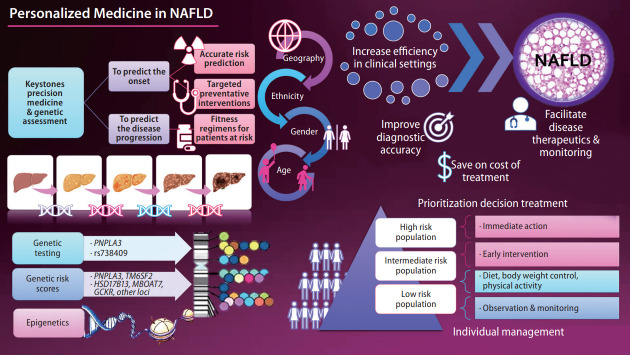
The keystone of precision medicine in complex traits is to predict the onset and progression of the disease. While there are different paths toward precision medicine [1], genetic assessment may be among the top strategies. The current knowledge of the complete human genome sequence and the availability of next-generation sequencing technologies [2] to decipher individual patients’ genetic make-up also invite fully integrated genomic medicine. Therefore, patients may be classified into high or low risk for disease onset or severity, and therapeutic interventions can be personalized. Most importantly, precision medicine must ensure health benefits across geography, ethnicity, age, and gender.
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease whose prevalence has reached global epidemic proportions in adults and children [3]. Once diagnosed, the treatment of NAFLD is complex and often requires pharmacological intervention to treat the liver disease and its associated risk factors, such as obesity, arterial hypertension, insulin resistance, and abnormal circulating lipid profiles [4]. Hence, in this complex scenario, NAFLD presents myriad challenges to physicians, researchers, and patients.
With advances in the genetic knowledge of NAFLD and nonalcoholic steatohepatitis (NASH) [1,5,6], it has become possible to use the information for clinical application. For example, genetic data could be leveraged to identify individuals who are at risk of NAFLD or estimate the risk of severe histological outcomes, including NASH-fibrosis, cirrhosis, and hepatocellular carcinoma [5].
The biological information gained through the years should be integrated with clinical data and translated into efficient, cost-effective, and reliable decision-making. This bioinformatics integration increases efficiency in the clinical setting and improves diagnosis, ultimately facilitating disease monitoring and therapeutics while also saving healthcare costs.
Major NAFLD-associated gene variants function in metabolic pathways, including substrate delivery for de novo lipogenesis, mitochondrial energy utilization, lipid droplet assembly, lipolytic catabolism, fatty acid compartmentalization very low density lipoprotein assembly, and secretion pathways [6]. However, variants in some genes, such as rs738409 in PNPLA3 and rs58542926 in TM6SF2, present pleiotropic associations that can even explain some systemic effects on emerging diseases, such as COVID-19 [7]. Pleiotropy analysis in a recent genome-wide association study of unexplained chronic alanine aminotransferase elevation as a proxy for NAFLD revealed that several replicated variants were jointly associated with metabolic and/or inflammatory traits, revealing a complex model of genetic architecture [8].
In addition, genetic knowledge integration, data modeling, and analysis are vital processes at the interface with drug discovery. However, drug discovery and precision medicine require a complete understanding of new technologies and gene and protein biology of the selected target [1].
A unique approach to advancing the identification of genetic factors associated with NAFLD is, for instance, the discovery of disease-specific eQTL (expression quantitative trait loci) [9]. This approach has shown that the rs2291702 variant located in alanine--glyoxylate aminotransferase 2 (AGXT2) gene protects against liver fibrosis in a genotype-dependent manner with the potential for therapeutic interventions [9].
Although the heritability of NAFLD is ~50%, as in other complex diseases, a significant proportion of the disease burden remains unexplained; the so-called “the missing heritability” [1,5]. The concept of the missing heritability includes not only the genetic and epigenetic modifiers [10], but the interaction with environmental exposure and diet [11], as well as highly interconnected and dynamic factors, such as the liver microbiome [12]. Some examples of the poorly explored heritability estimates of NAFLD and NASH that explain the disease variance are rare variants, which probably would have substantial effects on the phenotype, the exploration of structural variation, the assessment of interaction among the involved loci-also known as epistasis, and the genetic diversity of the mitochondrial DNA, among many others [5].
Recently, a human study demonstrated that the variants influencing the risk/protection against NAFLD-histological severity (PNPLA3-rs738409, TM6SF2-rs58542926, MBOAT7-rs641738, and HSD17B13-rs72613567) and a variant influencing macronutrient intake (FGF21-rs838133) may affect the liver microbial DNA composition [13]. The significant variants’ combined effect associated with a polygenic risk score (PRS) suggested a link between the liver metataxonomic profile, host genetics, and cardiovascular risk [13].
