Abstract
Background:
Two studies that examined the interaction between HLA-DRB1 and smoking in Parkinson’s disease (PD) yielded findings in opposite directions.
Objective:
To perform a large-scale independent replication of the HLA-DRB1×smoking interaction.
Methods:
We genotyped 182 SNPs associated with smoking initiation in 12424 cases and 9480 controls to perform a Mendelian randomization (MR) analysis in strata defined by HLA-DRB1.
Results:
At the amino-acid level, a valine at position 11 (V11) in HLA-DRB1 displayed the strongest association with PD. MR showed an inverse association between genetically-predicted smoking initiation and PD only in absence of V11 (odds ratio=0.74, 95% confidence interval=0.59-0.93, PInteraction=0.028). In silico predictions of the influence of V11 and smoking-induced modifications of alpha-synuclein on binding affinity showed consistent findings.
Conclusions:
Despite being one of the most robust findings in PD research, the mechanisms underlying the inverse association between smoking and PD remain unknown. Our findings may help better understand this association.
INTRODUCTION
Genome-wide association studies (GWAS) in Parkinson’s disease (PD) identified an association with the Human Leukocyte Antigen (HLA) region, in particular with HLA-DRB1. Hollenbach et al.1 reported an inverse association of PD with the shared epitope (SE), a combination of amino acids (AA) coded by HLA-DRB1, only in the presence of a valine at position 11 (V11). The strongest association in a cross-ethnic GWAS meta-analysis was an inverse association with a histidine at position 13 (H13) in HLA-DRB1, strongly correlated with V11.2 The latest study, with some overlap with the previous two, highlighted three AA (V11, H13, H33) encoded by HLA-DRB1 inversely associated with PD.3
Following studies showing interactions between smoking and HLA-DRB1 in other conditions,4–6 Chuang et al.7 genotyped one SNP in the HLA-DRB1 region whose minor G allele is inversely associated with PD (2056 cases, 2723 controls) and reported a significant positive interaction between self-reported smoking and rs660895-G: the inverse association between smoking and PD was stronger in carriers of the AA genotype compared to G-allele carriers.7 Based on a smaller selected sample (837 cases, 918 controls), the study that identified an inverse association of the SE and V11 combination (SE+V11+) with PD also showed an interaction with smoking, but in the opposite direction: the inverse association between smoking and PD was restricted to SE+V11+ carriers.1 The authors hypothesized that post-translational modifications of alpha-synuclein induced by smoking (citrullination/homocitrullination) explained this interaction.
We performed a large-scale independent replication of the HLA-DRB1×smoking interaction by performing a Mendelian randomization (MR) analysis using smoking predisposing genes as instrumental variables in strata defined by HLA-DRB1.
SUBJECTS AND METHODS
Courage-PD
The Courage-PD (COmprehensive Unbiased Risk Factor Assessment for Genetics and Environment in Parkinson’s Disease) consortium pooled individual-level data from 35 studies and used the Neurochip array to genotype participants (Supplementary Methods). Analyses are based on 26 studies with at least 50 cases or controls of European descent (12424 cases, 9480 controls; Supplementary Table 1). Additional methods on genotyping and imputation of HLA alleles/haplotypes/AA are available as Supplementary methods. All studies were approved by local ethical committees following procedures of each country.
Smoking initiation: two-sample Mendelian randomization
As self-reported smoking was not available in most studies, we used SNPs associated with smoking initiation to perform two-sample MR.8 Summary statistics for the association between SNPs and smoking initiation (182 SNPs independently associated at P<5×10−8) came from the GWAS and Sequencing Consortium of Alcohol and Nicotine use (n=1,232,091, European descent; Supplementary Methods),9 and those for associations with PD came from Courage-PD (Supplementary Table 2).
In silico prediction of binding affinity of HLA-DRB1 alleles to alpha-synuclein
We assessed the binding affinity (nM) of HLA-DRB1 alleles to alpha-synuclein derived peptides using NetMHCIIpan 4.0, and predicted whether peptides are strong, weak, or non-binders.10 After targeting 607 four-digit HLA-DRB1 alleles, we restricted our analyses to 34 alleles observed in Courage-PD. Of 126 alpha-synuclein derived peptides,1 we retained 96 peptides with lysine residues that can be homocitrullinated in order to examine the role of smoking-related post-translational modifications. We also performed analyses restricted to a single peptide (Tyrosine 39, Y39) that induces T cell responses in PD patients,11 and was previously used for binding affinity predictions.2
Statistical analyses
We used SAS9.4 (SAS Institute Inc, NC), STATA16 (StataCorp, TX), and R packages HIBAG12 and TwoSampleMR13 (R Foundation for Statistical Computing, Austria).
Interaction between genetically-predicted smoking initiation and HLA-DRB1
To perform an independent replication of the HLA-DRB1×smoking interaction, we excluded the French study that contributed to identify the interaction between smoking and rs660895 in PD.7
We used the random-effects inverse-variance weighted (IVW)8 approach to perform MR analyses for genetically-predicted smoking initiation in two strata defined by the presence of V11 encoded by HLA-DRB1 alleles (Supplementary Methods). We compared the two MR estimates using the statistic , where β1 and β2 are MR estimates in the two strata with variances SE(β1)2 and SE(β2)2; this difference represents the interaction between smoking and HLA-DRB1 and follows a normal distribution. In sensitivity analyses, we used other MR approaches that are less powerful but more robust to pleiotropy (weighted median-method and mode-based, MR-PRESSO, MR-Lasso);8 we also performed analyses after excluding 31 pleiotropic SNPs associated with alcohol drinking and/or body mass index (Supplementary methods).
