation both in vitro and in vivo. The competing endogenous RNA function of circ-RBM23 was evaluated by the RNA immunoprecipitation assay and found that circ-RBM23 shares miRNA response elements with RRM2. Overexpressed circ-RBM23 bound competitively to miR-139-5p, preventing the miRNA-mediated degradation of RRM2, activating the expression of eIF4G and AKT/mTOR, and facilitating liver regeneration. These results indicate that hPMSCs-EVs prevent hepatic dysfunction and improve liver regeneration in vivo and hepatocytes proliferation in vitro, potentially via circ-RBM23 delivery.
Keywords: Extracellular vesicles, circ-RBM23, placental stem cells, liver regeneration, ceRNA mechanism
Introduction
Liver regeneration is a compensatory process in which functional liver that has been lost to injury or disease is replaced.1,2 The mechanisms by which regeneration occurs are highly complex and involves multiple factors including the actions of cytokines, extracellular vesicles (EVs), growth factors, and signaling pathways.3–5 Liver possesses the extraordinary capacity to regenerate after partial hepatectomy (PH). However, subtotal hepatectomy often results in an insufficient future liver volume (FLV), which can lead to post-hepatectomy liver failure (PHLF).2 The available treatments mostly alleviate symptoms but do not cure the condition. Novel therapeutic options are thus required to enhance regeneration and improve hepatic failure.
Transplantation with MSCs can improve liver function in acute hepatic failure and end-stage liver disease.3,6–8 Human placental-derived mesenchymal stem cells (hPMSCs) exhibit self-renewing potential and can differentiate into multiple cell type lineages.9 The therapeutic potential of hPMSCs for tissue damage repair following ischemic disorders such as stroke or myocardial and cerebral infarctions has been demonstrated, indicating that hPMSCs-based therapies might be developed for future clinical application.10–14 Alternatively, stem cells may have beneficial effects through paracrine effects, which are mediated by the release of soluble factors. hPMSCs release EVs, growth factors and cytokines via paracrine signaling to regulate hemostasis, improve microcirculation, and inhibit inflammatory storms, thereby promoting liver regeneration.15 The latest evidence suggests that the cell-free hPMSCs-EVs treatment, which is not limited by ethical requirements, possesses approximately the same biological effect as hPMSCs.14 Discovering the key factors by which hPMSCs promote liver regeneration can better target and regulate liver regeneration.
Circular RNAs (circRNAs) are special types of long non-coding RNAs (lncRNAs) with stable properties, special structures, and conserved sequences in different biological tissues and cells.16 Some circRNAs increase the complexity of RNA regulatory networks by acting as microRNA (miRNA) sponges, thereby suppressing the functional activity of the miRNAs.17 Salmena et al.18 proposed a regulatory mechanism in the form of competing endogenous RNAs (ceRNAs), which act as molecular sponges.18 Recent studies have showed that circRNAs play an important role in the occurrence and development of liver disease and can serve as potential therapeutic targets. For instance, circRNA-14723 has been reported to promote cyclin D1 translation and enhance cell cycle progression, enhancing rat liver regeneration.19 Nevertheless, few studies have reported on the action of circRNAs in liver regeneration, especially following hepatectomy.
EVs are typically bilayer-enclosed spherical particles covered in phospholipids that are secreted by all MSCs and other cell types into the extracellular space.20 EVs can also be detected in all biological fluids such as blood, bile, urine, and ascites fluid, and carry complex cargo loads that may include active molecules such as proteins, DNA, and RNAs, which potentially influence multiple biological cellular processes.20–22 While the role of MSCs-derived soluble factors in effecting the paracrine effect have been investigated, the contribution of MSCs-derived EVs, especially hPMSCs-EVs circRNAs, and their function in liver regeneration have not been well characterized. Thus, we aimed to evaluate the effects of hPMSCs-EVs in experimental hepatectomy and thereby to determine the potential utility of these vesicles as therapeutic agents for tissue repair and functional restoration in liver regeneration.
This study is an evaluation of the effects of hPMSCs-EVs on murine/human hepatocytes (in vitro) and a 70% PH model (in vivo), as well as the underlying mechanisms.
Materials and methods
Human placenta mesenchymal stem cell preparation
The methods for preparing hPMSCs have been described previously.23 The sample collection and use were approved by the review board and ethics committee at Zhu Jiang Hospital (approval number: 2019-KY-015-02). All participants provided written informed consent prior to sample collection.
Ribonuclease R (RNase R) treatment
RNase R was utilized to degrade linear RNA. The total RNA (2 μg) was incubated at 37°C for 30 min with 0 or 20 units of RNase R. The RNA was then purified by phenol-chloroform extraction, retro-transcribed, and used for real-time polymerase chain reaction (RT-qPCR) using actin as a reference gene.
Isolation of EVs
EVs were harvested from primary hPMSCs. To extract EVs from the conditioned medium of cell cultures, 5 × 107 cells were seeded in 25 mL media supplemented with 10% exo-depleted FBS in a 150 cm2 tissue culture plate for 48 h. Standard differential centrifugation was used to isolate EVs from the conditioned medium, which was then subjected to sequential centrifugation of 200g for 10 min and 2000g for 15 min to eliminate excess cells and cellular debris. The supernatant was then centrifuged at 10,000g for 30 min and filtered using a 0.22 μm filter (Merck Millipore, Germany) to eliminate any remaining cell debris membranes and EVs. Ultracentrifugation was then conducted for 70 min at 100,000g to produce a pellet that was collected using a P28Ti fixed angle rotor (Himac CP100xn, Japan) before resuspension in 100 µL PBS for downstream analyses.
Characterization of EVs
The characterization of EVs was administrated according to the guidance of minimal information for studies of extracellular vesicles 2018 (MISEV2018). 20 µL EVs sample were added to a copper-mesh grid and was then negatively stained with 2% uranyl acetate solution for 4 min, dried off with filter paper, then observed under transmission electron microscopy (TEM; H-600 HITACHI microscope, Japan) at 80 keV.
The size and number of EVs were established and analyzed using nanoparticle tracking analysis (NTA) ZetaView (Particle Metrix, Germany). EVs were resuspended in 1 mL of PBS with filtered PBS as a control. Five microliter EVs samples were diluted with PBS to 30 µL, and a performance test conducted with a standard before samples were loaded. Testing was performed using the manufacturer’s instructions. ZetaView software 8.04.02 was used to analyze the EVs parameters. Analysis was started once the pictures of the particle on the screen become stable. The particle size and concentration information were obtained after completing the sample test. EVs purity was examined by Western blotting (see the after methods) to determine EVs surface markers (CD9, CD63, CD81, and HSP70), please refer to section on Western blotting.
