Abstract
Wastewater-based epidemiology (WBE) has gained increasing attention as a complementary tool to conventional surveillance methods with potential for significant resource and labour savings when used for public health monitoring. Using WBE datasets to train machine learning algorithms and develop predictive models may also facilitate early warnings for the spread of outbreaks. The challenges associated with using machine learning for the analysis of WBE datasets and timeseries forecasting of COVID-19 were explored by running Random Forest (RF) algorithms on WBE datasets across 108 sites in five regions: Scotland, Catalonia, Ohio, the Netherlands, and Switzerland. This method uses measurements of SARS-CoV-2 RNA fragment concentration in samples taken at the inlets of wastewater treatment plants, providing insight into the prevalence of infection in upstream wastewater catchment populations. RF's forecasting performance at each site was quantitatively evaluated by determining mean absolute percentage error (MAPE) values, which was used to highlight challenges affecting future implementations of RF for WBE forecasting efforts. Performance was generally poor using WBE datasets from Catalonia, Scotland, and Ohio with ‘reasonable’ or better forecasts constituting 0 %, 5 %, and 0 % of these regions' forecasts, respectively. RF's performance was much stronger with WBE data from the Netherlands and Switzerland, which provided 55 % and 45 % ‘reasonable’ or better forecasts respectively. Sampling frequency and training set size were identified as key factors contributing to accuracy, while inclusion of too many unnecessary variables (or e.g., flow data) was identified as a contributing factor to poor performance. The contribution of catchment population on forecast accuracy was more ambiguous. This study determined that the factors governing RF's forecast performance are complicated and interrelated, which presents challenges for further work in this space. A sufficiently accurate further iteration of the tool discussed within this study would provide significant but varying value for public health departments for monitoring future, or ongoing outbreaks, assisting the implementation of on-time health response measures.
Keywords: Wastewater-based epidemiology, COVID-19, Machine learning, Time series forecasting
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 virus, was first officially reported in December 2019. The global pandemic caused by this disease is ongoing and has caused detrimental physical and mental health impacts in addition to global economic, political, and environmental consequences (Hill et al., 2020; Ramelli and Wagner, 2020). Public health efforts have aimed to contain and mitigate the spread and associated impacts of COVID-19 through vaccine development, quarantine requirements, travel restrictions, and considerable allocation of resources to health departments.
COVID-19 diagnosis and case management using traditional epidemiological evidence is based on the detection of SARS-CoV-2 RNA in nasopharyngeal swabs or saliva samples using reverse transcription quantitative polymerase chain reaction (PCR) testing (Sasaki et al., 2022). This approach relies upon testing of symptomatic cases or close contacts of confirmed cases, introducing inaccuracies due to the frequency of asymptomatic COVID-19 cases. Sampling of entire populations is largely impractical and results in underreporting of positive cases, limiting the ability for authorities to make informed and timely health responses which ultimately reduces response efficacy (Sims and Kasprzyk-Hordern, 2020).
Wastewater-based epidemiology (WBE) offers an alternative to conventional surveillance practices and has gained recent attention as a supplementary tool for monitoring viral prevalence within the community. This approach facilitates detection of asymptomatic or previously undetected cases within monitored wastewater catchments while mitigating biases introduced through traditional surveillance methods, including spatial and temporal differences in health-seeking behaviours, testing capacities, and contact tracing capabilities (Larsen and Wigginton, 2020; Aberi et al., 2021; Zhu et al., 2021). Detection can occur prior to onset of symptoms or before detection through epidemiological methods, providing early warning capabilities that can facilitate pre-emptive or preventative actions by health departments (Hellmér et al., 2014; Ahmed et al., 2020; Sims and Kasprzyk-Hordern, 2020; Róka et al., 2021).
Faeces from infected individuals, including asymptomatic cases, contain SARS-CoV-2 viral genome fragments (Zhu et al., 2021; Zhang et al., 2022). Fragments are transported through sewers towards wastewater treatment plants (WWTPs), where samples can be collected and analysed to quantify viral prevalence. Since samples collected at WWTP inlets are composed of wastewater from across an entire catchment area, they can represent a region's entire population provided their sanitary systems are connected to sewerage networks. Inclusion of other measured parameters including volumetric flow rate and catchment population can then be used to calculate active community COVID-19 cases, facilitating effective allocation of medical resources and reducing pressure on health systems (Ahmed et al., 2020; Xagoraraki and O’Brien, 2020; Zhu et al., 2021).
Machine learning (ML) provides value for the analysis of large datasets due to its capability for self-improvement based on supplied data. While time series analysis is easier to model and use, root causes and factors are not taken into account. Specific ML models can be selected to learn from data, identify patterns, and make predictions with minimal human intervention (De Las Heras et al., 2020; Yadav, 2020; Abdalla et al., 2022; Truong, 2022). ML models have been widely applied to both COVID-19 forecasting and WBE data interpretation, however studies considering the former have largely utilised traditional epidemiological datasets. Riberio et al. (2020) evaluated autoregressive integrated moving average (ARIMA), cubist regression (CUBIST), random forest (RF), ridge regression (RIDGE), support vector regression (SVR), and stacking-ensemble learning to forecast the growth of COVID-19 cases in Brazil using cumulative case counts as a training parameter. Singh et al. (2020) also tested ARIMA for COVID-19 spread prediction. Chimmula and Zhang (2020) applied long short-term memory (LSTM) networks to predict future infection conditions and indicate a potential stopping time for COVID-19 outbreaks in Canada. Within WBE contexts historical data can be provided to an algorithm and used for future predictions of targeted water quality parameters or chemical/biological indicators within wastewater (Granata et al., 2017; Tomperi et al., 2017). These authors considered lead time of 1 to 6 days, however the current knowledge on lead time for pandemic early warning systems is very limited.