As secondary analyses, we ran MR analyses stratified by rs6608957 and HLA-DRB1*04,3 which are both inversely associated with PD and in linkage disequilibrium with V11. Analyses stratified by rs660895 have the advantage that they did not involve HLA imputation and are therefore based on a larger number of cases and controls than analyses that required HLA imputation.
In silico prediction of binding affinity
To examine the influence of V11 encoded by HLA-DRB1 alleles and homocitrullination (HC) of alpha-synuclein derived peptides on binding affinity, we described binding affinity for the four groups defined by the combination of V11 and HC. All 2×2 differences were tested using the Wilcoxon non-parametric test corrected for multiple comparisons.14 We compared the percentage of binding peptides in the four groups using multinomial logistic regression.
Data availability:
Results can be reproduced using the Supplementary material.
RESULTS
Supplementary Table 3 shows 19 SNPs from the HLA region associated with PD after accounting for multiple comparisons, of which 17 were located near HLA-DRB1 (including rs660895); none of them was associated with smoking initiation (P>0.05). Among 64 alleles of HLA class 2 genes (HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRB1), five were significantly and inversely associated with PD (HLA-DQA1*03:01, HLA-DQA1*03:03; HLA-DQB1*03:02; HLA-DRB1*04:01, HLA-DRB1*04:04; Supplementary table 4). The OR for the association of all HLA-DRB1*04 alleles with PD was of 0.84 (95% CI=0.78-0.91, P=3.9×10−6). Associations between DRB1~HLA-DQB1 haplotypes and PD are shown in Supplementary table 4.
Among 131 AA encoded by HLA-DRB1 and 116 by HLA-DQB1, 11 AA were associated (9 inversely, 2 positively) with PD and were all encoded by HLA-DRB1 (Supplementary Table 5). Two AA, V11 and S37, remained significantly associated with PD after a backward stepwise selection procedure, with a stronger association for V11 (OR=0.85, 95% CI=0.79-0.92, P=2.2×10−5) than S37 (OR=1.07, 95% CI=1.00-1.14, P=0.040). The association of H13 and H33 with PD was explained by V11 (Supplementary Table 6). We found no significant interaction between SE and V11 (P=0.29); only V11 remained associated with PD (OR=0.81, 95% CI=0.74-0.89, P<10-3) when both were included in the model (Supplementary table 7).
The overall association between genetically-predicted smoking initiation and PD was of 0.86 (95% CI=0.73-1.05, P=0.10) without evidence of heterogeneity (P=0.40). Compared with 26% (N=2212) of the controls, 22% (N=2531) of the cases carried at least one V11 residue. Genetically-predicted smoking initiation was inversely associated with PD in the absence of V11 (ORIVW=0.74, 95% CI=0.59-0.93, P=0.009) but not in its presence (ORIVW=1.25, 95% CI=0.83-1.87, P=0.29), with a positive and significant interaction (P=0.028; Table 1, Figure 1). There was no significant heterogeneity across SNPs and MR-Presso did not detect pleiotropy (all P>0.10). Results of pleiotropy-robust approaches were consistent with the IVW method, though CIs were generally larger. Similar conclusions were reached after excluding 31 pleiotropic SNPs (Figure 1, Supplementary Table 8). Results were similar in analyses stratified by rs660895 or HLA-DRB1*04.
Table 1.
Mendelian randomization analysis of the relation between genetically-predicted smoking initiation (182 SNPs) and Parkinson’s disease stratified by HLA-DRB1
| 0 allele or AA residue |
1/2 alleles or AA residues |
|||||||
|---|---|---|---|---|---|---|---|---|
| HLA-DRB1 | OR per 1-SD increase in the prevalence of ever smoking (95% CI) | P | P-het. | OR per 1-SD increase in the prevalence of ever smoking (95% CI) | P | P-het. | Interaction OR (95% CI)c | P |
| Valine 11 a | 6383 controls, 8812 cases | 2212 controls, 2531 cases | ||||||
| Inverse variance weighted | 0.74 (0.59-0.93) | 9.2×10−3 | 0.73 | 1.25 (0.83-1.87) | 0.29 | 0.40 | 1.68 (1.06-2.68) | 0.03 |
| Weighted median | 0.75 (0.53-1.07) | 0.11 | 1.14 (0.61-2.15) | 0.68 | 1.52 (0.75-3.11) | 0.26 | ||
| Weighted mode | 0.63 (0.30-1.31) | 0.22 | 1.72 (0.38-7.82) | 0.48 | 2.74 (0.51-14.77) | 0.24 | ||
| MR-Lasso | No invalid SNP (λ=0.20) | 1.30 (0.87-1.96) | 0.20d | 1.76 (1.10-2.81) | 0.02 | |||
| MR-PRESSO | 0.59 | 0.47 | ||||||
| rs660895-G b | 6498 controls, 8903 cases | 2982 controls, 3521 cases | ||||||
| Inverse variance weighted | 0.73 (0.59-0.91) | 4.8×10−3 | 0.84 | 1.33 (0.95-1.87) | 0.10 | 0.41 | 1.83 (1.22-2.74) | 3.5×10−3 |
| Weighted median | 0.72 (0.52-1.00) | 0.05 | 1.04 (0.62-1.73) | 0.89 | 1.45 (0.78-2.66) | 0.24 | ||
| Weighted mode | 0.68 (0.31-1.48) | 0.34 | 0.99 (0.23-4.26) | 0.99 | 1.46 (0.30-7.08) | 0.66 | ||
| MR-Lasso | No invalid SNP (λ=0.19) | 1.25 (0.89-1.75) | 0.20e | 1.71 (1.14-2.56) | 9.1×10−3 | |||
| MR-PRESSO | 0.83 | 0.40 | ||||||
| HLA-DRB1*04 a | 6563 controls, 9014 cases | 2032 controls, 2329 cases | ||||||
| Inverse variance weighted | 0.73 (0.59-0.92) | 6.8×10−3 | 0.77 | 1.29 (0.83-2.00) | 0.26 | 0.47 | 1.75 (1.07-2.87) | 0.03 |
| Weighted median | 0.70 (0.50-0.97) | 0.03 | 1.16 (0.59-2.29) | 0.66 | 1.67 (0.81-3.46) | 0.18 | ||
| Weighted mode | 0.67 (0.30-1.48) | 0.32 | 1.51 (0.34-6.66) | 0.59 | 2.26 (0.38-13.39) | 0.34 | ||
| MR-Lasso | No invalid SNP (λ=0.20) | 1.18 (0.76-1.83) | 0.46f | 1.61 (0.98-2.64) | 0.06 | |||
| MR-PRESSO | 0.67 | 0.57 | ||||||
OR, odds ratio; CI, confidence interval; AA, amino acid; P-het., P for heterogeneity; λ, tuning parameter for MR-Lasso.