EVs labeling and uptake
EVs were isolated from hPMSCs culture medium and labeled using a PKH26 red fluorescent labeling kit (Umibio, China) in line with standard protocols. Briefly, 100 μg of EVs (approximately 6.9 × 1010 EVs) and 4 μL of PKH26 dye were mixed with 1 mL Diluent C and incubated for 5 min, stopped the reaction with 1% BSA. Excess dye was removed by centrifugation at 200,000g for 30 min with a 30% sucrose/D2O density gradient. The sucrose was then removed using a 100-kD ultrafiltration tube to obtain PKH26-labeled EVs. PKH26-labeled EVs were co-cultured with cells for 3 h, and fixed with 4% paraformaldehyde at 25°C for 30 min, fixed cells were rinsed with PBS thrice, nuclear staining with DAPI for 10 min (Invitrogen, USA). Confocal laser scanning microscopy was applied to detect the cellular uptake and internalization of EVs.
XenoLight DiR (Perkin Elmer, USA) was used to track hPMSCs-EVs in vivo. The XenoLight DiR solution was diluted to 300 μM in PBS and the resultant XenoLight DiR solution was then added to 10 μg of hPMSCs-EVs in 1 mL PBS to obtain a final concentration of 2 μM. The cells were incubated with hPMSCs-EVs at 25°C for 30 min. The solution was then centrifuged for 3 min at 14,000 rpm. The pellet containing the fluorescently labeled hPMSCs-EVs was resuspended in 100 μL of PBS before 50 μg of hPMSCs-EVs were administered via the tail vein. IVIS analysis (Caliper Life Sciences, USA) was performed after 2 and 8 h.
High-throughput sequencing
High-throughput sequencing and OE Biotech (Shanghai, China) were used to determine the circRNAs expression profiles. Total RNA (2 μL) was extracted with TRIzol and the RNA purity and concentration (100–150 ng/uL) were determined, and a cDNA library was constructed following the removal of ribosomal RNA and RNase R digestion. The Illumina sequencer (HiSeqTM 2500) was used after passing a quality inspection. Then, circRNAs were confirmed and statistically analyzed.
Bioinformatics analysis
Gene expression profiles were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Data describing differentially expressed genes (DEGs) were analyzed via heatmap and clustering with R software (version 3.6.2) and limma, with |logFC| > 1 and p = <0.05 set as the cut-off for DEG identification. R software (version 3.6.2) was also applied for functional and pathway enrichment analyses of the DEGs, including the GO and KEGG pathways. Pathway enrichment analysis depended on references from the KEGG pathways. A false discovery rate (FDR) of <0.05 was applied as the cut-off.
Animals
Male C57BL/6 mice (6–8 weeks) were purchased from the animal center at Guangdong Medical Laboratory (Certificate of Conformity: SYXK (YUE) 2019-0215). The experimental protocols were approved by the Institutional Animal Care and Use Committee Center at Zhu Jiang Hospital (Southern Medical University, Guangzhou, China). The mice were housed in plastic cages at a controlled temperature of 22°C ± 1°C. Standard rodent chow and water were provided. All animals and samples were assigned a number that did not expose their treatment group so that blind analysis could be performed.
Murine 70% PH model
Anaesthetization was performed using 70 mg/kg pentobarbital prior to PH as described in Mitchell and Willenbring.24 The left, median, and posterior right lobes were ligated and excised, and 70% of the liver was removed.
RNA-binding protein immunoprecipitation (RIP) assay
Immunoprecipitation of circ-RBM23 that was bound to Ago2 was performed using a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore, Germany). A total of 2 × 107 L02 cells were harvested in RIP lysis buffer and lysates were stored at −80°C. Samples comprising 8 µg of anti-Ago2 (MA5-23515, Invitrogen, USA) or normal control IgG were then incubated with magnetic beads at 25°C for 2 h. RIP lysate (100 µL) was mixed with 900 µL RIP immunoprecipitation buffer and added to the bead-antibody complexes before incubation overnight at 4°C. The beads were then mixed with proteinase K buffer and incubated at 55°C for 30 min before RNA was extracted for PCR.
Dual-luciferase report assay
A dual-luciferase reporter assay kit (Promega, USA) was used to conduct the dual-luciferase assay. L02 cells were seeded on 24-well plates and cultured to 50% confluence before transfection with circ-RBM23-wt or circ-RBM23-mut plasmids, or negative control renilla luciferase plasmid with miR-139-5p for 48h. The same procedure was conducted with miR-139-5p and RRM2. Cells were lysed then luciferase and renilla substrates were added and left for 24 h. Luciferase analysis was then performed using the dual-luciferase reporter kit following the manufacturer’s instructions.
Cell culture
Murine hepatocyte cell line AML12 was purchased from the Procell Life Science & Technology Co., Ltd (Wuhan China). Cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 100 units/mL penicillin/streptomycin mixture (Gibco, USA). Human hepatocyte cell line L02 was purchased from Procell Life Science & Technology Co., Ltd (Wuhan China). Cells were cultured in RPMI-1640 medium supplemented with 10% FBS. Cell lines were incubated at 37°C under 100% humidity in 5% CO2.
Cell transfection
Circ-RBM23-knockdown vector, miR-139-5p inhibitor, and miR-139-5p mimics were obtained from GenePharma Co., Ltd (Shanghai, China). Gene-specific siRNAs, scrambled control siRNAs, miR-139-5p inhibitor, and miR-139-5p mimic were transfected into L02 cells using Lipofectamine 2000 (Invitrogen, USA) for 48 h before cells were collected for use in subsequent experiments.
Flow cytometry
Flow cytometry was used to analyze the characteristics of the cultured hPMSCs. Briefly, cells were incubated in 1% bovine serum albumin (BSA; Gibco, USA) in PBS for 30 min to block nonspecific antigens. Conjugated monoclonal antibodies were then used according to the manufacturer’s instructions. Samples were analyzed using an Epics xL flow cytometer (Beckman Coulter, High Wycombe, UK).
Cell proliferation assays
In vitro hepatocyte proliferation was determined by CCK8 assay. Hepatocytes were treated with hPMSCs-EVs for 24 h prior to the CCK8 assay (10 μL/well, APExBIO, USA). The results were normalized by the number of viable cells and compared with the control. The absorbance at 450 nm was then measured using a microplate absorbance reader (Thermo Scientific, USA).
Liver histological and serum aminotransferase analysis
The liver tissues were processed and sectioned into 4μm sections for paraffin embedding. The sections were then routinely stained with hematoxylin and eosin (H&E staining). For hepatocyte proliferation examination, tissue samples were fixed in 10% neutral-buffered formalin and embedded in paraffin. The sections were then stained for proliferating cell nuclear antigen (PCNA). PCNA staining was performed according to the manufacturer’s instructions (Ser10; dilution of 1:200, Cell Signaling Technology, USA). Sections were counterstained with hematoxylin. The positive nuclei were quantified, and the percentages calculated (400–600 hepatocytes from 4 to 6 fields at 200× magnification were counted for each mouse).
The serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured using a Hitachi 7600 automatic analyzer (Hitachi, Ltd., Japan).
Western blotting analysis (WB)
WB and the analysis of the blot results was conducted as previously reported. In brief, liver tissue or cultured cells were lysed in RIPA buffer containing 50 mM Tris-HCl pH 8, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), phosphatase inhibitors, and a protease inhibitor cocktail (Roche, Germany). Then centrifuged at 12,000g for 10 min at 4°C. The supernatants were denatured at 100°C for 5 min and mixed with a loading buffer (Beyotime, China). Solubilized proteins were collected by centrifugation and quantified using a BCA protein assay kit (Thermo Scientific, USA). Proteins were separated on SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, USA) for antibody blotting. Next, the PVDF membranes with the transferred proteins were blocked with 5% nonfat milk diluted in TBST for 2 h at 25°C and incubated with primary antibodies at 4°C for 12 h. Then the membrane was washed again with TBST and incubated with a 1:10,000 dilution of horseradish peroxidase (HRP)-labeled anti-rabbit IgG (Beyotime, China) for 1 h at 25°C. Finally, the respective protein bands were visualized using enhanced chemiluminescence reagents (BOSTER, China). The protein band positions were determined according to the protein molecular weight suggested by the antibody specification and the results of preliminary experiment. Protein bands were quantified using the grayscale analysis function in ImageJ software. The primary antibodies: CD9, CD63, CD81, HSP70, RRM2, AKT, p-AKT, eIF4G, p-mTOR, mTOR (Abcam, UK), and β-actin (Sigma-Aldrich, Germany).
Statistical analysis
Continuous variables were presented as mean ± the standard error of the mean (SEM). Analysis of variance (ANOVA) and post hoc Bonferroni analysis were conducted for multiple comparisons using GraphPad Prism 8.0. p < 0.05 was considered statistically significant.
Results
Identification of hPMSCs and hPMSCs-EVs
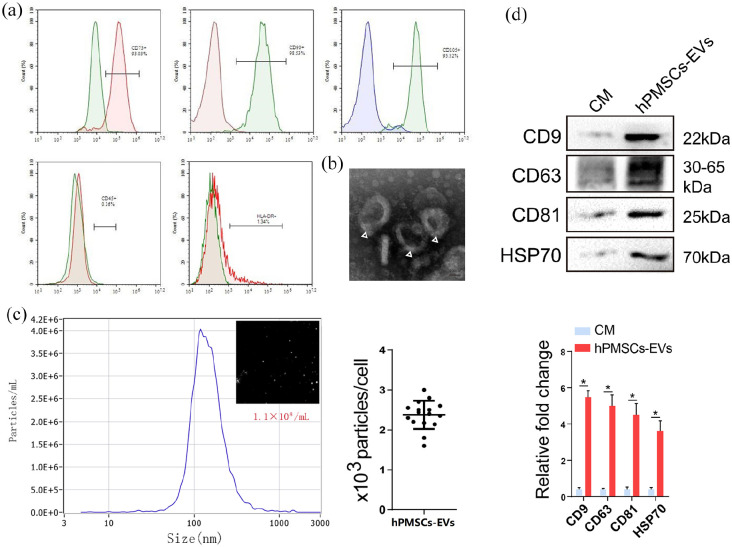
The obtained hPMSCs were found to have the same essential features of MSCs and were positive for the surface cell markers CD73, CD90, CD105 and negative for CD45 and HLA-DR (Figure 1(a)). EVs were isolated from hPMSCs supernatant as described previously.20 TEM revealed that hPMSCs-EVs were spherical in shape (Figure 1(b), Supplemental Figure S1F). NTA indicated hPMSCs-EVs diameters of 30–150 nm (Figure 1(c)). WB analysis demonstrated that the collected hPMSCs-EVs expressed precise surface markers including CD9, CD63, and CD81 (Figure 1(d)), and exhibit specific characteristics that are identical to those described previously.20
Figure 1.
Identification of hPMSCs and hPMSCs-EVs. (a) Flow cytometry analysis of surface markers of hPMSCs (the positive markers CD73, CD90, CD105 and the negative markers CD45, HLA-DR). (b) TEM image of hPMSCs-EVs (scale bar, 100 nm). (c) Particle size of EVs was detected with NTA. The X-axis shows the particle size inside of the sample, and the Y-axis shows the concentration of particles with a certain size (the EVs particle size mean: 130 nm). (d) Total protein was extracted from EVs and analyzed with Western blots. The specific surface markers CD9, CD63, CD81, and HSP70 were measured in hPMSCs-EVs. Western blots were quantified based on at least three replicates. CM: culture medium.
HPMSCs-EVs promoted liver regeneration in vivo and hepatocyte proliferation in vitro
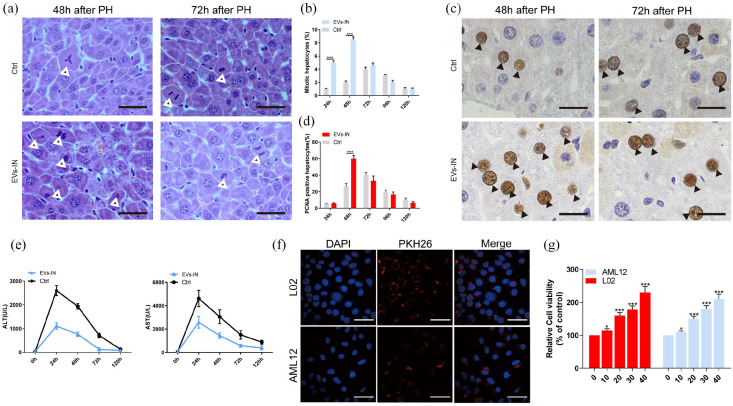
To determine whether hPMSCs-EVs facilitate hepatocyte proliferation in vivo, 1 × 108 particles/mL hPMSCs-EVs were administered 24 h before 70% PH surgery by tail vein injection. Relative to control mice, EVs treatment was found to significantly increase liver mitosis after 2 days as indicated by H&E staining (Figure 2(a) and (b)). Additionally, PCNA staining showed that EVs promoted cell proliferation after 2 days post PH (Figure 2(c) and (d)). Compared to EVs treated mice, untreated mice exhibited significantly higher levels of ALT and AST (Figure 2(e)), indicating that hPMSCs-EVs have a protective effect on the liver. Since the effective absorption of EVs into hepatocyte was one of the prior conditions for further treatment experiments, AML-12 and L02 cells were co-cultured with hPMSCs-EVs labeled with PKH26 (a cell membrane marker for EVs). Confocal analysis showed that EVs were directly absorbed by cells (Figure 2(f)), indicating uptake of hPMSCs-EVs by hepatocytes. Besides, in order to track the distribution of the hPMSCs-EVs in vivo, the XenoLight DiR and IVIS system was used, and the results indicated that most of the hPMSCs-EVs distributed in liver (Supplemental Figure S1G). Moreover, CCK8 assays showed elevated cell proliferation after 24 h treatment with hPMSCs-EVs (Figure 2(g)). These results indicating that hPMSCs-EVs showed a functionally protective role in PH mice and facilitated the proliferation of hepatocytes.
