Abstract
Chicken blue-greenish coloration (BGC) was known as a classic Mendel trait caused by a retrovirus (EAV-HP) insertion in the SLCO1B3 gene. Lueyang black-boned chicken (LBC) BGC is light and varies continuously, implying that LBC BGC may be controlled by a new molecular mechanism. The aim of this study was to provide an insight into the molecular basis of LBC BGC. The EAV-HP was detected in the BGC (n = 105) and non-BGC LBC (n = 474) using a PCR-based method. The association of SLCO1B3 expression in shell glands and sequence variants in a 1.6-kb region upstream from the transcription start site of SLCO1B3 with eggshell color and biliverdin (pigment for BGC) concentration was studied. Promoter activities of haplotypes in the 1.6-kb region were analyzed by luciferase reporter assay. This study did not found the EAV-HP in BGC and Non-BGC LBC, but detected a strong positive correlation between levels of SLCO1B3 expression in shell glands and biliverdin concentrations. A total of 31 SNP were found in the 1.6-kb region. Twenty-two of 31 SNP formed 42 types of haplotypes in the re-sequenced samples (n = 94). Haplotype 4 was present in higher frequency in the BGC (52%) than Non-BGC (3%). Haplotype 13 was significantly associated with Non-BGC (Non-BGC vs. BGC = 26% vs. 6%). In line with the above associations, Haplotype 4 showed higher (P < 0.05) levels of SLCO1B3 expression in shell glands, biliverdin concentration, and promoter activity than Haplotype 13. This study confirms that LBC BGC is not caused by the EAV-HP, but remains to be associated with the change of SLCO1B3 expression. Haplotype 4 accounts to some extents for the molecular basis of LBC BGC. The new molecular mechanism supports LBC BGC independently evolved.
Key words: Lueyang black-boned chicken, blue-greenish egg coloration, biliverdin, SLCO1B3, EAV-HP
INTRODUCTION
Eggshell color is an important component of avian reproductive system (Cassey et al., 2010). For open-nesting birds, it acts as a camouflage or an aposematism that dramatically reduces predation risk, and as a recognizing marker that offers protection against intraspecific and interspecific brood parasitism (Reynold et al., 2009). In addition to the visual functions, some other actions such antimicrobial effect (Ishikawa et al., 2010), filtering solar radiation (Lahti, 2008) and reinforcing eggshell (Gosler et al., 2005), were proposed to support the biological significance of eggshell color. Blue-greenish coloration (BGC) is an ancient and common eggshell color. It appears not only in nonavian dinosaurs (Wiemann et al., 2017), but in some modern avian species (Siefferman et al., 2006; Wang et al., 2013; Chen et al., 2020; Hamchand et al., 2020).
BGC is mainly caused by biliverdin IX, a product from oxidative degradation of heme (Kennedy and Vevers, 1976; Ryter and Tyrrel, 2000). With the change of biliverdin contents and the involvement of protoporphyrin IX, another eggshell pigment responsible for red-brown color (Kennedy and Vevers, 1976), BGC eggs show a wide spectrum of chroma from blue-greenish eggs laid by chicken and duck (Wang et al., 2013; Chen et al., 2020), deep blue eggs laid by eastern bluebird (Siefferman et al., 2006), olive-green eggs laid by elegant crested tinamou to blackish-green eggs laid by emu (Hamchand et al., 2020). BGC was hypothesized to signal female quality to male which allocates more parental care for their offspring depending on antioxidant properties of biliverdin (Stocker et al., 1987; Mereno and Osorno, 2003). Although the signaling function is negligible in poultry, BGC eggs are favorable because consumers believe that BGC eggs are more organic and nutritious than white and brown eggs laid by commercial hens. BGC is relevant to yolk ratio and contents of cholesterol, carotenoid and protein in the yolk (Somes et al., 1977; Wang et al., 2009; Butler and MeGraw, 2013). However, a combination of advantages (higher yolk/albumen ratio) and disadvantages (higher cholesterol contents in yolk and lower total protein contents) leaves BGC eggs as a “health eggs” inconclusive (Somes et al., 1977).
Chickens and ducks are two domestic fowls laying BGC eggs. It was reported that chicken and duck BGC belong to a classic Mendelian trait controlled by a dominant mutation (Punnett, 1933; Lai and Zhang, 1991). BGC of Dongxiang chickens was caused by a retrovirus (EAV-HP) insertion in 5’ flanking region of SLCO1B3 gene (Wang et al., 2013). The causative gene controlling duck BGC was ABCG2 (Chen et al., 2020). Lueyang black-boned chicken (LBC) is one of indigenous breeds laying BGC eggs in China. But differing from uniform BGC seen in the Dongxiang chicken and duck, LBC BGC varies in degree from light green to deep olive green, which seems to follow the characteristics of continuous variation. No evidence is available to support that there is a gene flow between LBC and other breeds laying blue eggs. The feature implies that LBC BGC may be controlled by a different molecular mechanism and independently evolve.
The aims of this study are to explore the molecular mechanisms controlling LBC BGC by detecting the EAV-HP and analyzing the association of SLCO1B3 expression in shell glands and sequence variants in a 1.6-kb region upstream of SLCO1B3 with LBC BGC and eggshell biliverdin concentration.
MATERIALS AND METHODS
Ethics Statement
Animal care, slaughter and experimental procedures were approved by Institutional Animal Care and Institutional Ethic Committee of Northwest A&F University (DK-2021020).
Management and Raising of LBC
LBC is a native breed from Lueyang city, Shannxi Province of China. Because of absence of selection for egg production, annual total number of eggs laid by LBC ranges from 150 to 230, peak production rate is 61%, and age at the first egg is 25 wk old (Wang et al., 2016). Majority of LBC lay light-brown and brown eggs. Approximate 10 % of LBC lay light-green to olive-green eggs. LBC used in this study were raised at the Shaanxi Longjia Agricultural Technology Development Co. from Dec. 2019 to Dec. 2020. These chickens were raised in battery cages with width × depth = 40 cm × 50 cm and 8-10 chicks per cage until 10 wk of age. Afterward they were transferred to a layer house and raised in individual cages. Temperature was 32 ± 1°C at the first week of brooding period, and gradually decreased every 2 to 3 d by 1°C. Artificial light was provided at a schedule of 18 h light and 6 h dark from 0 to 6 wk of age, 12 h light and 12 h dark from 7 to 20 wk of age, and 16 h light and 8 h dark at the egg-laying period. The chicks were fed a starter diet (ME, 3,050 kcal/kg; CP, 23% (wt/wt)) from 0 to 6 wk, a grower diet (ME, 3,210 kcal/kg; CP, 18%) from 7 to 18 wk, and a layer diet (ME, 3,260 kcal/kg; CP, 18%) at the egg-laying period. Each cage contained one drinking nipple with a cup underneath. All birds can eat and drink ad libitum.