Valine 11 amino-acids and HLA-DRB1*04 alleles were determined using imputation of HLA alleles and amino-acids based on SNPs from the HLA region.
Total number: 8595 controls, 11343 cases.
Total number: 9480 controls, 12424 cases.
The interaction OR represents the OR in carriers of 1/2 alleles or AA residues divided by the OR in carriers of 0 allele or AA residue.
Number of invalid SNPs=4; λ=0.17.
Number of invalid SNPs=4; λ=0.19.
Number of invalid SNPs=11; λ=0.19.
Figure 1.
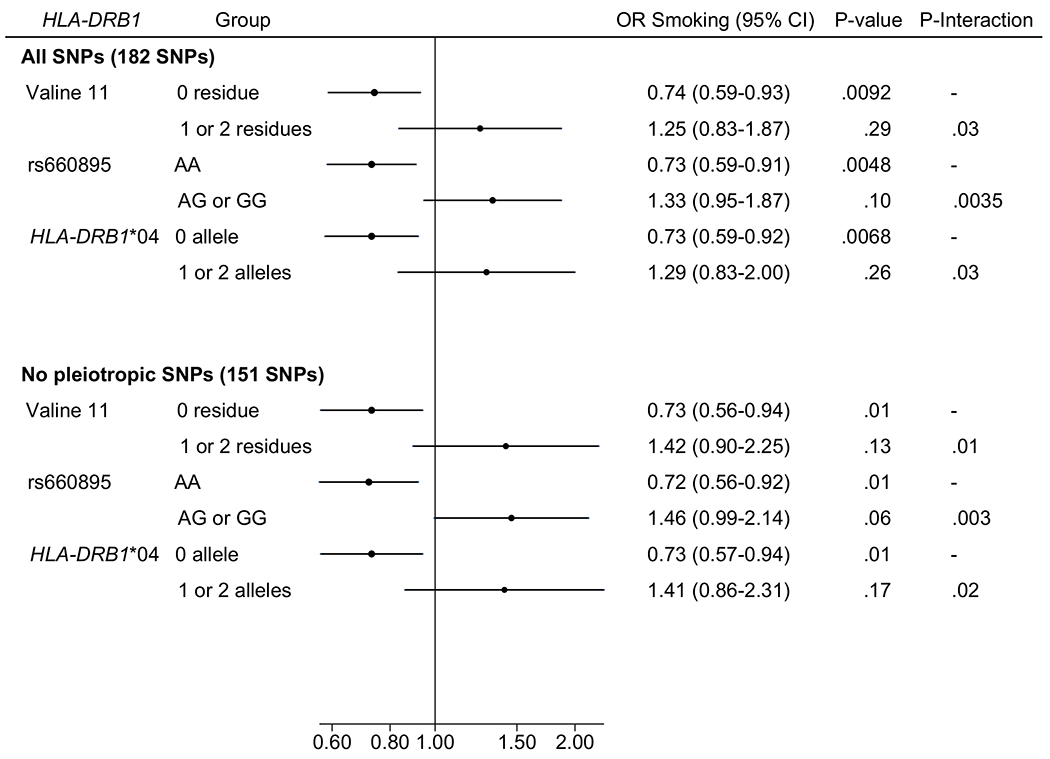
Forest plot of the association between genetically-predicted smoking initiation (inverse variance weighted estimate) and Parkinson’s disease stratified by HLA-DRB1
Compared to V11−HC−, V11+HC− and V11−HC+ were both associated with decreased binding affinity, with a stronger effect of HC+ than V11+ (Figure 2, Supplementary Table 9). Alternatively, in the presence of HC+, V11+ increased binding affinity (all peptides) or had no effect (Y39); HC+ had no effect on binding affinity in the presence of V11+. Analyses of binding and non-binding peptides paralleled these results (Supplementary Table 10).
Figure 2. Prediction of binding affinity (nM) according to the presence of a valine at position 11 (V11) coded by HLA-DRB1 alleles and homocitrullination (HC) of alpha-synuclein derived peptides.