Figure 2.
hPMSCs-EVs promote liver regeneration in vivo and ameliorates liver injury after PH, and facilitate hepatocyte proliferation in vitro. (a) Liver tissues were collected and followed by H&E staining of the liver postoperative 48 and 72 h. (b) The number of mitotic hepatocytes was counted. (c) After 2 and 3 days post PH, liver tissue sections were performed PCNA staining. (d) The percentages of PCNA-positive cells were analyzed. (e) Serum ALT and AST levels were analyzed to evaluate the liver injury. (f) PKH26-labeled hPMSCs-EVs were colocalized with L02 and AML12 cells. Immunofluorescent images of PKH26 (red) together with DAPI for nuclei (blue). (g) Cell viabilities were measured by CCK8 assay in AML12 and L02 cells treated with hPMSCs-EVs at the concentrations of 0%, 10%, 20%, 30%, and 40% for 24 h. Error bars represent the mean±standard error of the mean (SEM). *p < 0.05, ***p < 0.001. EVs-IN: hPMSCs-EVs treated.
Identification and characteristics of circ-RBM23 in hPMSCs-EVs
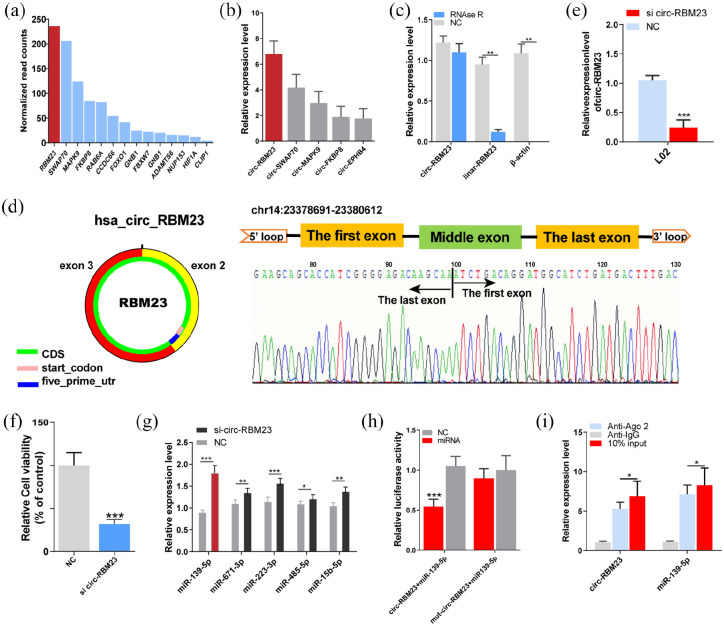
circRNAs expression in hPMSCs-EVs was profiled using high-throughput sequencing. Fourteen possible circRNAs were selected based on their higher expression (Figure 3(a)). Of these, circ-RBM23, circ-SWAP70, circ-MAPK9, circ-FKBP8, and circ-EPHB4 were highly expressed in hPMSCs-EVs. RT-qPCR revealed the highest expression for circ-RBM23 (Figure 3(b), circRNAs primers are given in Supplementary Table 1). Therefore, circ-RBM23 was selected for subsequent analysis. Linear RBM23 and circ-RBM23 primers are given in Supplemental Table 2. Compared to linear RBM23, circ-RBM23 was found more resistant to RNase R digestion (Figure 3(c)). Sanger sequencing and circPrimer 2.0 were used to verify the head-to-tail splicing and circular features and characterize circ-RBM23 expression in hPMSCs (Figure 3(d)). siRNA assay was used to knock down circ-RBM23 in L02 cells, which was confirmed by RT-qPCR assay (p < 0.01, Fig 3E). CCK8 assay also showed that downregulating circ-RBM23 inhibited the proliferation of L02 cells (Fig 3F).
Figure 3.
hPMSCs-EVs circ-RBM23 serve as a sponge for miR-139-5p. (A) High-throughput sequencing was used to obtain the top 14 expression levels of circ-RNA in hPMSCs-EVs, of which circ-RBM23 was highest. (b) Related RNA expression level (RT-qPCR) of the top five circ-RNAs in hPMSCs-EVs. (c) RT-qPCR analysis showed that linear RNA can be digested by RNase R and that β-actin expression decreased significantly while circ-RBM23 was not particularly affected. (d) Sanger sequencing and circPrimer2.0 were used to confirm the characteristics of circ-RBM23 in hPMSCs. (e) Related RNA expression level of the circ-RBM23 in L02 cells treated with circ-RBM23-siRNA for 72 h. (f) Cell viabilities were measured by CCK8 in L02 cells treated with circ-RBM23-siRNA for 72 h. (g) Relative RNA expression level of indicated miRNAs after L02 cells were co-transfected with circ-RBM23-siRNA. (h) Relative luciferase activity detected after L02 cells were co-transfection with miR-139-5p and circ-RBM23 promoter or circ-RBM23-siRNA. (i) RIP experiment using anti-Ago2 or lgG antibodies as immunoprecipitates; circ-RBM23 and miR-139-5p expressions measured by RT-qPCR. Error bars represent the SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Circ-RBM23 functions as a sponge for miR-139-5p
Previous studies have indicated that circRNAs primarily function as miRNA sponges. A rough miRNA target prediction was made for circ-RBM23 using the bioinformatics tool starBase2.0 (https://starbase.sysu.edu.cn/). The top-5 potential circ-RBM23 target miRNAs and the associated primers are listed in Supplemental Table 3. RT-qPCR analysis showed that circ-RBM23 silencing upregulated miRNA expression and that miR-139-5p was the most significantly upregulated (Figure 3(g)). Dual-luciferase reporter analysis identified that the co-transfection of wild circ-RBM23 and miR-139-5p hindered Rluc expression, indicating the sponge effect of circ-RBM23 (Figure 3(h)). RIP showed that circ-RBM23 was more enriched in Ago2-containing immunoprecipitates than actin or miR-139-5p (Figure 3(i)), suggesting the predicted function of circ-RBM23 as a sponge for miR-139-5p. Compared to sham mice, circ-RBM23 expression was found to increase in PH mice while miR-139-5p expression decreased (Supplemental Figure S1A, B).