Few examples of ML analysis of COVID-19 WBE data could be identified by the authors of this paper. Using time-series forecasting algorithms, future prevalence of SARS-CoV-2 RNA fragments within wastewater can be predicted to provide early warnings and vital time for the development of response strategies, with further development presenting a potentially valuable tool for management of future and ongoing epidemics (Chimmula and Zhang, 2020; Ribeiro et al., 2020; Aberi et al., 2021; Li et al., 2021; Abdalla et al., 2022; Daza-Torres et al., 2022). Koureas et al. (2021) examined the relationship between viral fragments and recorded COVID-19 cases in two Greek municipalities with two supervised ML models, Random Forest (RF) and Linear Regression (LR), which were trained and evaluated. RF exhibited superior performance within this study as evidenced by higher correlations and smaller mean absolute percentage error (MAPE) values relative to LR. Several additional ML algorithms are commonly used in timeseries forecasting and therefore demonstrate potential value within WBE studies, including Multilayer Perceptron (MLP), and Decision Tree (DT); furthermore, strengths and limitations for specific forecasting purposes must be considered (Hastie et al., 2009; Zhou et al., 2019).
Multilayer perceptron (MLP) is a feedforward artificial neural network that is trained through backpropagation. This algorithm is used widely for solving problems requiring supervised learning and for research in computational neuroscience and parallel distributed processing. It is capable of learning nonlinear relationships but is has high complexity due to its large number of parameters and is sensitive to feature scaling (Hastie et al., 2009). Linear regression (LR) is a simple model used to find the best fit linear line between two variables, however for many cases the relationship between variables is non-linear which results in low accuracy. In addition, LR is sensitive to outliers, which further disrupts model performance and accuracy; while non-linear regression techniques provide better performance for greater complexity and cost (Khamis et al., 2005; Yan and Su, 2009). Decision trees (DT) are a widely used data mining and machine learning method due to their relative simplicity and ease of implementation (Wu et al., 2008). A binary classification process splits analysed data at decision nodes into progressively more refined ‘branches’, ‘twigs’, and ‘leaves’ (Saloux and Candanedo, 2018). Each branching point represents a decision making point where the required output is provided at the leaf node (Suresan et al., 2021).
Random Forest (RF) is one of the most popular supervised learning algorithms due to its high flexibility and ease of implementation (Tyralis and Papacharalampous, 2017; Ray, 2019). RF is an ensemble algorithm that builds a ‘forest’ consisting of many decision trees and has a high accuracy due to the random selection of predictors which reduces variance and lowers correlation among trees (Suchetana et al., 2017). A greater number of trees within the forest tends to provide a more robust algorithm, however this also increases computational costs, and the relationship shows diminishing returns at scale (Oshiro et al., 2012; Tyralis and Papacharalampous, 2017). RF is also capable of handling datasets with missing values which improves performance when applied to real-world scenarios such as WBE datasets (Carranza and Laborte, 2015). Despite offering many advantages, RF has several limitations. Building the forest and training the model can be a time consuming and computationally intensive process. In addition, bias can be introduced in measures of variable importance when predictors are correlated (Buskirk, 2018).
The objective of this study is to explore the challenges associated with using machine learning algorithms for analysis of WBE datasets, specifically surveillance of SARS-CoV-2 RNA fragments. This involved a review of the performance and accuracy of machine learning algorithms for short forecasting periods (<7 days) based on WBE datasets, which led to the selection of RF for subsequent analysis. Data were collected from WWTPs within Scotland, Barcelona, Ohio, the Netherlands, and Switzerland to examine forecasting performance against local factors. This work intends to highlight the challenges associated with machine learning forecasting for further applications using WBE datasets, which may assist health responses to future epidemics. To the best of the authors' knowledge this work is a novel application of machine learning forecasting to WBE data across a larger scale with multiple examined sites.
2. Materials and methods
Dataset and site selection: WBE Datasets were accessed and downloaded from the Wastewater-Sphere (W-SPHERE) website, which is part of a larger wastewater surveillance project led by PATH and funded by the Bill & Melinda Gates Foundation and the Global Innovation Fund (Global Water Pathogens Project 2022, 2022).
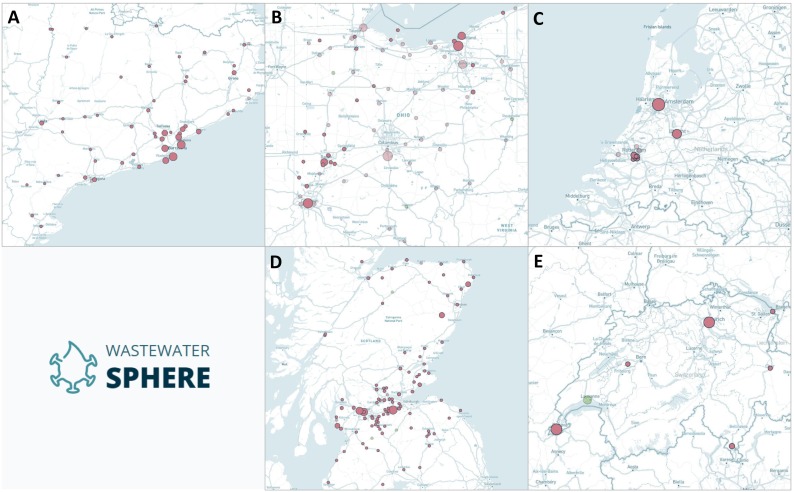
These data contained timeseries information on samples taken from WWTPs listed in Fig. 1 , sampling frequency, and populations of serviced catchments. Target concentrations measured nucleoprotein (N1 and N2) genome regions and polymerase IP4 genomic fragments of SARS-CoV-2 (Chavarria-Miró et al., 2021). Five regions (Fig. 1) were selected to provide datasets with varied population densities and sampling regimes, and diverse geographic distributions of WWTPs/sewerage catchment areas. This variability assists an initial evaluation of factors affecting prediction accuracy and informs parameters necessary within future studies. These unstructured datasets were also identified as high-quality examples, facilitating improved analysis within this preliminary assessment.
Fig. 1.
Datasets collected from W-SPHERE for machine learning analysis. Monitored regions were (A) Catalonia (https://sphere.waterpathogens.org/dataset/bbd61ae1-e40e-4660-af26-549cb00b3d03), (B) Ohio (https://sphere.waterpathogens.org/dataset/a29fcfe8-0c34-4e73-b08f-ede0520c1d4f), (C) the Netherlands (https://sphere.waterpathogens.org/dataset/5e4225bc-5edb-49cc-bf1f-84724c3152ef & https://sphere.waterpathogens.org/dataset/a3b51f71-ebea-408d-b100-06ac652f9d44), (D) Scotland (https://sphere.waterpathogens.org/dataset/634a33cd-9444-4f4a-8234-b7886e45d020), (E) Switzerland (https://sphere.waterpathogens.org/dataset/456690eb-8424-416f-8010-247179d67847).
Algorithm selection: Algorithms which had demonstrated value within relevant past studies were considered for trialling with W-SPHERE timeseries data. RF was identified as appropriate for this study as it is available on open-source training platforms, is relatively simple to train, and has demonstrated value within prior studies.
Software selection: Waikato Environment for Knowledge Analysis (WEKA) was selected as a platform for training/running algorithms and providing timeseries forecasts as it is simple and open-source. WEKA provides integrated data preparation, classification, and timeseries forecasting capabilities using pre-loaded algorithms and with a streamlined user interface. Original algorithms can also be modified using this platform if required (Vanam et al., 2021).
Data pre-processing: W-SPHERE data were pre-processed before being provided to WEKA. Original data contained within a single column were separated into “csv” format readable by WEKA and split into different sets according to its sampling site. Each site was given a unique label – i.e., the first site examined in Scotland was labelled sc1. Some regions recorded volumetric wastewater flowrate (m3/s) data, which were included to provide a parallel forecast to the same data in the absence of a volumetric input. Some sample results were absent due to the lack of sample taken on that day, which needed to be manually removed to prevent RF misinterpreting these results as zeros. RF is robust to below detection limit data points (Ray, 2019). Separated datasets were further split to form the training sets (80 %) and testing sets (20 %) based on frequently used splits in machine learning applications (Zhou et al., 2019; Brady et al., 2018; Chimmula and Zhang, 2020). As use of machine learning for WBE applications is not a mature practice commonly used split ratio from other machine learning applications was used. Separated dataset files within Excel were refined to remove all parameters not required by WEKA before being converted to an Attribute-Relation File Format (ARFF) appropriate for analysis. Furthermore, the performance of RF for forecasts using data from a larger geographic scale was assessed by training the algorithm with all COVID-19 WBE data held by W-SPHERE in a single training instance.
Random Forest analysis: Pre-prepared data were provided to WEKA with an estimated 100 random trees. Predictions were set to one step ahead, which is equivalent to the sampling frequency for each site and ranged from one day to one week depending on the region and specific WWTP. ‘Automatically detected’ periodicity was selected, and the outputted forecast values were recorded. This process was repeated for each datapoint in the 80 % training set, with all sampled values from earlier in the timeseries subsequently inputted as additional training values.
Forecast accuracy evaluation: Forecast accuracies and model performances were evaluated by calculating a Mean Absolute Percent Error (MAPE) for each forecasted dataset. MAPE is a common evaluation metric appropriate for trend evaluation as it gives all items the same weight and outputs results as a single easily comparable percentage (Lewis, 1982). MAPE was calculated according to Eq. (1):
| (1) |
where n is the number of iterations. MAPE values can range from 0 % to infinity (If the data contain zeros, the MAPE can be infinite as it will involve division by zero) where smaller values indicate a higher accuracy. Evaluation criteria outlined by Lewis (1982) was used in this study to assess model performance (Table 1 ). This allows direct comparison between different sites and regions.
Table 1.
A scale for the judgment of forecast accuracy.
| MAPE | Judgment of forecast accuracy |
|---|---|
| <10 % | Highly accurate |
| 10 % to 20 % | Good forecast |
| 21 % to 50 % | Reasonable forecast |
| 51 % or more | Inaccurate forecast |
3. Results and discussion
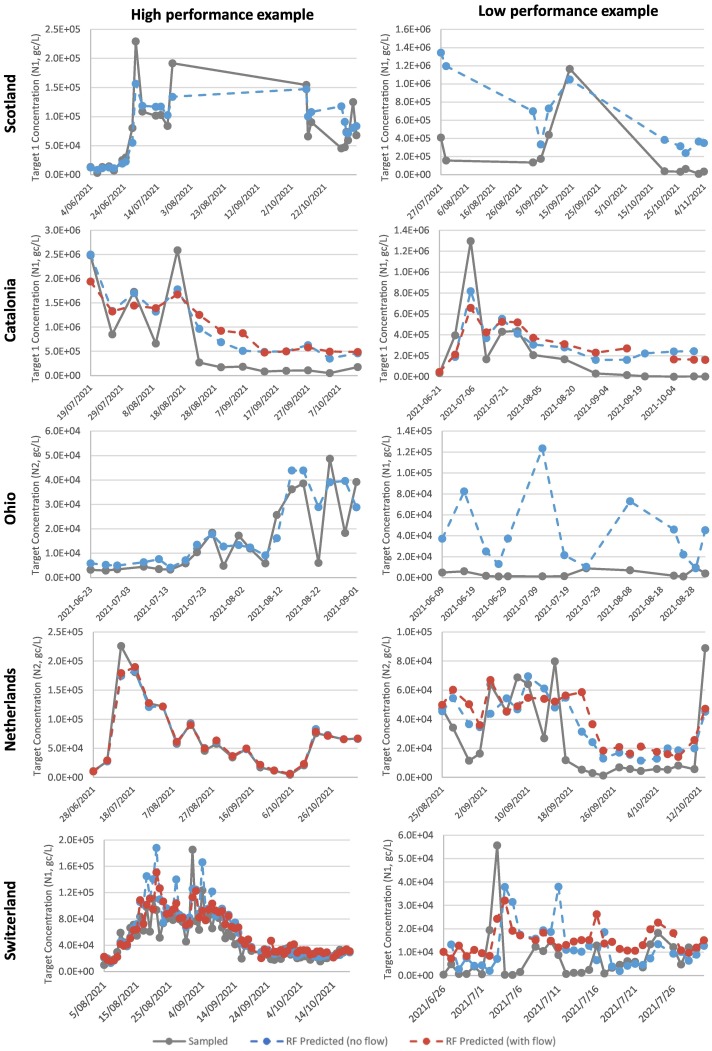
Random Forest forecasting performance: High and low performance examples (Fig. 2 ) illustrate the variability of RF's performance even within the same region. Timeseries in the left column of Fig. 2 were the best forecasts created in each region, which is reflected through generally accurate trend predictions (i.e. predicted increase or decrease in target concentration), predictions of peaks, and generally low magnitudes of differences between forecast and actual values. The opposite can generally be said for the low performance examples in the right column.
Fig. 2.
Random Forest forecasting one period ahead. Left column - examples with low MAPE, high performance. Right column - examples with high MAPE, low performance.
40 sites (Table S1) were analysed from Scotland data (summarised in Table 2 ), among which 16/40 (40 %) had a MAPE below 100 % and 2/40 (5 %) had a MAPE of 20–50 % so could be considered a reasonable forecast. Sampling intervals across examined Scottish sites ranged from three to thirteen days with an average of four. These relatively low and irregular sampling frequencies increased the pattern recognition challenge faced by RF, reducing forecast accuracy. Scotland's sites had the lowest average population per catchment, which was coupled with reasonable algorithm performance relative to other sites.
Table 2.
Averaged results from all sites for all sampling regions covering global data from five countries located in two continents varying life-style pandemic responses patterns.
| Region | Number of sites | Sampling frequency (samples per week) | Samples per site | Population | Percentage of data with a reasonable forecast (with flow data) | Percentage of data with a reasonable forecast (without flow data) |
|---|---|---|---|---|---|---|
| Scotland | 40 | 1.75 | 82 | 81,900 | N/A | 5 % |
| Catalonia | 4 | 1.00 | 59 | 829,000 | 0 % | 0 % |
| Ohio | 42 | 1.00 | 80 | 101,000 | N/A | 0 % |
| The Netherlands | 11 | 2.64 | 141 | 127,000 | 45 % | 55 % |
| Switzerland | 7 | 7.00 | 310 | 207,000 | 43 % | 43 % |
N/A = not applicable since flow data were not provided.
Of all examined regions, RF provided the most inaccurate forecast on Catalonian WBE data. All forecasts in Catalonia were inaccurate (Table 1) with a MAPE >100 % (summarised in Table 2). This finding used N1 and N2 target concentrations and was including or excluding flow data across 16 total forecasts (Table S2), which indicates the presence of factors disrupting RF's performance. A potential cause is low (weekly) sampling frequency, which required RF to make a prediction further into the future. Relative to daily sampling, variable factors governing viral spread are liable to compound over a week and will be difficult for RF to model accurately, highlighting machine learning forecasting challenges of WBE data including potential for data censoring. A low sampling frequency also limits the total dataset available for algorithm training since fewer total samples will have been taken since the beginning of WBE monitoring in that region. This site also had a considerably higher monitored population per catchment than other regions, indicating that the population may have some impact on algorithm performance as discussed within ‘Factors affecting prediction accuracy’. Interestingly, inclusion of flowrate data resulted in slightly poorer RF performance on average (Table S2).
Of the 42 sites (Table S3) investigated in Ohio, 6/40 (14 %) had a MAPE below 100 % and none were below 50 %, which indicates that RF provided inaccurate forecast with Ohio's WBE data (summarised in Table 2). Relative to Catalonia's results, RF performed better on Ohio's sites despite having the same sampling frequency. This is likely due to the greater average samples available per site and may also be influenced by the lower population per catchment in Ohio relative to Barcelona. As with Scotland, Ohio's data had no flow information which prevented comparison of RF performance with and without flow data.
Data from 11 sites (Table S4) in the Netherlands were used to train RF, providing forecasts both with and without flow data. RF provided slightly better forecast accuracy without access to flow data, with 6/11 (55 %) of forecasts made without flow data yielding a MAPE below 50 % compared to 5/11 (45 %) with flow data. Relative to other analysed regions, the Netherlands had a high sampling frequency with large training sets available (Table 2). This likely provided RF sufficient data to form reasonable forecasts, which is demonstrated through the low average MAPE values and high proportion below 50 %.
As with the Netherlands, Switzerland's data also contain flowrate information. RF provided good or reasonable forecast accuracy with most of the 7 Swiss sites (Table S5), with 3/7 (43 %) forecasts with and without flow achieving a MAPE below 50 % and a further 2/7 only slightly over 50 %. RF performed very poorly on two sites (sw5 and sw7), which raised the average MAPE considerably (Table 2). It is likely that the poor performance using data from these sites was due to frequent low/zero values for target concentrations, which impacted prediction performance. These results indicate that RF forecasting is most appropriate for regions with significant community transmission. Other than these outlier forecasts, Switzerland's data provided a good basis for reasonable RF modeling. This is likely due to these datasets having the highest sampling frequency and largest training sets by a considerable margin.
Factors affecting prediction accuracy: This study's results, together with insights gathered from the available literature, were investigated to discuss factors affecting prediction accuracy. This discussion focusses on parameters that were included within W-SPHERE's datasets to illustrate the challenge associated with creating accurate forecasts using real-world data. Data collected within this study suggest that the inclusion of flow data negatively impacts forecast accuracy. This result aligns with findings from previous studies with RF, which associated inclusion of unimportant variables with poorer model performance (Kuhn and Johnson, 2013; Tyralis and Papacharalampous, 2017). This demonstrates the importance of appropriate parameter selection.
Forecast results indicate that sampling frequency is a key parameter for improving RF's performance as discussed previously (Ahmed et al., 2020; Hill et al., 2020). The results of this study indicate that sampling more than once a week is necessary to yield reasonable forecasts, with greater frequency yielding greater accuracy. Viral incubation periods for COVID-19 may be partially responsible for this observation, which is estimated as 1–14 days but typically between 3 and 7 days (Hill et al., 2020). Weekly samples do not represent an average of viral concentration across the previous week and therefore lower sampling frequencies may miss peak viral loads in wastewater streams in addition to daily fluctuations. The impact of sampling frequency on forecast accuracy is likely governed by incubation periods, with shorter incubation periods necessitating increased sampling frequency. As increased sampling frequency adds costs and complexity, determining minimum required sampling frequencies based upon factors including average incubation period presents a future research direction.
Forecasting results also indicate that the standardisation of sampling procedures in sites with variable sampling intervals would improve predicted result reliability while also improving the reproducibility and comparability of the study. Furthermore, signal decay/persistence in sewage, in-sewer hydraulics, collection method, transient populations, analytical sensitivity and analytical turnaround times impact the data and consequently the forecasting (Ahmed et al., 2020; Hill et al., 2020; Wade et al., 2022).
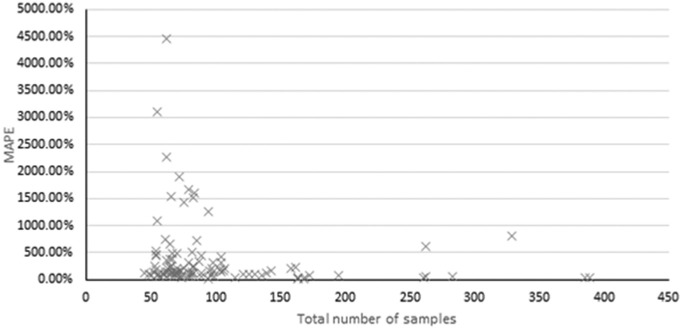
A greater number of samples per site were observed to generally improve model performance and deliver more consistent results (Fig. 3 ). This observation aligns with established machine learning algorithm training techniques which utilise larger training sets to improved pattern recognition capabilities (Ajiboye et al., 2015). Fig. 3 suggests that increasing the training set size reduces the frequency of high MAPE forecasts, however due to the limited number of sites with a large training set this evidence is not conclusive. It is also important to note that while a large sample training set appears to improve accuracy, accurate results were still attainable with smaller training sets. This highlights a further unknown for future forecasting efforts as ideally forecasts would be constructed with the smallest training set possible, allowing accurate forecasting early into an epidemic or with reduced costs.
Fig. 3.
MAPE versus total number of samples across all sites, excluding ‘with flow’ forecasts.
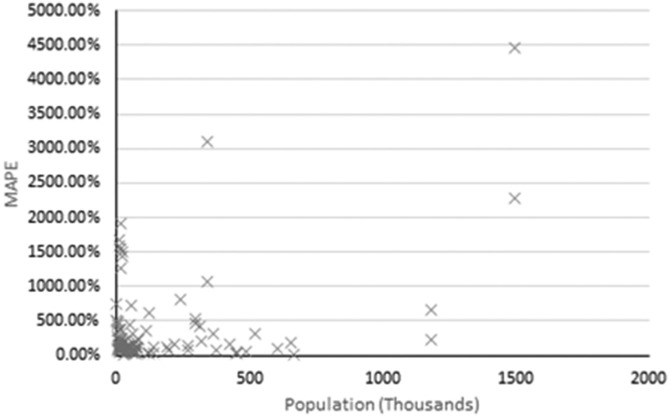
Population within each served catchment was not supplied as an input variable to RF, however forecast performances were considered within the context of respective populations to assess for noticeable impacts on performance. On an averaged regional scale, RF forecasting accuracy tended to decrease as monitored population size increased (Table 2). On a localised scale however this trend was not present, such as in the Netherlands where the largest catchment (ne1, population = 670,000) had the greatest accuracy (MAPE = 7.32 %) while the smallest catchment (ne10, population = 4880) had the weakest performance with RF (MAPE = 239 %). A linear regression analysis of all populations versus MAPEs of respective forecasts revealed no notable correlations (Fig. 4 ).
Fig. 4.
MAPE versus catchment population across all sites, excluding ‘with flow’ forecasts.
Despite the lack of clear correlation, population is likely to have considerable indirect impacts on forecast performance. Many higher-performing forecasts were from catchments with low populations, which may be caused by reduced complexity in these regions. Often larger population centres have disproportionately higher mobility and experience increased travel from regions to urban areas, which contributes to viral spread (Wade et al., 2022). This adds additional parameters governing viral spread and therefore complicates pattern recognition efforts for RF.
An additional observation is that RF often performed poorly when constructing forecasts with datasets containing many low/zero values, e.g. low performance examples. This indicates that forecasting with RF is useful only for regions with higher viral prevalence in the community and may also account for high MAPEs associated with very low population catchments such as ne10 and others in Fig. 4. It is likely that such catchments had periods of very low/zero COVID-19 prevalence in their communities, which disrupted RF's pattern recognition capabilities. The value of WBE is not limited to high prevalence periods where the relationship between disease and measurement is stronger and emergence of new outbreak can be detected at low concentrations over time. However, there is little publicly available that goes into depth on how to address low prevalence periods (Hill et al., 2020).
RF was unsuccessful in creating forecasts after being trained with the entire W-SPHERE dataset. The primary reason for this result is that different sites had different sampling frequencies, but another potential reason is that training with a global dataset prevented RF from learning the local factors that governed spread within specific sites examined within this study.
While the results from this study indicate that machine learning may provide value for future outbreak forecasting, improvements should be made within further studies.
■ RF's forecasting performance may be improved through structural changes appropriate for low-dimensional datasets, such as those proposed by Wang et al. (2018).Deeper understanding of fundamental knowledge of the system and processes is required for further improvement.
■ Algorithms such as others discussed within this paper should be trialled for efficacy.
■ Implementation of WBE for the purpose of forecasting should be done with high sampling frequency to provide adequate training sets and allow the algorithm to identify more patterns, which will thereby improve accuracy.
■ Forecasting accuracy may be improved through inclusion of data regarding population mobility, sampling methods, chemical composition and physicochemical properties of the wastewater samples, and environmental parameters which may be responsible for a portion of forecast uncertainties (Koureas et al., 2021).
■ As identified for flow rate data, additional data may also have opposite effects on accuracy and cause poorer forecast performance, necessitating further work to identify key parameters beyond those considered in this study.
■ Development of sufficiently accurate forecasts should prompt extension of forecast ranges, i.e. forecast to two sampling periods instead of one.
■ A further challenge that was not considered in this study is the impacts of changing restrictions and other public health responses on the parameters governing viral spread. It is hypothesised that a change in restrictions will negatively impact RF performance as this changes rates of infection. Further factors that were not included within this preliminary assessment include vaccination effects and the impacts of variants.
■ Implementation of forecasting for the qualitative identification of rising and falling target concentration may provide value for health departments by indicating whether an outbreak is expected to accelerate or shrink. This would provide valuable time for the preparation of health resources or indicate if current public health measures are effective.
■ Further exploration of the ‘Trailing W-SPHERE data on a larger geographic scale’ approach could use global datasets for ‘baseline training’, which is then tweaked according to local parameters and specific site data.
■ It is important to note that ML algorithms may be limited in value, being suitable in a specific context (training dataset) but can fail when applied to novel data. A single algorithm is not necessarily the most suitable to be applied across all datasets or for all use cases. Future studies exploring ML would need to test several algorithms and do comparator analyses.
4. Conclusion
This study explored challenges associated with implementing Random Forest for forecasting COVID-19 fragments in wastewater across multiple sites in five regions. MAPE was calculated to evaluate forecast accuracy and guide discussion on the factors affecting RF performance. RF performance was generally poor using WBE datasets from Catalonia, Scotland, and Ohio largely due to low sampling frequency across these sites. Of these sites, Scotland performed best and also had the highest sampling frequency. RF's performance was much stronger with WBE data from the Netherlands and Switzerland, likely due to generally higher sampling frequencies and total training dataset sizes. As identified in this study the factors governing forecast accuracy are complicated and interrelated, presenting challenges for the development of reliable and accurate forecasting using WBE data. Sufficient development of this tool may provide significant value for public health departments to monitor future, emerging, or ongoing outbreaks and provide information necessary to implement on-time health response measures.
CRediT authorship contribution statement
Liam Vaughan: Conceptualization, Methodology, Validation, Investigation, Writing – original draft, Writing – review & editing. Muyang Zhang: Methodology, Software, Validation, Investigation, Writing – original draft. Haoran Gu: Methodology, Software, Validation, Investigation, Writing – original draft. Joan B. Rose: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing – review & editing. Colleen C. Naughton: Conceptualization, Investigation, Data curation. Gertjan Medema: Conceptualization, Investigation, Data curation. Vajra Allan: Conceptualization, Investigation, Data curation. Anne Roiko: Conceptualization, Methodology, Validation, Investigation, Supervision. Linda Blackall: Conceptualization, Methodology, Validation, Investigation, Resources, Writing – review & editing, Supervision. Arash Zamyadi: Conceptualization, Methodology, Validation, Investigation, Resources, Writing – original draft, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge in-kind contribution from Wastewater-Sphere (W-SPHERE) website team, which is part of a larger wastewater surveillance project led by PATH.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159748.
Appendix A. Supplementary data
Supplementary tables
Data availability
Data will be made available on request.
References
- Abdalla W., Renukappa S., Suresh S. Managing COVID-19-related knowledge: a smart cities perspective. Knowl. Process. Manag. 2022;1–23 doi: 10.1002/kpm.1706. [DOI] [Google Scholar]
- Aberi P., Arabzadeh R., Insam H., Markt R., Mayr M., Kreuzinger N., Rauch W. Quest for optimal regression models in SARS-CoV-2 wastewater based epidemiology. Int. J. Environ. Res. Public Health. 2021;18(20):10778. doi: 10.3390/ijerph182010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajiboye A.R., Abdullah-Arshah R., Qin H., Isah-Kebbe H. Evaluating the effect of dataset size on predictive model using supervised machine learning technique. International Journal of Computer Systems & Software Engineering. 2015;1(1):75–84. doi: 10.15282/ijsecs.1.2015.6.0006. [DOI] [Google Scholar]
- Brady S., Magoni D., Murphy J., Assem H., Portillo-Dominguez A.O. Analysis of Machine Learning Techniques for Anomaly Detection in the Internet of Things. 2018 IEEE Latin American Conference on Computational Intelligence (LA-CCI) 2018:1–6. doi: 10.1109/LA-CCI.2018.8625228. [DOI] [Google Scholar]
- Buskirk T. Surveying the forests and sampling the trees: an overview of classification and regression trees and random forests with applications in survey research. Surv. Pract. 2018;11(1):1–13. doi: 10.29115/SP-2018-0003. [DOI] [Google Scholar]
- Carranza E.J.M., Laborte A.G. Random forest predictive modeling of mineral prospectivity with small number of prospects and data with missing values in Abra (Philippines) Comput. Geosci. 2015;74:60–70. doi: 10.1016/j.cageo.2014.10.004. [DOI] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona. Spain. Applied and Environmental Microbiology. 2021;87(7):e02750. doi: 10.1128/AEM.02750-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Torres M.L., Kim M., Olson R., Bess C.W., Rueda L., Susa M., Tucker L., Schmidt A.J., Naughton C., Pollock B.H., Shapiro K., Bischel H.N., Montesinos-Lopez J.C., Garcia Y.E., Nuno M. Model training periods impact estimation of COVID-19 incidence from wastewater viral loads. 2022;medRxiv:1–26. doi: 10.1016/j.scitotenv.2022.159680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Heras A., Luque-Sendra A., Zamora-Polo F. Machine learning technologies for sustainability in smart cities in the post-COVID era. Sustainability. 2020;12(22):9320. doi: 10.3390/su12229320. [DOI] [Google Scholar]
- Granata F., Papirio S., Esposito G., Gargano R., De Marinis G. Machine learning algorithms for the forecasting of wastewater quality indicators. Water. 2017;9(2):105. doi: 10.3390/w9020105. [DOI] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. Data Mining, Inference, and Prediction. (2nd Ed.). Springer; 2009. Elements of statistical learning. [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis a virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Zamyadi A., Deere D., Vanrolleghem P.A., Crosbie N.D. SARS-CoV-2 known and unknowns, implications for the water sector and wastewater-based epidemiology to support national responses worldwide: early review of global experiences with the COVID-19 pandemic. Water Qual. Res. J. 2020;56(2):57–67. doi: 10.2166/wqrj.2020.100. [DOI] [Google Scholar]
- Khamis A., Ismail Z., Khalid H., Mohammed A. The effects of outliers data on neural network performance. J. Appl. Sci. 2005;5(8):1394–1398. [Google Scholar]
- Koureas M., Amoutzias G.D., Vontas A., Kyritsi M., Pinaka O., Papakonstantinou A., Dadouli K., Hatzinikou M., Koutsolioutsou A., Mouchtouri V.A., Speletas M., Tsiodras S., Hadjichristodoulou C. Wastewater monitoring as a supplementary surveillance tool for capturing SARS-COV-2 community spread. A case study in two greek municipalities. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., Johnson K. Applied predictive modeling. Springer, New York. 2013 doi: 10.1007/978-1-4614-6849-3. [DOI] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.D. Butterworth-Heinemann; 1982. Industrial and business forecasting methods: a practical guide to exponential smoothing and curve fitting. [Google Scholar]
- Li X., Kulandaivelu J., Zhang S., Shi J., Sivakumar M., Mueller J., Luby S., Ahmed W., Coin L., Jiang G. Data-driven estimation of COVID-19 community prevalence through wastewater-based epidemiology. Sci. Total Environ. 2021;1:789–147947. doi: 10.1016/j.scitotenv.2021.147947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro T.M., Perez P.S., Baranauskas J.A. In: Machine Learning and Data Mining in Pattern Recognition. Perner P., editor. Springer; 2012. How many trees in a random Forest? pp. 154–168. [DOI] [Google Scholar]
- Riberio M.H.D.M., da Silva R.G., Mariani V.C., Coelho L.D.S. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelli S., Wagner A. In: Mitigating the COVID Economic Crisis: Act Fast and Do Whatever It Takes. Baldwin R., Mauro B.W.D., editors. CEPR Press; 2020. What the stock market tells us about the consequences of COVID-19. [Google Scholar]
- Ribeiro M.H.D.M., da Silva R.G., Mariani V.C., dos Coelho L., S. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos, Solitons & Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S. A Quick Review of Machine Learning Algorithms. 2019 International Conference on Machine Learning, Big Data. Cloud and Parallel Computing (COMITCon), IEEE. 2019:35–39. doi: 10.1109/COMITCon.2019.8862451. [DOI] [Google Scholar]
- Róka E., Khayer B., Kis Z., Kovács L.B., Schuler E., Magyar N., Málnási T., Oravecz O., Pályi B., Pándics T., Vargha M. Ahead of the second wave: early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloux E., Candanedo J.A. Forecasting district heating demand using machine learning algorithms. Energy Procedia. 2018;149:59–68. doi: 10.1016/j.egypro.2018.08.169. [DOI] [Google Scholar]
- Sasaki T., Inoue O., Ogihara S., Kubokawa K., Oishi S., Shirai T., Iwabuchi K., Suzuki-Inoue K. Detection of SARS-CoV-2 RNA using RT-qPCR in saliva samples and nasopharyngeal, lingual, and buccal mucosal swabs. Jpn. J. Infect. Dis. 2022;75(1):102–104. doi: 10.7883/yoken.JJID.2021.091. [DOI] [PubMed] [Google Scholar]
- Singh R.K., Rani M., Bhagavathula A.S., Sah R., Rodriguez-Morales A.J., Kalita H., Nanda C., Sharma S., Sharma Y.D., Rabaan A.A., Rahmani J., Kumar P. Prediction of the COVID-19 pandemic for the top 15 affected countries: advanced autoregressive integrated moving average (ARIMA) model. JMIR Public Health Surveillance. 2020;6(2) doi: 10.2196/19115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchetana B., Rajagopalan B., Silverstein J. Assessment of wastewater treatment facility compliance with decreasing ammonia discharge limits using a regression tree model. Sci. Total Environ. 2017;598:249–257. doi: 10.1016/j.scitotenv.2017.03.236. [DOI] [PubMed] [Google Scholar]
- Suresan A., P S., Venkatraman M., Suresh S. In: Image Processing and Capsule Networks. Chen J.I.-Z., Tavares J.M.R.S., Shakya S., Iliyasu A.M., editors. Springer International Publishing; 2021. Comparison of machine learning algorithms for smart license number plate detection system; pp. 63–75. [DOI] [Google Scholar]
- Tomperi J., Koivuranta E., Leiviskä K. Predicting the effluent quality of an industrial wastewater treatment plant by way of optical monitoring. Journal of Water Process Engineering. 2017;16:283–289. doi: 10.1016/j.jwpe.2017.02.004. [DOI] [Google Scholar]
- Truong T.C. The impact of digital transformation on environmental sustainability. Advances in Multimedia. 2022;20(22):1–12. doi: 10.1155/2022/6324325. [DOI] [Google Scholar]
- Tyralis H., Papacharalampous G. Variable selection in time series forecasting using random forests. Algorithms. 2017;10(4):114. doi: 10.3390/a10040114. [DOI] [Google Scholar]
- Vanam M.K., Amirali Jiwani B., Swathi A., Madhavi V. High performance machine learning and data science based implementation using Weka. Materials Today: Proceedings. 2021 doi: 10.1016/j.matpr.2021.01.470. [DOI] [Google Scholar]
- Wade M.J., Lo Jacomo A., Armenise E., Brown M.R., Bunce J.T., Cameron G.J., Fang Z., Farkas K., Gilpin D.F., Graham D.W., Grimsley J.M.S., Hart A., Hoffmann T., Jackson K.J., Jones D.L., Lilley C.J., McGrath J.W., McKinley J.M., McSparron C., Kasprzyk-Hordern B.… Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu T., Zheng X., Peng H., Xin J., Zhang B. Short-term prediction of groundwater level using improved random forest regression with a combination of random features. Appl Water Sci. 2018;8(5):125. doi: 10.1007/s13201-018-0742-6. [DOI] [Google Scholar]
- Wu X., Kumar V., Ross Quinlan J., Ghosh J., Yang Q., Motoda H., McLachlan G.J., Ng A., Liu B., Yu P.S., Zhou Z.-H., Steinbach M., Hand D.J., Steinberg D. Top 10 algorithms in data mining. Knowl. Inf. Syst. 2008;14(1):1–37. doi: 10.1007/s10115-007-0114-2. [DOI] [Google Scholar]
- Xagoraraki I., O’Brien E. In: Women in Water Quality: Investigations by Prominent Female Engineers. O’Bannon D.J., editor. Springer International Publishing; 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [DOI] [Google Scholar]
- Yadav R.S. Data analysis of COVID-2019 epidemic using machine learning methods: a case study of India. Int. J. Inf. Technol. 2020;12:1321–1330. doi: 10.1007/s41870-020-00484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Su X.G. World Scientific Publishing Co.; 2009. Linear regression analysis: theory and computing. [Google Scholar]
- Zhang D., Duran S.S.F., Lim W.Y.S., Tan C.K.I., Cheong W.C.D., Suwardi A., Loh X.J. SARS-CoV-2 in wastewater: from detection to evaluation. Materials Today Advances. 2022;13 doi: 10.1016/j.mtadv.2022.100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Zhang Q., Sornette D., Jiang L. Cascading logistic regression onto gradient boosted decision trees for forecasting and trading stock indices. Appl. Soft Comput. 2019;84 doi: 10.1016/j.asoc.2019.105747. [DOI] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables
Data Availability Statement
Data will be made available on request.