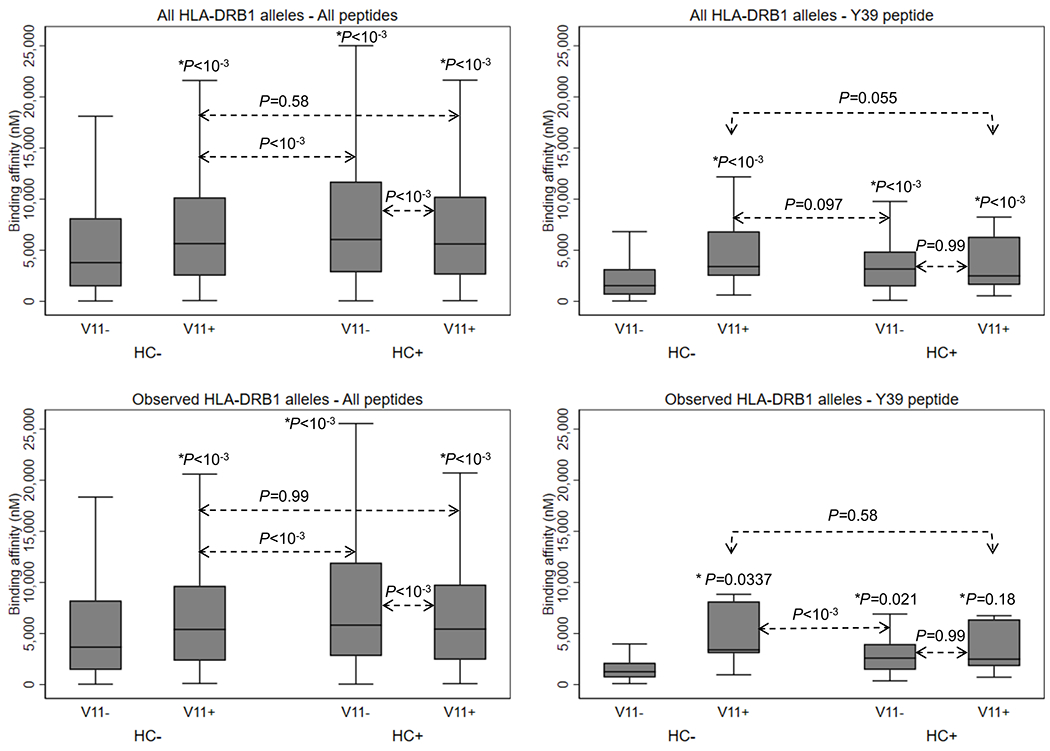
* P-values for the comparison versus the reference group (V11−HC−).
DISCUSSION
We replicate an interaction between HLA-DRB1 and smoking,7 according to which the inverse association between smoking and PD was only present in participants without protective HLA-DRB1 AA/alleles. In silico predictions of binding affinity were consistent with an interaction between V11 and post-translational smoking-induced modifications of alpha-synuclein derived peptides.
Recent MR studies showed an inverse association between genetically-predicted smoking and PD.15–18 These findings are in favour of a causal role of smoking in PD, but the underlying mechanisms remain unknown and gene-environment interactions analyses may contribute to their understanding. The interaction pattern we found is similar to the interaction between self-reported smoking and rs660895 reported by Chuang et al.7 Our study represents a fully independent replication using a different approach to define smoking (MR) and SNP-based imputation of HLA amino-acids which allowed us to examine this interaction at the AA level. Therefore, our findings contradict those from Hollenbach et al.1 who reported an interaction in the opposite direction based on a selected sample of smaller size.
Lower binding affinity for alpha-synuclein derived peptides is associated with a weaker immune response which may explain decreased PD risk.19 Our binding affinity analyses are consistent with the interaction pattern we identified. While V11 and HC both decreased binding affinity for alpha-synuclein derived peptides in the absence of each other, consistent with the inverse association of V11 and smoking with PD, there was a positive interaction between V11 and HC, whereby both V11 and HC had a weaker or no effect in the presence of each other; this pattern is consistent with the lack of association between smoking and PD in V11 carriers that we found.
We used MR to define genetically-predicted smoking initiation, rather than self-reported smoking; MR has the advantage that, provided that a set of assumptions are met, smoking-PD association estimates are less likely to be biased by confounding or reverse causation than association estimates based on self-reported smoking.8 Another strength of our study compared to Chuang et al.7 is that rather than using a single SNP, we used genome-wide data to impute AA encoded by HLA-DRB1. Finally, using an independent dataset, we report similar associations with HLA alleles and AA as previous studies.2, 3 One limitation of our HLA-DRB1×smoking interaction analyses is that the approach we used allowed us to estimate the association between smoking initiation and PD stratified by HLA-DRB1, but did not allow us to estimate the association between HLA-DRB1 and PD stratified by smoking.
Despite being one of the most robust findings in PD, the mechanisms underlying its inverse association with smoking remain unknown. This work represents the first example of large-scale replication of a gene-environment interaction in PD, and allows to propose a biological mechanism to explain the inverse smoking-PD association, in the context of a larger body of work on the relationship between the immune system and PD.19
Supplementary Material
Acknowledgements:
We thank the GSCAN consortium (GWAS and Sequencing Consortium of Alcohol and Nicotine use) for providing summary statistics for this analysis. Additional Courage-PD investigators are: Sophia N Pchelina (Saint Petersburg, Russia), Thomas Brücke (Wien, Austria), Marie-Anne Loriot (Paris, France), Claire Mulot (Paris, France), Yves Koudou (Villejuif, France), Alain Destée (Lille, France), Georgia Xiromerisiou (Larissa, Greece), Christos Koros (Athens, Greece), Matina Maniati (Athens, Greece), Maria Bozi (Athens, Greece), Micol Avenali (Pavia, Italy), Margherita Canesi (Milan, Italy), Giorgio Sacilotto (Milan, Italy), Michela Zini (Milan, Italy), Roberto Cilia (Milan, Italy), Francesca Del Sorbo (Milan, Italy), Nicoletta Meucci (Milan, Italy), Rosanna Asselta (Milan, Italy), Radha Procopio (Catanzaro, Italy), Clara Hellberg (Lund, Sweden), Manabu Funayama (Tokyo, Japan), Aya Ikeda (Tokyo, Japan), Takashi Matsushima (Tokyo, Japan), Yuanzhe Li (Tokyo, Japan), Hiroyo Yoshino (Tokyo, Japan), Zied Landoulsi (Luxembourg, Luxembourg), Rubén Fernández-Santiago (Barcelona, Spain), Nicholas Wood (London, UK), Huw R Morris (London, UK).