Circ-RBM23 regulated miR-139-5p/RRM2 in cell proliferation
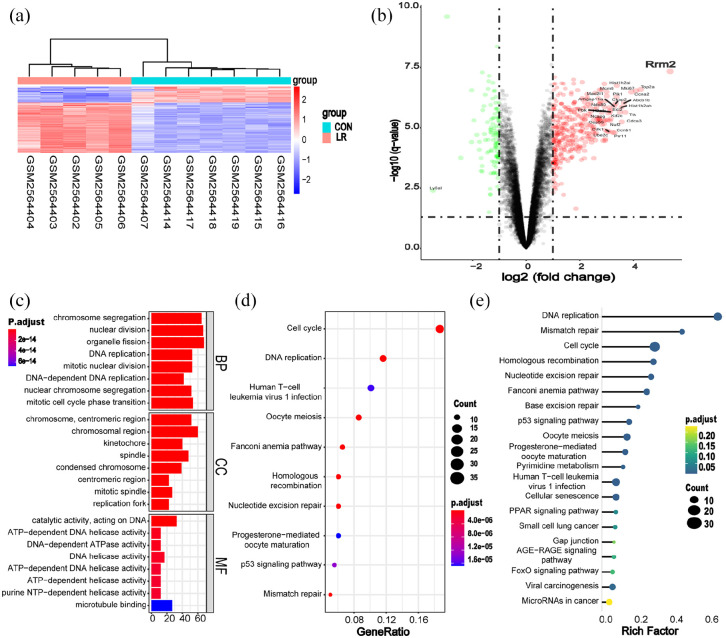
To identify the important genes involved in liver regeneration, R software was utilized to analyze dataset GSE97429 from GEO for the genes that are involved in liver regeneration following PH. Gene expression profiles from 12 samples were extracted from the GEO database. The samples were then divided into control and LR (liver resection, 70% PH) groups (Figure 4(a)). Volcano plot analysis was used to visualize DEGs (Figure 4(b)). Functional enrichment analysis was conducted to analyze the functions of the DEGs. Gene Ontology (GO) analysis showed that the DEGs were implicated in both “DNA replication” and “nuclear division” (Figure 4(c)). KEGG pathway analysis indicated that the DEGs have significant involvement in cell cycle signaling and DNA replication pathways (Figure 4(d) and (e)). Among the DEGs, RRM2 (Ribonucleotide Reductase Regulatory Subunit M2) elicited attention. RRM2 is one of the targets for miR-139-5p that provides DNA synthesis precursors and accelerates the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides. Huang et al.25 reported that RRM2 bound with miR-139-5p in a dual luciferase reporter assay of non-small cell lung cancer cells. Our data also showed that RRM2 was the target gene for miR-139-5p, and that binding occurred in L02 cells (Supplemental Figure S1C). Therefore, we hypothesized that circ-RBM23 might regulate RRM2 via miR-139-5p.
Figure 4.
The key gene involved in liver regeneration following PH. (a) Hierarchical cluster analysis. Analysis correctly classified five PH (red lines) and seven control samples (blue lines). (b) Volcano plot for differentially expressed genes. Red and green points represent DEGs that are statistically significant (p < 0.05) or show fold change >1, respectively. RRM2, in the red area, represents an upregulated gene. (c) GO term analysis for all DEGs, with top eight GO terms indicated (p < 0.05). BP: biological process; CC: cellular component; MF: molecular function. (d) KEGG pathway analysis for all DEGs. Top 10 pathways are showed in the figure (p < 0.05). (e) Bubble plot of signaling pathways analyses for DEGs. The rich factor indicates the ratio of DEG numbers to total genes annotated in this pathway group. A larger rich factor represents a larger degree of pathway enrichment, with FDR ⩽ 0.05 indicating significant pathway enrichment. DEGs: differentially expressed genes; GO: gene ontology; FDR: false discovery rate.
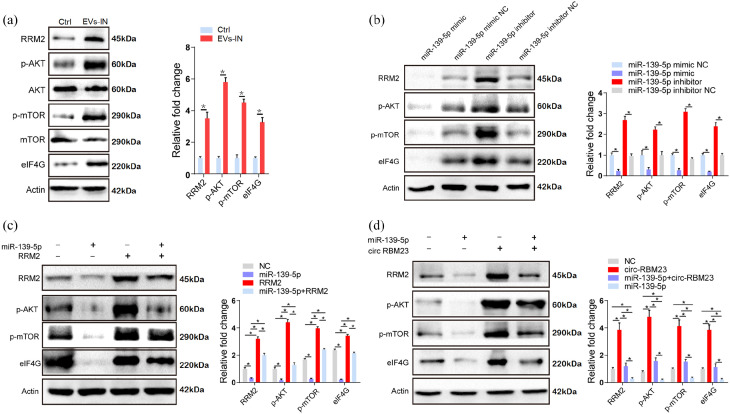
The administration of hPMSCs-EVs upregulated the expression of RRM2, p-AKT, p-mTOR, and eIF4G in L02 cells, indicating that hPMSCs-EVs might regulate cell proliferation via the AKT/mTOR pathway (Figure 5(a)). Further, we found that the miR-139-5p mimic downregulated RRM2 expression while the miR-139-5p inhibitor upregulated RRM2 expression. A similar expression pattern was observed in p-AKT, p-mTOR, and eIF4G (Figure 5(b)). Finally, the results indicated that miR-139-5p inhibits the expression of AKT, mTOR, and eIF4G, and that this inhibition is blocked by RRM2 overexpression in L02 cells (Figure 5(c)). circ-RBM23 upregulated the expression levels of RRM2, p-AKT, p-mTOR, and eIF4G; however, this upregulation was inhibited by miR-139-5p in L02 cells (Figure 5(d)). Binding sites for circ-RBM23, miR-139-5p, and RRM2 are summarized in Supplemental Table 4.
Figure 5.
circ-RBM23 regulates hepatocytes proliferation via the miR-139-5p/RRM2/AKT/mTOR pathway in vivo and in vitro. (a) Western blot analysis and mRNA expression levels of RRM2 and AKT/mTOR-associated protein in murine liver samples with or without hPMSCs-EVs treatment (n = 10). (b) mRNA and protein levels of AKT/mTOR-associated protein in L02 cells with miR-139-5p mimic or mimic NC (negative control) and miR-139-5p inhibitor or inhibitor NC (negative control). (c) mRNA and protein levels of RRM2 and AKT/mTOR-associated protein after transfection of miR-139-5p mimics or mimic NC in L02 cells with RRM2 overexpression or NC. (d) mRNA and protein levels of RRM2 and AKT/mTOR associated protein after transfection of miR-139-5p mimics or mimic NC in L02 cells with circ-RBM23 overexpression or NC. Western blots were quantified based on at least three replicates. Error bars represent the SEM. *p < 0.05.