The ultimate goal of precision medicine is to estimate the patient’s risk of developing a particular disease. In this regard, PRS is a valuable instrument for stratifying the population risk according to the individual’s genetic profile. However, some challenges remain, including the missing heritability, discovery of rare variants [14,15], and reproducibility of GWAS findings in diverse populations [1]. On the other hand, there is the advantage of potentially distinguishing patients with high or low risk of severe disease [1]. Although outside of the focus of this snapshot, the advances in transcriptomics, proteomics, and metabolomics integrated with genomics in a multi-omics approach [16] are presenting a fascinating scenario for the personalized medicine of NAFLD and comorbidities.
In conclusion, the newly acquired genetic information should be used to improve the understanding of disease pathogenesis, which may allow the early identification of at-risk patients, who would benefit most from early treatment. Potential avenues for future research should focus on the prevention, surveillance, and prognosis assessment of NAFLD and NASH. The ultimate role of precision therapy relies on defining the appropriate prediction strategies for implementing patient-based therapies.
Abbreviations
- AGXT2
alanine--glyoxylate aminotransferase 2
- eQTL
expression quantitative trait loci
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- PRS
polygenic risk score
Footnotes
Authors’ contributions
C.J.P Concept of the work, manuscript writing and approval. S.S. Concept of the work, manuscript writing and approval.
Conflicts of Interest
The authors have no conflicts to disclose.
REFERENCES
- 1.Sookoian S, Pirola CJ. Precision medicine in nonalcoholic fatty liver disease: new therapeutic insights from genetics and systems biology. Clin Mol Hepatol. 2020;26:461–475. doi: 10.3350/cmh.2020.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salatino A, Sookoian S, Pirola CJ. Computational pipeline for next-generation sequencing (NGS) studies in genetics of NASH. Methods Mol Biol. 2022;2455:203–222. doi: 10.1007/978-1-0716-2128-8_16. [DOI] [PubMed] [Google Scholar]
- 3.Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sookoian S, Pirola CJ. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis. 2019;39:124–140. doi: 10.1055/s-0039-1679920. [DOI] [PubMed] [Google Scholar]
- 6.Sookoian S, Pirola CJ, Valenti L, Davidson NO. Genetic pathways in nonalcoholic fatty liver disease: insights from systems biology. Hepatology. 2020;72:330–346. doi: 10.1002/hep.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirola CJ, Sookoian S. PNPLA3 and COVID-19 outcomes: thinking outside the box might explain the biology behind pleiotropic effects of rs738409 on the immune system. Liver Int. 2021;41:2801–2804. doi: 10.1111/liv.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vujkovic M, Ramdas S, Lorenz KM, Guo X, Darlay R, Cordell HJ, et al. A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. Nat Genet. 2022;54:761–771. doi: 10.1038/s41588-022-01078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo T, Joo SK, Kim HJ, Kim HY, Sim H, Lee J, et al. Disease-specific eQTL screening reveals an anti-fibrotic effect of AGXT2 in non-alcoholic fatty liver disease. J Hepatol. 2021;75:514–523. doi: 10.1016/j.jhep.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Pirola CJ, Sookoian S. Metabolic dysfunction-associated fatty liver disease: advances in genetic and epigenetic implications. Curr Opin Lipidol. 2022;33:95–102. doi: 10.1097/MOL.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 11.Vilar-Gomez E, Pirola CJ, Sookoian S, Wilson LA, Belt P, Liang T, et al. Impact of the association between PNPLA3 genetic variation and dietary intake on the risk of significant fibrosis in patients with NAFLD. Am J Gastroenterol. 2021;116:994–1006. doi: 10.14309/ajg.0000000000001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sookoian S, Salatino A, Castaño GO, Landa MS, Fijalkowky C, Garaycoechea M, et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69:1483–1491. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 13.Pirola CJ, Salatino A, Quintanilla MF, Castaño GO, Garaycoechea M, Sookoian S. The influence of host genetics on liver microbiome composition in patients with NAFLD. EBioMedicine. 2022;76:103858. doi: 10.1016/j.ebiom.2022.103858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baselli GA, Jamialahmadi O, Pelusi S, Ciociola E, Malvestiti F, Saracino M, et al. Rare ATG7 genetic variants predispose patients to severe fatty liver disease. J Hepatol. 2022;77:596–606. doi: 10.1016/j.jhep.2022.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Pirola CJ, Flichman D, Dopazo H, Fernández Gianotti T, San Martino J, Rohr C, et al. A rare nonsense mutation in the glucokinase regulator gene is associated with a rapidly progressive clinical form of nonalcoholic steatohepatitis. Hepatol Commun. 2018;2:1030–1036. doi: 10.1002/hep4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirola CJ, Sookoian S. Multiomics biomarkers for the prediction of nonalcoholic fatty liver disease severity. World J Gastroenterol. 2018;24:1601–1615. doi: 10.3748/wjg.v24.i15.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]