Relevant conflicts of interest/Financial disclosures
ABS reports grants from Department of Defense, during the conduct of the study; grants from Michael J Fox Foundation, outside the submitted work.
WP reports personal fees from Grünenthal, personal fees from AbbVie, personal fees from AOP Orphan, personal fees from Zambon, personal fees and other from Boehringer Ingelheim, personal fees from Stada, personal fees from UCB Pharma, outside the submitted work.
AEL reports personal fees from AbbVie, personal fees from AFFiRis, personal fees from Janssen, personal fees from Biogen, personal fees from Merck, personal fees from Sun Pharma, personal fees from Corticobasal Solutions, personal fees from Sunovion, personal fees from Paladin, personal fees from Lilly, personal fees from Medtronic, personal fees from Theravance, personal fees from Lundbeck, personal fees from Retrophin, personal fees from Roche, personal fees from PhotoPharmics, outside the submitted work.
AB reports grants from France Parkinson + FRC, grants from ANR - EPIG - Agence nationale de recherche, grants from ANR - JPND - Agence nationale de recherche, grants from RDS (Roger de Spoelberch Foundation), grants from France Alzheimer, grants from Institut de France, grants from ANR - EPIG, grants from FMR (maladies rares), outside the submitted work.
JCC reports grants from the Michael J Fox Foundation, Sanofi, and served in advisory boards for Air Liquide, Biogen, Denali, Ever Pharma, Idorsia, Prevail Therapeutic, Theranexus, UCB, outside the submitted work.
MCCH reports grants from France Parkinson, grants from ANR –Agence nationale de recherche, (MetDePaDi, Synapark), grant from ANR – JPND (TransNeuro), Agence nationale de recherche, Grant; grants from Fondation de France, grants from the Michael J Fox Foundation, outside the submitted work.
KB reports grants from MJFF, grants from BMBF, personal fees from Zambon, UCB, Abbvie, grants from University of Tuebingen, outside the submitted work.
LS over the past year has received the following grants: PPMI2 (supported by the Michael J. Fox Foundation), IMPRIND-IMI2 Number 116060 (EU, H2020), “Transferring autonomous and non-autonomous cell degeneration 3D models between EU and USA for development of effective therapies for neurodegenerative diseases (ND) - CROSS NEUROD” (H2020-EU 1.3.3., Grant Number778003), «Chaperone-Mediated Autophagy in Neurodegeneration» (Hellenic Foundation for Research and Innovation Grant HFRI-FM17–3013), and “CMA as a Means to Counteract alpha-Synuclein Pathology in Non-Human Primates” Grant by the Michael J. Fox Foundation (Collaborator). He is co-Head and PI at the NKUA of the General Secretariat of Research and Technology (GSRT)-funded Grant “National Network of Precision Medicine for Neurodegenerative Diseases”. He has served on an Advisory Board for Abbvie, ITF Hellas and Biogen and has received honoraria from Abbvie and Sanofi. There are no specific disclosures related to the current work.
EMV serves as Associate Editor of Journal of Medical Genetics, Section Editor of Pediatric Research, Member of the Editorial Board of Movement Disorders Clinical Practice; grants from the Italian Ministry of Health, CARIPLO Foundation, Pierfranco and Luisa Mariani Foundation, Telethon Foundation Italy, outside the submitted work.
NH reports grants from - Japan Agency for Medical Research and Development (AMED), grants from - Japan Society for the Promotion of Science (JSPS), grants from - Ministry of Education Culture,Sports,Science and Technology Japan; Grant-in-Aid for Scientific Research on Innovative Areas, personal fees and other from Dai-Nippon Sumitomo Pharma Co.,Ltd, personal fees and other from Takeda Pharmaceutical Co.,Ltd., personal fees and other from Kyowa Kirin Co.,Ltd., personal fees and other from GSK K.K, personal fees and other from Nippon Boehringer Ingelheim,Co.,Ltd, personal fees and other from FP Pharmaceutical Corporation, personal fees and other from Eisai Co.,Ltd., personal fees and other from Kissei Pharmaceutical Company, personal fees and other from Nihon Medi-physics Co.,Ltd, personal fees and other from Novartis Pharma K.K, personal fees and other from Biogen Idec Japan Ltd, personal fees and other from AbbVie, from Medtronic, Inc., other from Boston Scientific Japan, personal fees and other from Astellas Pharma Inc., grants and other from Ono Pharmaceutical Co.,Ltd, other from Nihon Pharmaceutical Co., Ltd, other from Asahi Kasei Medical Co.,Ltd, other from Mitsubishi Tanabe Pharma Corporation, personal fees and other from Daiichi Sankyo Co., other from OHARA Pharmaceutical Co.,Ltd, other from Meiji Seika Pharma, personal fees from Sanofi K.K., personal fees from Pfizer Japan Inc., personal fees from Alexion Pharmaceuticals, personal fees from Mylan N.V, personal fees from MSD K.K, personal fees from Lund Beck Japan, other from Hisamitsu Pharmaceutical Co.,Inc, outside the submitted work.
KN reports grants from - Japan Society for the Promotion of Science (JSPS), outside the submitted work.
PK reports other from Centre Hospitalier de Luxembourg; University of Luxembourg, grants from Fonds National de Recherche (FNR), from null, outside the submitted work.
BPCW reports grants from ZonMW, grants from Hersenstichting, grants from uniQure, other from uniQure, grants from Gossweiler Fund, grants from Radboud university medical centre, outside the submitted work.
BRB reports grants from Netherlands Organization for Health Research and Development, grants from Michael J. Fox Foundation, grants from Parkinson Vereniging, grants from Parkinson Foundation, grants from Gatsby Foundation, grants from Verily Life Sciences, grants from Horizon 2020, grants from Topsector Life sciences and Health, grants from Stichting Parkinson Fonds, grants from UCB, grants from Abbvie, during the conduct of the study; personal fees from Biogen, personal fees from Abbvie, personal fees from Walk with Path, personal fees from UCB, personal fees from Abbvie, personal fees from Zambon, personal fees from Bial, personal fees from Roche, outside the submitted work; and Serves as editor-in-chief of the Journal of Parkinson’s Disease and serves on the editorial board of Practical Neurology and Digital Biomarkers.
MT (M.Toft) reports grants from Research Council of Norway, during the conduct of the study; grants from South-Eastern Norway Regional Health Authority, grants from Michael J. Fox Foundation, outside the submitted work.
LP reports grants from Norwegian Health Association, grants from South-Eastern Norway Regional Health Authority, outside the submitted work.
JJF reports grants from GlaxoSmithKline, grants from Grunenthal, grants from Fundação MSD (Portugal), grants from TEVA, grants from MSD, grants from Allergan, grants from Novartis, grants from Medtronic, grants from GlaxoSmithKline, grants from Novartis, grants from TEVA, grants from Lundbeck, grants from Solvay, grants from BIAL, grants from Merck-Serono, grants from Merz, grants from Ipsen, grants from Biogen, grants from Acadia, grants from Allergan, grants from Abbvie, grants from Sunovion Pharmaceuticals, personal fees from Faculdade de Medicina de Lisboa, personal fees from CNS - Campus Neurológico Sénior, personal fees from BIAL, personal fees from Novartis, outside the submitted work.
ET received honoraria for consultancy from TEVA, Bial, Prevail Therapeutics, Boehringer Ingelheim, Roche and BIOGEN and has received funding for research from Spanish Network for Research on Neurodegenerative Disorders (CIBERNED)- Instituto Carlos III (ISCIII), and The Michael J. Fox Foundation for Parkinson’s Research(MJFF)
KW reports grants from Swedish Research Council, during the conduct of the study.
NLP reports grants from Swedish Research Council, during the conduct of the study.
AP reports grants from Parkinsonfonden (The Swedish Parkinson Foundation), grants from ALF (Swedish Government), grants from Region Skåne, Sweden, grants from Skåne University Hospital, grants from Hans-Gabriel och Trolle Wachtmeister Stiftelse för Medicinsk Forskning, Sweden, grants from Multipark – a strategic research environment at Lund University, during the conduct of the study; personal fees from Elsevier, outside the submitted work.
EYR reports grants from ALF (Swedish Government), grants from Hans-Gabriel och Trolle Wachtmeister Stiftelse för Medicinsk Forskning, Sweden, and grants from Demensfonden (all in Sweden).
MT (M. Tan) reports grants from Parkinson’s UK, other from Michael J Fox Foundation, other from University College London, outside the submitted work.
RK reports grants from Fonds National de Recherche (FNR), grants from German Research Council (DFG), non-financial support from Abbvie, Zambon, during the conduct of the study; personal fees from University of Luxembourg; Luxembourg Institute of Health; Centre Hospitalier de Luxembourg, grants from Fonds National de Recherche, Luxembourg (FNR), grants from Fonds National de Recherche, Luxembourg (FNR), grants from Fonds National de Recherche (FNR), Luxembourg/German Research Council (DFG), grants from Fonds National de Recherche, Luxembourg (FNR), personal fees from Desitin/Zambon, personal fees from Abbvie GmbH, personal fees from Medtronic GmbH, outside the submitted work.
TG reports personal fees from UCB Pharma, personal fees from Novartis, personal fees from Teva, personal fees from MedUpdate, grants from The Michael J Fox Foundation for Parkinsońs Research, grants from Bundesministerium für Bildung und Forschung (BMBF), grants from Deutsche Forschungsgemeinschaft (DFG), other from “Joint Programming for Neurodegenerative Diseases”(JPND) program, funded by the European Commission, outside the submitted work; in addition, Dr. Gasser has a patent Patent Number: EP1802749 (A2) KASPP (LRRK2) gene, its production and use for the detection and treatment of neurodegenerative disorders issued.
AE reports grants from Agence nationale de recherche (ANR), Michael J Fox foundation, Plan Ecophyto (French ministry of agriculture), and France Parkinson, outside the submitted work.
Funding agencies
This study used data from the Courage-PD consortium, conducted under a partnership agreement between 35 studies. The Courage-PD consortium is supported by the EU Joint Program for Neurodegenerative Disease research (JPND; https://www.neurodegenerationresearch.eu/initiatives/annual-calls-for-proposals/closed-calls/risk-factors-2012/risk-factor-call-results/courage-pd/).
CD is the recipient of a doctoral grant from Université Paris-Saclay, France.
PM was funded by the Fonds National de Recherche (FNR), Luxembourg as part of the National Centre of Excellence in Research on Parkinson’s disease (NCER-PD, FNR11264123) and the DFG Research Units FOR2715 (INTER/DFG/17/11583046) and FOR2488 (INTER/DFG/19/14429377).
ABS, DGH, and CE are funded by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project ZO1 AG000949.
ER is funded by the Canadian Consortium on Neurodegeneration in Aging.
SK is funded by MSWA.
PT is the recipient of an Estonian Research Council Grant PRG957.
EMV is funded by the Italian Ministry of Health (Ricerca Corrente 2021).
SB and JC are supported by grants from the National Research Foundation of South Africa (Grant Number: 106052); the South African Medical Research Council (Self-Initiated Research Grant); and Stellenbosch University, South Africa; they also acknowledge the support of the NRF-DST Centre of Excellence for Biomedical Tuberculosis Research; South African Medical Research Council Centre for Tuberculosis Research; Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town.
PP and MDF have received funding from the Spanish Ministry of Science and Innovation (SAF2013-47939-R).
KW and NLP are funded by the Swedish Research Council, grant numbers K2002-27X-14056-02B, 521-2010-2479, 521-2013-2488, 2017-02175.
NLP is funded by the National Institutes of Health, grant numbers ES10758 and AG 08724.
CR is funded by the Märta Lundkvist Foundation, Swedish Brain Foundation, Karolinska Institutet Research Fund.
ACB from the Swedish Brain Foundation, Swedish Research Council, Karolinska Institutet Research Funds.
MT is funded by the Parkinson’s UK.
MS was supported by the grants from the German Research Council (DFG/SH 599/6-1), MSA Coalition, and Michael J Fox Foundation (USA Genetic Diversity in PD Program: GAP-India Grant ID: 17473).
PG GEN sample collection was funded by the MRC and UK Medical Research Council (CEC, KEM).
The sponsors had no role in the study design, data collection, data analysis, data interpretation, the writing of the report, or the decision to submit the paper for publication.
Author’s roles
Cloé Domenighetti had a major role in the acquisition of data; conceptualized the study; designed, analyzed, and interpreted the data; drafted, and revised the manuscript; is the guarantor.
Venceslas Douillard conceptualized the study; designed and interpreted the data; drafted, and revised the manuscript.
Pierre-Emmanuel Sugier had a major role in the acquisition of data; analyzed and interpreted the data; revised the manuscript.
Ashwin Ashok Kumar Sreelatha had a major role in the acquisition of data; analyzed and interpreted the data; revised the manuscript.
Claudia Schulte had a major role in the acquisition of data, revised the manuscript.
Sandeep Grove had a major role in the acquisition of data, revised the manuscript.
Patrick May had a major role in the acquisition of data, revised the manuscript.
Dheeraj R. Bobbili had a major role in the acquisition of data, revised the manuscript.
Milena Radivojkov-Blagojevic had a major role in the acquisition of data, performed the genotyping, revised the manuscript.
Peter Lichtner had a major role in the acquisition of data, performed the genotyping, revised the manuscript.
Andrew B. Singleton performed the genotyping, revised the manuscript.
Dena G. Hernandez performed the genotyping, revised the manuscript.
Connor Edsall performed the genotyping, revised the manuscript.
Pierre-Antoine Gourraud conceptualized the study; drafted, and revised the manuscript.
George D. Mellick had a major role in the acquisition of data, revised the manuscript.
Alexander Zimprich had a major role in the acquisition of data, revised the manuscript.
Walter Pirker had a major role in the acquisition of data, revised the manuscript.
Ekaterina Rogaeva had a major role in the acquisition of data, revised the manuscript.
Anthony E. Lang had a major role in the acquisition of data, revised the manuscript.
Sulev Koks had a major role in the acquisition of data, revised the manuscript.
Pille Taba had a major role in the acquisition of data, revised the manuscript.
Suzanne Lesage had a major role in the acquisition of data, revised the manuscript.
Alexis Brice had a major role in the acquisition of data, revised the manuscript.
Jean-Christophe Corvol had a major role in the acquisition of data, revised the manuscript.
Marie-Christine Chartier-Harlin had a major role in the acquisition of data, revised the manuscript.
Eugénie Mutez had a major role in the acquisition of data, revised the manuscript.
Kathrin Brockmann had a major role in the acquisition of data, revised the manuscript.
Angela B. Deutschländer had a major role in the acquisition of data, revised the manuscript.
Georges M. Hadjigeorgiou had a major role in the acquisition of data, revised the manuscript.
Efthimos Dardiotis had a major role in the acquisition of data, revised the manuscript.
Leonidas Stefanis had a major role in the acquisition of data, revised the manuscript.
Athina Maria Simitsi had a major role in the acquisition of data, revised the manuscript.
Enza Maria Valente had a major role in the acquisition of data, revised the manuscript.
Simona Petrucci had a major role in the acquisition of data, revised the manuscript.
Stefano Duga had a major role in the acquisition of data, revised the manuscript.
Letizia Straniero had a major role in the acquisition of data, revised the manuscript.
Anna Zecchinelli had a major role in the acquisition of data, revised the manuscript.
Gianni Pezzoli had a major role in the acquisition of data, revised the manuscript.
Laura Brighina had a major role in the acquisition of data, revised the manuscript.
Carlo Ferrarese had a major role in the acquisition of data, revised the manuscript.
Grazia Annesi had a major role in the acquisition of data, revised the manuscript.
Andrea Quattrone had a major role in the acquisition of data, revised the manuscript.
Monica Gagliardi had a major role in the acquisition of data, revised the manuscript.
Hirotaka Matsuo had a major role in the acquisition of data, revised the manuscript.
Yusuke Kawamura had a major role in the acquisition of data, revised the manuscript.
Nobutaka Hattori had a major role in the acquisition of data, revised the manuscript.
Kenya Nishioka had a major role in the acquisition of data, revised the manuscript.
Sun Ju Chung had a major role in the acquisition of data, revised the manuscript.
Yun Joong Kim had a major role in the acquisition of data, revised the manuscript.
Pierre Kolber had a major role in the acquisition of data, revised the manuscript.
Bart PC van de Warrenburg had a major role in the acquisition of data, revised the manuscript.
Bastiaan R. Bloem had a major role in the acquisition of data, revised the manuscript.
Jan Aasly had a major role in the acquisition of data, revised the manuscript.
Mathias Toft had a major role in the acquisition of data, revised the manuscript.
Lasse Pihlstrøm had a major role in the acquisition of data, revised the manuscript.
Leonor Correia Guedes had a major role in the acquisition of data, revised the manuscript.
Joaquim J. Ferreira had a major role in the acquisition of data, revised the manuscript.
Soraya Bardien had a major role in the acquisition of data, revised the manuscript.
Jonathan Carr had a major role in the acquisition of data, revised the manuscript.
Eduardo Tolosa had a major role in the acquisition of data, revised the manuscript.
Mario Ezquerra had a major role in the acquisition of data, revised the manuscript.
Pau Pastor had a major role in the acquisition of data, revised the manuscript.
Monica Diez-Fairen had a major role in the acquisition of data, revised the manuscript.
Karin Wirdefeldt had a major role in the acquisition of data, revised the manuscript.
Nancy L. Pedersen had a major role in the acquisition of data, revised the manuscript.
Caroline Ran had a major role in the acquisition of data, revised the manuscript.
Andrea C. Belin had a major role in the acquisition of data, revised the manuscript.
Andreas Puschmann had a major role in the acquisition of data, revised the manuscript.
Emil Ygland Rödström had a major role in the acquisition of data, revised the manuscript.
Carl E. Clarke had a major role in the acquisition of data, revised the manuscript.
Karen E. Morrison had a major role in the acquisition of data, revised the manuscript.
Manuela Tan had a major role in the acquisition of data, revised the manuscript.
Dimitri Krainc had a major role in the acquisition of data, revised the manuscript.
Lena F. Burbulla had a major role in the acquisition of data, revised the manuscript.
Matt J. Farrer had a major role in the acquisition of data, revised the manuscript.
Rejko Krüger obtained funding and supervised the study, had a major role in the acquisition of data, revised the manuscript.
Thomas Gasser obtained funding and supervised the study, had a major role in the acquisition of data, revised the manuscript.
Manu Sharma obtained funding and supervised the study, had a major role in the acquisition of data, revised the manuscript.
Nicolas Vince conceptualized the study; designed and interpreted the data; drafted, and revised the manuscript.
Alexis Elbaz obtained funding and supervised the study; had a major role in the acquisition of data; conceptualized the study, designed, analyzed, and interpreted the data; drafted, and revised the manuscript; is the guarantor.
REFERENCES
- 1.Hollenbach JA, Norman PJ, Creary LE, et al. A specific amino acid motif of HLA-DRB1 mediates risk and interacts with smoking history in Parkinson’s disease. Proc Natl Acad Sci U S A 2019;116:7419–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naito T, Satake W, Ogawa K, et al. Trans-Ethnic Fine-Mapping of the Major Histocompatibility Complex Region Linked to Parkinson’s Disease. Mov Disord 2021;36:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu E, Ambati A, Andersen MS, et al. Fine mapping of the HLA locus in Parkinson’s disease in Europeans. NPJ Parkinsons Dis 2021;7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedström AK, Hillert J, Brenner N, et al. DRB1-environment interactions in multiple sclerosis etiology: results from two Swedish case-control studies. J Neurol Neurosurg Psychiatry 2021;92:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlson EW, Chang SC, Cui J, et al. Gene-environment interaction between HLA-DRB1 shared epitope and heavy cigarette smoking in predicting incident rheumatoid arthritis. Ann Rheum Dis 2010;69:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baecklund F, Foo JN, Askling J, et al. Possible Interaction Between Cigarette Smoking and HLA-DRB1 Variation in the Risk of Follicular Lymphoma. Am J Epidemiol 2017;185:681–687. [DOI] [PubMed] [Google Scholar]
- 7.Chuang YH, Lee PC, Vlaar T, et al. Pooled analysis of the HLA-DRB1 by smoking interaction in Parkinson disease. Ann Neurol 2017;82:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess S, Davey Smith G, Davies NM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res 2019;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M, Jiang Y, Wedow R, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019;51:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynisson B, Barra C, Kaabinejadian S, et al. Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J Proteome Res 2020;19:2304–2315. [DOI] [PubMed] [Google Scholar]
- 11.Sulzer D, Alcalay RN, Garretti F, et al. T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 2017;546:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Shen J, Cox C, et al. HIBAG—HLA genotype imputation with attribute bagging. The Pharmacogenomics Journal 2014;14:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas CE, Michael FA. On distribution-free multiple comparisons in the one-way analysis of variance. Communications in Statistics - Theory and Methods 1991;20:127–139. [Google Scholar]
- 15.Grover S, Lill CM, Kasten M, et al. Risky behaviors and Parkinson disease: A mendelian randomization study. Neurology 2019;93:e1412–e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilbron K, Jensen MP, Bandres-Ciga S, et al. Unhealthy Behaviours and Risk of Parkinson’s Disease: A Mendelian Randomisation Study. J Parkinsons Dis 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez-Baleon C, Ong JS, Scherzer CR, et al. Understanding the effect of smoking and drinking behavior on Parkinson’s disease risk: a Mendelian randomization study. Sci Rep 2021;11:13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domenighetti C, Sugier PE, Sreelatha AAK, et al. Mendelian Randomisation Study of Smoking, Alcohol, and Coffee Drinking in Relation to Parkinson’s Disease. J Parkinsons Dis 2022;12:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan EK, Chao YX, West A, et al. Parkinson disease and the immune system - associations, mechanisms and therapeutics. Nat Rev Neurol 2020;16:303–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Results can be reproduced using the Supplementary material.