Circ-RBM23-EVs regulate RRM2 via the AKT/mTOR pathway
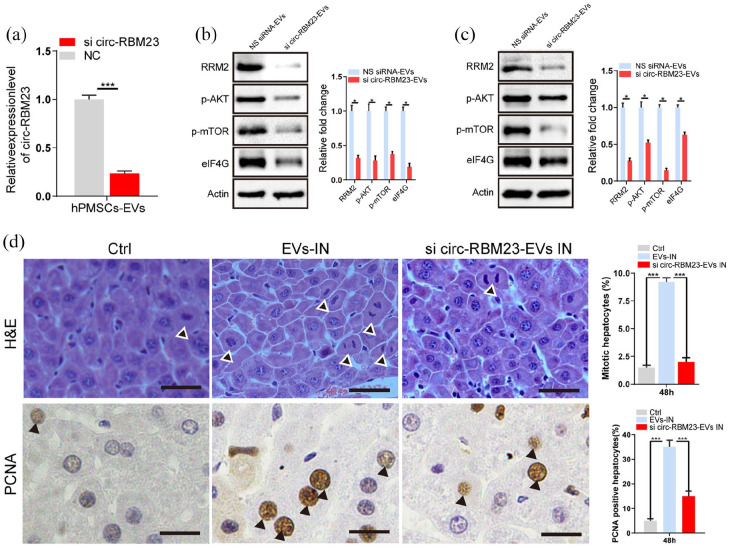
The EVs of stable si-circ-RBM23 cells showed low circ-RBM23 expression; thus, these cells were used to establish whether EVs circ-RBM23 regulates RRM2 in L02 cells (Figure 6(a)). After co-culturing si-circ-RBM23-EVs with L02 cells for 24 h, RT-qPCR analysis showed that si-circ-RBM23-EVs activate miR-139-5p expression (Supplemental Figure S1D), while WB analysis indicated that si-circ-RBM23-EVs inhibit the expression of RRM2, p-AKT, p-mTOR, and eIF4G (Figure 6(b)).
Figure 6.
Knockdown of hPMSCs-EVs circ-RBM23 inhibits hepatocyte proliferation. (a) Related expression level (RT-qPCR) of the circ-RBM23 in hPMSCs-EVs treated with circ-RBM23-siRNA for 72 h. (b) mRNA and protein levels of RRM2 and AKT/mTOR associated protein after co-culture with circ-RBM23-siRNA EVs or circ-RBM23-siRNA NS in L02 cells. (c) mRNA and protein levels of RRM2 and AKT/mTOR associated protein after PH with circ-RBM23-siRNA EVs or circ-RBM23-siRNA NS treatment in liver tissues. Western blots were quantified based on at least three replicates. (d) After 2 days post PH, liver tissue sections from hPMSCs-EVs, circ-RBM23-siRNA EVs or control treatment were performed H&E staining and PCNA staining, the number of mitotic hepatocytes and percentages of PCNA-positive cells were counted. Error bars represent the SEM. *p < 0.05, ***p < 0.001. NS siRNA: non-specific siRNA.
Si-circ-RBM23-EVs were injected into the tail veins of mice 24 h prior to 70% PH. RT-qPCR analysis showed si-circ-RBM23-EVs activated miR-139-5p expression (Supplemental Figure S1E) and WB analysis showed similar results, with downregulated RRM2, p-AKT, p-mTOR, and eIF4G expression observed in the si-circ-RBM23-EVs treated liver (Figure 6(c)). H&E staining revealed that the mitotic counts were significantly lower in the hepatocellular area of si-circ-RBM23-EVs treatment as compared to hPMSCs-EVs treatment. PCNA staining also showed a similar trend in si-circ-RBM23-EVs treated mice as compared to hPMSCs-EVs treated mice. Interestingly, no differences were observed in terms of liver mitosis and PCNA staining for si-circ-RBM23-EVs treated mice as compared to the control (Figure 6(d)).
Discussion
This study described a novel and significant function of hPMSCs-EVs. The results show that hPMSCs-EVs fuse directly with hepatocytes, promoting their proliferation both in vitro and in vivo. hPMSCs-EVs circ-RBM23 regulate hepatocyte proliferation by sponging miR-139-5p via RRM2/AKT/mTOR signaling. The results of this study improve our current understanding of hPMSCs-EVs and highlight the therapeutic potential of these vesicles against liver disease.
circRNA is a special type of non-coding RNA (ncRNA) with a stable structure and conserved sequence in different biological tissues and cells.16,17 hPMSCs-EVs contain circRNAs, of which circ-RBM23, circ-SWAP70, circ-MAPK9, circ-FKBP8, and circ-EPHB4 were more enriched in EVs. Of these, only circ-RBM23 was found to promote hepatocyte proliferation both in vitro and in vivo. hPMSCs-EVs with fewer circ-RBM23 were found to significantly decrease hepatocyte proliferation as compared to normal EVs. These results suggest that this type of circRNA could stimulate the growth of hepatic cells. Previous studies have indicated that circRNA plays a critical role in various liver diseases, including metabolic-associated fatty liver disease, hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC).26 Zhu et al.27 showed that exosomes derived from circ_0000623-modified adipose-derived MSCs can prevent liver fibrosis. Previous reports have described several functions for RBM23, including the regulation of HCC behaviors and autism neuronal conduction disorders. Han et al.28 revealed that RBM23 promotes the angiogenesis properties of HCC via the NF-κB signaling pathway. Thus, the function of RBM23 in normal hepatocytes and HCC cells may differ. However, no study has yet reported a link between circ-RBM23 and liver regeneration. The therapeutic approach of circ-RBM23 in liver disease therefore requires more thorough investigation. Here, we discovered that circ-RBM23 is potentially a key circRNA in hPMSCs-EVs, and that it regulates hepatocyte cell proliferation.
ceRNA is one of the mechanisms by which circRNAs regulation occurs.18 To further validate the relationship between circ-RBM23 and miR-139-5p and RRM2, we examined the protein expression of RRM2 with or without treatment with hPMSCs-EVs. The luciferase reporter assay indicated an interaction between miR-139-5p and circ-RBM23 in L02 cells. RIP assay showed that circ-RBM23 interacts with miR-139-5p. Bioinformatic analysis revealed that RRM2, which is targeted by miR-139-5p, regulates the development and promotes liver proliferation following PH. These findings suggested that circ-RBM23 could sponge miR-139-5p to inhibit its function in hepatocytes during the PH. Xiong et al.29 indicated that RRM2 activates the AKT pathway in renal cell carcinoma, and Jiao et al.30 found that RRM2 may regulate doxorubicin-induced cardiotoxicity through the AKT/mTOR signaling pathway. Huang et al.25 reported that AFAP1-AS1 upregulates RRM2 expression by sponging miR-139-5p in NSCLC cells. The comprehensive analysis of qRT-PCR confirmation and the subsequent luciferase reporter, RIP assay in this study revealed that circ-RBM23 enhances hepatocyte proliferation through the upregulation of RRM2 by inhibiting miR-139-5p expression. The WB results indicate that the action of circ-RBM23 in sponging miR-139-5p upregulates RRM2 through the AKT/mTOR signaling pathway.
