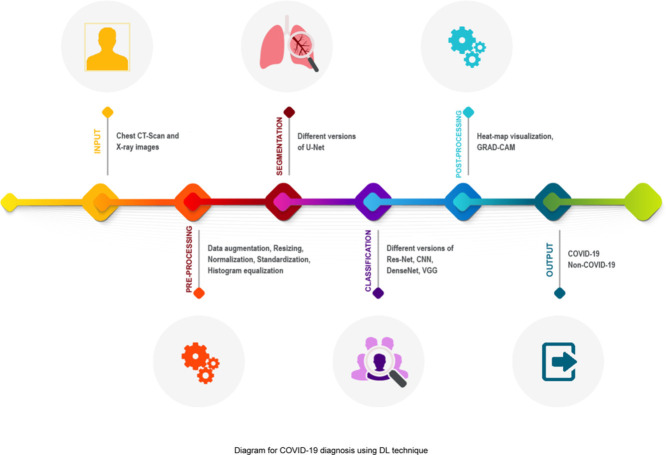
Abstract
The infectious disease known as COVID-19 has spread dramatically all over the world since December 2019. The fast diagnosis and isolation of infected patients are key factors in slowing down the spread of this virus and better management of the pandemic. Although the CT and X-ray modalities are commonly used for the diagnosis of COVID-19, identifying COVID-19 patients from medical images is a time-consuming and error-prone task. Artificial intelligence has shown to have great potential to speed up and optimize the prognosis and diagnosis process of COVID-19. Herein, we review publications on the application of deep learning (DL) techniques for diagnostics of patients with COVID-19 using CT and X-ray chest images for a period from January 2020 to October 2021. Our review focuses solely on peer-reviewed, well-documented articles. It provides a comprehensive summary of the technical details of models developed in these articles and discusses the challenges in the smart diagnosis of COVID-19 using DL techniques. Based on these challenges, it seems that the effectiveness of the developed models in clinical use needs to be further investigated. This review provides some recommendations to help researchers develop more accurate prediction models.
Keywords: Artificial intelligence, Classification, Segmentation, CT-Scan, X-ray
Graphical abstract
List of Abbreviations
- Abbreviation
Definition
- Acc
Accuracy
- AUC
the Area Under the ROC Curve
- CNN
Convolutional neural network
- COVID-19
Coronavirus disease 2019
- CT
Computed Tomography
- DL
Deep Learning
- GAN
Generative Adversarial Network
- GRAD-CAM
Gradient-weighted Class Activation Mapping
- HIS
Hyperspectral Imaging
- LRN
Local Response Normalization
- LUS
Lung Ultrasound
- PET
Positron Emission Tomography
- Pre
Precision
- ResNet
Residual Network
- ROI
Region of Interest
- RT–PCR
Reverse Transcriptase-Polymerase Chain Reaction
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SEM
Scanning Electron Microscopy
- Sen
Sensitivity
- Spe
Specificity
- SVM
Support Vector Machine
- Te
Test
- Tr
Training
- V
Validation
- WHO
World Health Organization
1. Introduction
In early December 2019, the first case of COVID-19, the disease caused by the virus Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), was confirmed by the authorities in Wuhan, China. COVID-19 developed rapidly into a global outbreak and spread all across the world. The World Health Organization (WHO) described the outbreak as a Public Health Emergency of International Concern on 30 January 2020 and declared it a pandemic on 11 March 2020. Globally, as of 6:06pm CEST, 19 October 2022, there have been 623,000,396 confirmed cases of COVID-19, including 6550,033 deaths, reported to WHO (https://covid19.who.int).
Although Reverse Transcriptase-Polymerase Chain Reaction (RT–PCR) is the gold standard for diagnosing COVID-19 [1], the sensitivity of the test is relatively poor, and thus even in patients with a negative RT–PCR result, COVID-19 infection cannot be entirely excluded [2,3]. Therefore, medical imaging, especially chest computed tomography (CT) scan and X-ray, is often used to complement the RT–PCR test to achieve more diagnostic certainty. These imaging techniques have a higher sensitivity than RT–PCR and play a critical role not only in the early diagnosis and treatment of COVID-19 patients but also in monitoring the progress of the disease [2,3]. However, the accuracy of COVID-19 diagnosis using CT and X-ray chest images depends on radiological expertise, and some radiologists may fail to interpret accurately the results of these images, hence leading to reduced sensitivity [4,5].
With recent advances in machine learning techniques, particularly deep learning (DL), and the success of these techniques in medical image processing, scientists and clinicians hope to improve the accuracy of COVID-19 diagnosis by applying deep learning methods to chest medical images. These methods have the potential in providing decision support for clinicians and reducing medical errors. In a short period, we have witnessed a large number of DL models developed for a very broad range of COVID-19-related applications. The purpose of the present review is to give more insight into deep learning applications in the diagnostics of COVID-19. We survey the literature on diagnosing patients infected by COVID-19 using DL techniques for a period from January 2020 to October 2021. Herein, our main focus is on classification and segmentation models proposed based on CT and X-ray chest images. Our review only includes peer-reviewed, well-documented articles and summarizes the technical details of models developed in these articles. We also discuss the challenges of using deep learning for the smart diagnosis of COVID-19. The challenges show that the performance of the models developed in the reviewed articles is probably optimistic and these models are not of potential for clinical use. We finally provide some recommendations that can help researchers develop more accurate and practical models for the COVID-19 diagnosis.
2. Methods
2.1. Data sources and search strategy
This search was intended to address the question: “What deep learning techniques have been developed for the COVID-19 diagnosis using CT and X-ray images?”. For a period from January 2020 to October 2021, we conducted systematic searches of the following on-line databases in order to identify relevant works: IEEE Xplore, ScienceDirect, Springer, PubMed, and Google Scholar.
2.2. Search terms
The keywords used for the literature search were “COVID-19; Coronavirus; Diagnosis; Detection; Artificial Intelligence; Machine Learning; Deep Learning; Medical Imaging; CT-Scan; X-ray.’’ They were connected using “and’’, or “or’’ to identify the articles that deal with the diagnosis of COVID-19 using DL techniques on medical images.
2.3. Selection criteria
The studies with the following criteria were included in the review: (1) Articles that employed machine learning or deep learning techniques for the COVID-19 diagnosis, (2) Articles that utilized techniques to analyze radiographic images (CT scan, and/or X-ray), (3) Articles that applied classification and segmentation models.
The following exclusion criteria were used to eliminate studies from consideration: (1) Articles that were not in the English language, (2) Articles that were not published in peer-reviewed journals, (3) Review Articles, (4) Articles that did not use ML/DL approaches for diagnosing COVID-19 based on medical images, (5) Articles that did not provide a clear explanation of the implemented model and its results.
3. Technical background
This review paper aims to survey different DL models developed in the literature for the smart diagnosis of COVID-19 in people with suspected infection. We focus on classification and/or segmentation techniques using CT and X-ray chest images. For this purpose, this section briefly discusses various DL architectures applied in the reviewed articles.
3.1. Classification models
In this section, the different deep neural networks for COVID-19 classification based on CT and X-ray chest images are discussed.
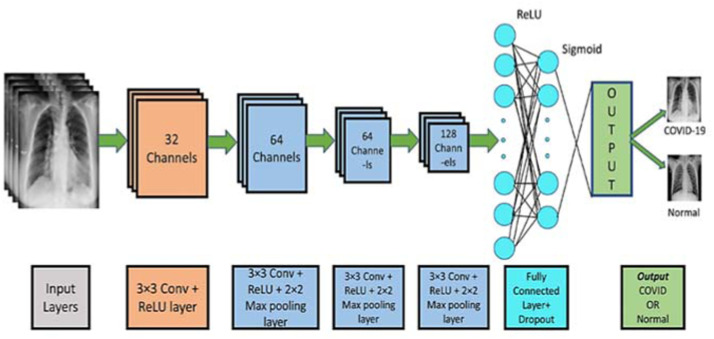
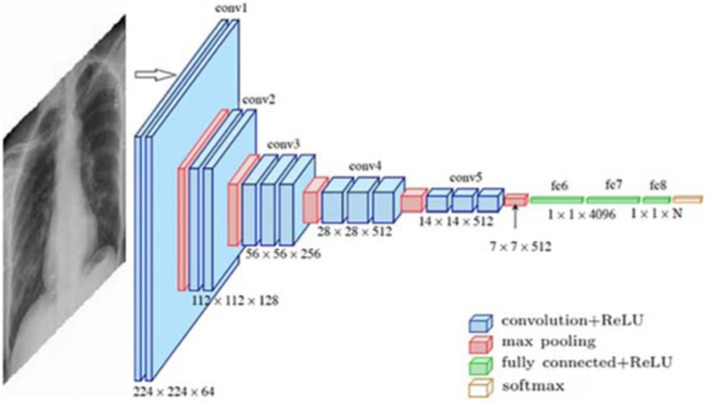
Standard CNN. The architecture of convolutional neural networks (CNN) is inspired by human and animal brains. The main advantage of CNN is detecting relevant features automatically and without any human supervision. CNN tries to overcome the overfitting problem by using convolutional layers. Fig. 1 depicts the architecture of standard deep CNN. The different parts of this architecture are described below:
-
•
The convolutional layer is the main building block of a CNN and applies a convolution operation to the input.
-
•
The pooling layer reduces the dimensions of the feature maps.
-
•
The fully connected layer is usually placed at the end of CNN architecture to do the classification task.
-
•
The loss function calculates the prediction error. Through network learning, this value is optimized.
-
•
Various techniques such as dropout, batch normalization, and data augmentation that are applied for better learning.
Fig. 1.
A CNN model for COVID-19 Detection [6].
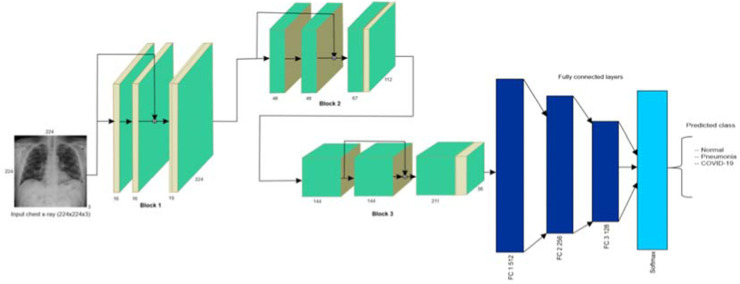
AlexNet. AlexNet is one of the earliest and most widely cited deep neural networks and a leading architecture for any object-detection task. It first introduced two new concepts named Local Response Normalization (LRN) and dropout to help the deep neural network learn better [7]. As a normalization layer, LRN implements the idea of lateral inhibition. Dropout is one of the regularization approaches to avoid the overfitting problem. It randomly skips some neurons during training and forces the other neurons in the layer to pick up the slack. The architecture of AlexNet is shown in Fig. 2 .
Fig. 2.
AlexNet Architecture used for COVID-19 classification [8].
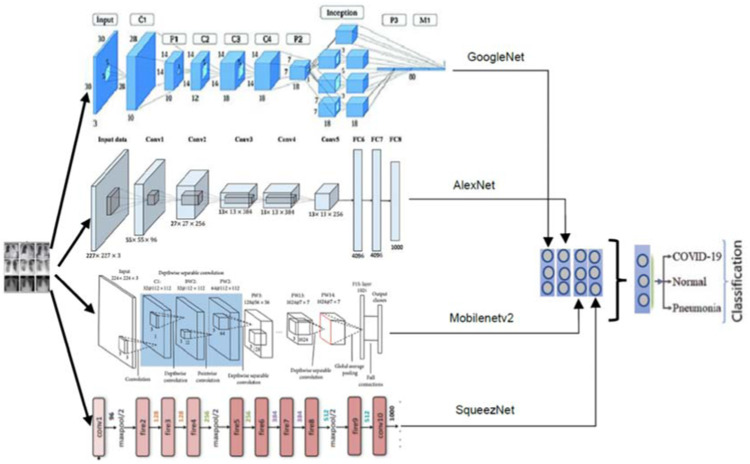
SqueezeNet. SqueezeNet is a convolutional neural network that utilizes design strategies to reduce the number of weights and is considered a more compact replacement for AlexNet [9]. On the ImageNet dataset, it achieves accuracies comparable to AlexNet while performing 3x faster and containing up to 50x fewer parameters. ImageNet (http://www.image-net.org) is a large-scale hierarchical dataset of annotated images for computer vision and machine learning research [10]. Fig. 3 depicts a combined architecture of AlexNet, SqueezeNet, GoogleNet, and MobileNetV2.
Fig. 3.
A combined architecture of AlexNet, SqueezeNet, GoogleNet, and MobileNetV2 for COVID-19 detection [11].