Measurement of LBC Eggshell Color
LBC hens (n = 846) at the 31 wk of age were selected from the 4th generation of egg-laying LBC breeding population. Three consecutive eggs were collected from each hen. Eggshell color at the blunt end was measured with the L*a*b color space where L represents lightness (value between 0 = black and 100 = white), a is the red-green scale (<0 = green, >0 = red), and b is the blue-yellow scale (<0 = blue, >0 = yellow). L, a, and b values were measured with a MiniScan EZ 4000 portable spectrophotometer (Hunter Associates Laboratory, Inc, Reston, VA). The illuminant D65 and the observer angle 10° were used during measuring.
Detection of EAV-HP in the SLCO1B3
A PCR-based method was developed to detect the EAV-HP in the LBC with reference to the published EAV-HP and genomic sequences at two sides of insertion site (Wang et al., 2013). Blood was collected from BGC (n = 105) and Non-BGC (n = 474) samples using wing venipuncture, and anticoagulated with acid citrate dextrose (ACD) solution (blood: ACD = 3:1). The ACD solution contains citric acid 4.8 g/L, sodium citrate 13.2 g/L, and dextrose 14.7 g/L. Genomic DNA was extracted using a FlexGen Blood DNA Kit (CWBIO, Beijing, China) according to the manufacturer's instructions. PCR mixture consists of 1 uL of DNA (50 ng), 1 uL of forward primer (10 μmol/L), 1 uL of reverse primer (10 μmol/L), 10 uL of Dye-added 2 × Es TaqMaster Mix (CWBIO) and 7 uL of ddH2O. PCR was conducted in a Bio-Rad T100 Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA), applying the thermal program: 95°C for 5 min, 33 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 60 s, followed by 72°C for 5 min. Four primers which were named test-nor-up, test-nor-down, test-eav-up, and test-eav-down were used to detect the EAV-HP. Primer sequences were 5’-TGTTTGTTGAGCACCCACTT-3’ for test-nor-up, 5’- GTTAATCCAATCGCCTTGTT-3’ for test-nor-down, 5’- CTGTAAGAAGGCAGAGGGTT-3’ for test-eav-up and 5’-AAGATACGGGTGGAAGATGA-3’ for test-eav-down. Test-nor-up, test-eav-up, and test-nor-down were located at 2 sides of the insertion site, and test-eav-down was located within the EAV-HP. If there is the EAV-HP in 5’ flanking region of SLCO1B3 gene, PCR will produce a 1,086-bp product with test-eav-up and test-eav-down. Otherwise, PCR will fail to amplify with this pair of primers, but can produce an 861-bp product with test-nor-up and test-nor-down. Three Dongxiang chickens that carried the EAV-HP were used as positive control to validate the PCR-based diagnosis. All samples were detected twice (1st: test-eav-up and test-eav-down, and 2nd: test-nor-up and test-nor-down) to eliminate diagnostic errors from PCR failure. PCR products were separated on a 1.5% (w/v) agarose gel containing 0.1 μg/mL ethidium bromide at 110 v for 30 min. Bands were visualized using a Gel Doc XR+ Imaging System (Bio-Rad Laboratories, Inc.).
Resequencing of a 1.6-kb Region Upstream of SLCO1B3 and Reconstruction of Haplotypes
A 1.6-kb region (−1,639/+18) upstream from the transcription start site of SLCO1B3 was re-sequenced using LBC (n = 94) with eggshell color records. The transcription start site (Chr1:65167287) was defined with reference to the chicken reference genome (Galgal 6.0) in the UCSC database. PCR was conducted using the abovementioned reaction mixture and thermal cycle protocol. The primer sequences were 5’- GAAAGTGGCACAGTTAGTGG-3’ and 5’-TTTTAATCTTGTCTCCTCCC-3’. After the specificity and the amount of PCR products were assessed by 1.5% (w/v) agarose gel electrophoresis, PCR products were bi-directionally sequenced using the Sanger sequencing. SNP calling was conducted by multiple sequence alignment using the Chromaspro v.1.33. Missing genotypes were imputed using a Bayesian approach in the BIMBAM software (Scheet and Stephens, 2006). Haplotypes in the 1.6-kb region were re-constructed using the PHASE v2.1.1 software with default parameters (Stephens et al., 2001).
Detection of SLCO1B3 Expression in Shell Glands
LBC (n = 24) with regular oviposition intervals at the 40 wk of age were used to expression analysis. These samples consisted of hens laying BGC (n = 8), light brown (n = 8), and brown (n = 8) eggs. Oviposition times of each hens were recorded. Hens were euthanized with CO2 3–5 h prior to oviposition to ensure tissue collection at the peak of pigment deposition. Shell glands (uterine part of oviduct) were opened at the ventral intermediate part. An approximately 500 mg of tissue was immersed into the RNAstore overnight at 4°C and then stored at −80°C. Total RNA was extracted from 100 mg of tissue using the TRNzol Universal (TIANGEN, Beijing, China). Concentrations of RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The integrities of RNA were evaluated by separating 100 ng of RNA on a 1.0% (w/v) agarose gel. One μg of RNA was reverse-transcribed using SumOnetube RT Mixture Ⅲ (SUMMER BIOTECH Inc., Beijing, China) according to the manufacturer's protocol. Real-time PCR mixture consisted of 1 μL of cDNA template, 10 μL of 2 × FastHotstart SYBR QPCR Mixture (SUMMER BIOTECH Inc., Beijing, China), 0.4 μL of forward primer (10 μmol/L), 0.4 μL of reverse primer (10 μmol/L) and 8.2 μL of ddH2O. GAPDH was used as the endogenous reference. Primer sequences were 5’-TTTTGGGGCACTGATTGA-3’ and 5´- AAGCCTGGAGTTCTTATT-3´ for SLCO1B3 and 5´-ATACACAGAGGACCAGGTTG-3´ and 5´-AAACTCATTGTCATACCAGG-3´ for GAPDH. Quantitative PCR was run in a LightCycler 96 System (Roche, Basel, Switzerland) applying the following conditions: 95°C for 2 min, followed by 40 cycles of amplification (95°C for 10 s, 60°C for 10 s and 60°C for 30 s). Melting curve analysis was performed by first heating to 95°C for 1 min, cooling to 40°C for 1 min, heating to 65°C, and then melting with continuous acquisition (15 readings/°C) of fluorescence signal until 97°C. Amplification for each sample was replicated 3 times. The ΔCq (CqGAPDH-CqSLCO1B3) was used to assess levels of SLCO1B3 expression in shell glands. The larger the ΔCq value is, the higher levels of SLCO1B3 expression is.