Several recent studies have demonstrated that MSCs-EVs exhibit hepato-protective effect.31,32 Our results revealed that ALT and AST were significantly decreased after hPMSCs-EVs administration, indicating that hPMSCs-EVs ameliorate liver injuries in the early stages following PH. Liver function recovery is associated with hepatocyte proliferation, and MSCs-EVs have been reported to increase hepatocyte proliferation after liver injury via carbon tetrachloride.6,33 Similarly, we observed a proliferative effect of hPMSCs-EVs in mice following PH. These results indicate a possible treatment that can enhance liver regeneration following post-hepatectomy-associated liver failure.
MSCs-EVs carry complex cargo, including nucleic acids, proteins, and lipids.20 Ultracentrifugation and NTA assay were used for the isolation and characterization of hPMSCs-EVs, and the fluorescent dye PKH26 was utilized for EV labeling. PKH26 is readily incorporated into any lipid structure and has been used widely for labeling lipoproteins and proteins. Takov found that the ultracentrifugation of plasma leads to the co-isolation of EVs and lipoproteins owing to their physically associated structure, and that density gradient centrifugation cannot be used to separate EVs from HDLs (high density lipoproteins) because of their similar densities.34 Thus, irrespective of the method used, the complete separation of lipoproteins from EVs is unlikely to be achieved. After careful consideration, we selected circRNAs for further research because the circular structure is more stable than that of linear nucleic acid, which is likely to enhance the reliability of the results. Previous research indicated that EVs cargos can play important role in modulating the recipient cell behavior in liver disease. Ichinohe et al.35 indicated that miR-146a-5p is involved in the production of EVs by BM-MCs, may play a major role in accelerating liver regeneration. Similarly, our results showed that the downregulated hPMSCs-EVs circ-RBM23 suppresses hepatocyte proliferation. These findings suggested that hPMSCs-EVs promote hepatocyte proliferation, which is in line with the results of previous studies. The hPMSCs-EVs that are involved in regulating liver regeneration and the role of circ-RBM23 require further clinical investigation.
However, the other main functional substance in hPMSCs-EVs and their regulatory mechanisms are still ambiguous and have attracted more attention. In addition, the pathogenesis of liver regeneration involves many kinds of cells, such as Kupffer cells and hepatic stellate cells. The multi-omics bioinformatics analysis of hPMSCs-EVs in different participant cells and the exchange of information between cells need to be further explored.
Conclusion
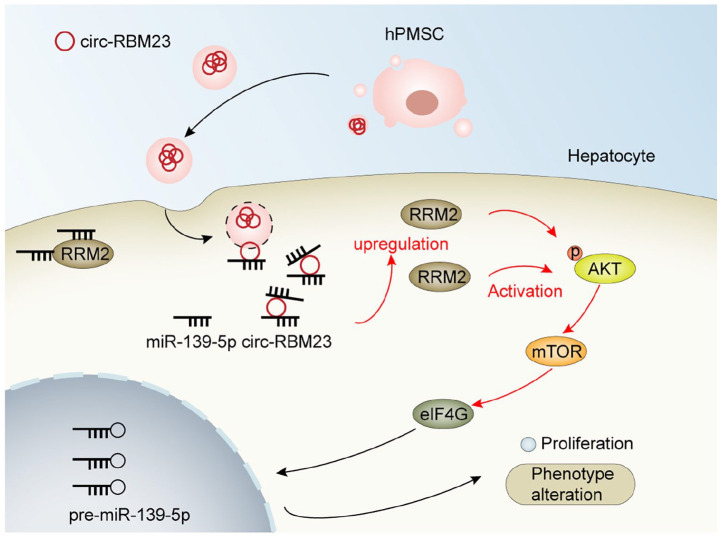
In summary, for the first time, the proliferative and protective impacts that hPMSCs-EVs have on hepatocytes following PH were described, and the hPMSCs-EVs circ-RBM23 serve as efficient miR-139-5p sponges and regulate liver regeneration of mouse after 70% PH via the RRM2/AKT/mTOR pathway (Figure 7). Our findings suggest a novel mechanism for liver regeneration and provides a potential therapeutic approach against liver disease following transplantation. This highlighted the importance of the ceRNA mechanism in pathogenesis and the cell-free treatment with hPMSCs-EVs as a possible therapeutic development for liver regeneration in the future.
Figure 7.
Schematic diagram of the mechanism by which hPMSCs-EVs circ-RBM23 leads to liver regeneration. Released hPMSCs contained circ-RBM23 hPMSCs-EVs, which were endocytosed and taken up by hepatocytes. The circ-RBM23 hPMSCs-EVs activate the miR-139-5p/RRM2/AKT/mTOR pathway to promote liver regeneration.
Supplemental Material
Supplemental material, sj-docx-1-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-jpg-2-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-rar-3-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-rar-4-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-tif-5-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Acknowledgments
We appreciate each participant providing assistance in this study.
Footnotes
Author contributions: All authors contributed to the study conception and design. The research was designed by Guolin He, Yi Gao, and Ting Li. Material preparation and data collection were performed by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, and Yizhou Zheng. Data analysis was performed by Shunjun Fu, Ting Li, Yang Li, Shao Li, Lin Zhou, Lujia Wang, and Lei Feng. The first draft of the manuscript was written by Guolin He and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by China Postdoctoral Science Foundation (Grant Number F121ZJ0216); the National Key R&D Program of China (2018YFC1106400; 2018YFA0108200); the National Natural Science Foundation of China (31972926); Guangdong Basic and Applied Basic Research Foundation (2020A1515111111); Medical Research Foundation of Guangdong Province (B2022086).
Ethics approval and consent to participate: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license Certificate of Conformity: SYXK (YUE) 2019-0215) granted by The institutional animal care and use committee center of Zhu Jiang Hospital, Southern Medical University, Guangzhou, China, in compliance with China national guidelines for the care and use of animals.