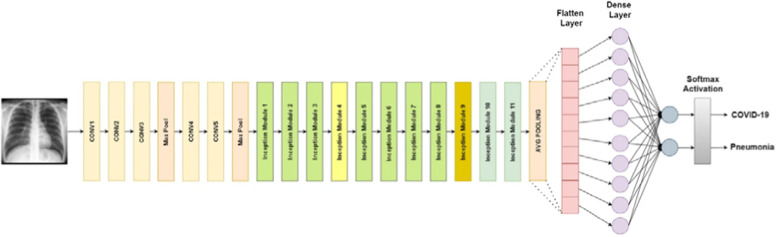
VGGNet. VGG is a classical CNN architecture. It consists of a few convolutional layers that apply the RELU activation function [7]. The number of convolution layers differs in various versions of VGG. A single max-pooling layer and three fully connected layers are placed following these activation layers, As shown in Fig. 4 . This architecture uses the Softmax classifier.
Fig. 4.
The structure of VGG16 for COVID-19 detection [12].
GoogleNet. GoogleNet, also called InceptionNet-V1, proposed the novel Inception blocks that improve recognition accuracy. The distinctive features of this architecture (see Fig. 3) are the large convolution masks, the less dimensional space, and reduced computational complexity. GoogleNet is deeper with much fewer parameters compared to its ancestors AlexNet and VGG [7].
MobileNet. The main characteristic of MobileNet (see Fig. 3) is using depth-wise separable convolution that consists of two layers named depth-wise convolution and point-wise convolution. Using these layers, MobileNet provides an intensive reduction in model size and decreases computational cost by about 8–9 times compared to standard convolutions. A RELU and batch normalization are placed after both of these layers. MobileNet is simple but efficient for mobile and embedded machine vision applications [13].
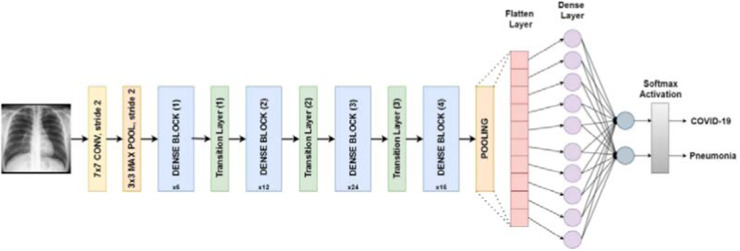
DenseNet. As a type of convolutional neural network, DenseNet utilizes dense connections between convolutional layers where each layer is connected to every other layer. DenseNet was introduced to overcome the issue of vanishing gradient. With effective usage of feature reuse, the network parameters decrease dramatically [14]. Fig. 5 depicts the architecture of a DenseNet network.
Fig. 5.
DenseNet architecture designed for COVID-19 classification [15].
ResNet. The core idea of the Residual network (ResNet) is introducing a technique called skip connection (or Shortcut Connection) that skips some of the layers in the network and feeds the output of a layer as the input to later layers, as shown in Fig. 6 . There are different variants of ResNet architecture composed of a different number of layers. The most popular type of ResNet network is ResNet50 including 49 convolutional layers followed by one fully connected layer [14].
Fig. 6.
ResNet architecture for COVID-19 image classification [16].
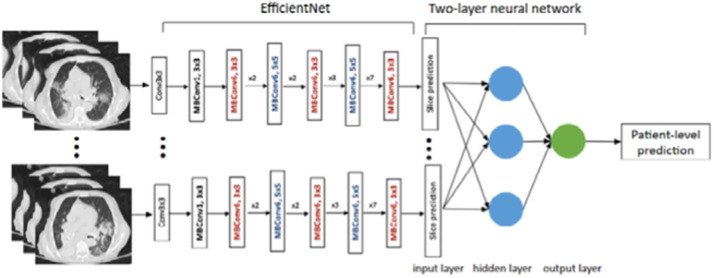
EfficientNet. The EfficientNet network relies on a compound scaling method that uniformly scales all three dimensions of depth/width/resolution while maintaining a balance between all network dimensions. using the compound scaling technique, the authors scaled the baseline network EfficientNet-B0 to get different variations including EfficientNet-B1 to B7. In comparison to squeeze-and-excitation networks, EfficientNet is 7.7 x smaller and 10 x faster [17]. Fig. 7 describes the architecture of an EfficientNet model proposed for COVID-19 image classification.
Fig. 7.
EfficientNet model for COVID-19 classification [18].
InceptionNet. An inception network is composed of repeating components referred to as Inception modules. InceptionNet employs the idea of auxiliary classifiers to address the gradient vanishing problem and improve the convergence of very deep networks. It also applies different convolution kernels of various sizes in parallel. This architecture helps to extract similar features in different sizes simultaneously. The popular versions of InceptionNet are InceptionV1 to V3 (as shown in Fig. 8 ) and Inception-ResNet.
Fig. 8.
InceptionV3 architecture for COVID-19 classification [15].
3.2. Segmentation models
The most common deep learning model developed for image segmentation, U-Net, has been widely utilized for lung region segmentation to diagnose COVID-19 patients:
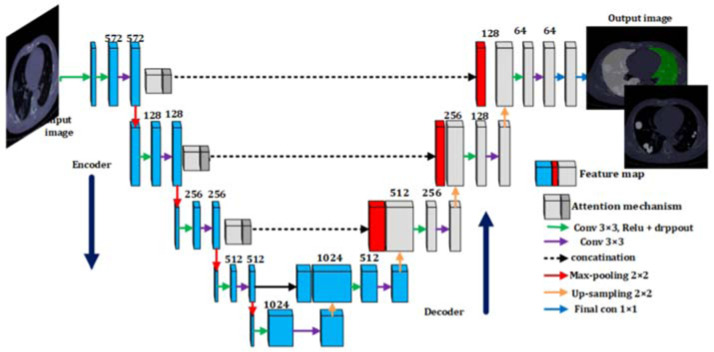
U-NET. U-Net architecture is primarily designed for segmentation applications. Its good performance in segmenting medical images makes it the primary tool for segmentation in this field. The U-Net architecture is an almost symmetrical u-shape network that consists of two paths called decoder and encoder. It is much faster than other segmentation networks, because of its context-based learning [19]. The architecture of U-Net is described in Fig. 9 .
Fig. 9.
The architecture of U-Net for COVID-19 segmentation [20].
3.3. Generative adversarial networks
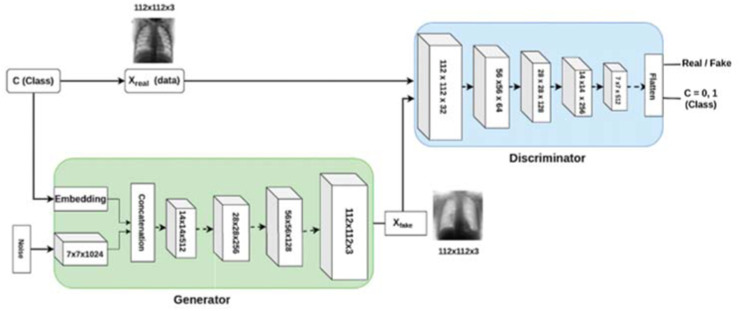
Generative Adversarial Network (GAN) is an unsupervised approach and a powerful method to produce unseen data samples with the same statistics as the training set. This way, it can overcome overfitting and data shortage problems and improve deep network performance. GAN architecture (as shown in Fig. 10 ) is composed of two sub-models named generator model and discriminator model contesting each other in a game. The generator model tries to generate new plausible images similar to real data. The discriminator model tries to distinguish between real and fake samples. This process continues until the generator samples become close to the actual input samples [7].
Fig. 10.
A GAN architecture for COVID-19 detection [21].
4. Results
From all the databases considered for finding relevant works (see Section 2.1 for more information), we identified 15,417 articles that satisfied our search terms, of which, 6209 articles were preprints (not peer-reviewed) and excluded from this review. For duplicate studies (157 articles), we ensured that the latest version of the article was considered. The titles, abstracts, and full texts were screened for relevance and eligibility. We removed 8983 articles that were irrelevant or not documented with enough detail to allow other researchers to reliably reproduce the results. Finally, our investigation retained 68 peer-reviewed, well-documented articles for consideration in this review. This section presents all the technical details of DL models developed in these articles for COVID-19 diagnosis in summary and explains the advantages and disadvantages of the used techniques. Table 1 summarizes the reviewed articles and reports informative details for them.
Table 1.
Summary of recent DL techniques for COVID-19 diagnosis.
| Refs. | Modality | Dataset | # Cases per Class | Test Method | Validation Method | Transfer Learning | ML Approach | Preprocessing | Classification | Segmentation | DNN | Classifier | Postprocessing | Performance Criteria (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [67] | X-ray | Combination of Two Different DBs | 142 COVID-19, 142 Normal Img. | 70%:30% | Hold-out | ImageNet | Supervised | Resizing, Data Augmentation | COVID-19 from Non-COVID-19 | NA | NCOVnet (Based on VGG-16) | Softmax | NA | Acc=97.62, Sen=97.62, Spe=78.57 |
| [68] | X-ray | Combination of Two Different DBs | 295 COVID-19, 65 Normal, 98 Pneumonia Img. | 70%:30% | 5-fold | NA | Supervised | Fuzzy Color Method, Image Stacking Technique | COVID-19 from Other Pneumonia | NA | MobileNetV2, SqueezeNet | SVM | Social Mimic Optimization Method | Acc=99.27 |
| [69] | X-ray | Combination of Two Different DBs | 105 COVID-19, 11 SARS, 80 Normal Img. | 70%:30% | Hold-out | ImageNet | Supervised | Data Augmentation, Histogram, Feature Extraction using AlexNet, PCA, K-means | COVID-19 from Other Pneumonia | NA | DeTraC (Based on ResNet18), AlexNet (Feature Extraction) | Softmax | Composition Phase | Acc=95.12, Sen=97.91, Spe=91.87 |
| [70] | X-ray | COVIDx | 76 COVID-19, 1583 Normal, 4290 Pneumonia Img. | 80%:10%:10% | Hold-out | NA | Supervised | Data Augmentation, RGB format, Normalizing | COVID-19 from Other Pneumonia | NA | COVIDiag-nosis-Net (Based on Deep SqueezeNet with Bayes Optimization) | Decision-Making System | Class Activation Mapping Visualization (Heat Map) | Acc=98.3, Spe=99.13, F1-Score=98.3 |
| [71] | X-ray | Combination of Two Different DBs | 127 COVID-19, 500 No-Findings, 500 Pneumonia Img. | 80%:20% | 5- fold | NA | Supervised | NA | COVID-19 from Non-COVID-19 | NA | DarkCovidNet (Based on CNN) | Linear | Heatmaps Visualization | Acc=98.08, Spe=95.3, Sen=95.13, Pre=98.03, F1-Score=96.51 |
| [72] | X-ray | RYDLS-20 | 90 COVID-19, 10 MERS, 11 SARS, 10 Varicella, 12 Streptococcus, 11 Pneumocystis Img. | 70%:30% | Hold-out | ImageNet | Supervised | Different Features, Early Fusion, Late Fusion, Different Resampling Algorithms | COVID-19 from Other Pneumonia | NA | Inception-V3 | Clus-HMC Framework | Friedman Statistical Test for Ranking | F1-Score=88.89 |
| [73] | X-ray | Combination of Two Different DBs | 284 COVID-19, 310 Normal, 330 Pneumonia Bacterial, 327 Pneumonia Viral Img. | 75%:25% | 4-fold | ImageNet | Supervised | Rescaling | COVID-19 from Other Pneumonia | NA | CoroNet (Based on Xception Architecture) | Softmax | NA | Acc=89.5, Pre=97, F1-Score=98 |
| [74] | X-ray | Combination of Four Different DBs | 455 COVID-19, 2109 Non-COVID Img. | 90%:10% | 10-fold | NA | Supervised | Rescaling, Data Augmentation | COVID-19 from Non-COVID-19 | NA | MobileNet V2 | NA | NA | Acc=99.18, Sen=97.36, Spe=99.42 |
| [21] | X-ray | Combination of Three Different DBs | 403 COVID-19, 721 Non-COVID Img. | 932–192 | Hold-out | NA | Supervised | Resizing, Normalization, Data Augmentation using CovidGAN Based AC-GAN | COVID-19 from Non-COVID-19 | NA | VGG16 | Softmax | PCA Visualization | Acc=95, Sen=90, Spe=97 |
| [75] | X-ray | Combination of Three Different DBs | 69 COVID-19, 79 Normal, 79 Pneumonia Viruses, 79 Pneumonia Bacterial Img. | 72%:18%:10% | Hold-out | ImageNet | Supervised | Data Augmentation using GAN | COVID-19 from Non-COVID-19 | NA | GoogleNet | Softmax | NA | Acc=100, Pre=100,F1-Score=100 |
| [76] | X-ray | Combination of Three Different DBs | 224 COVID-19, 504 Healthy, 400 Bacteria, 314 Viral Pneumonia Img. | 90%:10% | 10-fold | ImageNet | Supervised | Resizing | COVID-19 from Non-COVID-19 | NA | MobileNet | NA | NA | Acc=96.78, Sen=98.66, Spe=96.46 |
| [47] | X-ray | Pediatric cxr,Twitter COVID-19 cxr, Montreal COVID-19 cxr DBs | 7595 Normal, 6012 Pneumonia of Unknown Type, 2780 Bacterial, 313 COVID-19 Img. | 72%:18%:10% | Hold-out | ImageNet | Supervised | Pixel Rescaling, Median Filtering for Noise Removal and Edge Preservation, Normalization, Standardization for Identical Feature Distribution | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | U-Net (Segmentation), Ensemble of Pruned Models: VGG-16, VGG-19, and Inception-V3 (Classification) | Softmax | Grad-CAM | Acc=99.01, Sen=99.01, Pre=99.01,F1-Score=99.01, AUC=99.72, MCC=98.2 |
| [77] | X-ray | Combination of Two Different DBs | 180 COVID-19, 6054 Pneumonia, 8851 Normal Img. | 3784–11,302 | 5-fold | ImageNet | Supervised | Data Augmentation | COVID-19 from Other Pneumonia | NA | Concatenation of the Xception and ResNet50V2 | Softmax | NA | Acc=91.4 |
| [78] | X-ray | Combination of Two Different DBs | 181 COVID-19, 364 Healthy Img. | 80%:20%:20% | Hold-out | ImageNet | Supervised | Normalization, Resizing | COVID-19 from Non-COVID-19 | NA | VGG-19 | Softmax | NA | Acc=96.3 |
| [37] | X-ray | Combination of Three Different DBs | 250 COVID-19, 2753 Other Pulmonary Diseases, 3520 Healthy Img. | 2000–803–1100 | Hold-out | ImageNet | Supervised | Resizing, Data Augmentation | COVID-19 from Other Pneumonia | NA | VGG-16 | Softmax | Grad-CAM | Acc=98, Sen=87, Spe=94,F1-Score=89 |
| [51] | X-ray | Combination of three Clinical DBs | 610 COVID-19, 1493 Non-COVID-19, 1888 Normal, 305 Pneumonia Viral, 3085 Bacterial Pneumonia Img. | 80%:20% | 5-fold | Clinical | Supervised | Reshaping to Different Resolutions, Normalization | COVID-19 from Non-COVID-19 | NA | Stacked Multi-Resolution CovXNet | Stacking using Meta-Learner | Grad-CAM | Acc=97.4, Spe=94.7, F1-Score=97.1, Recall=97.8, Pre=96.3, AUC=96.9 |
| [79] | X-ray | Combination of Different DB | 162 COVID-19, 2003 Healthy, 4280 Viral and Bacterial Pneumonia, 400 Tuberculosis Img. | 90%:10% | 10-fold | ImageNet | Supervised | Resizing, Normalization, Data Augmentation using CovidGAN Based AC-GAN | COVID-19 from Other Pneumonia | NA | Truncated InceptionNet V3 | Softmax | Activation Maps Generation | Acc=98.77, Sen=95, Spe=99, Pre=99, F1-Score=97 |
| [80] | X-ray | Combination of Five Different DBs | 180 COVID-19, 191 Normal, 54 Bacterial Pneumonia, 20 Viral Pneumonia Img. | 70%:10%:20% | Hold-out | ImageNet | Supervised | Standard Preprocessing, Segmentation, Data Type Casting, Histogram Equalization, Gamma Correction, Resizing | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | ResNet18 (Classification), FC-DenseNet103 (segmentation) | Majority Voting | Probabilistic Grad-CAM Saliency Map Visualization | Sen=100, Pre=76.9 |
| [81] | X-ray | COVIDx, Clinical DB | 9466 Normal, 9501 Non-COVID Pneumonia, 283 COVID-19 Img. | 90%:10% | 10-fold | NA | Supervised | NA | COVID-19 from Non-COVID-19 | NA | Faster R–CNN | NA | NA | Acc=97.36,F1-Score=98.46, Pre=99.29, Spe=95.48, Sen=97.65 |
| [38] | X-ray | Combination of Two Different DBs | 1428 COVID-19, 700 Bacterial Pneumonia, 504 Healthy Img. | 70%:30% | Hold-out | ImageNet | Supervised | Data Augmentation | COVID-19 from Non-COVID-19 | NA | VGG-16 | Softmax | Grad-CAM | Acc=96 |
| [82] | X-ray | Combination of Three Different DBs | 585 Abnormal, 585 Normal Img. | 1000–170 | Hold-out | ImageNet | Supervised | Resizing, Data Augmentation | COVID-19 from Non-COVID-19 | NA | ResNet18 | Softmax | NA | Acc=98,Sen=99, Spe=97,Pre=97, AUC=98 |
| [83] | X-ray | Combination of Three Different DBs | 137 COVID-19, 150 Pneumonia Img. | 80%:20% | 5-fold | ImageNet | Supervised | Resizing, Data Augmentation, Normalization | COVID-19 from Other Pneumonia | NA | Inception V3 | Softmax | Grad-CA, resize | AUC=1,Acc=100, Sen=99,Spe=100 |
| [48] | X-ray | BIMCV-COVID19+, PadChest, Clinical DB | 2589 COVID-19, 4337 Normal Img. | 80%:20% | 5-fold | NA | Supervised | Histogram Matching Process, Rib Shadows Suppression, Convert to Grayscale, Contrast Enhancement by Contrast Limited Adaptive Histogram Equalization, Data Augmentation, Random Rotations, Width and Height Shift, Shear, Zoom, Horizontal Flips | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | U-Net (Segmentation), COVID-Xnet (Based on CNN, Classification) | Softmax | Grad-CAM | Acc=94.43, AUC=98.8, F1-score=93.14, Pre=93.76, Spe=96.3, Sen=92.53 |
| [84] | X-ray | Combination of Four Different DBs | 219 COVID-19, 1341 Normal, 1345 Viral Pneumonia Img. | 70%:30% | Hold-out | NA | Supervised | Data Augmentation | COVID-19 from Other Pneumonia | NA | CNN | SVM | NA | Acc=98.97, Sen=89.39, Spe= 99.75,F1-Score=96.72 |
| [6] | X-ray | Clinical | 659 Normal, 295 COVID-19 Img. | 80%:20% | Hold-out | NA | Supervised | NA | COVID-19 from Non-COVID-19 | NA | CNN | Sigmoid | NA | Acc=97.5,F1-Score=97.5, Pre=97.5, Recall=97.5, ROC=0.975 |
| [85] | X-ray | Combination of Two Different DBs | 202 COVID-19, 300 Normal, 300 Pneumonia Img. | 80%:20% | 5-fold | ImageNet | Supervised | Intensity Normalization, Class-label Encoding, Data Augmentation by CycleGAN, Width and Height Shift, Random Rotation, Horizontal Flips | COVID-19 from Other Pneumonia | NA | EfficientNetB0 | Softmax | Grad-CAM | Acc=96.8 |
| [86] | X-ray | Combination of Two Different DBs | 224 COVID-19, 700 Bacterial, 504 Healthy: Patients | 70%:30% | Hold-out | ImageNet | Supervised | Data Augmentation, Resizing | COVID-19 from Non-COVID-19 | NA | VGG-16 | Softmax | Grad-CAM (to highlight the regions of interest) | Acc=96, spec=97.27, Sen=92.64 |
| [87] | X-ray | Combination of Different DBs | 2665 COVID-19, 3692 Pneumonia, 3692 Normal | 9149- 450- 450 | NA | ImageNet | Supervised | Data Augmentation, Resizing, Lung Segmentation, CLAHE histogram equalization, Denoising | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | VGG-19 (Classification), U-Net (Segmentation) | Naive Bayes | Grad-CAM visualization | Acc=98.67,Kappa score=0.98,F1-Score=100 |
| [88] | X-ray | Cohen and Kaggle | 455 Normal, 457 Bacterial Pneumonia, 470 Viral Pneumonia (Stage1), 480 Viral Pneumonia (Stage2), 440 COVID-19 Img. | 80%:20% | 5-fold | ImageNet | Supervised | Resizing, Normalization, Data Augmentation | COVID-19 from Other Pneumonia | NA | Resnet101 | Softmax | Grad-CAM visualization | Acc=98.93, Sen=98.93, Spec=98.66, Pre=96.39, F1-score=98.15 |
| [5] | X-ray | COVIDx | 8066 Normal, 5521 Pneumonia, 183 COVID-19 Img. | 13,569–231 | NA | ImageNet | Supervised | Intensity Normalization, Resizing, Data Augmentation | COVID-19 from Other Pneumonia | NA | EfficientNet | Swish | NA | Acc=93.9,Sen=96.8, PPV=100 |
| [89] | X-ray | Kaggle | 219 COVID-19, 1341 Normal, 1345 Viral Pneumonia Img. | 70%:10%:20% | 5-fold | NA | Supervised | Cropping, Resizing | COVID-19 from Other Pneumonia | NA | CVDNet | Softmax | NA | Acc=96.69 |
| [90] | X-ray | Combination of Six Different DBs | 900 COVID-19, 900 Normal, 900 Pneumonia Img. | 70%:30% | Hold-out | ImageNet | Supervised | Resizing, Converting to Color Image | COVID-19 from Other Pneumonia | NA | E‑DiCoNet | ELM | NA | Acc=94.07, Sen=98.15, Spec=91.48 |
| [91] | X-ray | Combination of Three Different DBs | 543 COVID_19, 600 Normal, 600 Pneumonia Img. | 1220–523 | Hold-out | ImageNet | Supervised | Resizing | COVID-19 from Other Pneumonia | NA | AlexNet, ReliefF | SVM | NA | Acc=98.64, Spec=98.64, Sen=98.64, F-score=98.63 |
| [92] | X-ray | chest X-ray (CXR) dataset | 27 Normal, 220 SARS, 17 Streptococcus Img. | 80%:20% | 5-fold | ImageNet | Supervised | Noise Removal by Wiener Filtering | COVID-19 from Other Pneumonia | NA | FM-CNN | MLP | NA | Acc=98.06, Spec=98.29, Sen=97.22, F-score=97.93 |
| [93] | X-ray | Combination of Different DBs | 423 COVID-19, 1341 Normal, 1345 Viral PNA Img. | 80%:20% | 5-fold | ImageNet | Supervised | Image Resize, CLAHE Image Enhancement, Image Augmentation | COVID-19 from Other Pneumonia | NA | VGG-19 | LMPL (Large margin piecewise linear) | Heatmap Visualization | Acc=99.39, F1-score=99.45, Pre=99.47, Sen=99.42 |
| [94] | CT-Scan | Clinical | 219 COVID-19, 224 Other Pneumonia, 175 Healthy Img. | 85.4%: 14.6% | Hold-out | NA | Supervised | Segmentation, Data Augmentation | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | ResNet-18 (Classification), VNET-IR-RPN (Segmentation) | Voting Strategy | Total Infection Confidence Score Calculation using Probability Formula of the Noisy-or Bayesian Function | Acc=86.7, Pre=81.3, F1-Score=83.9 |
| [95] | CT-Scan | COVID-19 DB | 1262 Positive COVID-19, 1230 Negative COVID-19 Img. | 68%:17%:15% | Hold-out | ImageNet | Supervised | Data Augmentation | COVID-19 from Non-COVID-19 | NA | DenseNet201 | Sigmoid | NA | Pre=96.29, Recall=96.29, F1-Score=96.29, Spe=96.21,Acc=96.25 |
| [96] | CT-Scan | Clinical | 368 COVID-19 Patients, 127 Patients with Other Pneumonia | 395–50–50 | Hold-out | NA | Supervised | Segmentation using Threshold Segmentation and Morphological Optimization Algorithms, Rescaling, Multi-view Fusion | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | ResNet50 | Dense Layer | NA | Acc=76, Sen=81.1, Spe=61.5 |
| [97] | CT-Scan | Combination of Different DBs | 1029 COVID-19, 1695 Non-COVID Img. | 1059–328–1337 | Hold-out | NA | Supervised | Clipping, Cropping, Data Augmentation by Image Intensity and Contrast Adjustment, Random Gaussian Noise, Flipping, and Rotation | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | DensNet-121 (Classification), AH—Net (Segmentation, Based on ResNet50) | NA | Grad-CAM | Acc=90.8, Sen=84, Spe=93, ppv=79.4, npv=94.8, Auc=94.9 |
| [98] | CT-Scan | Clinical | 108 COVID-19, 86 Non-COVID Patients | 80%:20% | Hold-out | ImageNet | Supervised | Different Methods | COVID-19 from Non-COVID-19 | NA | ResNet-101, Xception | Softmax | NA | Sen=98.04, Spe=100, Acc=99.02 |
| [99] | CT-Scan | Clinical | 3389 COVID-19, 1593 Non-COVID Img. | Separate DB | 5-fold | NA | Supervised | Standard Preprocessing, VB-Net Toolkit for Segmentation, Lung Mask Generation | COVID-19 from Non-COVID-19 | Lung Lesion-Oriented Method | 3D ResNet34 (Classification), VB-Net (Segmentation) | Ensemble Learning | Grad-CAM | Acc=87.5, Sen=86.9, Spe=90.1, AUC=94.4, F1-Score=82.0 |
| [100] | CT-Scan | Clinical | 146 COVID-19, 149 Non-COVID Img. | 135–20–140 | Hold-out | NA | Supervised | Data Augmentation | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | DenseNet (Classification) | Softmax | CAM | Acc=92, Sen=97, Spe=87, F1-Score=93 |
| [101] | CT-Scan | Clinical | 558 COVID-19 Patients | 60%:20%:20% | 5-fold | NA | Weakly-supervised learning | NA | COVID-19 from Other Pneumonia | NA | AD3D-MIL (Based on CNN) | NA | Proposed Loss Function | Dice=80.72, RVE=15.96 |
| [102] | CT-Scan | Combination of Two Different DBs | 1118 COVID-19, 96 Pneumonia, 107 Healthy Img. | 70%:30% | Hold-out | NA | Supervised | Binary Mask, Lung Segmentation using Histogram Thresholding, Morphological Operation (Dilation, Hole Filling), and Removing All Small Connected Objects | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | CNN | LSTM | NA | Acc=99.68 |
| [103] | CT-Scan | Clinical | 151 COVID-19, 498 Non-COVID Patients | 490–82–77 | Hold-out | NA | Supervised | Resizing, Padding, Data Augmentation | COVID-19 from Non-COVID-19 | NA | CNN | Softmax | Visual Interpretation | AUC=70 |
| [104] | CT-Scan | Combination of Two Different DBs | 413 COVID-19, 439 Normal or Pneumonia Img. | 50%:10%:40% | 10-fold | ImageNet | Supervised | NA | COVID-19 from Non-COVID-19 | NA | ResNet | Softmax | NA | Acc=93.01, Spe=94.77, Sen=91.45, Pre=95.18 |
| [105] | CT-Scan | Clinical | 73 COVID-19 Patients | NA | 20-fold | NA | Supervised | Hyper-Parameter Tuning using MODE Algorithm | COVID-19 from Non-COVID-19 | NA | CNN | NA | NA | Acc=92, F1-Score=90, Sen=90, Spe=90 |
| [106] | CT-Scan | Clinical | 98 COVID-19, 103 Non-COVID Patients | 80%:10%:10% | Hold-out | NA | Supervised | Visual Inspection | COVID-19 from Non-COVID-19 | NA | BigBiGAN | Linear | NA | AUC=97.2, Sen=92, Spe=91 |
| [107] | CT-Scan | COVID-19 Set | 470 COVID-19 Suspects | 370–100 | Hold-out | COPDGene | Supervised | Standardization, Re-scaling, Down Sampling, Zero padding | NA | Lung Region-Oriented Method | RTSU-Net (Based on RU-Net) | Sigmoid, Softmax | Upsampling | IOU=92.2, ASSD=86.6 |
| [44] | CT-Scan | Clinical | 79 COVID-19, 100 Common Pneumonia, 130 Patients without Pneumonia | 378–50–130 | 5-fold | NA | Semi-supervised | Data Augmentation | NA | Lung Lesion-Oriented Method | AD3D-MIL (Based on CNN) | Bernouli Distribution | CAM | Acc=97.9, AUC=99, F1-Score=97.9, Pre=97.9, Recall=97.9 |
| [108] | CT-Scan | COVID-19 Clinical DB | 150 COVID-19, 150 CAP, 150 NP Img. | 80%:20% | 5-fold | TCIA | Weakly-supervised | Data Augmentation, Fixed-Sized Sliding Window, Segmentation | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | CNN (Classification), U-Net (Segmentation) | Softmax | Multi-Window Voting, Sequential Information Attention Module, CAM (Class Activation Maps), Categorical- Specific Joint Saliency | Acc=96.2, Pre=97.3, Sen=94.5, Spe=95.3, AUC=97 |
| [45] | CT-Scan | COVID-SemiSeg | 150 COVID-19, 150 CAP, 150 NP Img. | 45–5–50 | Hold-out | 1600 CT images with pseudo labels | Semi-supervised | Pseudo Label Generation, Resizing | NA | Lung Lesion-Oriented Method | Semi-Supervised Inf-Net | Sigmoid | NA | Dice=73.9, Sen=72.5, Spe=96, MAE=6.4 |
| [43] | CT-Scan | Clinical | 1315 COVID-19, 3342 Non-COVID Img. | 3997- 60 - 600 | 5-fold | NA | Weakly-supervised | Lobe Segmentation, Cropping, Resizing, Data Augmentation | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | 3D-ResNets (Classification), 3d-U-Net (Segmentation) | Softmax | Heatmap Visualization | Acc=93.3, Sen=87.6, Spe=95.5 |
| [50] | CT-Scan | Clinical | 296 COVID-19, 1735 CAP, 1325 Non-Pneumonia Img. | Separate DB | NA | NA | Supervised | Lung Segmentation using U-net | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | COVNet (Classification, Based on ResNet), U-Net (Segmentation) | Softmax | Grad-CAM | AUC=96, Sen=90, Spe=96 |
| [109] | CT-Scan | Clinical | 1266 COVID-19, 4106 Lung Cancer Patients | Separate DB | NA | Clinical | Supervised | Lung Segmentation, 3-Dimensional Bounding Box, Non-Lung Area Suppression Operation | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | DenseNet (Segmentation), COVID-19Net (Classification) | Sigmoid | Combining Feature Vectors, Multivariate Cox Proportional Hazard Model, Visualizations | Acc=81.24, AUC=0.90, Sen=78.93, Spe=89.93, F1-Score=86.92 |
| [18] | CT-Scan | Clinical | 521 COVID-19, 665 Non-COVID Patients | 70%:20%:10% | Hold-out | ImageNet | Supervised | Lung Segmentation using Thresholding and The Manual Active Contour Segmentation Method, Padding, Resizing, Lung windowing, Normalization, Standardization | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | EfficientNet B4 | Sigmoid | Generating Heat Map | Acc=96,Sen=95, Spec=96 |
| [110] | CT-Scan | Combination of Two Different DBs | 1272 COVID-19,1230 Non-COVID Img. | 70%:30% | 5-fold and 10-fold | NA | Semi-supervised | Normalization, Standardization. | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | PQIS-Net (Segmentation) | Majority Voting | Patch-Based Classification on Segmented Lung CT Images | Acc=93.1, Pre=89, Recall=83.5,F1-Score=82.6,AUC=98.2 |
| [111] | CT-Scan | Combination of Different DBs | 1194 COVID-19, 1357 Non-COVID Pneumonia, 1442 Nonpneumonia Img. | 80%:20% | Hold-out | ImageNet | Supervised | Converting to One-channel Grayscale PNG Images, Rescaling, Normalizing, Data Augmentation | COVID-19 from Other Pneumonia | NA | ResNet-50 | Softmax | Grad-CAM | Acc=99.87,Spec=100, Sen=99.58 |
| [46] | CT-Scan | Clinical DB, SPIE-AAPM-NCI | 111 COVID-19, 115 CAP, 109 Normal Patients | 300–15–20 | Hold-out | NA | Supervised | Generating Pseudo-Infection Anomalies using Perlin Noise, Resampling, Resizing, Normalization | COVID-19 from Other Pneumonia | Lung Lesion-Oriented Method | BCDU-Net (Segmentation), CNN (Classification) | NA | NA | Acc=86.66,Spec=100, Sen=90.91 |
| [112] | CT-Scan | Clinical | 255 COVID-19, 420 Typical Viral Pneumonia Img. | Separate DB | Hold-out | ImageNet | Supervised | Converting to Grayscale, Grayscale Binarization, Background Area Filling, Reverse Color, Cropping To Obtain ROI Images | COVID-19 from Non-COVID-19 | NA | M-inception (Based on GoogleNet Inception-V3) | Softmax | NA | Acc=79.3,Spec=83, Sen=67,AUC=81,F1-Score = 63 |
| [113] | CT-Scan | Clinical | 51 COVID-19 Patients, 55 Patients with Other Diseases | Separate DB | Hold-out | ImageNet | Supervised | NA | NA | Lung Lesion-Oriented Method | UNet++ (Based on ResNet-50) | NA | NA | Acc=96, Spec=94, Sen=98, PPV=94.23, NPV=97.92 |
| [114] | CT-Scan | Clinical | 777 COVID-19, 505 Bacteria Pneumonia, 708 Normal Img. | 60%:10%:30% | Hold-out | ImageNet | Supervised | Lung Detection, Filling the Blank Area of Image with Its Rotational Lung Areas |
COVID-19 from Other Pneumonia | NA | DRENet (Based on ResNet-50 and FPN) | NA | Grad-CAM | Pre=93, Recall=93, Acc=93, Spec=93, F1-Score=93 |
| [115] | CT-Scan | Mosmed Dataset | 1100 COVID-19, 1980 Normal Img. | 1980–1100 | NA | ImageNet | Supervised | Resizing, Random Crops | COVID-19 from Non-COVID-19 | NA | ResNet-50 | Majority Voting | NA | Acc=96, AUC=90, Sen=100, Spec=96 |
| [116] | CT-Scan | COVID-CTx | 324 COVID-19, 504 Normal Img. | 60%:20%:20% | 3-fold | NA | Supervised | Resizing, Lung Segmentation, K-means Clustering, Contrast Enhancement, Morphological Closing, Hole Filling, Data Augmentation | COVID-19 from Non-COVID-19 | Lung Region-Oriented Method | AM-SdenseNet (classification), U-Net (Segmentation) | Sigmoid | NA | Acc=99.18, Pre=99.32, Recall=98.97, F1-score=91.14 |
| [117] | CT-Scan | Combination of Two Different DBs | 1601 COVID-19, 1626 Non-COVID Img. | 80%:20% | Hold-out | NA | Supervised | NA | COVID-19 from Non-COVID-19 | NA | EfficientNet and Convolutinal Block Attention Module (CBAM) | SVM | NA | Acc=98, Pre=98, Sen=98, F1-score=98 |
| [118] | CT-Scan | Combination of Two Different DBs | 259 COVID-19, 171 Non-COVID-19 Patients | NA | 10-fold | DeepLesi LIDC-IDRI | Supervised | Image Resize, Image Augmentation (Resizing, Random Flipping, Random Cropping, Color Distortions) | COVID-19 from Non-COVID-19 | NA | Prototypical Network | Relu | NA | Acc=88.5, Pre=89.9, Recall=88.6, AUC=94.5 |
| [119] | CT-Scan, X-ray | Combination of Two Different DBs | 3065 COVID-19, 3065 Non-COVID Img. | 70%:30% | NA | NA | Supervised | Data Augmentation | COVID-19 from Non-COVID-19 | NA | ConvLSTM | NA | NA | Acc=98.45, F1-score=98.07, Mcc=96.81 |
| [49] | CT-Scan, X-ray | Combination of Two Different DBs | 2780 Bacterial Pneumonia, 1493 Viral Pneumonia, 231 COVID-19, 1583 Normal Img. | 80%:20% | NA | ImageNet | Supervised | Intensity Normalization, CLAHE Method, DA, Resizing | COVID-19 from Other Pneumonia | Lung Region-Oriented Method | U-Net (Segmentation), Inception ResNetV2 (Classification) | MLP | NA | Acc=92.18, Sen=92.11, Spec=96.06, Pre=92.38, F1-Score=92.07 |
Generally, imaging modalities considered in COVID-19 diagnosis are X-ray, Computed Tomography (CT), and ultrasound. Although ultrasound imaging is a more widely available, cost-effective, safe, and real-time imaging technique [22], it has a relatively lower sensitivity in comparison to chest CT-Scan [23] and cannot usually detect lesions that are deep and intrapulmonary [24,25]. Since more deep learning-based COVID-19 detection and segmentation works focus on CT and X-ray modalities, we only reviewed articles that have used these modalities. The column Imaging Modality in the table indicates which imaging modalities have been used as the input to the DL network developed in each article. Chest radiological imaging, such as CT and X-ray, has a vital role in the early diagnosis and treatment of COVID-19 disease [26].
The second column Dataset refers to image datasets collected by or utilized in the reviewed articles. These datasets contain CT and/or X-ray images from healthy, COVID-19, and Non-COVID patients. Since COVID-19 is a new disease, there is no dataset of appropriate size for its diagnosis purpose. Therefore, many works combine images from several different publicly available datasets, gather chest images from multiple institutions, or use a combination of their clinical images and public datasets.
The column # Cases Per Class indicates the number of images/patients in each class. In other words, the value of each cell in this column shows which classes have been considered in the corresponding article, and how many images or patients there are in each class. COVID-19 datasets are strongly imbalanced which can impact the model accuracy. Hence, several studies have proposed different class balancing techniques to overcome the data imbalance issue [27].
The column Test Method shows which technique has been used for splitting data into three non-overlapping parts including the Training (Tr) set, Validation (V) set, and Test (Te) set. The division of input data into these three subsets is crucial in the creation of robust prediction models to avoid overfitting, as well as to increase generalization. Overfitting is one of the main issues in the training of machine learning algorithms [28]. It occurs when the training error is low, and the generalization error is high. Although a small training dataset is the main reason for the overfitting problem, the complexity of the model can be another reason [29]. Many works apply a validation set to identify the overfitting problem during the model training by controlling the model complexity [30]. It can help the model determine when to stop the training process. For simplicity, we use two different patterns for reporting the test method. The first pattern is Tr%: V%: Te% which indicates the percent of data assigned to each part, and the second pattern is Tr-V-Te showing the number of data assigned to each part. We use the phrase “Separate DB” for indicating the studies which have utilized separate datasets for training and testing their DL models.
The column Validation Method refers to the validation method used to evaluate the efficiency of DL models. The applied validation methods contain K-fold cross-validation and the holdout method. The K-fold cross-validation technique ensures that every observation from the original dataset has the chance of appearing in the training and test sets [31]. In this approach, at first, the data is randomly split into K equal-sized folds (the K value is usually selected in the range of five to ten depending on the data size). Then, the model is trained on the K-1 folds and validated on the remaining fold. This process is repeated K times, with each of the K folds used exactly once as the test set. This technique is one of the best validation approaches if there are not enough instances to train on [32]. On the other hand, the holdout method partitions data randomly into two sets, training, and test. The size of the test set is typically smaller than that of the training set. Since the method involves a single run, results depend on how the data is split into these two sets [33]. The holdout method is not effective for comparing multiple models and tuning their hyper-parameters. Therefore, another very popular form of this method is usually utilized which splits data into three separate sets including training, validation, and test sets [34]. The validation set which is a holdout subset from the training set is used to tune various hyper-parameters and choose the best performing model. The final model is evaluated by the test set and the found error is considered the generalization error.
The dataset used for the transfer learning process is reported in the column Transfer Learning. The philosophy behind transfer learning is that people can intelligently utilize knowledge learned in the past to solve new problems faster or more efficiently [35]. Transfer learning is an approach in machine learning where knowledge learned in one or more tasks is transferred to improve learning in another related task [36]. The main goal of this technique is to decrease training task time, improve generalization performance [37], and deal with small input dataset problems [38]. Transfer learning has been widely adopted in many COVID-19 studies to compensate for the scarcity of large-scale public datasets for this disease.
The column Learning Method can contain the values Supervised, Unsupervised, Semi-supervised, and Weakly-supervised, showing which machine learning approach has been used for training the deep neural network. Supervised learning refers to a class of methods that learn on a labeled dataset, where each training sample is a pair of inputs and its desired output [39]. In an unsupervised learning model, in contrast, the algorithm tries to extract features and patterns from unlabeled data. In other words, the algorithm does not receive any feedback from the environment [40]. Clustering, association, and dimensionality reduction are three main tasks in unsupervised learning. A weakly-supervised model utilizes training data with incomplete, inexact, and inaccurate annotation [41]. Finally, semi-supervised learning describes a class of algorithms that seek to learn from both unlabeled and labeled samples typically assumed to be sampled from the same or similar distributions [42].
Since labeling lesion annotations is time-consuming and impractical for clinicians [43], Noisy labels exist widely for the segmentation of large-scale 3D medical images. It can be either due to challenges for accurate annotation, such as low contrast, ambiguous boundaries, and complex appearances of the target, or caused by low-cost inaccurate annotations such as annotations provided by non-experts, human-in-the-loop strategies [44], and some algorithms generating pseudo labels [45,46]. For this reason, some COVID-19 studies have applied weakly supervised or semi-supervised learning frameworks in their segmentation tasks.
The pre-processing functions applied to input images are shown in the column Pre-processing. In image processing, the pre-processing phase aims to process raw input images to improve the quality of images, produce a more understandable format for the algorithm, enhance accuracy, increase the number of data, reduce the processing time, or standardize data acquired from multiple devices [47,48]. There are several important steps in the pre-processing phase, such as data augmentation, histogram equalization, image normalization, morphological operators, segmentation, standardization, etc.
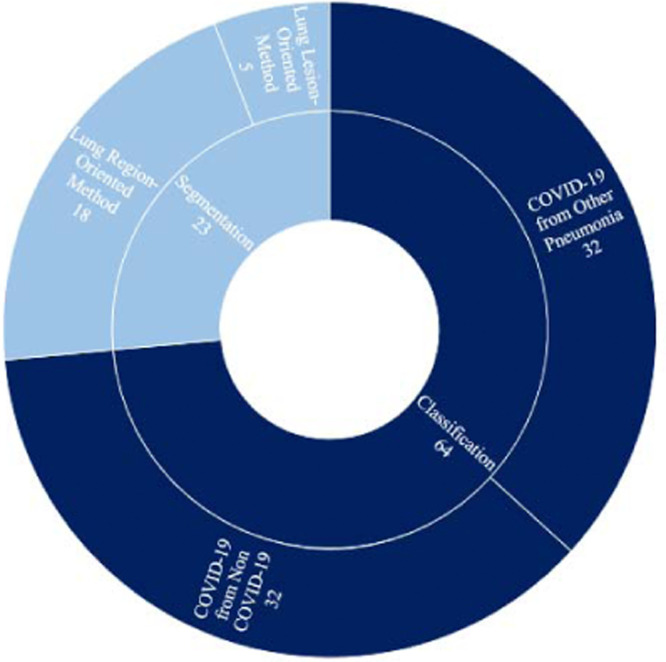
The column Classification categorizes COVID-19 classification models into two main groups: COVID-19 from Non-COVID and COVID-19 from Other Pneumonia. The former indicates a two-class classification where the class Non-COVID includes common pneumonia cases and non-pneumonia cases, and the latter shows a multi-class classification that distinguishes among COVID-19, other types of pneumonia, and healthy cases.
The column Segmentation classifies COVID-19 segmentation models into two categories. The first category, called the Lung-Region-Oriented method [47,49,50], is a prerequisite step in COVID-19 applications and separates the whole lung region and lung lobes from other regions in input images. The second category, called the Lung-Lesion-Oriented method [44,51], aims to separate lesions or metal and motion artifacts from the lung regions which is a challenging detection task.
The column DNN, which stands for deep neural network, shows the networks utilized for classification and segmentation tasks. In addition to well-known models, some new DNNs have been also introduced in the reviewed articles to detect COVID-19 infection in CT and X-ray chest images.
The value of the column Classifier indicates the classifier algorithm used in deep learning models for COVID-19 diagnosis purposes. These functions are applied in the last layer of a neural network. Some of the most used classifiers are introduced as follows. The Softmax function [52] normalizes the real values of the input vector into a probability distribution consisting of probabilities. The output values are in the interval (0,1). The support vector machine (SVM) [53] constructs a set of hyperplanes that provide the maximum margin distance to generate the least generalization error. The Sigmoid function [52], also called the logistic function, is a very popular activation function for artificial neural networks. The input value to this function is transformed into a probability that is between 0 and 1. The Majority Voting classifier [54] merges different classification rules to produce a classifier that is superior to any of the individual ones. It classifies each input value in the class that obtains a large number of votes.
The column Post-processing shows post-processing procedures applied to the output of a deep neural network. Some of the most common post-processing techniques include generating heat-map, heat-map visualization, calculating infection confidence, and GRAD-CAM (Gradient-weighted Class Activation Mapping).
Finally, the column Performance Criteria reports the performance of deep learning models developed in the studies. The common performance criteria contain Accuracy (Acc), Sensitivity (Sen), Specificity (Spe), F1-score, Precision (Pre), Recall, and the area under the ROC curve (AUC).
It is worth mentioning that some of the reviewed articles have proposed several DL models for COVID-19 diagnosis purposes or evaluated their models in two-class and multi-class modes. In these cases, for each study, we only provide the details of the model which has obtained the best result that may be the most helpful to researchers.
5. Discussion
During the recent coronavirus pandemic, RT-PCR frequently produced false-negative findings that increased over time [55]. Due to high false-negative rates, this test sometimes considers COVID-19 patients to be healthy people, which has severe consequences. The probability of a false-negative result on one day a patient has contracted the virus is 100% and falls to 67% over the four days of infection. On day five, the typical time of symptom onset, the chance of a false-negative result is 38% and drops to 20% on day eight. Then it begins to increase again, from 21% on day nine to 66% on day 21 [56]. As a result, an earlier diagnosis tool is necessary to reduce the spread of the disease.
Chest imaging could be a part of COVID-19 diagnosis in suspected or portable COVID-19 patients in the absence of an RT-PCR test, or where the RT-PCR test results are delayed or are negative in the early infection stage in the presence of symptoms indicative of COVID-19 [57]. Benmalek et al. [58] reported the typical imaging features for confirmed COVID-19 pneumonia cases which can help in the early screening and tracking of the disease.
The chest CT scan has been found to have a sensitivity value of 98% and is one of the most precise methods for diagnostics of this disease [59]. Hence, a chest CT may be viewed as a reliable complementary diagnostic measure for RT-PCR to help physicians assess patients more perfectly [18]. The report by Li et al. [60] showed that an initial CT scan performed as early as 5 days after the onset of symptoms would allow for a confident separation of severe and non-severe patients. Lung abnormalities on chest CT peak within 6–13 days after the initial onset of symptoms reaching the highest severity score [61], [62], [63]. On the other hand, X-ray has a reported sensitivity of 69% for COVID-19 diagnosis [64]. However, compared to CT, it is faster, less harmful, easy to perform, and costs less [65]. The radiographic findings detected on chest X-ray in COIVD-19 patients show the greatest severity on days 5–10 of symptom onset [66].
The work carried out by Shazia et al. [15] states that applying deep learning techniques to radiological images for novel coronavirus identification has the potential to reduce the workload of medical practitioners and increase the accuracy and efficiency of COVID-19 diagnosis. In what follows, we discuss the reviewed articles on developing DL models for the early diagnosis of COVID-19 using medical images, and attempt to answer these research questions: What are the most commonly used DL techniques for COVID-19 detection using medical images? What challenges may we face during the deep neural network training process on chest images, and how can we cope with them? What are the advantages and disadvantages of each imaging modality in classification and segmentation tasks?
5.1. Deep neural networks for COVID-19 diagnosis
A revolution in image processing and machine vision has taken place with the advent of deep learning techniques [120]. Deep neural networks have been used in the domain of image processing to solve difficult problems including image classification, detection, and segmentation [121]. These networks eliminate the need for manual feature extraction. They have introduced the concept of end-to-end learning by receiving annotated images as input and discovering the underlying patterns in each image class automatically [122]. A convolutional neural network can have hundreds of layers where each layer learns to detect different image features. DNNs have also achieved great success in analyzing medical images by providing high accuracy, stability, scalability, and efficiency [123]. They can provide Health professionals with more details about the internal organs and patient's tissues, and help physicians diagnose many types of disease, such as pneumonia, brain injuries, cancer, internal bleeding, and pneumonia, brain injuries, cancer, internal bleeding, etc. [124]. Not only in medical image analysis for the diagnosis of such diseases but also during the recent COVID-19 pandemic, deep learning has proved its success [23]. Gaur et al. [125] give evidence on the successful application of deep learning techniques for COVID-19 infection detection. Their work confirms that deep machine vision models can be implemented in the healthcare sector to screen and detect COVID-19 from chest X-rays. Sethi et al. [126] also utilize different deep CNN architectures to provide doctors with diagnosis recommendations for COVID-19. They show that CNN-based architectures have the potential to diagnose COVID-19 disease using chest X-ray Images.
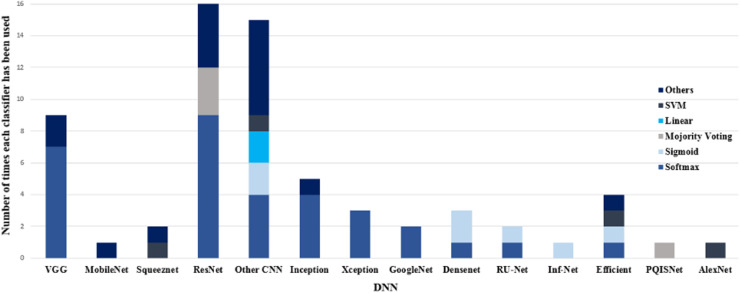
A variety of deep neural networks developed for COVID-19 diagnosis purposes is shown in Fig. 11 . This figure also shows which classifiers and how many times have been used in each DNN. Due to using different datasets with different sizes and properties, a direct comparison between deep networks considering performance metrics is not possible. However, based on our review, Res-Net and VGG are the most common networks used in classification tasks, and for segmentation tasks, various versions of U-Net deep neural networks have been utilized.
Fig. 11.
Number of times that each pair of DNNs and classifiers has been used in reviewed papers for COVID-19 diagnosis.
Different experimental studies have proved good results of the state-of-the-art deep architectures VGG and ResNet for challenging recognition and localization problems such as image segmentation [127]. The main advantage of VGG architecture is its good generalization ability in new datasets [128]. In the 2014-ILSVRC competition, VGG became famous, despite being in 2nd place, due to its simplicity, homogenous topology, and increased depth [127]. On the other hand, ResNet can prevent the degradation problem of deep CNNs. This problem occurs when the accuracy gets saturated and quickly degrades with network depth increasing [129]. ResNet has shortcut connections that skip one or more layers and facilitate deep network training without adding extra parameters or computational complexity. ResNet also lets feature maps from the initial layers that usually include fine details easily propagate to the deeper layers [130].
It is clear also from Fig. 11 that the majority of works have utilized the Softmax classifier in their models. As mentioned before, this classifier represents a probability distribution over class labels, and its output can be directly displayed. Besides, since output values are between 0 and 1, they can be directly fed into any other model without the need for normalization.
It is worth mentioning that using different pre-processing approaches, diverse classifiers, applying the transfer learning method, and other techniques employed for compensating for data shortage could directly affect the results of DNNs. A sufficient number of clinically annotated data is critical for the training step. However, there are very few clinical datasets on COVID-19 which are publicly available and these datasets contain a limited number of COVID-19 cases. Several reviewed studies [18,21,[79], [80], [81], [82], [83], [84], [85],103,37,45,51,[74], [75], [76], [77], [78]] have applied transfer learning and image preprocessing techniques such as image augmentation or used a combination of different datasets to overcome the COVID-19 data leakage and data imbalance problems and enhance their performance. However, these approaches may lead to high variance estimation of deep learning models’ performance during the test phase [131]. Since many of the COVID-19 prediction models are poorly reported, and at high risk of bias and model overfitting [132], more investigation will be needed to ensure the performance of these models in clinical use.
5.2. Classification and segmentation techniques for COVID-19 diagnosis
Fig. 12 shows the total number of studies conducted in the field of segmentation and/or classification for diagnosing COVID-19 patients. Based on the results of this figure, most studies have only focused on the classification task. As mentioned before, we categorized COVID-19 classification approaches into two main classes named COVID-19 from Non-COVID, and COVID-19 from Other Pneumonia, which respectively refer to two-class and multi-class classifications. Although the differentiation of COVID-19 from other types of pneumonia is sometimes challenging, an equal number of studies have modeled the COVID-19 diagnosis problem as a two-class and multi-class classification task. Some researchers have also taken advantage of both classification and segmentation techniques to improve model accuracy. These models first extract the ROI (Region of Interest) of the image in the pre-processing phase and then search within the ROI region instead of the whole region. However, most classification works have preferred to process the whole of the image rather than the ROI region. The main reason is that annotating COVID-19 medical images for a segmentation task is difficult and time-consuming for clinicians.
Fig. 12.
Total number of research works conducted in COVID-19 classification and segmentation, and number of studies done in each sub-class.
5.3. Imaging modalities for COVID-19 diagnosis
The most common imaging modalities for COVID-19 infection identification are CT scan, Lung Ultrasound (LUS), and Chest X-ray radiography. The chest X-ray is low cost, easy to perform, and has low radiation. It also is fast and immediately available for radiologist analysis; the last feature has made it one of the first imaging modalities during the COVID-19 pandemic [133]. Nevertheless, it has limited sensitivity in detecting lung lesions in the early stages of infections [134]. While the Chest CT, being a rapid imaging procedure, may be more sensitive and accurate for the early diagnosis of COVID-19, it exposes patients to more radiation than typical X-rays. Moreover, it is time-consuming for radiologists to diagnose COVID-19 from CT-scan images (about 21.5 min for experienced ones). After each patient, CT scanners will need to be sanitized which is a time-consuming, tedious, and expensive process [135,136].
Compared to the other two modalities, chest ultrasound is low-cost and radiation-free. It can accurately detect the location of objects in real-time and be applied to different lung diseases [137]. Ultrasound has a low false-negative rate in COVID-19 diagnosis [138]. All COVID-19 abnormalities which are visible in CT scan images are also presented on ultrasound images just as clearly [139]. Its accuracy in identifying lung pathologies is better than Chest X-ray, it also is portable, safe, non-invasive, repeatable, and easy to use [134]. However, it shows low sensitivity in comparison to chest X-rays, and it cannot detect deep and intrapulmonary lesions [140].
In the absence of a CT scan, a chest MRI could be recommended for suspected or confirmed COVID-19 patients. Although MRI has low anatomic resolution and is in danger of inevitable artifacts because of breathing motion, its ability to visualize variations in lung structure is constantly evolving [141].
Some other imaging techniques have also been utilized for the COVID-19 diagnosis such as Hyperspectral Imaging (HSI), Scanning Electron Microscopy (SEM), and Positron Emission Tomography (PET). HSI is a medical imaging technique that offers noninvasive disease diagnosis. HSI, also known as Imaging Spectroscopy, can capture spectral information for multiple wavelengths at each image pixel [142]. The maximum contrast in hyperspectral imaging is related to the maximum particle concentration [143]. The main disadvantages of hyperspectral imaging are its cost and complexity. To analyze hyperspectral data, fast computers, sensitive detectors, and large data storage capacities are required.
SEM is a powerful tool for infectious disease diagnosis and scans the sample surface morphology by bright focused electron beam emission [144]. It can produce high-resolution and high-quality images that reveal complex and delicate structures. However, it has several cons. In this technique, samples must always be in a vacuum, so this technique cannot be used for live specimens. Electron microscopes are large and need plenty of space in a laboratory. They also are high-sensitive and could be affected by magnetic fields and vibrations of other lab equipment. Artifacts may be present in SEM images and need to be avoided [145].
PET is a type of nuclear medicine imaging. It applies a small amount of radioactive material to visualize and measure variations in metabolic processes. This non-invasive approach can display very early changes in the cells. Despite the advantages of the PET approach, it is not a routine examination for COVID-19 diagnosis because of its low global accessibility, high inspection cost, complex scanning procedures, and concurrent risks of virus spreading.
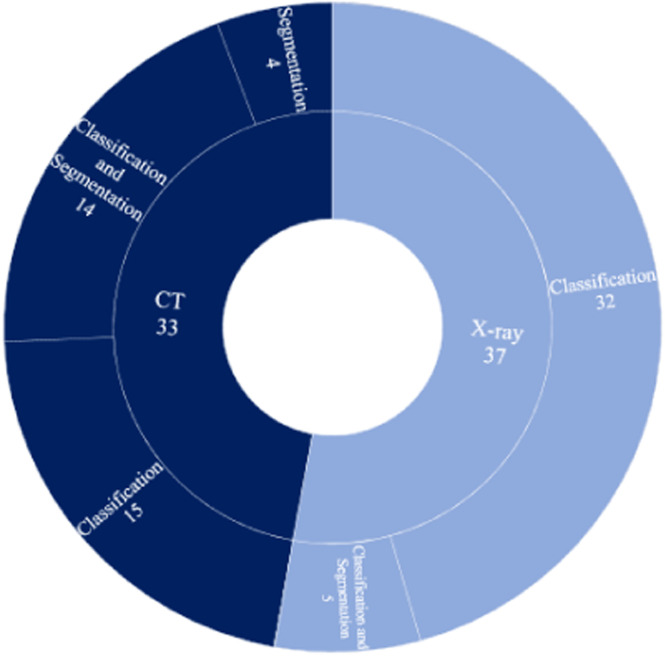
Considering the importance and the role of CT and X-ray chest images in diagnosing COVID-19 patients, our review has investigated machine vision-based techniques developed in the literature for the early diagnostics of COVID-19 and other types of pneumonia using these imaging modalities. Fig. 13 illustrates the total number of research works reviewed for each one. It also shows the number of classification and segmentation tasks conducted for each modality. As one can see, most researchers have selected to work on the X-ray modality. Also, the combination of CT-Scan and segmentation in the diagnosis of COVID-19 patients is more common than X-ray and segmentation. The reason is that CT-Scan produces a 3D view of the chest region and allows for a more detailed view compared to X-ray. Most X-ray modality-based studies have focused on classification for COVID-19 diagnosis, which is a less challenging task.
Fig. 13.
Total number of studies conducted on each modality, and number of classification and segmentation works done for each modality. .
6. Challenges
Deep learning approaches require a large amount of data to build a robust predictive model. However, COVID-19 is a new infectious disease, and the lack of large-scale labeled data is the main challenge in this area. It might lead to overfitting and reduce the accuracy of DL-based models. To overcome this challenge, a majority of works [21,37,51,[74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85]] have combined several public datasets to compensate for the scarcity of well-annotated datasets of appropriate size. However, duplication of images across these datasets is a potential risk during data combination tasks.
Transfer learning is another technique to cope with overfitting and data shortage problems. It is more likely to have a more accurate model by fine-tuning the pre-trained model than training the model from scratch on a small dataset [77,78,146]. Some studies [18,45,47,49,80,113] have used transfer learning to effectively train models on relatively small labeled datasets in COVID-19 segmentation tasks. However, they have mostly applied deep neural networks pre-trained on the ImageNet dataset which includes images that are entirely different from chest images. A small number of segmentation tasks have also used chest images with pseudo labels for the pre-training stage [45]. Preparing noisy labels for the segmentation task is much easier than obtaining clean annotations at the pixel level. Consequently, some research works have attempted to develop a framework to learn from noisy label images [44].
Class imbalance is another main issue in COVID-19 diagnosis since the number of data related to COVID-19 is much less than other pneumonia diseases. Several works [21,37,67,69,70,100,103] applied data augmentation methods to compensate for the small COVID-19 datasets and avoid unbalancing data problems. Also, some researchers [21,75,79,85] used the Generative Adversarial Network (GAN) framework in the pre-processing phase to generate synthetic images to deal with this problem.
Most of the COVID-19 images compress into Non-DICOM formats leading to quality loss. Since the quality of public health information is essential for health monitoring, the lack of image quality remains a challenge in the COVID-19 diagnosis problem. The work carried out by Harmon et al. [97] has reconstructed CT images by super-resolution technique to achieve better accuracy. This technique improves the accuracy of algorithms by making enhanced images with higher contrast.
Generally, lung segmentation can improve the accuracy of a classification task and reduce misdiagnosis [147]. As mentioned before, we grouped the COVID-19 segmentation approaches into two main categories, named the lung-region-oriented methods and the lung-lesion-oriented methods. Lesion detection is a challenging task in the medical imaging area as the lesion size is small in comparison to the whole lung size. In addition, the shape of lesions and their texture and location are very diverse. Most works [108,[148], [149], [150]] applied U-Net architecture or Attention-based U-Net to segment lesions and lung regions in COVID-19 automatic detection tasks. Another challenging task is to discriminate COVID-19 from other types of viral pneumonia since COVID-19 is similarly caused by an infective agent. Several DL-based models have been developed in the literature to distinguish between them [51,68].
7. Recommendations
The awareness and knowledge of the different clinical features of COVID-19 are essential in the early diagnosis and management of the recent pandemic. Some studies were conducted to compare the clinical presentations of COVID-19 patients versus patients infected with other types of pneumonia [151], [152], [153], [154], [155]. Researchers found that considering a combination of information such as background and clinical findings of patients, the duration of the symptoms, ancillary imaging findings, and follow-up CT-Scan imaging when needed would be helpful in the differential diagnosis [152]. Based on this, we strongly recommend developing DL-based models that utilize clinical text data, as well as CT and X-ray images to provide a more accurate diagnosis of COVID-19. While many DL-based techniques have been proposed in the literature for detecting COVID-19 patients based on X-ray images, CT-scan images, or both, less work is being done on diagnosis and prediction using clinical text data. Moreover, to the best of our knowledge, there is no research work in the literature focusing on COVID-19 diagnosis based on both medical images and textual data.
As we mentioned before, dealing with small-size datasets is the main challenge in the COVID-19 diagnosis area which can lead to overfitting and reduce the model generalization performance. Researchers may address this problem through various model enhancement and data generation techniques. Since more complex models with more hyper-parameters are more overfitting-prone than shallower ones, the adaptation of model complexity to the complexity of data can help researchers overcome the issue of overfitting [156]. Another effective way to have more accurate predictions is combining the results of multiple models, which is called ensemble learning [157]. This approach involves several individual models combined in some way such as weighted averaging, and voting. The ensemble model obtains better generalization performance compared to any of the individual models.
Data imbalance is another main issue in diagnosing COVID-19 patients which can undermine model predictability. The oversampling methods can alleviate both data scarcity and data imbalance problems [158]. One approach to oversampling is generating synthetic samples using augmentation techniques such as GAN.
8. Conclusion
COVID-19 has spread dramatically in a short time around the world. The SARS-CoV-2 infection causes symptoms from weak to severe in different people, and to date, more than 6.5 million deaths have been reported to WHO (https://covid19.who.int). Early diagnosis of COVID-19 can help to prevent the spread of this pneumonia, and save many lives. Although the standard test for COVID-19 diagnosis is RT-PCR, this test is time-consuming, and it is possible to show false-negative results [159]. Therefore, many researchers applied deep learning models to detect COVID-19 in the early stage using X-ray and CT images. Chest imaging has a useful role in the detection and management of COVID-19, even in the early stage of the disease. It helps in suspected patients’ identification, Breaking the disease transmission chain, and preventing the further spread of infection [113]. Deep learning approaches can extract rich features from chest images. They are capable of interpreting medical images and distinguishing different types of pneumonia. Physicians can predict COVID-19, estimate its severity, and extract and interpret the infected region by the data extracted from these networks [65].
During the course of this study, we have undertaken a comprehensive review of some 60 papers that have appeared in the literature on the subject of deep learning-based COVID-19 segmentation and classification using X-ray and CT images. We reported the informative details of each model proposed in the reviewed papers. We also listed the public datasets for diagnosing COVID-19 patients and provided several charts summarizing various techniques used in the different phases of developing a DL-based diagnostic model for COVID-19.
Finally, we discussed the challenges in the research on COVID-19 diagnosis based on medical imaging. We found that the reviewed models are at high risk of bias especially because of the overfitting problem, and their generalization performance is therefore unreliable. Consequently, none of these models are recommended for clinical use. Since the scarcity of large-scale public annotated datasets is one of the main challenges leading to overfitting, the compilation, curating, and sharing of well-annotated datasets is urgently needed in order to develop and validate more accurate models for diagnosing COVID-19.
Funding
None.
Ethical approval
Not required.
Declaration of Competing Interest
None declared.
References
- 1.Mei X., Lee H.C., Diao K., Huang M., Lin B., Liu C., et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020;30:4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., et al. Performance of Radiologists in Differentiating COVID-19 from Non-COVID-19 Viral Pneumonia at Chest CT. Radiology. 2020;296:E46–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luz E., Silva P., Silva R., Silva L., Guimarães J., Miozzo G., et al. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res Biomed. Eng. 2021 doi: 10.1007/s42600-021-00151-6. [DOI] [Google Scholar]
- 6.Haque K.F., Abdelgawad A. A deep learning approach to detect COVID-19 patients from chest X-ray images. AI. 2020;1:418–435. doi: 10.3390/ai1030027. [DOI] [Google Scholar]
- 7.Alom M.Z., Taha T.M., Yakopcic C., Westberg S., Sidike P., Nasrin M.S., et al. A state-of-the-art survey on deep learning theory and architectures. Electronics (Basel) 2019;8 doi: 10.3390/electronics8030292. [DOI] [Google Scholar]
- 8.Kaur M., Kumar V., Yadav V., Singh D., Kumar N., Das N.N. Metaheuristic-based deep COVID-19 screening model from chest X-ray images. J. Healthc. Eng. 2021;2021 doi: 10.1155/2021/8829829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. ArXiv Prepr ArXiv160207360; 2016. SqueezeNet: AlexNet-level accuracy With 50x Fewer Parameters and < 0.5MB Model Size. [Google Scholar]
- 10.Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. Proceedings of the Seventh International Conference on Advances in Pattern Recognition. IEEE; 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 11.Zabbah I., Layeghi K., Ebrahimpour R. Improving the Diagnosis of COVID-19 by using a combination of deep learning models. J Electr Comput Eng Innov. 2022;10:411–424. doi: 10.22061/jecei.2022.8200.491. [DOI] [Google Scholar]
- 12.Yang D., Martinez C., Visuña L., Khandhar H., Bhatt C., Carretero J. Detection and analysis of COVID-19 in medical images using deep learning techniques. Sci. Rep. 2021;11:19638. doi: 10.1038/s41598-021-99015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha D., El-Sharkawy M. Proceedings of the IEEE 10th IEEE Annual Ubiquitous Computing, Electronics & Mobile Communication Conference (UEMCON) 2019. Thin MobileNet: an enhanced MobileNet architecture; pp. 280–285. [DOI] [Google Scholar]
- 14.Alzubaidi L., Zhang J., Humaidi A.J., Al-Dujaili A., Duan Y., Al-Shamma O., et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J. Big Data. 2021;8:53. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shazia A., Xuan T.Z., Chuah J.H., Usman J., Qian P., Lai K.W. A comparative study of multiple neural network for detection of COVID-19 on chest X-ray. EURASIP J. Adv. Signal Process. 2021;2021:50. doi: 10.1186/s13634-021-00755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim M., Döhmen T., Rebholz-Schuhmann D., Decker S., Cochez M., Beyan O. ArXiv Prepr ArXiv200404582; 2020. DeepCOVIDExplainer: Explainable COVID-19 Diagnosis Based On Chest X-ray Images. [Google Scholar]
- 17.Pachade S., Porwal P., Kokare M., Giancardo L., Mériaudeau F. NENet: nested EfficientNet and adversarial learning for joint optic disc and cup segmentation. Med. Image Anal. 2021;74 doi: 10.1016/j.media.2021.102253. [DOI] [PubMed] [Google Scholar]
- 18.Bai H.X., Wang R., Xiong Z., Hsieh B., Chang K., Halsey K., et al. Artificial Intelligence Augmentation of Radiologist Performance in Distinguishing COVID-19 from Pneumonia of Other Origin at Chest CT. Radiology. 2020;296:E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddique N., Paheding S., Elkin C.P., Devabhaktuni V. U-net and its variants for medical image segmentation: a review of theory and applications. IEEE Access. 2021;9:82031–82057. doi: 10.1109/ACCESS.2021.3086020. [DOI] [Google Scholar]
- 20.Ahmed I., Chehri A., Jeon G. A sustainable deep learning-based framework for automated segmentation of COVID-19 infected regions: using U-net with an attention mechanism and boundary loss function. Electronics. 2022;11 doi: 10.3390/electronics11152296. (Basel) [DOI] [Google Scholar]
- 21.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. CovidGAN: data augmentation using auxiliary classifier GAN for improved COVID-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imaging. 2020;39:2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 23.Lu W., Zhang S., Chen B., Chen J., Xian J., Lin Y., et al. A Clinical Study of Noninvasive Assessment of Lung Lesions in Patients with Coronavirus Disease-19 (COVID-19) by Bedside Ultrasound TT - Nicht-invasive Beurteilung von pulmonalen Läsionen bei Patienten mit Coronavirus-Erkrankung (COVID-19) durch Ultrasch. Ultraschall Med. 2020;41:300–307. doi: 10.1055/a-1154-8795. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y., Wang S., Liu Y., Zhang Y., Zheng C., Zheng Y., et al. A preliminary study on the ultrasonic manifestations of peripulmonary lesions of non-critical novel coronavirus pneumonia (COVID-19) 2020. 10.21203/rs.2.24369/v1.
- 25.Sofia S., Boccatonda A., Montanari M., Spampinato M., D'ardes D., Cocco G., et al. Thoracic ultrasound and SARS-COVID-19: a pictorial essay. J. Ultrasound. 2020;23:217–221. doi: 10.1007/s40477-020-00458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zu Z.Y., Di Jiang M, Xu P.P., Chen W., Ni Q.Q., Lu G.M., et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punn N.S., Agarwal S. Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Appl Intell. 2020 doi: 10.1007/s10489-020-01900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from Overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 29.Hawkins D.M. The problem of overfitting. J. Chem. Inf. Comput. Sci. 2004;44:1–12. doi: 10.1021/ci0342472. [DOI] [PubMed] [Google Scholar]
- 30.Ng A.Y. Preventing “overfitting” of cross-validation data. Proceedings of the Fourteenth International Conference Machine Learning; San Francisco, CA, USA; Morgan Kaufmann Publishers Inc.; 1997. pp. 245–253. [Google Scholar]
- 31.Bengio Y., Grandvalet Y. No unbiased estimator of the variance of K-fold cross-validation. J Mach Learn Res. 2004;5:1089–1105. [Google Scholar]
- 32.Yadav S., Shukla S. Proceedings of the IEEE 6th International Conference on Advanced Computing (IACC) 2016. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification; pp. 78–83. [DOI] [Google Scholar]
- 33.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI. 1995;14:1137–1145. [Google Scholar]
- 34.Bryll R., Gutierrez-Osuna R., Quek F. Attribute bagging: improving accuracy of classifier ensembles by using random feature subsets. Pattern Recognit. 2003;36:1291–1302. doi: 10.1016/S0031-3203(02)00121-8. [DOI] [Google Scholar]
- 35.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010;22:1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 36.Torrey L., Shavlik J., editors. Transfer Learning. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques. edited by Olivas, Emilio Soria and José David Martín Guerrero, Marcelino Martinez-Sober, Jose Rafael Magdalena-Benedito, and Antonio José Serrano López. IGI Global; Hershey, PA: 2010. pp. 242–264. [DOI] [Google Scholar]
- 37.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalid P.M., Amin B.S. SARS n-CoV2-19 detection from chest X-ray images using deep neural networks. Int. J. Pervasive Comput. Commun. 2020;16:419–427. doi: 10.1108/IJPCC-06-2020-0060. [DOI] [Google Scholar]
- 39.Cunningham P., Cord M., Delany S.J., Cord M., Cunningham P, eds. Heidelberg: Springer Berlin Heidelberg; Berlin: 2008. Supervised Learning BT - Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval; pp. 21–49. [DOI] [Google Scholar]
- 40.Ghahramani Z., Bousquet O, von Luxburg U, Rätsch G. eds. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004. Unsupervised learning BT - Advanced Lectures on Machine Learning: ML Summer Schools 2003, Canberra, Australia, February 2 - 14, 2003, Tübingen, Germany, August 4 - 16, 2003, Revised Lectures; pp. 72–112. [DOI] [Google Scholar]
- 41.Zhou Z.H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018;5:44–53. doi: 10.1093/nsr/nwx106. [DOI] [Google Scholar]
- 42.Zhai X., Oliver A., Kolesnikov A., Beyer L. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. S4L: self-supervised semi-supervised learning; pp. 1476–1485. vol. abs/1905.0. [Google Scholar]
- 43.Wang J., Bao Y., Wen Y., Lu H., Luo H., Xiang Y., et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020;39:2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- 44.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., et al. A Noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39:2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan D.P, Zhou T., Ji G.P., Zhou Y., Chen G., Fu H., et al. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39:2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 46.Javaheri T., Homayounfar M., Amoozgar Z., Reiazi R., Homayounieh F., Abbas E., et al. CovidCTNet: an open-source deep learning approach to diagnose COVID-19 using small cohort of CT images. NPJ Digit. Med. 2021;4:29. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duran-Lopez L., Dominguez-Morales J.P., Corral-Jaime J., Vicente-Diaz S., Linares-Barranco A. COVID-XNet: a custom deep learning system to diagnose and locate COVID-19 in Chest X-ray images. Appl Sci. 2020;10:5683. doi: 10.3390/app10165683. [DOI] [Google Scholar]
- 49.El Asnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nwankpa C., Ijomah W., Gachagan A., Marshall S. Practice and Research for Deep Learning; 2018. Activation Functions: Comparison of Trends. [Google Scholar]
- 53.Noble W.S. What is a support vector machine? Nat. Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 54.G.M. James, Majority vote classifiers: Theory and applications. Ph.D. thesis, Department of Statistics, University of Stanford, CA, 1998.
- 55.MubarakAli D. Comprehensive Review on Rapid Diagnosis of New Infection COVID-19. Appl. Biochem. Biotechnol. 2022;194:1390–1400. doi: 10.1007/s12010-021-03728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020;173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organisation . COVID-19 a Rapid Advice Guide; 2020. COVID-19 Use of Chest Imaging. [Google Scholar]
- 58.Benmalek E., Elmhamdi J., Jilbab A. Comparing CT scan and chest X-ray imaging for COVID-19 diagnosis. Biomed. Eng. Adv. 2021;1 doi: 10.1016/j.bea.2021.100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li K., Liu X., Yip R., Yankelevitz D.F., Henschke C.I., Geng Y., et al. Early prediction of severity in coronavirus disease (COVID-19) using quantitative CT imaging. Clin. Imaging. 2021;78:223–229. doi: 10.1016/j.clinimag.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwee T.C., Kwee R.M. Chest CT in COVID-19: what the radiologist needs to know. Radiographics. 2020;40:1848–1865. doi: 10.1148/rg.2020200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin. Imaging. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swapnarekha H., Behera H.S., Nayak J., Naik B. Role of intelligent computing in COVID-19 prognosis: a state-of-the-art review. Chaos, Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rousan L.A., Elobeid E., Karrar M., Khader Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20:245. doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos, Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2020 doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ucar F., Korkmaz D. COVIDiagnosis-Net: deep bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Costa Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan A.I., Shah J.L., Bhat M.M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Progr. Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 Biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. 2020;40:462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loey M., Smarandache F., Khalifa N.E.M. Within the Lack of Chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 2020;12:651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 76.Apostolopoulos I.D., Mpesiana T.A. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rahimzadeh M., Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inf. Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaid S., Kalantar R., Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int. Orthop. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das D., Santosh K.C., Pal U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys. Eng. Sci. Med. 2020;43:915–925. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 81.Shibly K.H., Dey S.K., Islam M.T.U., Rahman M.M. COVID faster R–CNN: a novel framework to diagnose novel coronavirus disease (COVID-19) in X-ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoo S.H., Geng H., Chiu T.L., Yu S.K., Cho D.C., Heo J., et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020;7:427. doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsiknakis N., Trivizakis E., Vassalou E.E., Papadakis G.Z., Spandidos D.A., Tsatsakis A., et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp. Ther. Med. 2020;20:727–735. doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and bayesian optimization. Appl. Soft Comput. 2020 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zebin T., Rezvy S. COVID-19 detection and disease progression visualization: deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 2021;51:1010–1021. doi: 10.1007/s10489-020-01867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pandit M.K., Banday S.A., Naaz R., Chishti M.A. Automatic detection of COVID-19 from chest radiographs using deep learning. Radiography. 2021;27:483–489. doi: 10.1016/j.radi.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh R.K., Pandey R., Babu R.N. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput Appl. 2021 doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect COVID-19 coronavirus with X-ray images. Biocybern Biomed Eng. 2020;40:1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (COVID-19) from chest X-ray images. Chaos, Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murugan R., E-DiCoNet Goel T. Extreme learning machine based classifier for diagnosis of COVID-19 using deep convolutional network. J. Ambient Intell. Humaniz Comput. 2021;12:8887–8898. doi: 10.1007/s12652-020-02688-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin W., Dong S., Dong C., Ye X. Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shankar K., Mohanty S.N., Yadav K., Gopalakrishnan T., Elmisery A.M. Automated COVID-19 diagnosis and classification using convolutional neural network with fusion based feature extraction model. Cogn. Neurodyn. 2021 doi: 10.1007/s11571-021-09712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azouji N., Sami A., Taheri M., Müller H. A large margin piecewise linear classifier with fusion of deep features in the diagnosis of COVID-19. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butt C., Gill J., Chun D., Babu B.A. RETRACTED ARTICLE: deep learning system to screen coronavirus disease 2019 pneumonia. Appl. Intell. 2020 doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020;0:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 96.Wu X., Hui H., Niu M., Li L., Wang L., He B., et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur. J. Radiol. 2020;128 doi: 10.1016/j.ejrad.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39:2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 100.Yang S., Jiang L., Cao Z., Wang L., Cao J., Feng R., et al. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann. Transl. Med. 2020;8:450. doi: 10.21037/atm.2020.03.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., et al. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging. 2020;39:2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 102.Hasan A.M., AL-Jawad M.M., Jalab H.A., Shaiba H., Ibrahim R.W., AL-Shamasneh A.R. Classification of COVID-19 coronavirus, pneumonia and healthy lungs in CT scans using Q-deformed entropy and deep learning features. Entropy. 2020;22:517. doi: 10.3390/e22050517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pu J., Leader J., Bandos A., Shi J., Du P., Yu J., et al. Any unique image biomarkers associated with COVID-19? Eur. Radiol. 2020;30:6221–6227. doi: 10.1007/s00330-020-06956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. IRBM; 2020. Deep Transfer Learning Based Classification Model for COVID-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh D., Kumar V., Vaishali Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song J., Wang H., Liu Y., Wu W., Dai G., Wu Z., et al. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur. J. Nucl. Med. Mol. Imaging. 2020;47:2516–2524. doi: 10.1007/s00259-020-04929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie W., Jacobs C., Charbonnier J.P., van Ginneken B. Relational modeling for robust and efficient pulmonary lobe segmentation in CT scans. IEEE Trans. Med. Imaging. 2020;39:2664–2675. doi: 10.1109/TMI.2020.2995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., et al. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 109.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020 doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Konar D., Panigrahi B.K., Bhattacharyya S., Dey N. Auto-diagnosis of COVID-19 using lung CT images with semi-supervised shallow learning network 2020. 10.21203/rs.3.rs-34596/v1.
- 111.Ko H., Chung H., Kang W.S., Kim K.W., Shin Y., Kang S.J., et al. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J. Med. Internet Res. 2020;22:e19569. doi: 10.2196/19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. 2021 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:19196. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans Comput Biol Bioinforma. 2021:1. doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Serte S., Demirel H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Q., Ning J., Yuan J., Xiao L. A depthwise separable dense convolutional network with convolution block attention module for COVID-19 diagnosis on CT scans. Comput. Biol. Med. 2021;137 doi: 10.1016/j.compbiomed.2021.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Canayaz M. C+EffxNet: a novel hybrid approach for COVID-19 diagnosis on CT images based on CBAM and EfficientNet. Chaos, Solitons Fractals. 2021;151 doi: 10.1016/j.chaos.2021.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen X., Yao L., Zhou T., Dong J., Zhang Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021;113 doi: 10.1016/j.patcog.2021.107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sedik A., Hammad M., Abd El-Samie F.E., Gupta B.B., Abd El-Latif A.A. Efficient deep learning approach for augmented detection of Coronavirus disease. Neural Comput. Appl. 2021 doi: 10.1007/s00521-020-05410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goswami T., Anguera J., Satapathy S.C., Bhateja V., Sunitha K.V.N. eds. Singapore: Springer Singapore; 2018. Impact of Deep Learning in Image Processing and Computer Vision BT - Microelectronics, Electromagnetics and Telecommunications; pp. 475–485. [Google Scholar]
- 121.Anwar S.M., Majid M., Qayyum A., Awais M., Alnowami M., Khan M.K. Medical image analysis using convolutional neural networks: a review. J. Med. Syst. 2018;42:226. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 122.O'Mahony N., Campbell S., Carvalho A., Harapanahalli S., Hernandez G.V., Krpalkova L., et al. In: Adv. Comput. Vis. editors. Arai K., Kapoor S., editors. Cham: Springer International Publishing; 2020. Deep learning vs. traditional computer vision; pp. 128–144. [Google Scholar]
- 123.Liu X., Gao K., Liu B., Pan C., Liang K., Yan L., et al. Advances in deep learning-based medical image analysis. Health Data Sci. 2021;2021 doi: 10.34133/2021/8786793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sharma P., Bhatt D.P. Approaches Applications Deep Learning. Virtual Med. Care; 2022. Importance of deep learning models in the medical imaging field; pp. 1–23. [DOI] [Google Scholar]
- 125.Gaur L., Bhatia U., Jhanjhi N.Z., Muhammad G., Masud M. Medical image-based detection of COVID-19 using deep convolution neural networks. Multimed Syst. 2021 doi: 10.1007/s00530-021-00794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sethi R., Mehrotra M., Sethi D. Proceedings of the Second International Conference on Inventive Research in Computing Applications (ICIRCA) 2020. Deep learning based diagnosis recommendation for COVID-19 using Chest X-rays images; pp. 1–4. [DOI] [Google Scholar]
- 127.Khan A., Sohail A., Zahoora U., Qureshi A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020;53:5455–5516. doi: 10.1007/s10462-020-09825-6. [DOI] [Google Scholar]
- 128.Simonyan K., Zisserman A. ArXiv Prepr ArXiv; 2014. Very Deep Convolutional Networks For Large-Scale Image Recognition. 14091556. [Google Scholar]
- 129.Nguyen H.G., Pica A., Hrbacek J., Weber D.C., La Rosa F., Schalenbourg A., et al. Proceedings of the International Conference Medical Imaging with Deep Learning. PMLR; 2019. A novel segmentation framework for uveal melanoma in magnetic resonance imaging based on class activation maps; pp. 370–379. [Google Scholar]
- 130.Wang T., Lei Y., Fu Y., Wynne J.F., Curran W.J., Liu T., et al. A review on medical imaging synthesis using deep learning and its clinical applications. J. Appl. Clin. Med. Phys. 2021;22:11–36. doi: 10.1002/acm2.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Acar E., Şahin E., Yılmaz İ. Improving effectiveness of different deep learning-based models for detecting COVID-19 from computed tomography (CT) images. Neural Comput. Appl. 2021;33:17589–17609. doi: 10.1007/s00521-021-06344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dhont J., Wolfs C., Verhaegen F. Automatic coronavirus disease 2019 diagnosis based on chest radiography and deep learning {\textendash} Success story or dataset bias? Med. Phys. 2022;49:978–987. doi: 10.1002/mp.15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zargari Khuzani A., Heidari M., Shariati S.A. COVID-Classifier: an automated machine learning model to assist in the diagnosis of COVID-19 infection in chest X-ray images. Sci. Rep. 2021;11:9887. doi: 10.1038/s41598-021-88807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aljondi R., Alghamdi S. Diagnostic value of imaging modalities for COVID-19: scoping review. J. Med. Internet Res. 2020;22:e19673. doi: 10.2196/19673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu Y.H., Gao S.H., Mei J., Xu J., Fan D.P., Zhang R.G., et al. JCS: an explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 136.Axiaq A., Almohtadi A., Massias S.A., Ngemoh D., Harky A. The role of computed tomography scan in the diagnosis of COVID-19 pneumonia. Curr. Opin. Pulm. Med. 2021;27:163–168. doi: 10.1097/MCP.0000000000000765. [DOI] [PubMed] [Google Scholar]
- 137.El-Rashidy N., Abdelrazik S., Abuhmed T., Amer E., Ali F., Hu J.W., et al. Comprehensive survey of using machine learning in the COVID-19 pandemic. Diagnostics. 2021;11 doi: 10.3390/diagnostics11071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tung-Chen Y., Martí de Gracia M., Díez-Tascón A., Alonso-González R., Agudo-Fernández S., Parra-Gordo M.L., et al. Correlation between chest computed tomography and lung ultrasonography in patients with coronavirus disease 2019 (COVID-19) Ultrasound Med. Biol. 2020;46:2918–2926. doi: 10.1016/j.ultrasmedbio.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rehouma R., Buchert M., Chen Y.P.P. Machine learning for medical imaging-based COVID-19 detection and diagnosis. Int. J. Intell. Syst. 2021;36:5085–5115. doi: 10.1002/int.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Khalili N., Haseli S., Iranpour P. Lung Ultrasound in COVID-19 Pneumonia: prospects and Limitations. Acad. Radiol. 2020;27:1044–1045. doi: 10.1016/j.acra.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vasilev Y.A., Sergunova K.A., Bazhin A.V., Masri A.G., Vasileva Y.N., Semenov D.S., et al. Chest MRI of patients with COVID-19. Magn. Reson. Imaging. 2021;79:13–19. doi: 10.1016/j.mri.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu G., Fei B. Medical hyperspectral imaging: a review. J. Biomed. Opt. 2014;19:1–24. doi: 10.1117/1.JBO.19.1.010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alafeef M., Moitra P., Dighe K., Pan D. Hyperspectral mapping for the detection of SARS-CoV-2 using nanomolecular probes with yoctomole sensitivity. ACS Nano. 2021;15:13742–13758. doi: 10.1021/acsnano.1c05226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brahim Belhaouari D., Fontanini A., Baudoin J.P., Haddad G., Le Bideau M., Bou Khalil J.Y., et al. The strengths of scanning electron microscopy in deciphering SARS-CoV-2 infectious cycle. Front Microbiol. 2020;11:2014. doi: 10.3389/fmicb.2020.02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Inkson B.J. In: Materials Characterization Using Nondestructive Evaluation (NDE) Methods. eds. Hübschen G., Altpeter I., Tschuncky R., Herrmann H.G., editors. Woodhead Publishing; 2016. 2 - Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) for materials characterization; pp. 17–43. [DOI] [Google Scholar]
- 146.Han T., Liu C., Wu R., Jiang D. Deep transfer learning with limited data for machinery fault diagnosis. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107150. [DOI] [Google Scholar]
- 147.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126 doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., et al. MedRxiv; 2020. AI-assisted CT Imaging Analysis For COVID-19 Screening: Building and Deploying a Medical AI System in Four Weeks. 2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gaál G., Maga B., Lukács A. ArXiv E-Prints; 2020. Attention U-Net Based Adversarial Architectures for Chest X-ray Lung Segmentation. arXiv:2003.10304. [Google Scholar]
- 150.Oktay O., Schlemper J., Le Folgoc L, Lee M.C.H., Heinrich M.P., Misawa K., et al. CoRR; 2018. Attention U-Net: Learning Where to Look for the Pancreas. abs/1804.0. [Google Scholar]
- 151.Dai W.C., Zhang H.W., Yu J., Xu H.J., Chen H., Luo S.P., et al. CT imaging and differential diagnosis of COVID-19. Can Assoc Radiol J = J l'Association Can Des Radiol. 2020;71:195–200. doi: 10.1177/0846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Duzgun S.A., Durhan G., Demirkazik F.B., Akpinar M.G., Ariyurek O.M. COVID-19 pneumonia: the great radiological mimicker. Insights Imaging. 2020;11:118. doi: 10.1186/s13244-020-00933-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li Y., Wang H., Wang F., Du H., Liu X., Chen P., et al. Comparison of hospitalized patients with pneumonia caused by COVID-19 and influenza A in children under 5 years. Int. J. Infect. Dis. 2020;98:80–83. doi: 10.1016/j.ijid.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., et al. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging. 2020;101:263–268. doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao D., Yao F., Wang L., Zheng L., Gao Y., Ye J., et al. A comparative study on the clinical features of coronavirus 2019 (COVID-19) pneumonia with other pneumonias. Clin. Infect. Dis. 2020;71:756–761. doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bejani M.M., Ghatee M. A systematic review on overfitting control in shallow and deep neural networks. Artif. Intell. Rev. 2021 doi: 10.1007/s10462-021-09975-1. [DOI] [Google Scholar]
- 157.Polikar R., Zhang C., Ma Y. eds. Springer US; Boston, MA: 2012. Ensemble Learning BT - Ensemble Machine Learning: Methods and Applications; pp. 1–34. [DOI] [Google Scholar]
- 158.Chen Z., Zhang Y., Simsek M., Kantarci B. ICC IEEE International Conference on Communications. 2020. Deep Belief network-based fake task mitigation for mobile crowdsensing under data scarcity; pp. 1–7. [DOI] [Google Scholar]
- 159.Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., et al. Use of Chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295:22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]