Measurement of Eggshell Biliverdin Concentration of LBC Eggs
Three eggs were collected from each of hens (n = 24) which were also used in the expression experiment. Eggshell biliverdin concentration was measured with reference to the reported method (Wang et al., 2009). Eggs were rinsed with deionized water, and dried overnight. Eggshell membrane was removed. Eggshell (0.25 g) was placed into 4 mL of solvent (methanol: concentrated HCl = 2:1), and vortex-shaked overnight in darkness until dissolved completely. The eggshell solution was centrifuged at 3,000 rpm for 45 min. Absorbance of the supernatant solution was measured with a Synergy H1 Hybrid Multi-Mode Reader (BioTek Instruments, Winoosk, VT) at wavelength of 670 nm. The standard solution of biliverdin was prepared by dissolving 4 mg of biliverdin hydrochloride (Sigma-Aldrich, St. Louis, MI) into 4 mL of solvent (methanol: concentrated HCl = 2:1). Concentration (1 mg/mL) of the standard solution decreased to 0.043 mg/mL by adding 87 μL of standard solution into 1,913 μL of solvent. A series of 2-fold dilutions that range from 1/2 to 1/256 were made by mixing 1/2 of the previous dilution and 1/2 of solvent. Biliverdin concentrations were calculated according to regression relationship between absorbance values and concentrations of serial dilutions, and expressed as μg biliverdin/g eggshell.
Conservation Analysis of Sequences in the 1.6-kb Region Upstream of SLCO1B3
Sequence conservation in the 1.6-kb region upstream of SLCO1B3 was analyzed by alignment of genome sequences of 52 birds and chicken using the Multiz tool in the UCSC database (Blanchette et al., 2004). Evolutionary conservation was quantitatively measured using the PhyloP program in the UCSC database (Siepel and Haussler, 2005). The PhyloP program computes p-values of conservation for each base under a null hypothesis of neutral evolution. The PhyloP score is defined as log p-values. Bases predicted to be conserved are assigned positive scores, while bases predicted to be fast-evolving are assigned negative scores.
Prediction of Transcription Factor Motifs
Putative motifs of CCAAT-enhancer binding protein (CEBPβ), TATA-binding protein (TBP) and 2 hepatocyte nuclear factors (HNF1α and HNF3β) in the re-sequenced region were predicted using the JASPAR database with relative profile score threshold set as 80% (Castro-Mondragon et al., 2021). There are 3 entries (MA0466.1, MA0466.2 and MA0466.3) for CEBPβ, two (MA0108.1 and MA0108.2) for TBP, two (MA0046.1 and MA0046.2) for HNF1α and three (MA0047.1, MA0047.2, and MA0047.3) for HNF3β (Foxa2) deposited in the JASPAR. All of 11 entries were selected to predict effect of SNP in the 1.6-kb region on these candidate transcription factors (TF) motifs.
Construction of Reporter Gene Plasmids
The 1.6-Kb region was PCR-amplified using genomic DNA from Haplotype 4 and Haplotype 13 homozygotes, primers and Dye-added 2 × Es TaqMaster Mix (CWBIO) in a 50 μL of reaction mixture. The forward and reverse primer sequences were 5’-GGGGGTACCGAAAGTGGCACAGTTAGTGG-3’ and 5’- GTTCACTCGAGTTTTAATCTTGTCTCCTCCC-3’. The forward primers contained an internal KpnI restriction site (underlined letters), and the reverse primers contained an internal XhoI site (underlined letters). PCR products were purified using Universal DNA Purification Kit (TIANGEN) according to the manufacture's instructions. Purified PCR products and pNL1.1 plasmid were digested with KpnI and XhoI (New England BioLabs Inc., MA) for 1 h at 37°C. Digested products were purified from agarose gels using Universal DNA Purification Kit (TIANGEN) according to the manufacture's instructions. Target fragments were ligated into multiple cloning site of Nanoluc reporter gene of pNL1.1 plasmid (Promega, Madison, WI) using DNA Ligation Kit (Takara, Dalian, China), yielding Haplotype 4 and Haplotype 13-containing plasmid constructions. These plasmids were transformed into DH5α competent cells (Weidi Biotechnology Co., Shanghai, China) by heat shock according to the manufacturer's instructions. The transformants were incubated overnight at 37°C on LB agar plates containing 100 μg/mL ampicillin (Sangon Biotech, Xi'an, China). Plasmid DNA was extracted from bacterial cultures from single colony using GoldHi EndoFree Plasmid Midi Kit (CWBIO) according to the manufacture's instructions. Concentrations of plasmid DNA were analyzed by 0.8% (w/v) agarose gel electrophoresis and measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Sequences of plasmid constructions were verified by the Sanger sequencing.
Cell Culture, Transfection, and Dual-Luciferase Reporter Assay
The chicken DF-1 embryonic fibroblast cell line, gifted by Dr. Y. N. Sun at the Northwest A&F University, was used for transient transfection of reporter plasmids. DF-1 cells were seeded in a 12-well culture plate at 3 × 104 cells/well and cultured in high-glucose DMEM (Solarbio, Beijing, China) supplemented with 1% of chicken serum (Solarbio), 10% of fetal bovine serum (EVERY GREEN, Zhejiang, China) and 1% of L-Glutamine-Penicillin-Streptomycin (Sigma-Aldrich). When cell growth reaches about 80% confluency, target and pGL3-control (Promega) plasmids were co-transfected into DF-1. One hundred μL of DNA/liposome mix was prepared for each well by diluting pNL1.1 constructs (1 μg) and pGL3-control plasmid (100 ng), 3 μL of Lipofectamine 2000 into DMEM without phenol red (Meilun, Dalian, China). After gentle mixed and incubated for 5 min at room temperature, DNA/liposome mix was added into culture medium.
Dual-Luciferase reporter assays were performed 24 h after transfection using a Nano&Firefly-Glo Luciferase Reporter Assay Kit (Meilun) according to the manufacture's instructions. Luminescence intensity was detected using an Infinite M200 PRO Multimode Microplate Reader (Tecan Austria GmbH, Grödig, Austria). Empty pNL1.1 plasmid served as negative control. The pGL3 plasmid was used as the constitutively expressed reporter. Luminescence intensity of the pNL1.1 construction was divided by that of the co-transfected pGL3-control plasmid to normalize experimental variations from transfection efficiency, cell number and cell viability etc. Luminescence intensities of each sample were detected 8 times.
Statistical Analysis
Extreme data that exceeded inner fence (low quartile − 1.5 × Interquartile range, upper quartile + 1.5 × Interquartile range) was removed before statistical analysis. Difference of means of L, a and b values between BGC and Non-BGC was tested by the two-tailed t test. Correlation of levels of SLCO1B3 expression in shell glands and biliverdin concentrations was calculated using the Hmisc package in R. Differences in SLCO1B3 expression, biliverdin concentration and L, a, b values between haplotypes were analyzed using one-way ANOVA. Multiple comparisons were done using the Duncan's multiple range test. The significance level was set at 0.05. Statistical analysis was conducted using the SAS University Edition.