ORCID iDs: Ting Li  https://orcid.org/0000-0002-5341-2485
https://orcid.org/0000-0002-5341-2485
Guolin He  https://orcid.org/0000-0001-8032-1434
https://orcid.org/0000-0001-8032-1434
Supplemental material: Supplemental material for this article is available online.
References
- 1. Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 2006; 43: S45–S53. [DOI] [PubMed] [Google Scholar]
- 2. Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol 2013; 3: 485–513. [DOI] [PubMed] [Google Scholar]
- 3. Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology 2009; 137: 466–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guerin DJ, Kha CX, Tseng KA. From cell death to regeneration: rebuilding after injury. Front Cell Dev Biol 2021; 9: 655048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 2021; 18: 40–55. [DOI] [PubMed] [Google Scholar]
- 6. Jiang W, Tan Y, Cai M, et al. Human umbilical cord MSC-derived exosomes suppress the development of CCl4-induced liver injury through antioxidant effect. Stem Cells Int 2018; 2018: 6079642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu C, Wu Z, Li L. Mesenchymal stromal cells promote liver regeneration through regulation of immune cells. Int J Biol Sci 2020; 16: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yao Y, Xia Z, Cheng F, et al. Human placental mesenchymal stem cells ameliorate liver fibrosis in mice by upregulation of Caveolin1 in hepatic stellate cells. Stem Cell Res Ther 2021; 12: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abumaree MH, Abomaray FM, Alshehri NA, et al. Phenotypic and functional characterization of mesenchymal stem/multipotent stromal cells from decidua parietalis of human term placenta. Reprod Sci 2016; 23: 1193–1207. [DOI] [PubMed] [Google Scholar]
- 10. Liu H, Honmou O, Harada K, et al. Neuroprotection by PlGF gene-modified human mesenchymal stem cells after cerebral ischaemia. Brain 2006; 129: 2734–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J, Shehadah A, Pal A, et al. Neuroprotective effect of human placenta-derived cell treatment of stroke in rats. Cell Transplant 2013; 22: 871–879. [DOI] [PubMed] [Google Scholar]
- 12. Trounson A, McDonald C. Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell 2015; 17: 11–22. [DOI] [PubMed] [Google Scholar]
- 13. Bai L, Sun L, Chen W, et al. Evidence for the existence of CD34(+) angiogenic stem cells in human first-trimester decidua and their therapeutic for ischaemic heart disease. J Cell Mol Med 2020; 24: 11837–11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barzegar M, Wang Y, Eshaq RS, et al. Human placental mesenchymal stem cells improve stroke outcomes via extracellular vesicles-mediated preservation of cerebral blood flow. eBioMedicine 2021; 63: 103161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao H, Yang J, Yu J, et al. Therapeutic potential of transplanted placental mesenchymal stem cells in treating Chinese miniature pigs with acute liver failure. BMC Med 2012; 10: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019; 20: 675–691. [DOI] [PubMed] [Google Scholar]
- 17. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 2018; 187: 31–44. [DOI] [PubMed] [Google Scholar]
- 18. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo X, Xi L, Li L, et al. CircRNA-14723 promotes hepatocytes proliferation in rat liver regeneration by sponging rno-miR-16-5p. J Cell Physiol 2020; 235: 8176–8186. [DOI] [PubMed] [Google Scholar]
- 20. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 2016; 36: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018; 19: 213–228. [DOI] [PubMed] [Google Scholar]
- 22. Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. New Engl J Med 2018; 379: 958–966. [DOI] [PubMed] [Google Scholar]
- 23. Yi X, Chen F, Liu F, et al. Comparative separation methods and biological characteristics of human placental and umbilical cord mesenchymal stem cells in serum-free culture conditions. Stem Cell Res Ther 2020; 11: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 2008; 3: 1167–1170. [DOI] [PubMed] [Google Scholar]
- 25. Huang N, Guo W, Ren K, et al. LncRNA AFAP1-AS1 supresses miR-139-5p and promotes cell proliferation and chemotherapy resistance of non-small cell lung cancer by competitively upregulating RRM2. Front Oncol 2019; 9: 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fu LY, Wang SW, Hu MY, et al. Circular RNAs in liver diseases: mechanisms and therapeutic targets. Life Sci 2021; 264: 118707. [DOI] [PubMed] [Google Scholar]
- 27. Zhu M, Liu X, Li W, et al. Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum Exp Toxicol 2020; 39: 1619–1627. [DOI] [PubMed] [Google Scholar]
- 28. Han H, Lin T, Fang Z, et al. RBM23 drives hepatocellular carcinoma by activating NF-kappaB signaling pathway. BioMed Res Int 2021; 2021: 6697476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiong W, Zhang B, Yu H, et al. RRM2 regulates sensitivity to sunitinib and PD-1 blockade in renal cancer by stabilizing ANXA1 and activating the AKT pathway. Adv Sci (Weinh) 2021; 8: e2100881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiao Y, Li Y, Zhang J, et al. RRM2 alleviates doxorubicin-induced cardiotoxicity through the AKT/mTOR signaling pathway. Biomolecules 2022; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao M, Liu S, Wang C, et al. Mesenchymal stem cell-derived extracellular vesicles attenuate mitochondrial damage and inflammation by stabilizing mitochondrial DNA. ACS Nano 2021; 15: 1519–1538. [DOI] [PubMed] [Google Scholar]
- 32. Zheng J, Lu T, Zhou C, et al. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells protect liver ischemia/reperfusion injury by reducing CD154 expression on CD4+ T cells via CCT2. Adv Sci (Weinh) 2020; 7: 1903746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tan CY, Lai RC, Wong W, et al. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res Ther 2014; 5: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takov K, Teng IJ, Mayr M. Isolation of circulating extracellular vesicles by high-performance size-exclusion chromatography. Methods Mol Biol (Clifton, NJ) 2022; 2504: 31–40. [DOI] [PubMed] [Google Scholar]
- 35. Ichinohe N, Ishii M, Tanimizu N, et al. Extracellular vesicles containing miR-146a-5p secreted by bone marrow mesenchymal cells activate hepatocytic progenitors in regenerating rat livers. Stem Cell Res Ther 2021; 12: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-jpg-2-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-rar-3-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-rar-4-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering
Supplemental material, sj-tif-5-tej-10.1177_20417314221132093 for A new cell-free therapeutic strategy for liver regeneration: Human placental mesenchymal stem cell-derived extracellular vesicles by Ting Li, Yu Fu, Zeyi Guo, Honglei Zhu, Hangyu Liao, Xiaoge Niu, Lin Zhou, Shunjun Fu, Yang Li, Shao Li, Lujia Wang, Yizhou Zheng, Lei Feng, Yi Gao and Guolin He in Journal of Tissue Engineering