Abstract
The definitive diagnosis and early treatment of many immune-mediated inflammatory diseases (IMIDs) is hindered by variable and overlapping clinical manifestations. Psoriatic arthritis (PsA), which develops in ~30% of people with psoriasis, is a key example. This mixed-pattern IMID is apparent in entheseal and synovial musculoskeletal structures, but a definitive diagnosis often can only be made by clinical experts or when an extensive progressive disease state is apparent. As with other IMIDs, the detection of multimodal molecular biomarkers offers some hope for the early diagnosis of PsA and the initiation of effective management and treatment strategies. However, specific biomarkers are not yet available for PsA. The assessment of new markers by genomic and epigenomic profiling, or the analysis of blood and synovial fluid/tissue samples using proteomics, metabolomics and lipidomics, provides hope that complex molecular biomarker profiles could be developed to diagnose PsA. Importantly, the integration of these markers with high-throughput histology, imaging and standardized clinical assessment data provides an important opportunity to develop molecular profiles that could improve the diagnosis of PsA, predict its occurrence in cohorts of individuals with psoriasis, differentiate PsA from other IMIDs, and improve therapeutic responses. In this review, we consider the technologies that are currently deployed in the EU IMI2 project HIPPOCRATES to define biomarker profiles specific for PsA and discuss the advantages of combining multi-omics data to improve the outcome of PsA patients.
Keywords: psoriatic diseases, psoriatic arthritis, psoriasis, multi-omics, data integration
1. Introduction
1.1. Psoriasis and Psoriatic Arthritis
Psoriasis is a chronic, immune-mediated inflammatory disease (IMID) of the skin, which affects 0.91–8.5% of the population, varying by age, region and ethnicity [1]. The most common manifestation is plaque psoriasis (psoriasis vulgaris), which accounts for ~80% of cases and typically involves the formation of erythematous and scaly plaques on the head, ears, elbows and knees, as well as gluteal and umbilical areas. These skin changes are often highly conspicuous, and the resulting stigmatization can lead to psychosocial issues. There is also a high rate of comorbidities, including cardiovascular disease and obesity [2]. Approximately 30% of psoriasis patients go on to develop psoriatic arthritis (PsA) [3], a mixed-pattern IMID characterized by the inflammation of mainly entheseal and synovial musculoskeletal structures [4]. Predisposition to the development of PsA has a strong genetic basis [3] and correlates with the severity of psoriatic skin lesions, including nail involvement (pitting, cracking, separation or nail loss). However, in a minority of cases, the symptoms of PsA develop alongside psoriasis or even before it. Various environmental and lifestyle factors also increase the risk of PsA at the population level, including a high body mass index and smoking [5,6,7], although paradoxically, smoking is negatively associated with progression to PsA at the level of individual psoriasis patients [8]. There is also increasing evidence that dietary factors influence the risk of progressing to PsA [9,10]. PsA can lead to structural damage and loss of function of the joints due to bone erosion, new bone formation and cartilage loss [11]. It has diverse presentations including asymmetric oligo-articular forms of arthritis, polyarticular disease, dactylitis and spinal inflammation [12].
1.2. Current Diagnostic Practices and Disease Management Strategies
A diagnosis of psoriasis is usually based on the appearance of the skin [13]. Blood tests or other diagnostic procedures are generally unnecessary [14]. If clinical diagnosis is uncertain, psoriasis can be differentiated from visually similar conditions (such as certain forms of eczema) by skin biopsy, which will confirm epidermal thickening interdigitating with the dermis, changes to the stratum granulosum, the presence of nuclei in the superficial layer, and the presence of infiltrating T cells [15]. In contrast, there is no definitive diagnosis for PsA because the clinical manifestations overlap with other arthritic diseases, including rheumatoid arthritis (RA), osteoarthritis and inflammatory bowel disease (IBD)-associated arthritis. Current diagnostic practice is based on rheumatologic assessment involving physical examination, medical history, blood tests and imaging. More definitive diagnosis is generally dependent on the presence of inflammation and musculoskeletal damage, which makes early intervention much more challenging. The identification of early and specific biomarkers of PsA would facilitate immediate treatment with the most appropriate drugs, therefore offering a much better prognosis for PsA patients and even preventing disease progression in its early stages [16]. In addition to the need for early diagnosis so that treatment can improve patient outcomes, the management of chronic disease plays an important role with a focus on individualized and personalized treatment strategies. Even following the initiation of appropriate immunosuppressive therapy, up to ~40% of patients may not respond or experience adverse effects [17]. There is an urgent medical need for biomarkers that facilitate the early differentiation of PsA and allow the prediction and monitoring of therapeutic responses during the chronic disease stage, thus helping to normalize function and improve outcomes and quality of life.
1.3. The Promise of Omics and Multi-Omics Technology
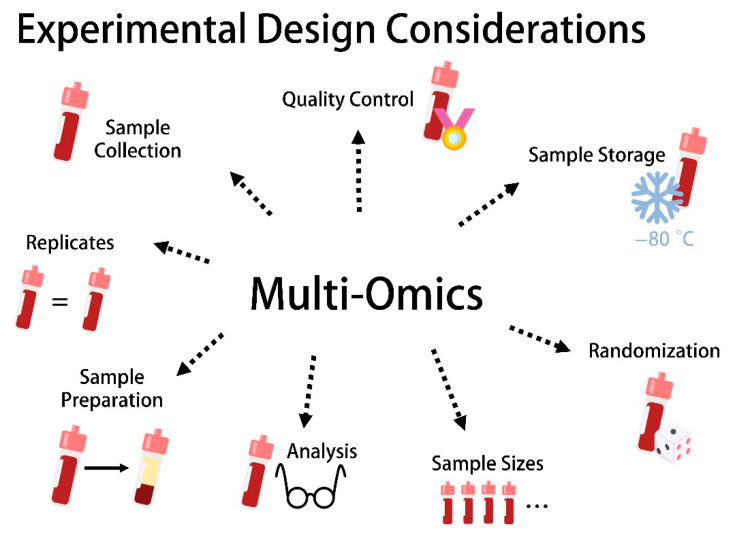
Biomarkers that are distinct for specific groups of patients can be used for the early diagnosis of diseases because they often correspond to qualitative or even quantitative indicators of biological and pathological processes [18,19]. The genomics revolution in the 2000s identified a large panel of new genetic markers that are associated with particular disease phenotypes, but the potential of biomarkers expanded enormously as omics technology broadened to encompass the global analysis of DNA modifications (epigenomics), RNA (transcriptomics), proteins (proteomics) and metabolites (metabolomics). Furthermore, it is reasonable to differentiate between the analysis of polar metabolites and the analysis of lipids (lipidomics) because the physicochemical properties of these compounds are quite distinct and optimized methods for analyzing these groups are necessary. The advent of proteomics and metabolomics/lipidomics in particular has raised the possibility of using combinations of markers to differentiate between diseases or disease stages in a quantitative manner, which is not possible with genetic markers outside the field of oncology. As the corresponding technologies have become increasingly sophisticated, sensitive and automated, the cost of analysis has fallen and more ambitious studies are possible, including the correlation of multiple omics biomarker profiles across large groups of patients. This requires stringent quality control standards to be applied during sample collection, storage, preparation and analysis, including due attention to sample sizes and replicates, as well as appropriate randomization (Figure 1).
Figure 1.
Experimental design considerations for the utilization of multi-omics data.
The EU-funded HIPPOCRATES project (https://hippocrates-imi.eu, accessed on 20 September 2022) is an ambitious collaboration that considers the potential of multiple molecular marker types across the spectrum of omics technology and seeks to combine them with conventional clinical diagnostic methods (imaging, medical records and physical examinations) for PsA. The value of omics technologies in the clinical care of PsA patients has been explored in a recent review article, including transcriptomics (which is not part of the HIPPOCRATES project) [20]. HIPPOCRATES aims to extend the concept by combining marker profiles for the differential diagnosis of psoriasis and PsA, as well as prognosis and the monitoring of treatment responses. In this review, we focus on the main objectives of the HIPPOCRATES project by considering the advantages and disadvantages of different omics technologies for the discovery of biomarkers for psoriasis and PsA, the potential of multi-omics approaches that combine different technologies to take advantage of synergies and how the diverse data formats may be combined and interrogated using advanced data evaluation tools (e.g., tools based on artificial intelligence) to identify patterns with diagnostic or prognostic value.
2. Genomics
2.1. Brief Overview of Relevant Genomics Technologies
Genomics is the branch of biology that deals with the analysis of genomes. In the context of psoriasis and PsA, genomics can be used to identify and characterize the genes, and more importantly the gene variants (alleles), that are associated with each disease. Many of the genes identified as associated with psoriasis have also been found to be associated with PsA when compared to population controls, highlighting their shared genetic basis. Susceptibility loci associated with PsA alone have also been identified, including several HLA-B alleles and IL23R [21,22]. The detection of pathological gene variants can be used to assist diagnosis and also to predict the age of onset, severity and likely symptoms of the disease. However, the multiple genes that distinguish between psoriasis and PsA may also be shared with other arthritic diseases, such as RA or ankylosing spondylitis.
The fundamental technology underlying the field of genomics is the genome-wide genotyping array, the contents of which are routinely enhanced by imputation, which provides the structure and sequence of key disease-associated genes and allows causative allelic variants to be identified. Genome-wide association studies (GWAS) and gene chip experiments have identified more than 20 additional loci outside the HLA system that are associated with PsA [23,24], some of which are exclusive (i.e., not also associated with psoriasis) [25]. The advent of next-generation sequencing platforms that are faster, cheaper and easier to automate than classic Sanger sequencing will enable researchers to amass a large body of sequence data from various patient cohorts, and this allows the comparison of patient groups to identify relevant alleles, in particular for rare variants not captured on genotyping arrays or by imputation.
2.2. Applications for Early Diagnosis, Prognosis and Treatment Monitoring
PsA is known to have a strong genetic component, which means that certain allelic variants are likely to be more prevalent among PsA patients than controls (or other disease cohorts). Because such genetic variation is present from conception, it should be possible to detect disease-causing alleles before the onset of symptoms and commence treatment as early as possible. Similarly, it should be possible to detect PsA-associated alleles in cohorts of psoriasis patients and thus identify those at the greatest risk of progression. Although many different alleles are associated with psoriasis, PsA or both, GWAS can be used to screen for large panels of variants in a single test, which is generally based on array hybridization or multiplex PCR [26,27]. The detection of one or more informative variants can therefore provide data to indicate causality. Other markers may be useful for the assessment of therapy, and to determine which subcomponents are heritable, and therefore more predictable [28]. Accordingly, prospective studies are needed in psoriasis patients, ideally recruited from primary care before disease-modifying therapy commences, to assess the ability of genetic variants to predict the onset of PsA.
2.3. Case Studies/Examples in Psoriasis and PsA
The primary genetic factors that distinguish PsA from psoriasis map to the HLA-B locus [29,30]. The alleles HLA-B*39, HLA-B*07, HLA-B*38 and in particular HLA-B*27 have been described as specific risk factors for PsA [31]. Although gene mapping is consistent across different studies, resolution to a precise allelic variant is conflicting when the reported index associations point to amino acid positions 45 or 97 (Table 1). Outside the HLA region, there is convincing evidence for a PsA-specific effect at the IL23R locus independent of the known psoriasis risk variant [32,33,34]. Other genes associated with PsA but not psoriasis include KIR2D [35], IL4 and KIF3A [36], B3GNT2 [37] and PTPN22 [25].
Table 1.
Genetic variants with evidence to support their ability to distinguish between PsA and cutaneous-only psoriasis.
3. Epigenomics
3.1. Brief Overview of Relevant Epigenomics Technologies
Epigenomics is the large-scale analysis of epigenetic phenomena, which include DNA methylation and histone modification as regulators of the 3D configuration of the genome, and the expression of small regulatory RNAs. Epigenetic mechanisms play a key role in the regulation of gene expression, and specific epigenetic markers can be associated with diseases such as psoriasis and PsA. Various technologies can be used to monitor genome-wide epigenetic phenomena, including chromatin immunoprecipitation (ChIP) followed by detection on microarrays (ChIP-chip) [38] or by sequencing (ChIP-Seq) [39], the detection of methylated DNA using bisulfite sequencing or (directly) by nanopore sequencing or SMRT sequencing [40], and enzyme-based chromatin accessibility assays [41]. The detection of chromosome conformation signatures (sequences that are likely to control the 3D structure of the genome) can also be used to pinpoint abnormal chromosome structures that are associated with diseases or responses to treatment. For example, the Oxford Biodynamics EpiSwitch platform is based on the testing of more than 10,000 samples in 30 disease indications, enabling the screening, evaluation, validation and monitoring of 3D genomic biomarkers [42].
3.2. Applications for Early Diagnosis, Prognosis and Treatment Monitoring
The EpiSwitch platform facilitates the discovery of stable and heriTable 3D genomic markers and the development of highly sensitive clinical assays based on non-invasive blood readouts. In the case of PsA, it can assist with a definitive diagnosis and prognosis in the context of comorbidities and overlapping symptoms, without resorting to biopsy. This technique has already delivered biomarkers that predict the response to methotrexate treatment in RA patients [43], that predict the response to immune checkpoint inhibitors in cancer [44], and that are prognostic for severe outcomes of COVID-19 based on individual patient immune health profiling [45]. The markers profiled by EpiSwitch technology are governed by all forms of genetic and epigenetic variation, and their combined influence has a major impact on the regulation of gene expression by controlling access to chromatin. Therefore, such markers are powerful high-level integrators of multi-omic signals [46]. In order to utilize the full potential of EpiSwitch, a representative cohort of whole blood samples with clinical annotations is required, representing extreme clinical outcomes. That spectrum will define the quality of the EpiSwitch biomarkers and their correlation with other modalities.
3.3. Case Studies/Examples in Psoriasis and PsA
Although chromosome conformation signatures for psoriasis and PsA are not yet available, the promise of the technique has been demonstrated in early RA patients commencing methotrexate treatment [43]. Using blood samples from responders, non-responders and healthy controls, a custom biomarker discovery array was refined to a five-marker chromosome conformation signature that could discriminate between responders and non-responders. Markers were validated using a blinded, independent cohort of 19 early RA patients (9 responders and 10 non-responders) and the corresponding loci were mapped to a RA-specific expression quantitative trait locus (eQTL). Finally, a five-marker chromosome conformation signature was found that could identify, at baseline, responders and non-responders to methotrexate. It consisted of binary chromosome conformations in the genomic regions of IFNAR1, IL-21R, IL-23, CXCL13 and IL-17A. When tested on a cohort of 59 RA patients the marker provided a negative predictive value of 90% for methotrexate response. When tested on a blinded independent validation cohort of 19 early RA patients, the signature demonstrated a true negative response rate of 86%, and 90% sensitivity for the detection of non-responders. Only conformations in responders mapped to the RA-specific eQTL.
4. Proteomics
4.1. Brief Overview of Relevant Proteomics Technologies
Proteomics can be defined as the large-scale analysis of proteins. In the context of PsA, it has been applied mainly to identify biomarkers that can be detected in blood, synovial fluid or skin samples for the early diagnosis of PsA and its differentiation from psoriasis [47,48,49]. The proteome is much more complex and dynamic than the genome because there are an estimated ~20,000 protein-encoding genes in the human genome [50], but these give rise to multiple variants by alternative transcription, splicing and processing of RNA, post-translational modification and protein–protein interactions. About 10% of the human proteome lacks experimental evidence, and the combined effect of differential protein abundance, protein modifications, sequence variation and interactions further complicate the task of measuring all proteins in every sample [50].
The technologies used to interrogate the proteome can be broadly divided into untargeted methods that attempt to consider all proteins in a sample, and targeted methods that focus on specific proteins or classes of proteins. Mass spectrometry (MS) is a key platform in both approaches because it is a sensitive, high-throughput technology that is relatively easy to automate. Proteins are digested into peptides using a protease with known specificity such as trypsin, and the mass of each peptide, and its fragments generated inside a collision cell, is correlated with values in databases to achieve peptide and hence protein identification. Untargeted methods are based on the analysis of complex, uncharacterized peptide mixtures from multiple proteins. These are generally fractionated by liquid chromatography before injection into the mass spectrometer (LC-MS) and/or by multiple rounds of MS. In the latter case, data-dependent acquisition (DDA) involves the selection of specific peptides during the first round of MS for further fragmentation in subsequent rounds, whereas data-independent acquisition (DIA) involves the fragmentation and further analysis of all peptides from the first round [51]. Targeted methods involve the selection of one or a relatively small number of proteins from a sample for quantitative analysis [52]. Targeted analysis can be undertaken using MS-based methods as exemplified in the Atturos platform or methods that rely on highly specific affinity reagents. In the latter case, the production of high-quality data requires the use of validated binders (affinity reagents) that capture target proteins at low abundance [53]. Current affinity proteomics methods can detect more than 3000 proteins simultaneously by using different selectivity concepts, as well as the amplification capabilities of DNA-based readout methods. One relevant example is the Olink platform, a proximity extension assay that involves the recognition of proteins by antibodies linked to protein-specific DNA barcodes that can be amplified by qPCR or sequencing [54]. This may have a broader dynamic range and greater sensitivity than LC-MS and can simultaneously detect 3000 human proteins in plasma samples [55]. The use of slow off-rate DNA aptamers, provided by SomaLogic, has enabled large-scale studies of 10,000 donors targeting 4000 circulating proteins across human diseases [56].
4.2. Applications for Early Diagnosis, Prognosis and Treatment Monitoring
For the proteomic analysis of body fluids, particularly blood, further challenges arise due to the broad concentration range of different proteins, dynamic changes induced by disease processes, and analytical factors that influence protein detection [57]. Collectively, more than 4500 proteins have been detected in plasma samples by discovery-driven MS [58]. Many abundant plasma constituents are secreted by the liver, whereas other secreted proteins, such as inflammatory cytokines, are often elevated only transiently [59]. Accordingly, differences have been observed between individuals and between molecular profiles at longitudinal study time points [60]. When searching for protein biomarkers in healthy individuals, as well as psoriasis and PsA patients, the heterogeneity of signatures from circulating proteins should be expected.
Multiple candidate biomarkers of PsA have been reported in serum and plasma, in addition to a smaller number found in synovial fluid/tissue and skin biopsies [47,48]. Most of the biomarker candidates are proposed for the detection of PsA [61,62], differentiation between mild and severe forms [63,64], measuring disease activity [65], or predicting which psoriasis patients are likely to develop PsA [66]. However, others have been proposed to distinguish PsA from other arthritic diseases such as RA [49,67] or to monitor responses to therapy [68,69,70,71]. For example, the label-free MS analysis of synovial fluid from PsA patients revealed 12 candidate PsA markers including the injury marker MMP3, as well as the inflammatory proteins S100A9 and CRP [62]. A subsequent study using LC-MS identified periostin, which is related to cell-adhesion proteins, and the angiogenesis marker PGK1 [72]. More recently, a systematic search of five bibliographic databases for clinical, laboratory and genetic markers was used to determine the level of evidence for each marker and its association with concomitant/developing PsA [73]. These have been converted into proteomic biomarkers in Table 2. For the prediction of PsA in psoriasis patients, highly characterized cohorts of patients are needed with each disease, minimizing the proportion of undiagnosed subclinical PsA patients in the psoriasis group. Alternatively, longitudinal observation and sample collection in the psoriasis group may directly identify those progressing to PsA, allowing the retrospective analysis of early samples to look for predictive biomarkers.
Table 2.
Proteomic markers with evidence to support their ability to distinguish between PsA and cutaneous-only psoriasis. A gene-centric table of candidates was created by using the biomarkers listed by Mulder et al. [73]. The proteins and mRNAs were converted into gene-centric entries using the Human Protein Atlas portal (www.proteinatlas.org (accessed on 5 July 2022)), and were annotated for secretion location, tissue expression and biological functional based on the recent clustering of single-cell expression data [75].
| Gene Name | Biomarker | UniProt ID | Category | Secretion | Tissue Expression | Biological Function |
|---|---|---|---|---|---|---|
| ADIPOQ | Adiponectin | Q15848 | Lipid | Blood | Adipose tissue | ECM organization |
| APOA1 | ApoA | P02467 | Lipid | Blood | Liver | Metabolism |
| APOB | ApoB | P04114 | Lipid | Blood | Liver | Metabolism |
| CMC2 | C16ORF61 | Q9NRP2 | Skin | N/A | Non-specific | Mitochondria |
| COL2A1 | C2C | P02458 | Bone | ECM | Epididymis | Unknown function |
| CCL1 | CCL1 | P22362 | mRNA | Blood | T cells | Adaptive immune response |
| CCL20 | CCL20 | P78556 | mRNA | Blood | Smooth muscle tissue | Mixed function |
| CCL7 | CCL7 | P80098 | mRNA | Blood | Neutrophils | Humoral immune response |
| CD5L | CD5L | O43866 | Serum | Blood | Macrophages | Immune response |
| COMP | COMP | P49747 | Bone | ECM | Skin | Epidermis development |
| C9 | Complement C9 | P02748 | Serum | Blood | Liver | Hemostasis and lipid |
| COL2A1 | CPII | P02458 | Bone | ECM | Epididymis | Unknown function |
| CPN2 | CPN2 | P22792 | Skin | Blood | Liver | Hemostasis |
| CRP | CRP | P02741 | Inflammation | Blood | Liver | Hemostasis |
| COL1A1 | CTX | P02452 | Bone | ECM | Fibroblasts | ECM organization |
| CX3CL1 | CX3CL1 | P78423 | mRNA | Blood | Adipose tissue | ECM organization |
| CXCL10 | CXCL10 | P02778 | Cytokines | Blood | Immune cells | Immune response |
| CXCL12 | CXCL12 | P48061 | Skin | Blood | Fibroblasts | ECM organization |
| CXCL2 | CXCL2 | P19875 | mRNA | Blood | Liver | Metabolism |
| CXCL5 | CXCL5 | P42830 | mRNA | Blood | Salivary gland | Salivary secretion |
| DKK1 | DKK-1 | O94907 | Bone | Other | Adipose tissue | ECM organization |
| ESR1 | ESR | P03372 | Inflammation | N/A | Fibroblasts | ECM organization |
| FHL1 | FHL1 | Q13642 | Skin | N/A | Striated muscle | Muscle contraction |
| GSN | Gelsolin | P06396 | Serum | Blood | Fibroblasts | ECM organization |
| GPS1 | GPS1 | Q13098 | Skin | N/A | Non-specific | Mitochondria |
| HAT1 | HAT1 | O14929 | mRNA | N/A | Non-specific | Ribosome |
| IFI16 | IFI16 | Q16666 | Serum | N/A | Immune cells | Immune response |
| IL12A | IL-12/23 p40 | P29459 | Cytokines | Blood | Brain and skin | Unknown function |
| IL12B | IL-12/23 p40 | P29460 | Cytokines | Blood | Non-specific | Cell cycle regulation |
| IL9 | IL-12/23 p40 | P15248 | Cytokines | Blood | N/A | N/A |
| IL17A | IL-17 | Q16552 | Cell culture secretion | Blood | Immune cells | Immune response |
| IL17C | IL-17C | Q9P0M4 | mRNA | Blood | Testis | DNA repair |
| IL17F | IL-17F | Q96PD4 | mRNA | Blood | B cells | Humoral immune response |
| IL2 | IL-2 | P60568 | Cell culture secretion | Blood | N/A | N/A |
| IL23 | IL-23 | Q9NPF7 | Cytokines | Blood | B cells | Humoral immune response |
| IL23R | IL23R | Q5VWK5 | Skin | N/A | Intestine | Brush border |
| IL3 | IL-3 | P08700 | mRNA | Blood | N/A | N/A |
| IL33 | IL-33 | O95760 | Cytokines | Blood | Fibroblasts | ECM organization |
| IL34 | IL-34 | Q6ZMJ4 | Cytokines | Blood | Macrophages | Immune response |
| EBI3 | IL-35 | Q14213 | Cytokines | Blood | Placenta | Pregnancy |
| IL12A | IL-35 | P29459 | Cytokines | Blood | Brain and skin | Unknown function |
| IL36A | IL-36a | Q9UHA7 | Cytokines | Blood | Esophagus | Epithelial cell function |
| IL1F10 | IL-38 | Q8WWZ1 | Cytokines | Blood | Skin | Cornification |
| IL6 | IL-6 | P05231 | Cytokines, mRNA | Blood | Adipose tissue | ECM organization |
| CXCL8 | IL-8 | P10145 | mRNA | Blood | Neutrophils | Humoral immune response |
| INS | Insulin | P01308 | Lipid | Blood | Pancreas | Digestion |
| ISG20 | ISG20 | Q96AZ6 | mRNA | N/A | Immune cells | Immune response |
| ITGB5 | ITGB5 | P18084 | Serum | N/A | Adipose tissue | ECM organization |
| ITGB5 | ITGB5 | P18084 | Skin | N/A | Adipose tissue | ECM organization |
| KRT17 | K17 | Q04695 | Serum | N/A | Skin | Epidermis development |
| LEP | Leptin | P41159 | Lipid | Blood | Adipose tissue | ECM organization |
| LGALS3BP | M2BP | Q08380 | Serum | Blood | Stomach | Digestion |
| CSF1 | M-CSF | P09603 | Cytokines | Blood | Non-specific | Angiogenesis |
| MMP3 | MMP3 | P08254 | Bone, mRNA | ECM | Salivary gland | Salivary secretion |
| MPO | MPO | P05164 | Serum | Membrane | Neutrophils | Humoral immune response |
| NOTCH2NLA | NOTCH2NL | Q7Z3S9 | mRNA | Blood | Testis | DNA repair |
| TNFRSF11B | OPG | O00300 | Bone | Other | Kidney | Transmembrane transport |
| POSTN | POSTN | Q15063 | Skin | ECM | Skin | Epidermis development |
| PTPA | PPP2R4 | Q15257 | Skin | N/A | Non-specific | Mitochondria |
| PRL | PRL | P01236 | Serum | Blood | Pituitary gland | Hormone signaling |
| TNFSF11 | RANKL | O14788 | Bone | Blood | Immune cells | Immune response |
| SETD2 | SETD2 | Q9BYW2 | mRNA | N/A | Non-specific | Transcription |
| IL2RA | sIL2R | P01589 | Serum | N/A | Immune cells | Immune response |
| IL2RB | sIL2R | P14784 | Serum | N/A | Immune cells | Immune response |
| IL2RG | sIL2R | P31785 | Serum | N/A | T cells | Adaptive immune response |
| SNCA | SNCA | P37840 | Skin | Membrane | Brain and bone marrow | Chromatin organization |
| SRP14 | SRP14 | P37108 | Skin | N/A | Non-specific | Mitochondria |
| SRPX | SRPX | P78539 | Skin | Unknown | Adipose tissue | ECM organization |
| STAT3 | STAT3 | P40763 | mRNA | N/A | Non-specific | Mitochondria and proteasome |
| STAT6 | STAT6 | P42226 | mRNA | N/A | Macrophages | Immune response |
| STIP1 | STIP1 | P31948 | Serum | N/A | Non-specific | Unknown function |
| SYK | SYK | P43405 | mRNA | N/A | Non-specific | Transcription |
| TBX21 | TBX21 | Q9UL17 | mRNA | N/A | Immune cells | Immune response |
| TNF | TNF-alpha | P01375 | Cytokines | Blood | Neutrophils | Inflammatory response |
| VCP | VCP | P55072 | Serum | N/A | Non-specific | Mitochondria |
| FLT4 | VEGFR-3 | P35916 | Serum | Blood | Non-specific | Transcription |
| CHI3L1 | YKL-40 | P36222 | Serum | Blood | Liver | Metabolism |
4.3. Case Studies/Examples in Psoriasis and PsA
In a recent study, a set of 951 circulating proteins was analyzed in serum samples to interrogate possible differences between patients with PsA, psoriasis and healthy controls [74]. Sixty-eight differentially expressed proteins were identified when comparing PsA patients and healthy controls, but no differentially expressed proteins were identified when comparing PsA and psoriasis patients. This led the authors to propose a “shared serum proteomic signature” between psoriasis and PsA. However, the cohorts were very small and subclinical PsA in the psoriasis group could not be excluded. Indeed, no information was provided about patient inclusion/exclusion criteria or the criteria used for the differentiation of PsA from psoriasis, which is necessary in such studies. In conclusion, the authors recommended that future studies focus on skin and synovial tissue to find differences between PsA and psoriasis patients.
5. Metabolomics
5.1. Brief Overview of Relevant Metabolomics Technologies
Metabolomics can be defined as the investigation of changes in the populations of endogenous and exogenous low-molecular-weight metabolites (<1500 Da), representing a shift from single metabolite monitoring to complex profiling and pattern recognition [76]. This is a considerable analytical challenge that involves the identification and quantification of a broad spectrum of molecules in biological matrices such as human plasma or urine, which contain hundreds or thousands of metabolites with diverse chemical and physical properties across a wide dynamic range of concentrations. The most widely used techniques include nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry in combination with gas chromatography (GC-MS) or liquid chromatography (LC-MS). Advanced bioinformatics and statistical tools are used to maximize the recovery of information from the resulting metabolomic datasets.
5.2. Applications for Early Diagnosis, Prognosis and Treatment Monitoring
Low-molecular-weight metabolites are important indicators and even integrators of phenotypes, reflecting the biochemical activity of cells and tissues. Metabolomics recognizes that changes in cell function are most evident at the level of small-molecule metabolism and can provide a coherent view of the response of individuals to a variety of genetic and environmental influences [77]. The abnormal cellular processes associated with disease often disrupt the composition of low-molecular-weight metabolites. Perturbations in metabolite abundance and temporal profiles in readily accessible body fluids may provide an index of disease severity through the direct measurement of biochemical changes. As such, metabolomics has the potential to identify biomarkers of PsA that may improve diagnostic accuracy and predict disease progression as well as defining patient responses to specific therapeutic interventions. Similarly, metabolomics may offer additional insight into the metabolic pathways that drive the chronic, immune-mediated processes that are characteristic of PsA, opening routes to potential new drug targets.
5.3. Case Studies/Examples in Psoriasis and PsA
Researchers are increasingly using metabolomics for the clinical assessment of PsA [78,79,80]. Several studies have reported alterations in the metabolomes of PsA patients in comparison to healthy controls or individuals with related inflammatory diseases such as psoriasis or RA. The serum levels of various amino acids are modified in PsA patients relative to RA cohorts [81,82]. Changes in the levels of circulating glucuronic acid and α-ketoglutaric acid were detected among psoriasis patients with or without PsA [83] and a correlation was made between serum levels of the choline metabolite trimethylamine N-oxide (TMAO) and inflammation in PsA patients [84]. A more recent study used untargeted metabolomics to characterize the metabolic changes in the transition from psoriasis to PsA, revealing differences in the abundance of bile acids (particularly glycoursodeoxycholic acid sulfate) and butyrate to differentiate between psoriasis patients who did or did not progress to PsA [85].
Metabolite profiles in other matrices can also provide a window of opportunity to elucidate the metabolic changes in PsA. It was recently reported that α/β-turmerone, glycerol 1-hexadecanoate, dihydrosphingosine, pantothenic acid and glutamine may act as fecal biomarkers for PsA [86]. In addition, a metabolomic study focusing on urinary metabolites revealed lower levels of citrate, alanine, methylsuccinate and trigonelline in PsA patients compared to unaffected individuals [87]. Metabolomic approaches have also been used to evaluate PsA patient responses to anti-TNF therapy. For example, histamine, glutamine, phenylacetic acid, xanthine, xanthurenic acid and creatinine levels were elevated in urine samples from patients who responded to TNF antagonists, whereas ethanolamine, p-hydroxyphenylpyruvic acid and phosphocreatine levels were depleted [88].
6. Lipidomics
6.1. Brief Overview of Relevant Lipidomics Technologies
Recent technological improvements in LC-MS enable comprehensive lipidomic analysis in clinical studies, analyzing extensive sample sets for different lipids and lipid mediators. Depending on the specific lipids, targeted or untargeted LC-MS may be most appropriate. The untargeted approach is based on high-resolution mass spectrometry (HRMS) and can potentially examine the whole lipidome in a single run, but focuses on the more abundant lipids because the dynamic range is not sufficient to detect scarce molecules such as lipid mediators alongside abundant lipids such as triglycerides. Scarce lipids are analyzed using targeted approaches based on tandem mass spectrometry (MS/MS), which has greater sensitivity and selectivity. However, targeted methods cannot display the whole lipidome, so an approach combining untargeted and targeted methods is used for comprehensive lipidomics analysis, searching for lipids and lipid mediators relevant in the context of psoriatic diseases.
6.2. Applications for Early Diagnosis, Prognosis and Treatment Monitoring
Lipids and lipid mediators play a fundamental role in the immune system and changes in homeostatic status are closely related to IMIDs [89,90,91,92,93] such as RA [94], IBD [95,96,97,98,99] and psoriatic diseases [100,101,102,103]. Lipids are involved in many different processes and are also essential building blocks of membranes and key components in energy metabolism. Lipid mediators such as oxylipins and endocannabinoids, which are present at very low concentrations, are signaling molecules implicated in diverse physiological and pathological processes. Therefore, lipid profiles might be used as biomarkers for early diagnosis, prognosis of disease progression or the development of comorbidities, and to guide the selection of the most promising therapeutic approach.
Biomarker discovery in the field of lipidomics is challenging due to strict procedural requirements for sampling, sample preparation and analysis. This is partly due to the variable concentration of lipids in different biological matrices, the broad spectrum of isomeric compounds and the special procedures required to ensure lipid stability at all stages, including pre-analytical sample handling. On the other hand, lipidomics covers a field of up to several thousand different molecules and one of its key advantages is the close temporal linkage between these markers and individual clinical phenotypes or disease states [77,104].
6.3. Case Studies/Examples in Psoriasis and PsA
The close link between lipid profiles and IMIDs was recently demonstrated in PsA patients, where oxylipins [102,103,105,106], endocannabinoids [103,107], fatty acids [105,106,107] and phospholipids [106] were found to be potentially pathophysiologically relevant. A recent study also found that the level of inflammatory lipid mediators in psoriasis patients increased following a PsA diagnosis, particularly leukotriene B4 [85]. However, a comprehensive study is required to compare the lipid profiles of patients with psoriasis and PsA, and this will be the first step toward the identification of lipid biomarkers that improve the diagnosis and treatment of PsA.
7. Complementary Technologies—Multiple Sequential Immunohistochemistry
Several multiple immunohistochemistry systems have been developed that allow the staining of tissue sections with a theoretically unlimited number of antibodies. The technology makes use of directly labeled antibodies carrying a fluorophore or heavy metal ion. The antibodies are applied to the sample in an automated process, which includes a short incubation period, washing steps, imaging and signal removal. The latter involves either bleaching or chemical inactivation, and is followed by the addition of the next antibody [108,109]. This process can be repeated as often as necessary, and typically creates image stacks representing 30–50 antibodies. Recent developments include software that combines single-cell phenotyping and localized information about neighboring cells, facilitating a quantitative “tissue FACS analysis” (FACS = fluorescence-activated cell sorting) with the description of disease-specific immunological neighborhoods within inflamed tissues [108].
One of the key benefits of multiple immunohistochemistry systems in the context of PsA is single-cell phenotyping in different patient groups using 40–50 antibody probes in automated cycles. By detecting and quantifying a large panel of corresponding markers, it is possible to identify nearly all immune cells and their subtypes, and to characterize their cellular neighborhood to quantify and visualize cellular networks (information that is lost during FACS analysis). The comparison of samples from psoriasis and PsA cohorts can therefore identify differences between the patient groups and generate information about biomarkers and immune cell networks/interactions that may lead to new therapeutic options.
8. Data Management/Integration and Artificial Intelligence
To benefit from the wealth of methods used to mine multi-omics data, it is essential to align the data and verify their quality before integration. Data should be formatted according to international standards, including standard bioinformatics file formats (such as FASTA, FASTQ, SAM/BAM, VCF and GFF), and “minimum information” standards for omics experiments [110], including MIGS/MIMS for genomics [111] and MIAPE for proteomics [112]. The data must be checked to ensure they include the same annotation references (e.g., genome version, standard gene and protein names). This is challenging with lipidomics and metabolomics data where there are currently no widely accepted standards, although efforts are ongoing to establish equivalent minimum information standard such as MIAMET [113] as well as standards for lipidomics analysis [114,115]. Following de-identification, clinical data are standardized using the OMOP common data model (Observational Health Data Sciences and Informatics, OHDSI program, available at https://ohdsi.org/ (accessed on 20 June 2022) and aligned to standard dictionaries to ensure interoperability. Once formatted and standardized, data are stored and accessed via a secure data management infrastructure specifically designed to protect sensitive clinical and biomedical data [116].
Multiple processing steps should be considered to ensure data integrity, including missing value imputation, normalization, transformations, aggregation and batch effect correction [117,118,119]. Unsupervised multivariate analysis methods such as common dimensions [120] can be used to assess overall variability, trends and potential biases across multiple integrated layers of multi-omics and clinical data before further data exploration by supervised multivariate analysis methods such as OPLS-DA [121] or artificial intelligence-based approaches such as machine learning. A wide range of computational methods can be applied depending on the study design and research aim. In addition to classical statistical analysis, machine learning can be used to evaluate data in an unsupervised manner for preliminary exploration and dimensional reduction (e.g., clustering approaches such as DBSCAN or k-means algorithms, or dimensional reduction such as PCA or TSNE). Batch effects in dimensional reduction and clustering approaches can reveal outliers [118]. In a clinical setting, supervised machine learning often tackles classification problems rather than regression. Due to the high dimensionality of multi-omics data and the so-called “curse of dimensionality” (low number of subjects and high number of features), feature selection algorithms such as LASSO or ridge can be applied to enhance the results of supervised learning [118,122]. In addition to feature selection, class imbalances are common challenges in multi-omics analysis, but can be solved by sampling or cost-sensitive learning [122]. Commonly used algorithms such as random forests, support vector machines and the k-nearest neighbor algorithm can provide insight into the underlying structures of datasets and can be developed into powerful models for the support of clinical decision making [123,124,125]. In order to develop further hypotheses and integrate data with the literature, pathway analysis can embed the data in a broader context [126].
9. The Advantage of Multi-Omics Evaluation
As discussed above, several markers have been identified that commonly occur in PsA patients, but no single marker stands alone as a specific indicator of the disease. Even the most reliable markers are also present in other IMIDs, which therefore makes it difficult if not impossible to achieve a definitive diagnosis. In other fields, the lack of definitive qualitative markers has been addressed by (a) seeking quantitative markers, whose abundance rather than presence/absence correlates with a disease, and (b) profiles based on combinations of several markers that are more informative than single molecules, a strategy first applied to ovarian cancer [127]. Indeed, this approach has also been successful in RA, where the proteomic analysis of serum and synovial fluid has revealed the elevation of multiple biomarkers representing processes such as joint inflammation and injury (e.g., MMP3, IL-12, IL-15 and IL-18), cartilage integrity and bone or connective tissue degradation (e.g., MMP13 and neoepitopes of collagen) [128]. Considering that such panels of RA markers have been assembled based solely on proteomics data, it is clear that the combination of proteomics with orthogonal omics datasets plus more diverse data can increase the power of this approach exponentially, both for diagnosis/prognosis [129] and the monitoring of drug responses [130]. In one recent study, metabolomics and lipidomics analysis revealed that a combination of the bile acid conjugate glycoursodeoxycholic acid sulfate and lipid mediator leukotriene B4 provided a sensitive and specific predictor of progression from psoriasis to PsA [85]. However, adding new features will also require the careful evaluation of added value, both for discovery in basic research and translation to the clinic. Models that incorporate more markers may be more sensitive and specific, but the cost of acquiring the data in routine clinical practice may be prohibitive, although this may not always be the case [131]. A risk remains that expensive and large datasets merely report already known aspects, such as the effect of the body mass index or inflammation on disease progression. The field should also strive to identify causal markers rather than solely correlative observations without a direct biochemical connection to the phenotype. As the amount and complexity of the data increase, it becomes more difficult for humans to identify consistent patterns that correlate with certain diseases, but machine learning algorithms either supervised to assign samples to known categories or devising categories de novo using unsupervised analysis have the power to reveal these hidden patterns and then to apply the same approach when analyzing data from new patients, greatly improving the accuracy of the resulting predictions.
10. Conclusions and Outlook
The definitive diagnosis and early treatment of PsA are not yet possible because the clinical manifestations and associated biomarkers are not, on an individual basis, able to distinguish PsA from other IMIDs or predict those individuals with psoriasis who will progress to PsA. However, the combination of biomarker profiles based on data from multi-omics technologies and classical sources, such as imaging data and clinical evaluations, could provide the body of information required for early diagnosis and the initiation of effective treatment before symptoms emerge. Combinations of different types of biomarkers have proven effective in other fields, particularly oncology, but such biomarker profiles are often mainly based on a single method. The power of biomarker profiles may increase with the number of complementary modalities that can be tested simultaneously. For PsA, combining information from disease-associated alleles and chromatin structures, the levels of proteins, lipids and other metabolites, multimodal image analysis, histology and classical phenotyping will provide an important step forward. Ultimately, the use of multiple orthogonal technologies that embed machine learning will lead to the generation of unique molecular and clinical fingerprints that can be used for PsA diagnosis, prognosis and therapeutic monitoring. However, research on the identification of biomarker profiles/fingerprints using different omics technologies is still in the discovery phase with much work to be conducted to turn the anticipated results of these analyses into assays which are applicable in routine clinical settings. The HIPPOCRATES project is therefore strategically important because it combines expertise from all relevant fields with access to comprehensive cohorts, technologies and translational research experience. This will ultimately improve the quality of life for those living with PsA or at risk of developing PsA.
Acknowledgments
R.G., M.K., L.H., S.K., E.R., S.R., K.S., G.G. and F.B. acknowledge support from the Fraunhofer Cluster of Excellence Immune Mediated diseases (CIMD). A.B. is an NIHR Senior Investigator. A.B. and J.B. receive support from Versus Arthritis (grant ref. 21754) and the NIHR Manchester Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, National Institute for Health and Care Research or the Department of Health. The authors would like to thank Daniel Kratz and Samuel Rischke for support in creating the figures.
Author Contributions
All authors contributed to the conceptualization, writing, reviewing and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors, including Richard M. Twyman, who is the director of the company TRM, declare no conflict of interest.
Funding Statement
HIPPOCRATES has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement no. 101007757. The JU receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Damiani G., Bragazzi N.L., Karimkhani Aksut C., Wu D., Alicandro G., McGonagle D., Guo C., Dellavalle R., Grada A., Wong P., et al. The Global, Regional, and National Burden of Psoriasis: Results and Insights from the Global Burden of Disease 2019 Study. Front. Med. 2021;8:743180. doi: 10.3389/fmed.2021.743180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong A.W., Read C. Pathophysiology, Clinical Presentation, and Treatment of Psoriasis: A Review. JAMA. 2020;323:1945–1960. doi: 10.1001/jama.2020.4006. [DOI] [PubMed] [Google Scholar]
- 3.Ritchlin C.T., Colbert R.A., Gladman D.D. Psoriatic Arthritis. N. Engl. J. Med. 2017;376:957–970. doi: 10.1056/NEJMra1505557. [DOI] [PubMed] [Google Scholar]
- 4.Simon D., Watad A., Rodrigues-Manica S., Perricone C. Editorial: Early Origins of Psoriatic Arthritis. Front. Med. 2021;8:794229. doi: 10.3389/fmed.2021.794229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumthekar A., Ogdie A. Obesity and Psoriatic Arthritis: A Narrative Review. Rheumatol. Ther. 2020;7:447–456. doi: 10.1007/s40744-020-00215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogdie A., Harrison R.W., McLean R.R., Lin T.-C., Lebwohl M., Strober B.E., Zhuo J., Patel V., Mease P.J. Prospective Cohort Study of Psoriatic Arthritis Risk in Patients With Psoriasis in a Real-World Psoriasis Registry. J. Am. Acad. Dermatol. 2022. in press . [DOI] [PubMed]
- 7.Gisondi P., Bellinato F., Maurelli M., Geat D., Zabotti A., McGonagle D., Girolomoni G. Reducing the Risk of Developing Psoriatic Arthritis in Patients with Psoriasis. Psoriasis. 2022;12:213–220. doi: 10.2147/PTT.S323300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pezzolo E., Naldi L. The relationship between smoking, psoriasis and psoriatic arthritis. Expert Rev. Clin. Immunol. 2019;15:41–48. doi: 10.1080/1744666X.2019.1543591. [DOI] [PubMed] [Google Scholar]
- 9.Owczarczyk-Saczonek A., Purzycka-Bohdan D., Nedoszytko B., Reich A., Szczerkowska-Dobosz A., Bartosiñska J., Batycka-Baran A., Czajkowski R., Dobrucki I.T., Dobrucki L.W., et al. Pathogenesis of psoriasis in the “omic” era. Part III. Metabolic disorders, metabolomics, nutrigenomics in psoriasis. Postepy Dermatol. Alergol. 2020;37:452–467. doi: 10.5114/ada.2020.98284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kocic H., Damiani G., Stamenkovic B., Tirant M., Jovic A., Tiodorovic D., Peris K. Dietary compounds as potential modulators of microRNA expression in psoriasis. Ther. Adv. Chronic Dis. 2019;10:2040622319864805. doi: 10.1177/2040622319864805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher J.U., Ogdie A., Merola J.F., Ritchlin C. Preventing psoriatic arthritis: Focusing on patients with psoriasis at increased risk of transition. Nat. Rev. Rheumatol. 2019;15:153–166. doi: 10.1038/s41584-019-0175-0. [DOI] [PubMed] [Google Scholar]
- 12.Ogdie A., Schwartzman S., Husni M.E. Recognizing and managing comorbidities in psoriatic arthritis. Curr. Opin. Rheumatol. 2015;27:118–126. doi: 10.1097/BOR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 13.Weigle N., McBane S. Psoriasis. Am. Fam. Physician. 2013;87:626–633. [PubMed] [Google Scholar]
- 14.Raychaudhuri S.K., Maverakis E., Raychaudhuri S.P. Diagnosis and classification of psoriasis. Autoimmun. Rev. 2014;13:490–495. doi: 10.1016/j.autrev.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Johnson M.A.N., Armstrong A.W. Clinical and histologic diagnostic guidelines for psoriasis: A critical review. Clin. Rev. Allergy Immunol. 2013;44:166–172. doi: 10.1007/s12016-012-8305-3. [DOI] [PubMed] [Google Scholar]
- 16.Pennington S.R., FitzGerald O. Early Origins of Psoriatic Arthritis: Clinical, Genetic and Molecular Biomarkers of Progression from Psoriasis to Psoriatic Arthritis. Front. Med. 2021;8:723944. doi: 10.3389/fmed.2021.723944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri S.P., Wilken R., Sukhov A.C., Raychaudhuri S.K., Maverakis E. Management of psoriatic arthritis: Early diagnosis, monitoring of disease severity and cutting edge therapies. J. Autoimmun. 2017;76:21–37. doi: 10.1016/j.jaut.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Ghasemi M., Nabipour I., Omrani A., Alipour Z., Assadi M. Precision medicine and molecular imaging: New targeted approaches toward cancer therapeutic and diagnosis. Am. J. Nucl. Med. Mol. Imaging. 2016;6:310–327. [PMC free article] [PubMed] [Google Scholar]
- 19.Vargas A.J., Harris C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer. 2016;16:525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grivas A., Fragoulis G., Garantziotis P., Banos A., Nikiphorou E., Boumpas D. Unraveling the complexities of psoriatic arthritis by the use of -Omics and their relevance for clinical care. Autoimmun. Rev. 2021;20:102949. doi: 10.1016/j.autrev.2021.102949. [DOI] [PubMed] [Google Scholar]
- 21.Caputo V., Strafella C., Termine A., Dattola A., Mazzilli S., Lanna C., Cosio T., Campione E., Novelli G., Giardina E., et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J. Cell. Mol. Med. 2020;24:13554–13563. doi: 10.1111/jcmm.15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Generali E., Scirè C.A., Favalli E.G., Selmi C. Biomarkers in psoriatic arthritis: A systematic literature review. Expert Rev. Clin. Immunol. 2016;12:651–660. doi: 10.1586/1744666X.2016.1147954. [DOI] [PubMed] [Google Scholar]
- 23.O’Rielly D.D., Jani M., Rahman P., Elder J.T. The Genetics of Psoriasis and Psoriatic Arthritis. J. Rheumatol. Suppl. 2019;95:46–50. doi: 10.3899/jrheum.190119. [DOI] [PubMed] [Google Scholar]
- 24.Shi C., Rattray M., Barton A., Bowes J., Orozco G. Using functional genomics to advance the understanding of psoriatic arthritis. Rheumatology. 2020;59:3137–3146. doi: 10.1093/rheumatology/keaa283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowes J., Loehr S., Budu-Aggrey A., Uebe S., Bruce I.N., Feletar M., Marzo-Ortega H., Helliwell P., Ryan A.W., Kane D., et al. PTPN22 is associated with susceptibility to psoriatic arthritis but not psoriasis: Evidence for a further PsA-specific risk locus. Ann. Rheum. Dis. 2015;74:1882–1885. doi: 10.1136/annrheumdis-2014-207187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortes A., Brown M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budu-Aggrey A., Bowes J., Barton A. Identifying a novel locus for psoriatic arthritis. Rheumatology. 2016;55:25–32. doi: 10.1093/rheumatology/kev273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez J., Fernández-Sueiro J.L., López-Mejías R., Montilla C., Arias M., Moll C., Alsina M., Sanmarti R., Lozano F., Cañete J.D. FCGR2A/CD32A and FCGR3A/CD16A variants and EULAR response to tumor necrosis factor-α blockers in psoriatic arthritis: A longitudinal study with 6 months of followup. J. Rheumatol. 2012;39:1035–1041. doi: 10.3899/jrheum.110980. [DOI] [PubMed] [Google Scholar]
- 29.Bowes J., Ashcroft J., Dand N., Jalali-Najafabadi F., Bellou E., Ho P., Marzo-Ortega H., Helliwell P.S., Feletar M., Ryan A.W., et al. Cross-phenotype association mapping of the MHC identifies genetic variants that differentiate psoriatic arthritis from psoriasis. Ann. Rheum. Dis. 2017;76:1774–1779. doi: 10.1136/annrheumdis-2017-211414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y., Han B., Tsoi L.C., Stuart P.E., Ellinghaus E., Tejasvi T., Chandran V., Pellett F., Pollock R., Bowcock A.M., et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 2014;95:162–172. doi: 10.1016/j.ajhg.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Rielly D.D., Rahman P. Genetic, Epigenetic and Pharmacogenetic Aspects of Psoriasis and Psoriatic Arthritis. Rheum. Dis. Clin. N. Am. 2015;41:623–642. doi: 10.1016/j.rdc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Bowes J., Budu-Aggrey A., Huffmeier U., Uebe S., Steel K., Hebert H.L., Wallace C., Massey J., Bruce I.N., Bluett J., et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat. Commun. 2015;6:6046. doi: 10.1038/ncomms7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budu-Aggrey A., Bowes J., Loehr S., Uebe S., Zervou M.I., Helliwell P., Ryan A.W., Kane D., Korendowych E., Giardina E., et al. Replication of a distinct psoriatic arthritis risk variant at the IL23R locus. Ann. Rheum. Dis. 2016;75:1417–1418. doi: 10.1136/annrheumdis-2016-209290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuart P.E., Nair R.P., Tsoi L.C., Tejasvi T., Das S., Kang H.M., Ellinghaus E., Chandran V., Callis-Duffin K., Ike R., et al. Genome-wide Association Analysis of Psoriatic Arthritis and Cutaneous Psoriasis Reveals Differences in Their Genetic Architecture. Am. J. Hum. Genet. 2015;97:816–836. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandran V., Bull S.B., Pellett F.J., Ayearst R., Pollock R.A., Gladman D.D. Killer-cell immunoglobulin-like receptor gene polymorphisms and susceptibility to psoriatic arthritis. Rheumatology. 2014;53:233–239. doi: 10.1093/rheumatology/ket296. [DOI] [PubMed] [Google Scholar]
- 36.Cascella R., Strafella C., Ragazzo M., Manzo L., Costanza G., Bowes J., Hüffmeier U., Potenza S., Sangiuolo F., Reis A., et al. KIF3A and IL-4 are disease-specific biomarkers for psoriatic arthritis susceptibility. Oncotarget. 2017;8:95401–95411. doi: 10.18632/oncotarget.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aterido A., Cañete J.D., Tornero J., Ferrándiz C., Pinto J.A., Gratacós J., Queiró R., Montilla C., Torre-Alonso J.C., Pérez-Venegas J.J., et al. Genetic variation at the glycosaminoglycan metabolism pathway contributes to the risk of psoriatic arthritis but not psoriasis. Ann. Rheum. Dis. 2019;78:355–364. doi: 10.1136/annrheumdis-2018-214158. [DOI] [PubMed] [Google Scholar]
- 38.Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Park P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schadt E.E., Banerjee O., Fang G., Feng Z., Wong W.H., Zhang X., Kislyuk A., Clark T.A., Luong K., Keren-Paz A., et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 2013;23:129–141. doi: 10.1101/gr.136739.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Pugh B.F. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merelli I., Tordini F., Drocco M., Aldinucci M., Liò P., Milanesi L. Integrating multi-omic features exploiting Chromosome Conformation Capture data. Front. Genet. 2015;6:40. doi: 10.3389/fgene.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carini C., Hunter E., Ramadass A.S., Green J., Akoulitchev A., McInnes I.B., Goodyear C.S. Chromosome conformation signatures define predictive markers of inadequate response to methotrexate in early rheumatoid arthritis. J. Transl. Med. 2018;16:18. doi: 10.1186/s12967-018-1387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter E., Dezfouli M., Koutsothanasi C., Wilson A., Santos F.C., Salter M., Westra J.W., Powell R., Dring A., Egan B., et al. Development and validation of blood-based predictive biomarkers for response to PD-(L)-1 checkpoint inhibitors: Evidence of a universal systemic core of 3D immunogenetic profiling across multiple oncological indications. medRxiv. 2021 doi: 10.1101/2021.12.21.21268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter E., Koutsothanasi C., Wilson A., Santos F.C., Salter M., Powell R., Dring A., Brajer P., Egan B., Westra J.W., et al. 3D genomic capture of regulatory immuno-genetic profiles in COVID-19 patients for prognosis of severe COVID disease outcome. bioRxiv. 2021 doi: 10.1101/2021.03.14.435295. [DOI] [Google Scholar]
- 46.Tordini F., Aldinucci M., Milanesi L., Liò P., Merelli I. The Genome Conformation as an Integrator of Multi-Omic Data: The Example of Damage Spreading in Cancer. Front. Genet. 2016;7:194. doi: 10.3389/fgene.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahendran S.M., Chandran V. Exploring the Psoriatic Arthritis Proteome in Search of Novel Biomarkers. Proteomes. 2018;6:5. doi: 10.3390/proteomes6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi F., Tan Y., Yao A., Yang X., He Y. Psoriasis to Psoriatic Arthritis: The Application of Proteomics Technologies. Front. Med. 2021;8:681172. doi: 10.3389/fmed.2021.681172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mc Ardle A., Kwasnik A., Szentpetery A., Hernandez B., Parnell A., de Jager W., de Roock S., FitzGerald O., Pennington S.R. Identification and Evaluation of Serum Protein Biomarkers That Differentiate Psoriatic Arthritis From Rheumatoid Arthritis. Arthritis Rheumatol. 2022;74:81–91. doi: 10.1002/art.41899. [DOI] [PubMed] [Google Scholar]
- 50.Adhikari S., Nice E.C., Deutsch E.W., Lane L., Omenn G.S., Pennington S.R., Paik Y.-K., Overall C.M., Corrales F.J., Cristea I.M., et al. A high-stringency blueprint of the human proteome. Nat. Commun. 2020;11:5301. doi: 10.1038/s41467-020-19045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bateman N.W., Goulding S.P., Shulman N.J., Gadok A.K., Szumlinski K.K., MacCoss M.J., Wu C.C. Maximizing peptide identification events in proteomic workflows using data-dependent acquisition (DDA) Mol. Cell. Proteom. 2014;13:329–338. doi: 10.1074/mcp.M112.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludwig C., Gillet L., Rosenberger G., Amon S., Collins B.C., Aebersold R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suhre K., McCarthy M.I., Schwenk J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2021;22:19–37. doi: 10.1038/s41576-020-0268-2. [DOI] [PubMed] [Google Scholar]
- 54.Zhong W., Edfors F., Gummesson A., Bergström G., Fagerberg L., Uhlén M. Next generation plasma proteome profiling to monitor health and disease. Nat. Commun. 2021;12:2493. doi: 10.1038/s41467-021-22767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrera A., von Toerne C., Behler J., Huth C., Thorand B., Hilgendorff A., Hauck S.M. Multiplatform Approach for Plasma Proteomics: Complementarity of Olink Proximity Extension Assay Technology to Mass Spectrometry-Based Protein Profiling. J. Proteome Res. 2021;20:751–762. doi: 10.1021/acs.jproteome.0c00641. [DOI] [PubMed] [Google Scholar]
- 56.Pietzner M., Wheeler E., Carrasco-Zanini J., Cortes A., Koprulu M., Wörheide M.A., Oerton E., Cook J., Stewart I.D., Kerrison N.D., et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374:eabj1541. doi: 10.1126/science.abj1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ignjatovic V., Geyer P.E., Palaniappan K.K., Chaaban J.E., Omenn G.S., Baker M.S., Deutsch E.W., Schwenk J.M. Mass Spectrometry-Based Plasma Proteomics: Considerations from Sample Collection to Achieving Translational Data. J. Proteome Res. 2019;18:4085–4097. doi: 10.1021/acs.jproteome.9b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deutsch E.W., Omenn G.S., Sun Z., Maes M., Pernemalm M., Palaniappan K.K., Letunica N., Vandenbrouck Y., Brun V., Tao S.-C., et al. Advances and Utility of the Human Plasma Proteome. J. Proteome Res. 2021;20:5241–5263. doi: 10.1021/acs.jproteome.1c00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uhlén M., Karlsson M.J., Hober A., Svensson A.-S., Scheffel J., Kotol D., Zhong W., Tebani A., Strandberg L., Edfors F., et al. The human secretome. Sci. Signal. 2019;12:609. doi: 10.1126/scisignal.aaz0274. [DOI] [PubMed] [Google Scholar]
- 60.Tebani A., Gummesson A., Zhong W., Koistinen I.S., Lakshmikanth T., Olsson L.M., Boulund F., Neiman M., Stenlund H., Hellström C., et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat. Commun. 2020;11:4487. doi: 10.1038/s41467-020-18148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cretu D., Gao L., Liang K., Soosaipillai A., Diamandis E.P., Chandran V. Differentiating Psoriatic Arthritis From Psoriasis Without Psoriatic Arthritis Using Novel Serum Biomarkers. Arthritis Care Res. 2018;70:454–461. doi: 10.1002/acr.23298. [DOI] [PubMed] [Google Scholar]
- 62.Cretu D., Liang K., Saraon P., Batruch I., Diamandis E.P., Chandran V. Quantitative tandem mass-spectrometry of skin tissue reveals putative psoriatic arthritis biomarkers. Clin. Proteom. 2015;12:1–8. doi: 10.1186/1559-0275-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalmády S., Kiss M., Képíró L., Kovács L., Sonkodi G., Kemény L., Gyulai R. Higher levels of autoantibodies targeting mutated citrullinated vimentin in patients with psoriatic arthritis than in patients with psoriasis vulgaris. Clin. Dev. Immunol. 2013;2013:474028. doi: 10.1155/2013/474028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansson C., Eriksson C., Alenius G.-M. S-calprotectin (S100A8/S100A9): A potential marker of inflammation in patients with psoriatic arthritis. J. Immunol. Res. 2014;2014:696415. doi: 10.1155/2014/696415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alenius G.-M., Eriksson C., Rantapää Dahlqvist S. Interleukin-6 and soluble interleukin-2 receptor alpha-markers of inflammation in patients with psoriatic arthritis? Clin. Exp. Rheumatol. 2009;27:120–123. [PubMed] [Google Scholar]
- 66.Abji F., Pollock R.A., Liang K., Chandran V., Gladman D.D. Brief Report: CXCL10 Is a Possible Biomarker for the Development of Psoriatic Arthritis among Patients with Psoriasis. Arthritis Rheumatol. 2016;68:2911–2916. doi: 10.1002/art.39800. [DOI] [PubMed] [Google Scholar]
- 67.Grazio S., Razdorov G., Erjavec I., Grubisic F., Kusic Z., Punda M., Anticevic D., Vukicevic S., Grgurevic L. Differential expression of proteins with heparin affinity in patients with rheumatoid and psoriatic arthritis: A preliminary study. Clin. Exp. Rheumatol. 2013;31:665–671. [PubMed] [Google Scholar]
- 68.Chandran V., Shen H., Pollock R.A., Pellett F.J., Carty A., Cook R.J., Gladman D.D. Soluble biomarkers associated with response to treatment with tumor necrosis factor inhibitors in psoriatic arthritis. J. