Abstract
Simple Summary
In the period 2014–2021, the circulation of CDV in dogs of Southern Italy was investigated. In this time span a reduction in the circulation of CDV was observed, with a higher frequency of detection of the pathogen in imported dogs (18.4%) compared to stray (7.4%) and household (3.9%) animals. These results underline the effectiveness of the prophylaxis strategy on autochthonous dogs as well as the importance of continuous surveillance of CDV, especially in imported dogs.
Abstract
This study aims to investigate the presence of canine distemper virus (CDV) infection in 949 autochthonous or illegally imported dogs from Southern Italy, over a period of eight years (2014–2021). CDV RNA was detected in 6.8% (65/949) of the animals tested, with no detection of CDV in dogs sampled in 2020–2021. The frequency of CDV detection was higher in imported dogs (19/103, 18.3%) with respect to stray (27/365, 7.4%) and household dogs (19/481, 3.9%). On sequence and phylogenetic analyses of selected strains, the analyzed viruses belonged to the Arctic clade, which has already been reported in Italy and in Europe. The results of our study may suggest a reduction of CDV circulation in Southern Italy, while at the same time highlighting the need for strict controls on dog importation, in order to prevent the introduction of viruses from endemic countries.
Keywords: canine distemper virus, dog, animal importation, CDV Arctic-lineage, CDV H gene
1. Introduction
Canine distemper virus (CDV) is an enveloped, negative-sense, single-stranded RNA virus belonging to the family Paramyxoviridae, genus Morbillivirus [1]. It was first described by H. Carré in 1905 [2] and it can affect a very broad range of host species worldwide [3,4,5,6]. Canidae and Mustelidae families are the most affected, but CDV has also been detected in the Felidae, Viverridae, Procyonidae, and Ursidae families [7,8]. CDV infections are present in genetic diversity worldwide [9,10,11,12].
Its genome encodes for six structural proteins: a single envelope-associated matrix (M) protein, two membrane glycoproteins [hemagglutinin/attachment protein (H) and fusion protein (F)], two transcriptase-associated proteins [the phosphoprotein (P) and the large (L) polymerase protein] and the nucleo-capsid (N) protein, that encapsidates the viral RNA [13,14]. The heterogeneity within the H genes allows the molecular typing of CDV. H-glycoprotein mediates the binding of the virus to receptors on the host cell and is composed of a short N-terminal cytoplasmic tail followed by a transmembrane domain and a large C-terminal ectodomain [13,15]. Sequence variations may affect the virulence, host range, and neutralisation-epitopes of CDV, and play an essential role in cell tropism [16]. CDV infection can be prevented by an adequate host immune response against protein H [15]. The fusion of the cell membrane with the viral envelope is promoted by the F protein that also promotes membrane fusion between the host cells, with formation of syncytia [17].
Twelve genetic lineages have been evidenced by molecular studies (America-1 and -2, Arctic-like, Asia -1, -2, and -3, Europe Wildlife, Europe-1/South America-1, South America-2, South America-3, South Africa, Rockborn-like) [11,18]. The majority of CDV field strains cluster into six major genetic lineages, designated America-1 and -2, Asia-1 and -2, European and Arctic [19,20,21,22,23,24,25,26,27,28,29,30,31]. America-1 CDVs have not been detected over the last five decades and it is not known whether they are still circulating in the field. In Italy, the circulation of 3 distinct lineages have been reported: Europe (also called Europe-1/South America-1), Europe Wildlife, and Arctic-like lineages [18,28,29,32,33,34,35]. In animal hosts other than dogs, such as wolves, Arctic-like lineage CDV strains have also been detected [10,36,37,38] in badgers [33] and tigers [39]. Europe and Europe wildlife lineage CDV strains have been also described in foxes [28] and wolves [38], and in bears [40], respectively.
The CDV genetic lineages are variously distributed according to geographic patterns, although not to species of origin. Disease prevalence exhibits temporal fluctuations and increases during the cold season. In the environment, CDV is quickly inactivated. Transmission mainly occurs by direct animal-to-animal contact or by exposure to respiratory and ocular fluids and exudates, while other body excretions and secretions (e.g., urine and faeces) [7] could also contribute to viral shedding during the acute phase of infection. In domestic dogs, trans-placental infection has also been documented [41]. This highly contagious pathogen enters the new host and promptly starts replication [42], resulting in severe immunosuppression. Targets of infection are mainly mucous membranes and lymphoid tissues. This can result in either subclinical infection (50–70% of CDV infections) or severe, often fatal, systemic disease, mostly observed in immune-compromised animals [43].
The severity of clinical signs is influenced by strain virulence, environmental conditions, host age and immune status [11]. The infection may be prevented by passive or active immunization and is not age-restricted [44]. Young pups are protected by passive immunity and most adult dogs are protected by vaccine immunization. Age-related susceptibility to infection (3–6 months old pups are more susceptible than older dogs) correlates with the decline in maternally derived immunity. Individual variations among the various strains are responsible for the differences in virulence or tropism rather than properties inherent to a given CDV lineage [27,45].
This study aims to investigate the presence of CDV infections in dogs in Italy, over a period of eight years (2014–2021) in order to better understand the virus origin and distribution among animal populations. In order to elucidate the genetic divergences, selected CDV strains were sequenced and their sequences were compared with those available in the GenBank.
2. Materials and Methods
A total of 949 dogs and 6374 matrices were tested for canine distemper virus (CDV) over a period of 8 years (2014–2021) (Table 1) at the Istituto Zooprofilattico Sperimentale del Mezzogiorno (IZSM), statal public institution that operates within the National Health Service. The samples tested in this study in the framework within the diagnostic activity of IZSM. Out of the 949 tested dogs, 858 (90.4%) were dead and the analyzed samples (intestine, spleen, liver, lung, kidney, brain, heart) were collected at necropsy. A total of 91 (9.6%) animals were alive and the following matrices were analyzed: oropharyngeal swab, oculo-conjunctival swab, rectal swab or faeces. Three hundred and sixty five stray dogs (38.5%) and 481 (50.7%) household animals were collected from Calabria and Campania regions of Italy, whilst 103 (10.8%) dogs were imported from Eastern Europe (Hungary and Romania) (Table 2). The samples were subjected to extraction of nucleic acids by means of an automatic extractor (QIAsymphony, Qiagen, Hilden, Germany), using the “Virus/Pathogen” kit (Qiagen). Viral RNA was detected by a real time RT-PCR [46] and the positive samples were submitted to RT-PCR protocol for amplification of the hemagglutinin gene of CDV [47] and sequence characterization for determination of the CDV lineage. In addition, the extracts were also analyzed for the following canine viruses, to evaluate the presence of co-infections: coronavirus (CCoV) [48,49,50,51], adenoviruses (CAdVs) [52,53], herpesvirus (CaHV-1) [54], rotavirus (RVA) [55,56], parvovirus (CPV2) [57,58]. For CPV2, a PCR able to discriminate between field variants and the vaccine virus was used [59].
Table 1.
Dogs tested in the period 2014–2021, animals CDV+ and their relative frequency expressed as percentage.
| Year | Dogs | CDV+ (%) |
|---|---|---|
| 2014 | 186 | 37 (19.9) |
| 2015 | 156 | 20 (12.8) |
| 2016 | 111 | 5 (4.5) |
| 2017 | 130 | 1 (0.8) |
| 2018 | 88 | 1 (1.1) |
| 2019 | 82 | 1 (1.2) |
| 2020 | 99 | 0 (0) |
| 2021 | 97 | 0 (0) |
| Total | 949 | 65 (6.8%) |
Table 2.
Imported, stray and owned dogs analyzed by their geographical distribution and frequency distribution of CDV+ animals.
| Category | nr of Dogs (%) | Geographical Distribution (nr) | CDV+ (%) |
|---|---|---|---|
| Household | 481 (50.7%) | Campania, Italy (405) | 19 (3.9) |
| Calabria, Italy (76) | |||
| Stray | 365 (38.5%) | Campania, Italy (346) | 27 (7.4) |
| Calabria, Italy (19) | |||
| Imported | 103 (10.8%) | Eastern Europe (Hungary, Romania) (103) |
19 (18.4) |
| Total | 949 (100%) | Italy, Eastern Europe (Hungary and Romania) (949) |
65 (6.8) |
The PCR products underwent agarose gel electrophoresis at 50 V for 90 min. PCR amplicons were visualized on a Gel Doc™ EZ (Bio-Rad Laboratories SRL, Segrate, Italy), and subsequently subjected to purification. Direct Sanger sequencing was performed in both directions by Eurofins Genomics (Ebersberg, Germany).
The online tools Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov, accessed on 28 June 2022) and FASTA (http://www.ebi.ac.uk/fasta33, accessed on 28 June 2022) were used, employing the default values to find homologous hits. Sequence editing was carried out by Geneious Prime version 2021.2 (Biomatters Ltd., Auckland, New Zealand). The sequences were aligned with other CDV strains recovered from the GenBank database by Multiple Alignment using Fast Fourier Transform (MAFFT) [60]. The correct substitution model for performing phylogenetic analyses and evaluation of selection pressure on coding sequences was obtained using “Find the best protein DNA/Protein Models” of MEGA X version 10.0.5 software [61]. Maximum-likelihood method, Tamura-Nei 4-parameter model, a discrete gamma distribution with six categories with 1000 replicates as statistical support were used. Bayesian inference and neighbor joining methods for the phylogeny were also explored, exhibiting similar topologies; maximum-likelihood tree was finally kept.
3. Results
Overall, out of the 949 dogs tested in a period spanning from 2014 to 2021, 6.8% (65/949) animals were positive for CDV by real time RT-PCR. CDV was not detected in 2020 and 2021 (Table 1). CDV was unevenly identified in the lung, intestine, brain, spleen, liver, heart and kidney of the positive animals. The frequency of CDV detection was higher in imported dogs (n = 19, 18.4%) than in stray (27, 7.4%) and household (n = 19, 3.9%) dogs (Table 2). The highest number of stray (nr = 22) and imported (nr = 10) CDV+ dogs was identified in 2014 whilst CDV was detected with the highest frequency in household dogs in 2015. Identification of CDV occurred in stray dogs in the 2014–2016 period, whilst in household dogs CDV was identified from 2014 to 2019 with the exception of 2017. CDV RNA was retrieved in imported dogs from 2014 to 2017 with the exception of 2017 (Table 3).
Table 3.
Distribution over 2014–2021 of imported, stray and household CDV+ dogs.
| Year | Category | CDV+ Dogs (%) |
|---|---|---|
| 2014 | Household | 5 (13.5) |
| Stray | 22 (59.5) | |
| Imported | 10 (27.0) | |
| Total | 37 (100) | |
| 2015 | Household | 6 (30.0) |
| Stray | 6 (30.0) | |
| Imported | 8 (40.0) | |
| Total | 20 (100) | |
| 2016 | Household | 3 (60.0) |
| Stray | 2 (40.0) | |
| Imported | 0 | |
| Total | 5 (100) | |
| 2017 | Household | 0 (0.0) |
| Stray | 0 (0.0) | |
| Imported | 1 100) | |
| Total | 1 (100) | |
| 2018 | Household | 1 (100) |
| Stray | 0 (0.0) | |
| Imported | 0 (0.0) | |
| Total | 1 (100) | |
| 2019 | Household | 1 (100) |
| Stray | 0 (0.0) | |
| Imported | 0 (0.0) | |
| Total | 1 (100) | |
| 2020 | - | - |
| 2021 | - | - |
The 65 CDV positive dogs exhibited the following co-infections: 20 had a single CDV viral infection, 23 dual viral infections, 21 triple viral infections and 1 dog (78870/2016), had a quadruple viral infection (CDV + CPV2 + CCoV + CAdV). Among the 23 dogs with double infection, there were two co-infection patterns: 20 animals were co-infected by CDV + CPV2 and 3 dogs by CDV + CCoV. Among the 21 dogs with triple viral infections, there were two patterns: 17 dogs were co-infected by CDV + CPV2 + CCoV and 4 dogs by CDV + CPV2 + CAdV. CDV positive dogs imported from Eastern Europe were 18/65 (28%), with 3 being co-infections with the pantropic canine coronavirus pCCoV (acc. N. 103480/2014, 15910-3/2015 and 78870/2016) [62].
A total of 13 CDV-positive samples were selected based on the viral load as assessed by real time RT-PCR, with a threshold cycle (Ct) lower than 25. These samples were used for RT-PCR amplification of the complete hemagglutinin gene of CDV. A total of 8/13 samples could be amplified by RT-PCR yielding visible PCR products under gel visualization, while only 6 were suitable for direct sequencing based on DNA yield. One sequence was of poor quality. The sequenced CDV strains, ITA/2015/dog/24100, ITA/2015/dog/15952-1, ITA/2015/dog/15952-4, ITA/2015/dog/15952-5 and ITA/2015/dog/15952-6 were all characterized as Arctic lineage. For these dogs, the following information was assessed in detail: breed, age, origin, organs, co-infections, anatomopathological lesions (Table 4).
Table 4.
Information of dogs infected by CDV Arctic strain.
| ID | Year | Species and Breed | Age | Origin | Organs CDV+ |
Viral Co-Infections | Non-Viral Co-Infections | Injury |
|---|---|---|---|---|---|---|---|---|
| ITA/2015/dog/24100 | 2015 | DOG half-breed male |
young adult (over 12 month) |
Client-owner | Liver, lung, encephalus | absent | acanthosis nigricans, leishmania, tenia, hunting pellets, lead, neospora | encephalitis, pneumonia, enteritis |
| ITA/2015/dog/15952-1 | 2015 | DOG akita female |
puppy | Imported from Eastern Europe (Hungary) | lung, encephalus | CPV2a vaccinal and wild type |
coccidia | enteritis, lymphoid atrophy of the spleen, bronchopneumonia, encephalitis |
| ITA/2015/dog/15952-4 | 2015 | DOG chow chow female |
puppy | Imported from Eastern Europe (Hungary) | lung, encephalus | CPV2a vaccinal |
coccidia, klebsiella pneumoniae | enteritis, lymphoid hyperplasia of the milzam broncho-pneumonia, encephalitis |
| ITA/2015/dog/15952-5 | 2015 | DOG pincher female |
puppy | Imported from Eastern Europe (Hungary) | lung, encephalus | CPV2a vaccinal and wild type |
coccidia | hemorrhagic enteritis with atrophy of the intestinal villi, lymphadenomegaly of the meseric lymphnodes, lymphoid hyperplasia of the spleen, broncho-pneumonia, encephalitis |
| ITA/2015/dog/15952-6 | 2015 | DOG husky female |
puppy | Imported from Eastern Europe (Hungary) | lung, encephalus | CPV2a wild type |
coccidia | enteritis, lymphoid hyperplasia of the spleen, broncho-pneumonia, encephalitis |
All the five animals from Table 3 were puppies except the animal #24100, was 12-months old.
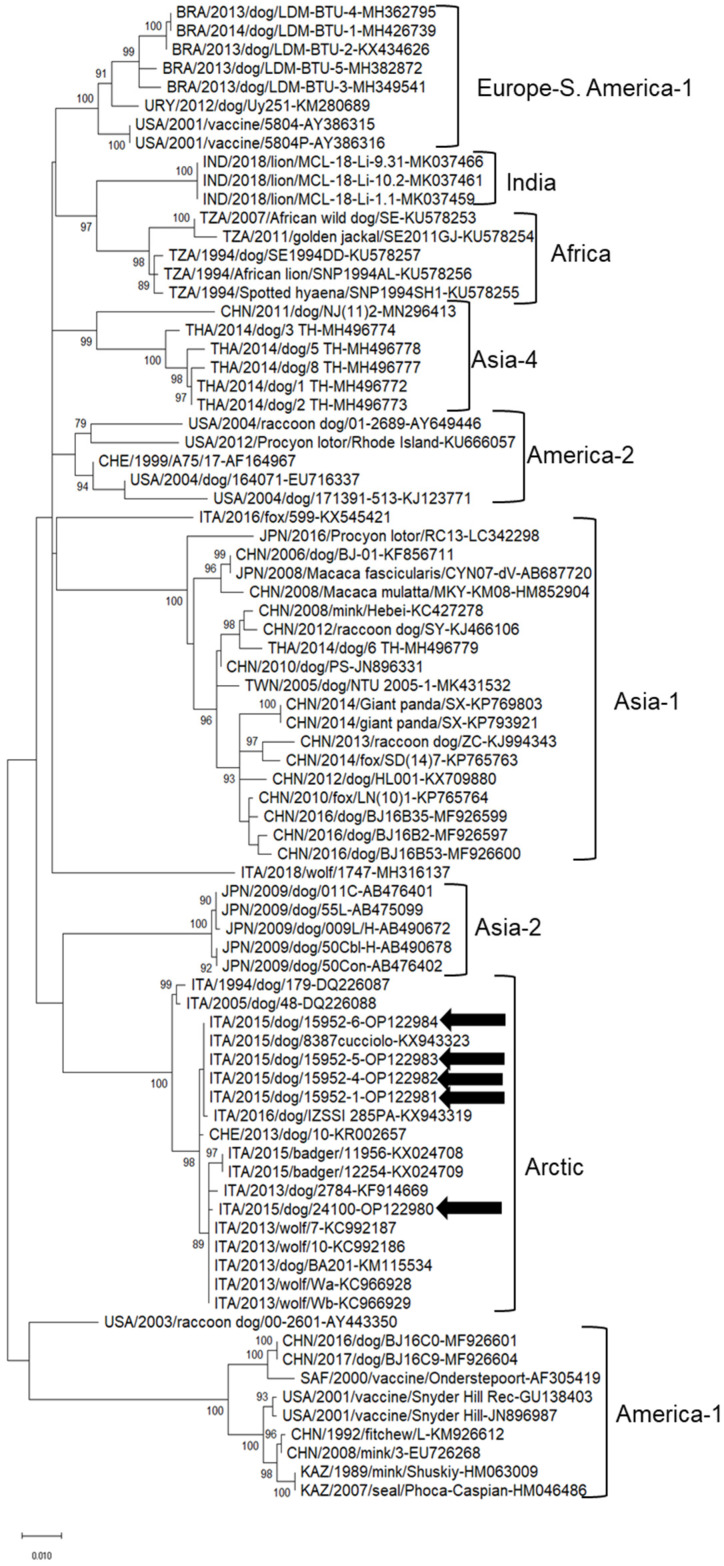
Four CDV strains (ITA/2015/dog/15952-1, ITA/2015/dog/15952-4, ITA/2015/dog/15952-5 and ITA/2015/dog/15952-6) showed the highest nt identities (99.8–100%) to the Italian strain ITA/2015/dog/8387cucciolo (KX943323) whilst the CDV strain ITA/2015/dog/24100 displayed 99.7% identity to strain ITA/2013/dog/BA201 (KM115534).