RESULTS
Quantitative Assessment of LBC Eggshell Color
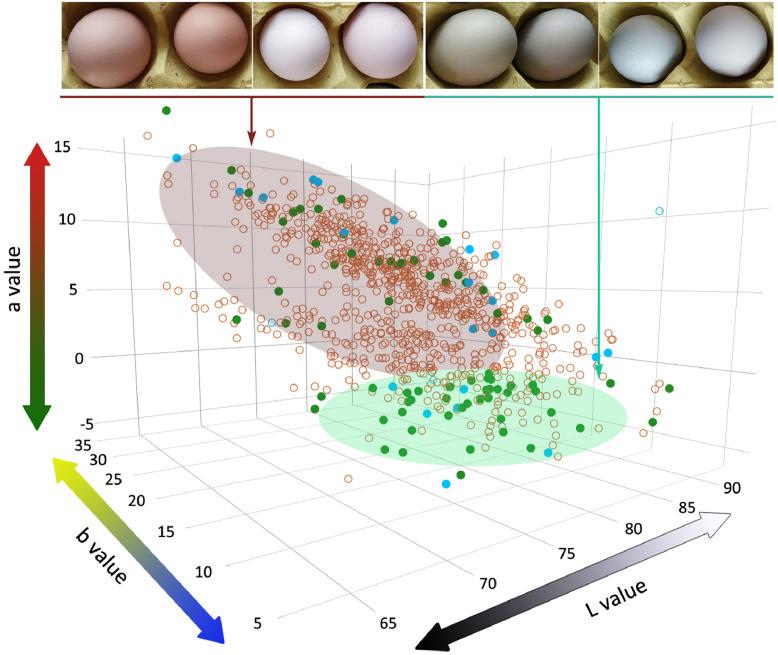
This study quantitatively assessed the change of LBC eggshell color (n = 846) using the L*a*b color space. L values of these samples varied from 63.0 to 92.5 with mean ± SD of 76.1 ± 4.4, a values ranged from −5.6 to 15.5 with mean ± SD of 4.4 ± 3.8, and b values ranged from 4.5 to 34.5 with mean ± SD of 15.8 ± 3.8 (Figure 1). This study selected 44 BGC and 50 Non-BGC samples based on subjective perception to assess the sensitivity of the L*a*b color space to color difference between BGC and Non-BGC. The 94 samples were also used in the following re-sequencing experiment. The L, a and b values had significant difference between BGC and Non-BGC (Table 1). The a value that had the largest between-group difference and coefficient of variation was the most sensitive index in response to the change of eggshell color among the L*a*b color space (Table 1).
Figure 1.
Distribution of L, a, and b values of Lueyang black-boned chicken eggs. Eggshell color of Lueyang black-boned chickens (n = 846) was quantitatively assessed by the L*a*b color space. Each point or open circle represents a sample used in this color-measured experiment. Green points indicate samples used in the re-sequencing experiment. Blue points indicate samples used in the re-sequencing and expression experiments. Blue open circles indicate samples that were only used in the expression experiments. Green transparent oval highlights color distribution of BGC eggs, and brown transparent oval for Non-BGC eggs. Abbreviation: BGC, blue-greenish coloration.
Table 1.
Comparison of L, a and b values between BGC and non-BGC samples.
| Egg color | N | L |
a |
B |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | C.V.2 | P value | Mean ± SD | C.V. | P value | Mean ± SD | C.V. | P value | ||
| BGC1 | 45 | 78.0 ± 4.2 | 5.4% | 0.0203 | −1.7 ± 1.5 | 88.2% | <0.0001 | 12.2 ± 2.6 | 21.3% | <0.0001 |
| Non-BGC1 | 50 | 75.5 ± 6.0 | 7.9% | 6.9 ± 3.4 | 49.3% | 16.8 ± 4.9 | 29.2% | |||
BGC, blue-greenish coloration, non-BGC, non-blue-greenish coloration.
C.V., coefficient of variation.
Detecting the EAV-HP Insertion and the Association of SLCO1B3 Expression With BGC in the LBC
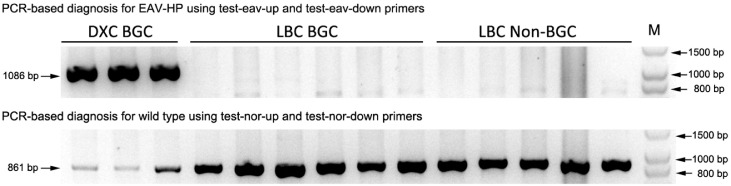
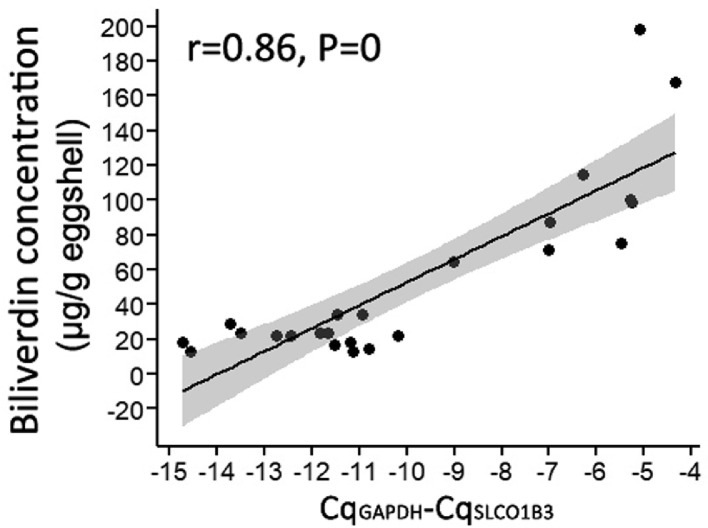
The EAV-HP insertion in the SLCO1B3 was recognized as the causative mutation for BGC in the DXC (Wang et al., 2013). We amplified a 1086-bp product using the EAV-HP diagnosis primers in the DXC, but failed to obtain the product not only in the Non-BGC samples, but in the BGC LBC (Figure 2). However, an 861-bp product which was derived from wild-type genomic sequence was successfully amplified in all LBC samples (Figure 2). These results confirmed there is no the EAV-HP in the homologous sequence of SLCO1B3 in the LBC. But levels of SLCO1B3 expression in shell glands of LBC showed a positive correlation with eggshell biliverdin concentration (Figure 3).
Figure 2.
A representing result of agarose gel electrophoresis of PCR amplified products using EAV-HP and wild-type diagnosis primer sets. The upper panel shows electrophoresis results of EAV-HP diagnosis products and the lower is the results of wild-type genome sequence in the SLCO1B3. Three DXC known to carry the EAV-HP insertion in the SLCO1B3 served as positive control in the EAV-HP diagnosis. PCR produced a 1086-bp amplicon in the DXC, but not for LBC. Only an 861-bp product was successfully amplified from wild-type genomic sequence in the LBC. Abbreviations: BGC, blue-greenish coloration; DXC, Dongxiang chicken; LBC, Lueyang black-boned chicken.
Figure 3.
Correlation of levels of SLCO1B3 expression with eggshell biliverdin concentration. Each point represents a sample. The black line is regression line. The gray band is 95% confidence band for the regression line. Numbers at the top are the correlation coefficient (r) and the P values of significance tests for r.
Association of Sequence Variants in the 1.6-kb Region Upstream of SLCO1B3 With LBC BGC
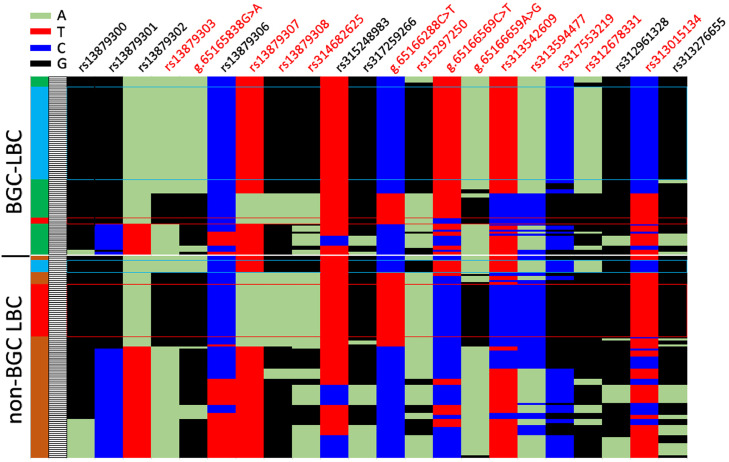
This study re-sequenced a1.6-kb region upstream of SLCO1B3. A total of 31 SNP were found in this region. There were no structural variants in this region. After excluding SNP with minor allele frequency < 5 % or Hardy-Weinberg Equilibrium P-value < 0.001, this study re-constructed haplotypes in the re-sequenced region using 22 SNP. A total of 42 types of haplotypes were found in the re-sequenced samples (Figure 4; Supplementary Table 1). Haplotype 4 that accounted for 52% (46/88) of haplotypes found in BGC samples (n = 44) was the predominant BGC-associated haplotype (Figure 4). In contrast, haplotype 13 that was present in higher frequency (26/100 = 26%) in Non-BGC (n = 50) than BGC (3/88 = 3.4%) was significantly associated with Non-BGC (Figure 4).
Figure 4.
Distribution of haplotypes in the 1.6-kb region upstream of SLCO1B3. Twenty-two SNP were used to construct haplotypes in the 1.6-kb region upstream of SLCO1B3. A total of 42 types of haplotypes were found in re-sequenced samples (n = 94). This figure shows distribution of 184 haplotypes. Each row represents one haplotype which was sorted according to haplotype type, and each column represents a SNP. The 94 samples were classified into BGC (n = 44) and non-BGC (n = 50) based on the subjective perception of eggshell color. Blue boxes indicate Haplotype 4 which is a predominant haplotype in BGC LBC, and red boxes indicate the haplotype 13 which is present in high frequency in the Non-BGC LBC. SNP shown in red has difference alleles between haplotype 4 and haplotype 13. Abbreviations: BGC, blue-greenish coloration; LBC, Lueyang black-boned chicken.
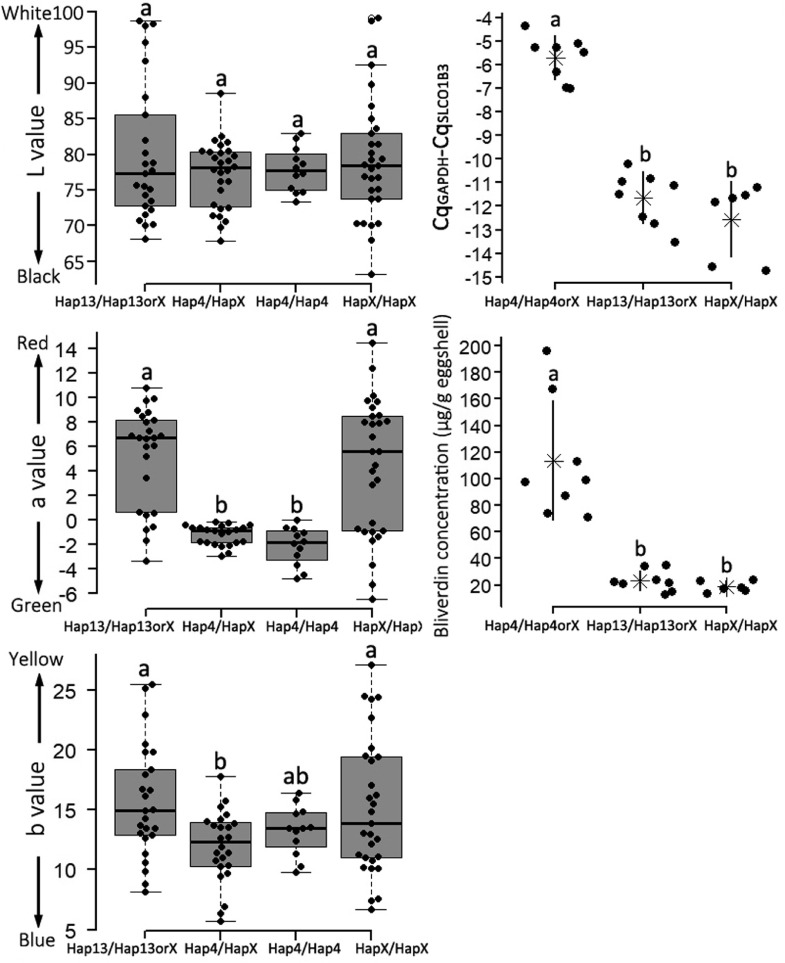
Comparisons of Eggshell Color, Levels of SLCO1B3 Expression, and Biliverdin Concentration Between Haplotypes
Haplotype 4 and 13 represented 2 promising candidate haplotypes that may affect SLCO1B3 expression in shell glands and LBC BGC. This study compared eggshell color, levels of SLCO1B3 expression, and eggshell biliverdin concentrations between haplotypes. There were 12 homozygotes (Hap4/Hap4) and 28 heterozygotes (Hap4/HapX) of haplotype 4, 3 homozygotes and 22 heterozygotes of haplotype 13, and 29 other genotypes (HapX/HapX) among the re-sequenced samples (Supplementary Table 1). The data from homozygotes and heterozygotes of haplotype 4 and 13 was pooled into Hap4/Hap4orX and Hap13/Hap13orX groups due to small sample size of homozygotes. The L values had no significant difference between haplotypes (Figure 5). Hap4/Hap4 and Hap4/HapX showed significantly lower a and b values than Hap13/Hap13orX and HapX/HapX (Figure 5). There was no significant difference between Hap4/Hap4 and Hap4/HapX (Figure 5), implying that haplotype 4 had no additive effect on BGC. In line with the association of haplotype 4 with BGC, Hap4/Hap4orX showed higher levels of SLCO1B3 expression and biliverdin concentration than Hap13/Hap13orX and HapX/HapX (Figure 5). There was no significant difference between Hap13/Hap13orX and HapX/HapX (Figure 5).
Figure 5.
Comparison of egg color, levels of SLCO1B3 expression, and eggshell biliverdin concentration between haplotypes. HapX represents any haplotypes other than Haplotype 4 and Haplotype 13. Hap4/Hap4 and Hap4/HapX represent homozygotes and heterozygotes of Haplotype 4, respectively. Hap4/Hap4orX represents individuals that carry one copy of Haplotype 4 at least. So is Hap13/Hap13orX. Lower letters on each column indicates the results of multiple comparison between haplotypes. The same letters represent nonsignificant (P > 0.05) difference between two groups, and different letters for significant (P < 0.05) difference. Asterisks and short lines at two sides of asterisk represent mean ± SD of data from each group.
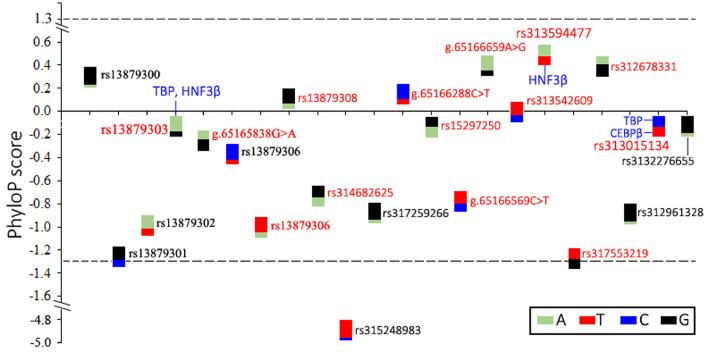
Conservation Analysis of Sequence Variants in the 1.6-kb Region Upstream of SLCO1B3
Cis-regulatory elements like coding sequence are conserved across evolution (Siepel et al., 2005). To provide an insight into cis-regulatory variants affecting SLCO1B3 expression in shell glands, this study analyzed conservation of 22 SNP loci by alignment of 52 birds and chicken genome sequences. The PhyloP scores of almost all SNP did not reach statistical significance except rs315248983 which belongs to a fast-evolving locus (Figure 6). Nevertheless, rs313594477 with the highest PhyloP score showed high conservation among birds (Figure 6). The T allele of rs313594477 was completely conserved among 51 birds except collared flycatcher (Ficedula albicollis) which has unalignable bases at this site (Supplementary Figure 1). The A allele of rs313594477 was uniquely present in chicken genome sequence (Supplementary Figure 1). Rs13879300, g.65166659A>G and rs312678331 also showed relatively high conservation. The G of rs13879300, the A of g.65166659A>G and the G allele of rs312678331 were found in 94 % (34/36), 94 % (46/49), and 96 % (49/51) of bird genomes regardless of bird species with genomic gap at the homogenous locus (Supplementary Figure 1).
Figure 6.
Conservation analysis results of 22 SNP loci located in upstream 1.6 kb region of SLCO1B3. Each double-color box represents one SNP. The ratio of colors represent allelic frequencies in re-sequenced samples (n = 94). Dash lines indicate significant level of the PhyloP score. SNP in red represent that different alleles are present in Haplotype 4 and Haplotype 13. Putative motif variant was indicated by short blue lines and transcription factors.
TF Prediction and Comparison of Promoter Activities Between Haplotypes
SLCO1B3 had significant expression difference between haplotype 4 and haplotype 13. Haplotype 4 differed from haplotype 13 at 14 SNP loci (Figure 4). This study predicted disruptive effect of these SNP on DNA binding motifs of TBP, CEBPβ, HNF1α and HNF3β that were known to be involved in expression regulation of human SLCO1B1 and SLCO1B3 gene (Jung et al., 2001; Vavricka et al., 2004). Of these SNP, 3 were predicted to disrupt binding motifs of these candidate TF. The G allele (harbored in the haplotype 13) of rs13879303 potentially destroys TBP and HNF3β motifs. The A allele (harbored in the Haplotype 4) of rs313594477 can disrupt HNF3β-DNA binding. Two alleles of rs313015134 can create a motif for TBP and for CEBPβ, respectively (Figure 6).
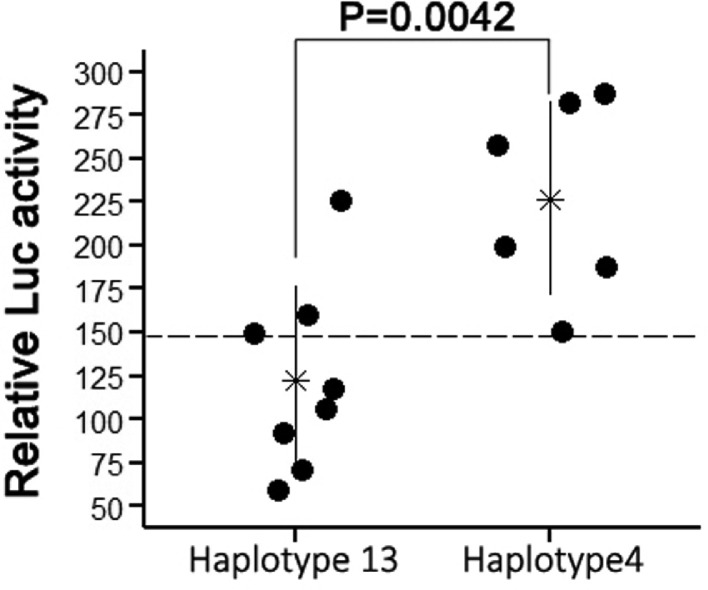
To elucidate whether or not these variants result in differential expression of SLCO1B3 between 2 haplotypes, this study assessed promoter activities of 2 haplotypes through reporter assay. Haplotype 4 showed a higher luciferase activity than Haplotype 13, supporting that these differential SNP loci between Haplotype 4 and 13 account to some extents for differential expression of SLCO1B3 and LBC BGC (Figure 7).
Figure 7.
Comparison of relative luciferase activities between Haplotype 4 and Haplotype 13. Empty pNL1.1 plasmid was used as negative control. Dash line indicates relative luciferase activity of the negative control. The number at the top is the P value of significant test. Asterisks and short lines at two sides of asterisk represent mean ± SD of 6-8 repeats.
DISCUSSION
Dongxiang and Araucana BGC were caused by the EAV-HP insertion in the SLCO1B3 (Wang et al., 2013). We did not detect the EAV-HP in BGC samples of LBC, but found a strong positive correlation between SLCO1B3 expression levels in shell glands and eggshell biliverdin concentration. Haplotype 4 that was associated with BGC and higher levels of SLCO1B3 expression and promoter activity accounted to some extents for molecular basis underlying LBC BGC. The findings confirm that LBC BGC is caused by a new molecular mechanism, but remains to be associated with the change of SLCO1B3 expression.
Independent Evolution of BGC Among Modern Birds
The identical types and deposition mode of eggshell pigments support homology of eggshell color between dinosaur and modern bird which can be traced back to a single evolutionary origin in eumaniraptorans (Wiemann et al., 2018). Not only was BGC early present in oviraptors (Wiemann et al., 2017), but it is commonly distributed in modern bird species (Siefferman et al., 2006; Wang et al., 2013; Chen et al., 2020; Hamchand et al., 2020). Different molecular mechanisms for chicken and duck BGC question the single evolutionary origin of BGC, and support that BGC should independently evolved among different avian species laying BGC eggs (Wang et al., 2013; Chen et al., 2020). Within the chicken species, EAV-HP were found in Chinese indigenous breeds (Dongxiang and Lushi chickens) and Chilean Araucana chickens laying BGC eggs (Wang et al., 2013; Wragg et al., 2013). But the EAV-HP inserted into 2 different genomic sites of SLCO1B3 in Dongxiang and Araucana chickens (Wang et al., 2013; Wragg et al., 2013). This study reported a distinct molecular mechanism in the LBC. This finding adds new evidence for the independent evolution hypothesis of BGC, and indicates that BGC independently evolved not only across avian species, but across breeds within the same species.
Biliverdin Transport Mechanisms May Be Conserved Among Birds Laying BGC Eggs
OATP1B3, the protein encoded by SLCO1B3, is a liver-specific transporter (Hagenbuch and Gui, 2008). In human, OATP1B3 is only expressed at the basolateral membrane of hepatocytes, and is involved in transports of multiple amphipathic organic compounds including bile salts (Hagenbuch and Gui, 2008). But in the blue-shelled chickens, SLCO1B3 was abnormally expressed in shell glands due to the EAV-HP insertion (Sacco et al., 2000; Wang et al., 2013). Given the role of OATP1B3 in bile salt transport, it was supposed that formation of chicken BGC was mostly associated with increased efficiency of biliverdin transport in shell glands (Wang et al., 2013). This study found positive correlation between levels of SLCO1B3 expression in shell glands and eggshell biliverdin concentration, which lends support to the biliverdin-transport mechanism. In duck, biliverdin concentration in uterus fluid from BGC was significantly higher than one from Non-BGC, but there was no difference in shell glands (Liu et al., 2010). The result implied that a transport mechanism that controls biliverdin transport from shell gland to uterus fluid is responsible for duck BGC (Liu et al., 2010). ABCG2, the causative gene for duck BGC, is also associated with transports of bile acid and protoporphyrin Ⅸ (Chen et al., 2020). Taken together, the biliverdin-transport mechanism may be conserved among birds laying BGC eggs although their genetic basis may differ from each other.
Potential Regulatory Mechanisms of SLCO1B3 Expression
Cross-species orthologous sequences that are significantly more similar than would be expected under a null hypothesis of neutral evolution are likely to have critical functional roles (Siepel et al., 2005). This provides a strategy for finding cis-regulatory variants by conservation analysis. SLCO1B3 as a causative gene for BGC was only confirmed in chicken to date (Wang et al., 2013). We hypothesize that 1) the causative mutations occurred within conserved cis-regulatory elements; 2) the wild-type alleles are conserved among birds, and the causative alleles should be embedded in the Haplotype 4. Of 22 SNP, 4 were located at conserved loci among 52 bird species and chicken. The G allele of rs13879300 is the conserved base across evolution. But the allele is shared by Haplotype 4 and Haplotype 13, suggesting that rs13879300 may not be the regulatory variant. The A allele of g.65166659A>G is conserved among birds and is present in Haplotype 4, which is contrary to the hypothesis (2). The A alleles of rs313594477 and rs312678331 harbored in the Haplotype 4 are only found in the genomes of chicken and duck, 2 known fowls laying BGC egg, and thus represent 2 promising candidate loci.
HNF1α and HNF3β are 2 well-known TF involved in regulation of liver-specific expression of transporters including OATP1B3 (Jung et al., 2001; Vavricka et al., 2004). HNF1α can activate transporter expression, however HNF3β represses OATP1B3 expression (Jung et al., 2001; Vavricka et al., 2004). In addition, some ubiquitously expressed TF, such as TBP and CEBPβ, are also involved in regulation of transporter expression (Jung et al., 2001). We found that 3 SNP potentially disrupt these TF motifs among 22 SNP. Given the results from conservation analysis and TF prediction, rs313594477 represents a promising cis-regulatory variant as the A allele was predicted to disrupt the binding site of HNF3β, a transcriptional repressor and associated with high expression of SLCO1B3.
CONCLUSIONS
This study identified a new molecular mechanism that single nucleotide variants at the promoter region of SLCO1B3 can alter gene expression and result in chicken BGC. Haplotype 4 is significantly associated with LBC BGC and serves as a useful marker for breeding of blue-shelled LBC. The findings broaden our understanding to the molecular basis of chicken BGC, and provide new evidence for the independent evolution of BGC that occurred not only across species, but across breeds.
Supplementary Table 1 Haplotypes of 94 re-sequenced samples
Supplementary Figure 1 Alignment of 53 bird genome sequences at 4 conserved SNP loci. The screen shots of UCSC Genome Browser shows alignment of genome sequences of 53 bird species at 4 conserved SNP loci. "−" represent no bases in the aligned species. "=" means that aligning species has one or more unalignable bases in the gap region. Two alleles of each SNP are highlighted in two colors. The upper base is the allele contained in the haplotype 4, and the lower in the haplotype 13.
ACKNOWLEDGMENTS
This study was funded by grants from Key Research and Development Program of Shaanxi Province (2021NY-028), Egg-laying Lueyang Black-boned Chicken Breeding Project (WJYJY-2021-3) and China Agriculture Research System of MOF and MARA(CARS-40-S20)
DISCLOSURES
The authors have no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2022.102223.
Appendix. Supplementary materials
REFERENCES
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Haussler D., Miller W. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.W., McGraw K. Eggshell coloration reflects both yolk characteristics and dietary carotenoid history of female mallards. Funct. Ecol. 2013;27:1176–1185. [Google Scholar]
- Cassey P., Portugal S.J., Maurer G., Ewen J.G., Boulton R.L., Hauber M.E., Blackburn T.M. Variability in avian eggshell colour: a comparative study of museum eggshells. PLoS One. 2010;5:e12054. doi: 10.1371/journal.pone.0012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Berhanu Lemma R., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N., Fornes O., Leung T.Y., Aguirre A., Hammal F., Schmelter D., Baranasic D., Ballester B., Sandelin A., Lenhard B., Vandepoele K., Wasserman W.W., Parcy F., Mathelier A. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic. Acids Res. 2021;30:gkab1113. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gu X., Huang X., Liu R., Li J., Hu Y., Li G., Zeng T., Tian Y., Hu X., Lu L., Li N. Two cis-regulatory SNPs upstream of ABCG2 synergistically cause the blue eggshell phenotype in the duck. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosler A.G., Higham J.P., Reynolds S.J. Why are birds’ eggs speckled? Ecol. Lett. 2005;8:1105–1113. [Google Scholar]
- Hagenbuch B., Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- Hamchand R., Hanley D., Prum R.O., Brückner C. Expanding the eggshell colour gamut: uroerythrin and bilirubin from tinamou (Tinamidae) eggshells. Sci. Rep. 2020;10:11264. doi: 10.1038/s41598-020-68070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S., Suzuki K., Fukuda E., Arihara K., Yamamoto Y., Mukai T., Itoh M. Photodynamic antimicrobial activity of avian eggshell pigments. FEBS Lett. 2010;584:770–774. doi: 10.1016/j.febslet.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Jung D., Hagenbuch B., Gresh L., Pontoglio M., Meier P.J., Kullak-Ublick G.A. Characterization of the human OATP-C (SLC21A6) gene promoter and regulation of liver-specific OATP genes by hepatocyte nuclear factor 1 alpha. J. Biol. Chem. 2001;276:37206–37214. doi: 10.1074/jbc.M103988200. [DOI] [PubMed] [Google Scholar]
- Kennedy G.Y., Vevers H.G. A survey of avian eggshell pigments. Comp. Biochem. Physiol. B. 1976;55:117–123. doi: 10.1016/0305-0491(76)90183-8. [DOI] [PubMed] [Google Scholar]
- Lahti D.C. Population differentiation and rapid evolution of egg color in accordance with solar radiation. Auk. 2008;125:796–802. [Google Scholar]
- Lai Y., Zhang S. Studies on the egg shell colour inheritance of the laying duck. Hereditas. 1991;13:4–5. [Google Scholar]
- Liu H.C., Hsiao M.C., Hu Y.H., Lee S.R., Cheng W.T.K. Eggshell pigmentation study in blue-shelled and white-shelled ducks. Asian-Aust. J. Anim. Sci. 2010;23:162–168. [Google Scholar]
- Moreno J., Osorno J.L. Avian egg colour and sexual selection: does eggshell pigmentation reflect female condition and genetic quality? Ecol. Lett. 2003;6:803–806. [Google Scholar]
- Punnett R.C. Genetic study in poultry-IX. The blue egg. Genetics. 1933;27:465–470. [Google Scholar]
- Reynolds S.J., Martin G.R., Cassey P. Is sexual selection blurring the functional significance of eggshell coloration hypotheses? Anim. Behav. 2009;78:209–215. [Google Scholar]
- Ryter S.W., Tyrrell R.M. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic. Biol. Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- Sacco M.A., Flannery D.M., Howes K., Venugopal K. Avian endogenous retrovirus EAV-HP shares regions of identity with avian leucosis virus subgroup J and the avian retrotransposon ART-CH. J. Virol. 2000;74:1296–1306. doi: 10.1128/jvi.74.3.1296-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M. A fast and exible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefferman L., Navara K.J., Hill G.E. Egg coloration is associated with female condition in eastern bluebirds (Sialia sialis) Behav. Ecol. Sociobiol. 2006;59:651–656. [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Weinstock G.M., Wilson R.K., Gibbs R.A., Kent W.J., Miller W., Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Haussler D. Pages 325–351 in Statistical Methods in Molecular Evolution. Springer; New York: 2005. Phylogenetic hidden Markov models. R. Nielsen, ed. [Google Scholar]
- Somes R.G.Jr, Francis P.V., Tlustohowicz J.J. Protein and cholesterol content of Araucana chicken eggs. Poult. Sci. 1977;56:1636–1640. doi: 10.3382/ps.0561636. [DOI] [PubMed] [Google Scholar]
- Stephens M., Smith N., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R., Yamamoto Y., McDonagh A.F., Glazer A.N., Ames B.N. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- Vavricka S.R., Jung D., Fried M., Grützner U., Meier P.J., Kullak-Ublick G.A. The human organic anion transporting polypeptide 8 (SLCO1B3) gene is transcriptionally repressed by hepatocyte nuclear factor 3beta in hepatocellular carcinoma. J. Hepatol. 2004;40:212–218. doi: 10.1016/j.jhep.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Wang X.L., Zheng J.X., Ning Z.H., Qu L.J., Xu G.Y., Yang N. Laying performance and egg quality of blue-shelled layers as affected by different housing systems. Poult. Sci. 2009;88:1485–1492. doi: 10.3382/ps.2008-00417. [DOI] [PubMed] [Google Scholar]
- Wang X.T., Zhao C.J., Li J.Y., Xu G.Y., Lian L.S., Wu C.X., Deng X.M. Comparison of the total amount of eggshell pigments in Dongxiang brown-shelled eggs and Dongxiang blue-shelled eggs. Poult. Sci. 2009;88:1735–1739. doi: 10.3382/ps.2008-00434. [DOI] [PubMed] [Google Scholar]
- Wang Z., Meng G., Li N., Yu M., Liang X., Min Y., Liu F., Gao Y. The association of very low-density lipoprotein receptor (VLDLR) haplotypes with egg production indicates VLDLR is a candidate gene for modulating egg production. Genet. Mol. Biol. 2016;39:380–391. doi: 10.1590/1678-4685-GMB-2015-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Qu L., Yao J., Yang X., Li G., Zhang Y., Li J., Wang X., Bai J., Xu G., Deng X., Yang N., Wu C. An EAV-HP insertion in 5′ Flanking region of SLCO1B3 causes blue eggshell in the chicken. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann J., Yang T.R., Norell M.A. Dinosaur egg colour had a single evolutionary origin. Nature. 2018;563:555–558. doi: 10.1038/s41586-018-0646-5. [DOI] [PubMed] [Google Scholar]
- Wiemann J., Yang T.R., Sander P.N., Schneider M., Engeser M., Kath-Schorr S., Müller C.E., Sander P.M. Dinosaur origin of egg color: oviraptors laid blue-green eggs. PeerJ. 2017;5:e3706. doi: 10.7717/peerj.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wragg D., Mwacharo J.M., Alcalde J.A., Wang C., Han J.L., Gongora J., Gourichon D., Tixier-Boichard M., Hanotte O. Endogenous retrovirus EAV-HP linked to blue egg phenotype in mapuche fowl. PLoS One. 2013;8:e71393. doi: 10.1371/journal.pone.0071393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.