In the phylogenetic tree based on the partial nucleotide sequence of the hemagglutinin gene, the CDV strains identified in this study clustered into the Arctic clades together with strains retrieved in Italy from 2013 to 2016 and Switzerland in 2013 (Figure 1). The nucleotide sequences of strains ITA/2015/dog/24100, ITA/2015/dog/15952-1, ITA/2015/dog/15952-4, ITA/2015/dog/15952-5 and ITA/2015/dog/15952-6 were deposited in the GenBank database under accession nrs. OP122980-OP122984.
Figure 1.
Phylogenetic tree based on the CDV partial nucleotide sequence of the hemagglutinin gene.
4. Discussion
The results of our study showed an active circulation of CDV in dogs in Southern Italy, either alone or in co-infection. Interestingly, we observed a marked reduction over the 8-year-long study period, with 37 cases in 2014, 20 in 2015, 5 in 2016, 1 in 2017, 1 in 2018 and 1 in 2019, with no cases in 2020 and 2021. CDV was identified in all the dog categories (household, stray, and imported animals) with peaks in 2014 and 2015 (Table 3). The higher frequency of detection of CDV in 2014 was mostly accounted for by infections in stray dogs (Table 3). A large distemper epidemic occurred in Italy during 2013 and involved primarily the Abruzzi region and neighboring regions, also affecting unvaccinated domestic and shepherd dogs, foxes (Vulpes vulpes), badgers (Meles meles), beech martens (Martes foina) and European polecats (Mustela putorius) [10,36]. The epidemic was sustained by an Arctic-lineage [34]. Therefore, it is possible that the peak of CDV activity observed in free-ranging dogs in Campania region in 2014 was somewhat linked to the epidemic observed in Central-Southern Italy in 2013.
Alternate periods of quiescence and peaks of activity of CDV in the population are likely linked to changes in population immunity. Since vaccination against CDV is a core recommendation for dogs, most individuals are expected to be vaccinated and protected, and population immunity should be high enough to keep CDV infection under control, with only sporadic cases occurring [63]. When for some reason, specific population immunity decreases, the patterns of CDV infection may shift from sporadic to epidemic, posing a serious threat to susceptible populations [64].
Interestingly, out of the 65 CDV-positive dogs, only in 20 animals was CDV the only pathogen identified, with CPV2 and CCoV being frequently detetced in co-infection. Co-infections are frequently found in dogs [65,66] and the extent of this phenomenon has been revealed by the adoption of syndromic diagnostics (i.e., multi-screening of samples against a recognized panel of pathogens) [67] and, more recently, by metagenomic investigations in domestic and wildlife animals [68,69]. We cannot rule out that concomitant changes in the epidemiology of viruses with immune-suppressive activity may also occur in the local canine population, amplifying the magnitude of the CDV epidemic. For instance, both canine parvovirus and canine coronavirus are able to trigger leukopenia in dogs [13,70,71]. Functional imbalance of TCD4 lymphocytes after infection by the pantropic canine coronavirus can persist for 40 days [72], even if coronavirus infection is asymptomatic.
Co-infection in CDV-infected animals could be facilitated by animal life conditions/habits. Interestingly, the prevalence of CDV was higher in imported dogs (18.4%) than in free-ranging dogs (7.4%) and in household dogs (3.9%). Movements for animals can be very stressful and can decrease the immunological defenses [62]. Also, illegal trading of dogs is usually associated with low hygiene and health standards with incomplete or unknown vaccination status of young pups. By interrogation of metadata and animal history when available, we noted that in most imported dogs the vaccination schedule was incomplete or altered using false certificates.
Several lineages or genotypes of CDV exist that are variously distributed throughout several continents. The detection of Arctic CDV strains, formerly believed to circulate exclusively in the Arctic ecosystem, was first reported in dogs in 2004 and 2005 in an Italian study (29), and subsequently in the United States [31] and other European areas [73,74,75], raising questions on the origin of these unusual strains. Legal or uncontrolled trading of animals, as observed for pCCoV [62], may be linked to the observed changes in CDV epidemiology, as a consequence of the introduction of novel strains into CDV-naïve areas, or accounting for the resurgence of CDV in countries where CDV spread had been effectively and successfully controlled by vaccine prophylaxis.
We were able to determine the partial sequence of the H gene of a selection of CDV strains. All the sequences clustered within the Arctic-like lineage, along with CDV sequences obtained from other Italian studies. Interestingly, a unique strain, 24100 (accession OP122980) fell into a distinct sub-cluster, suggesting the circulation of different epidemic strains.
5. Conclusions
Although vaccination against canine distemper has been used widely for many decades, this infection still represents an important disease for dogs. Several factors, including the high number of unvaccinated stray dogs present in Italy [76,77] and the illegal trading of dogs [62,78], may be related to the persistence and spread of CDV in the Italian canine population. Implementing controls on imported animals and enacting continuous surveillance plans, along with reinforced vaccination programs, would be necessary to defeat the threat of CDV.
Acknowledgments
We thank Manuela Sannino for her kind and excellent assistance.
Author Contributions
Conceptualization, F.A. and N.D. (Nicola Decaro); methodology, F.A., G.M., N.D. (Nicola D’Alessio), M.G.A. and M.S.L.; Investigation, F.A., A.G., C.A. and M.S.L.; writing—original draft preparation, F.A. and G.L.; writing—review and editing, M.G.L., G.F., N.D. (Nicola Decaro) and V.M.; supervision, G.F., E.D.C., N.D. (Nicola Decaro) and V.M.; project administration, G.F.; funding acquisition, F.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical review and approval were waived for this study, as samples were part of the diagnostic activity of Istituto Zooprofilattico Sperimentale del Mezzogiorno, statal public institution that operate within the national health service.
Informed Consent Statement
Informed consent was obtained from the owners of the animals for client-owned animals involved in this study.
Data Availability Statement
The data presented in this study are available in this manuscript. Sequence data presented in this study are openly available in the GenBank database.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by the Italian Ministry of Health, Ricerca Corrente 2019, IZS ME 04/19 RC: “Coronavirus emergenti dei carnivori domestici e selvatici ed implicazioni zoonosiche”, recipient Flora Alfano.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Vries R.D., Duprex W.P., de Swart R.L. Morbillivirus infections: An introduction. Viruses. 2015;7:699–706. doi: 10.3390/v7020699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carré H. Sur la maladie des jeunes chiens. Comptes Rendu De L’académie Des. Sci. 1905;140:689–690. [Google Scholar]
- 3.Carvalho O.V., Botelho C.V., Ferreira C.G.T., Scherer P.O., Soares-Martins J.A.P., Almeida M.R., Silva Júnior A. Immunopathogenic and neurological mechanisms of canine distemper virus. Adv. Virol. 2012;2012:163860. doi: 10.1155/2012/163860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu W., Zheng Y., Zhang S., Fan Q., Liu H., Zhang F., Wang W., Liao G., Hu R. Canine distemper outbreak in rhesus monkeys. China Emerg. Infect. Dis. 2011;17:1541–1543. doi: 10.3201/eid1708.101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trogu T., Canziani S., Salvato S., Bianchi A., Bertoletti I., Gibelli L.R., Alborali G.L., Barbieri I., Gaffuri A., Sala G., et al. Canine Distemper Outbreaks in Wild Carnivores in Northern Italy. Viruses. 2021;13:99. doi: 10.3390/v13010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshikawa Y., Ochikubo F., Matsubara Y., Tsuruoka H., Ishii M., Shirota K., Nomura Y., Sugiyama M., Yamanouchi K. Natural infection with canine distemper virus in a Japanese monkey (Macaca fuscata) Vet. Microbiol. 1989;20:193–205. doi: 10.1016/0378-1135(89)90043-6. [DOI] [PubMed] [Google Scholar]
- 7.Deem S.L., Spelman L.H., Yates R.A., Montali R.J. Canine distemper in terrestrial carnivores: A review. J. Zoo Wildl. Med. 2000;31:441–451. doi: 10.1638/1042-7260(2000)031[0441:CDITCA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Gutierrez M., Ruiz-Saenz J. Diversity of susceptible hosts in canine distemper virus infection: A systematic review and data synthesis. BMC Vet. Res. 2016;12:78. doi: 10.1186/s12917-016-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budaszewski R.F., Pinto L.D., Weber M.N., Caldart E.T., Alves C.D., Martella V., Ikuta N., Lunge V.R., Canal C.W. Genotyping of canine distemper virus strains circulating in Brazil from 2008 to 2012. Virus Res. 2014;180:76–83. doi: 10.1016/j.virusres.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Di Sabatino D., Lorusso A., Di Francesco C.E., Gentile L., Di Pirro V., Bellacicco A.L., Giovannini A., Di Francesco G., Marruchella G., Marsilio F., et al. Arctic lineage-canine distemper virus as a cause of death in Apennine wolves (Canis lupus) in Italy. PLoS ONE. 2014;9:e82356. doi: 10.1371/journal.pone.0082356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinal M.A., Díaz F.J., Ruiz-Saenz J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet. Microbiol. 2014;172:168–176. doi: 10.1016/j.vetmic.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Panzera Y., Calderón M.G., Sarute N., Guasco S., Cardeillac A., Bonilla B., Hernández M., Francia L., Bedó G., La Torre J., et al. Evidence of two co-circulating genetic lineages of canine distemper virus in South America. Virus Res. 2012;163:401–404. doi: 10.1016/j.virusres.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Martella V., Elia G., Buonavoglia C. Canine distemper virus. Vet. Clin. N. Am. Small Anim. Pract. 2008;38:787–789. doi: 10.1016/j.cvsm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 14.van Regenmortel H.V.M., Fauquet C.M., Bishop D.H.L., Carstens E., Estes M.K., Lemon S., Maniloff J., Mayo M.A., McGeoch D. Virus taxonomy. In: Pringle C.R., Wickner R.B., editors. Proceedings of the Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press; New York, NY, USA: 2000. [Google Scholar]
- 15.von Messling V., Zimmer G., Herrler G., Haas L., Cattaneo R. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 2001;75:6418–6427. doi: 10.1128/JVI.75.14.6418-6427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke G.M., Ho C.H., Chiang M.J., Sanno Duanda B., Chung C.S., Lin M.Y., Shi Y.Y., Yang M.H., Tyan Y.C., Liao P.C., et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet. Res. 2015;11:164. doi: 10.1186/s12917-015-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb R.A., Paterson R.G., Jardetzky T.S. Paramyxovirus membrane fusion: Lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balboni A., De Lorenzo Dandola G., Scagliarini A., Prosperi S., Battilani M. Occurrence of different Canine distemper virus lineages in Italian dogs. Vet. Ital. 2014;50:227–231. doi: 10.12834/VetIt.52.2173.2. [DOI] [PubMed] [Google Scholar]
- 19.Bolt G., Jensen T.D., Gottschalck E., Arctander P., Appel M.J.G., Buckland R., Blixenkrone-Møller M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J. Gen. Virol. 1997;78:367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter M.A., Appel M.J.G., Roelke Parker M.E., Munson L., Hofer H., East M., O’Brien S.J. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet. Immunol. Immunopathol. 1998;65:259–266. doi: 10.1016/S0165-2427(98)00159-7. [DOI] [PubMed] [Google Scholar]
- 21.Haas L., Martens W., Greiser-Wilke I., Mamaev L., Butina T., Maack D., Barrett T. Analysis of the haemagglutiningene of current wild-type canine distemper virus isolates from Germany. Virus Res. 1997;48:165–171. doi: 10.1016/S0168-1702(97)01449-4. [DOI] [PubMed] [Google Scholar]
- 22.Harder T.C., Kenter M., Vos H., Siebelink K., Huisman W., van Amerongen G., Orvell C., Barrett T., Appel M.J., Osterhaus A.D. Canine distemper virus from diseased large felids: Biological properties and phylogenetic relationships. J. Gen. Virol. 1996;77:397–405. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto M., Une Y., Mochizuki M. Hemagglutinin genotype profiles of canine distemper virus from domestic dogs in Japan. Arch. Virol. 2001;146:149–155. doi: 10.1007/s007050170198. [DOI] [PubMed] [Google Scholar]
- 24.Iwatsuki K., Miyashita N., Yoshida E., Gemma T., Shin Y.S., Mori T., Hirayama N., Kai C., Mikami T. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J. Gen. Virol. 1997;78:373–380. doi: 10.1099/0022-1317-78-2-373. [DOI] [PubMed] [Google Scholar]
- 25.Lan N.T., Yamaguchi R., Furuya Y., Inomata A., Ngamkala S., Naganobu K., Kai K., Mochizuki M., Kobayashi Y., Uchida K., et al. Pathogenesis and phylogenetic analyses of canine distemper virus strain 007Lm, a new isolate in dogs. Vet. Microbiol. 2005;110:197–207. doi: 10.1016/j.vetmic.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Lan N.T., Yamaguchi R., Inomata A., Furuya Y., Uchida K., Sugano S., Tateyama S. Comparative analyses of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet. Microbiol. 2006;115:32–42. doi: 10.1016/j.vetmic.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Lednicky J.A., Dubach J., Kinsel M.J., Meehan T.P., Bocchetta M., Hungerford L.L., Sarich N.A., Witecki K.E., Braid M.D., Pedrak C., et al. Genetically distant AmericanCanine distemper virus lineages have recently caused epizooticswith somewhat different characteristics in raccoons livingaround a large suburban zoo in the USA. Virol. J. 2004;2:1–2. doi: 10.1186/1743-422X-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martella V., Pratelli A., Cirone F., Zizzo N., Decaro N., Tinelli A., Foti M., Buonavoglia C. Detection and genetic characterization of canine distemper virus (CDV) from free-ranging red foxes in Italy. Mol. Cell Probes. 2002;16:77–83. doi: 10.1006/mcpr.2001.0387. [DOI] [PubMed] [Google Scholar]
- 29.Martella V., Cirone F., Elia G., Lorusso E., Decaro N., Campolo M., Desario C., Lucente M.S., Bellacicco A.L., Blixenkrone-Møller M., et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet. Microbiol. 2006;116:301–309. doi: 10.1016/j.vetmic.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 30.Mochizuki M., Hashimoto M., Hagiwara S., Yoshida Y., Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J. Clin. Microbiol. 1999;37:2936–2942. doi: 10.1128/JCM.37.9.2936-2942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardo I.D., Johnson G.C., Kleiboeker S.B. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J. Clin. Microbiol. 2005;43:5009–5017. doi: 10.1128/JCM.43.10.5009-5017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Francesco C.E., Di Francesco D., Di Martino B., Speranza R., Santori D., Boari A., Marsilio F. Detection by hemi-nested reverse transcription polymerase chain reaction and genetic characterization of wild type strains of Canine distemper virus in suspected infected dogs. J. Vet. Diagn. Investig. 2012;24:107–115. doi: 10.1177/1040638711425700. [DOI] [PubMed] [Google Scholar]
- 33.Di Sabatino D., Di Francesco G., Zaccaria G., Malatesta D., Brugnola L., Marcacci M., Portanti O., De Massis F., Savini G., Teodori L., et al. Lethal distemper in badgers (Meles meles) following epidemic in dogs and wolves. Infect. Genet. Evol. 2016;46:130–137. doi: 10.1016/j.meegid.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Marcacci M., Ancora M., Mangone I., Teodori L., Di Sabatino D., De Massis F., Camma C., Savini G., Lorusso A. Whole genome sequence analysis of the arctic-lineage strain responsible for distemper in Italian wolves and dogs through a fast and robust next generation sequencing protocol. J. Virol. Methods. 2014;202:64–68. doi: 10.1016/j.jviromet.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Monne I., Fusaro A., Valastro V., Citterio C., Pozza M.D., Obber F., Trevisiol K., Cova M., De Benedictis P., Bregoli M., et al. A distinct CDV genotype causing a major epidemic in Alpine wildlife. Vet. Microbiol. 2011;150:63–69. doi: 10.1016/j.vetmic.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Lorusso A., Savini G. Old diseases for new nightmares: Distemper strikes back in Italy. Vet. Ital. 2014;50:151–154. doi: 10.12834/VetIt.66.191.2. [DOI] [PubMed] [Google Scholar]
- 37.Di Francesco C.E., Smoglica C., Angelucci S. Infectious Diseases and Wildlife Conservation Medicine: The Case of the Canine Distemper in European Wolf Population. Animals. 2020;10:2426. doi: 10.3390/ani10122426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricci I., Cersini A., Manna G., Marcario G.A., Conti R., Brocherel G., Grifoni G., Eleni C., Scicluna M.T. A Canine Distemper Virus Retrospective Study Conducted from 2011 to 2019 in Central Italy (Latium and Tuscany Regions) Viruses. 2021;13:272. doi: 10.3390/v13020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seimon T.A., Miquelle D.G., Chang T.Y., Newton A.L., Korotkova I., Ivanchuk G., Lyubchenko E., Tupikov A., Slabe E., McAloose D. Canine distemper virus: An emerging disease in wild endangered Amur tigers (Panthera tigris altaica) mBio. 2013;4:e00410-13. doi: 10.1128/mBio.00410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Francesco C.E., Smoglica C., Di Pirro V., Cafini F., Gentile L., Marsilio F. Molecular Detection and Phylogenetic Analysis of Canine Distemper Virus in Marsican Brown Bear (Ursus arctos marsicanus) Animals. 2022;12:1826. doi: 10.3390/ani12141826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krakowka S., Hoover E.A., Koestner A., Ketring K. Experimental and naturally occurring transplacental transmission of canine distemper virus. Am. J. Vet. Res. 1977;38:919–922. [PubMed] [Google Scholar]
- 42.Appel M.J. Pathogenesis of canine distemper. Am. J. Vet. Res. 1969;30:1167–1182. [PubMed] [Google Scholar]
- 43.Loots A.K., Mitchell E., Dalton D.L., Kotzé A., Venter E.H. Advances in canine distemper virus pathogenesis research: A wildlife perspective. J. Gen. Virol. 2017;98:311–321. doi: 10.1099/jgv.0.000666. [DOI] [PubMed] [Google Scholar]
- 44.Appel M.J.G. Canine distemper virus. In: Horzinek M.C., editor. Virus Infections of Carnivores. Elsevier Science Publishers; Amsterdam, The Netherlands: 1987. pp. 133–159. [Google Scholar]
- 45.Summers B.A., Greisen H.A., Appel M.J. Canine distemper encephalomyelitis: Variation with virus strain. J. Comp. Pathol. 1984;94:65–75. doi: 10.1016/0021-9975(84)90009-4. [DOI] [PubMed] [Google Scholar]
- 46.Elia G., Decaro N., Martella V., Cirone F., Lucente M.S., Lorusso E., Di Trani L., Buonavoglia C. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol. Methods. 2006;136:171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J.J., Yan X.J., Chai X.L., Martella V., Luo G.L., Zhang H.L., Gao H., Liu Y.X., Bai X., Zhang L., et al. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet. Microbiol. 2010;140:34–42. doi: 10.1016/j.vetmic.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Decaro N., Pretelli A., Campolo M., Elia G., Martella V., Tempesta M., Buonavoglia C. Quantitation of canine coronavirus RNA in the feaces of dogs by TaqMan RT-PCR. J. Virol. Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Di Trani L., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for detection and quantitation of canine coronavirus type I and type II RNA in fecal sample of dogs. J. Virol. Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decaro N., Cordonnier N., Demeter Z., Egberink H., Elia G., Grellet A., Le Poder S., Mari V., Martella V., Ntafis V., et al. European surveillance for pantropic canine coronavirus. J. Clin. Microbiol. 2013;51:83–88. doi: 10.1128/JCM.02466-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratelli A., Martella V., Pistello M., Elia G., Decaro N., Buonavoglia D., Camero M., Tempesta M., Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linnè T. Differences in the E3 regions of the canine adenovirus type 1 and type 2. Virus Res. 1992;23:119–133. doi: 10.1016/0168-1702(92)90072-H. [DOI] [PubMed] [Google Scholar]
- 53.Peters I.E., Helps C.R., Batt R.M., Day M.J., Hall E.J. Quantitative real-time RT-PCR meascurement of mRNA encoding alpha-chain, pIgR and J-chain from canine doudenal mucosa. J. Immunol. Methods. 2003;275:213–222. doi: 10.1016/S0022-1759(03)00056-5. [DOI] [PubMed] [Google Scholar]
- 54.Decaro N., Amorisco F., Desario C., Lorusso E., Camero M., Bellacicco A.L., Sciarretta R., Lucente M.S., Martella V., Buonavoglia C. Development and validation of a real-time PCR assay for specific and sensitive detection of canid herpesvirus 1. J. Virol. Methods. 2010;169:176–180. doi: 10.1016/j.jviromet.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Logan C., O’Leary J.J., O’Sullivan N. Real time reverse transcription-PCR for detection of Rotavirus and Adenovirus as causative agents of acute viral gastroenteritis in children. J. Clin. Microbiol. 2006;44:3189–3195. doi: 10.1128/JCM.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotavirus. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 57.Decaro N., Elia G., Martella V., Desario C., Campolo M., Di Trani L., Tarsitano E., Tempesta M., Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet. Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Decaro N., Desario C., Lucente M.S., Amorisco F., Campolo M., Elia G., Cavalli A., Martella V., Buonavoglia C. Specific identification of feline panleukopenia virus and its rapid differentation from canine parvoviruses using minor grove probes. J. Virol. Methods. 2008;147:67–71. doi: 10.1016/j.jviromet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Decaro N., Elia G., Desario C., Roperto S., Martella V., Campolo M., Lorusso A., Cavalli A., Buonavoglia C. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J. Virol. Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alfano F., Fusco G., Mari V., Occhiogrosso L., Miletti G., Brunetti R., Galiero G., Desario C., Cirilli M., Decaro N. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound. Emerg. Dis. 2020;67:1991–1999. doi: 10.1111/tbed.13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rikula U., Nuotio L., Sihvonen L. Vaccine coverage, herd immunity and occurrence of canine distemper from 1990–1996 in Finland. Vaccine. 2007;25:7994–7998. doi: 10.1016/j.vaccine.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Ek-Kommonen C., Sihvonen L., Pekkanen K., Rikula U., Nuotio L. Outbreak off canine distemper in vaccinated dogs in Finland. Vet. Rec. 1997;141:380–383. doi: 10.1136/vr.141.15.380. [DOI] [PubMed] [Google Scholar]
- 65.Alves C.D.B.T., Granados O.F.O., Budaszewski R.D.F., Streck A.F., Weber M.N., Cibulski S.P., Pinto L.D., Ikuta N., Canal C.W. Identification of enteric viruses circulating in a dog population with low vaccine coverage. Braz. J. Microbiol. 2018;49:790–794. doi: 10.1016/j.bjm.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M., Yan N., Ji C., Wang M., Zhang B., Yue H., Tang C. Prevalence and genome characteristics of canine astrovirus in southwest China. J. Gen. Virol. 2018;99:880–889. doi: 10.1099/jgv.0.001077. [DOI] [PubMed] [Google Scholar]
- 67.Deng X., Zhang J., Su J., Liu H., Cong Y., Zhang L., Zhang K., Shi N., Lu R., Yan X. A multiplex PCR method for the simultaneous detection of three viruses associated with canine viral enteric infections. Arch Virol. 2018;163:2133–2138. doi: 10.1007/s00705-018-3828-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ergunay K., Mutinda M., Bourke B., Justi S.A., Caicedo-Quiroga L., Kamau J., Mutura S., Akunda I.K., Cook E., Gakuya F. Metagenomic Investigation of Ticks From Kenyan Wildlife Reveals Diverse Microbial Pathogens and New Country Pathogen Records. Front. Microbiol. 2022;13:932224. doi: 10.3389/fmicb.2022.932224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Worsley-Tonks K.E.L., Miller E.A., Gehrt S.D., McKenzie S.C., Travis D.A., Johnson T.J., Craft M.E. Characterization of antimicrobial resistance genes in Enterobacteriaceae carried by suburban mesocarnivores and locally owned and stray dogs. Zoonoses Public Health. 2020;67:460–466. doi: 10.1111/zph.12691. [DOI] [PubMed] [Google Scholar]
- 70.Carmichael L.E., Binn L.N. New enteric viruses in the dog. Adv. Vet. Sci. Comp. Med. 1981;25:1–37. [PubMed] [Google Scholar]
- 71.Buonavoglia C., Decaro N., Martella V., Elia G., Campolo M., Desario C., Castagnaro M., Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerg. Infect Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marinaro M., Mari V., Bellacicco A.L., Tarsitano E., Elia G., Losurdo M., Rezza G., Buonavoglia C., Decaro N. Prolonged depletion of circulating CD4+ T lymphocytes and acute monocytosis after pantropic canine coronavirus infection in dogs. Virus Res. 2010;152:73–78. doi: 10.1016/j.virusres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martella V., Elia G., Lucente M.S., Decaro N., Lorusso E., Banyai K., Blixenkrone-Møller M., Lan N.T., Yamaguchi R., Cirone F., et al. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet. Microbiol. 2007;122:32–42. doi: 10.1016/j.vetmic.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Mira F., Purpari G., Di Bella S., Vicari D., Schirò G., Di Marco P., Macaluso G., Battilani M., Guercio A. Update on canine distemper virus (CDV) strains of Arctic-like lineage detected in dogs in Italy. Vet. Ital. 2018;54:225–236. doi: 10.12834/VetIt.1455.7862.2. [DOI] [PubMed] [Google Scholar]
- 75.Demeter Z., Lakatos B., Palade E.A., Kozma T., Forgách P., Rusvai M. Genetic diversity of Hungarian canine distemper virus strains. Vet. Microbiol. 2007;122:258–269. doi: 10.1016/j.vetmic.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corrain R., Di Francesco A., Bolognini M., Ciucci P., Baldelli R., Guberti V. Serosurvey for CPV-2, distemper virus, ehrlichiosis and leishmaniosis in free-ranging dogs in Italy. Vet. Record. 2007;160:91–92. doi: 10.1136/vr.160.3.91. [DOI] [PubMed] [Google Scholar]
- 77.Verardi A., Lucchini V., Randi E. Detecting introgressive hybridization between free-ranging domestic dogs and wild wolves (Canis lupus) by admixture linkage disequilibrium analysis. Mol. Ecol. 2006;15:2845–2855. doi: 10.1111/j.1365-294X.2006.02995.x. [DOI] [PubMed] [Google Scholar]
- 78.Alfano F., Dowgier G., Valentino M.P., Galiero G., Tinelli A., Decaro N., Fusco G. Identification of Pantropic Canine Coronavirus in a Wolf (Canis lupus italicus) in Italy. J. Wildl. Dis. 2019;55:504–508. doi: 10.7589/2018-07-182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in this manuscript. Sequence data presented in this study are openly available in the GenBank database.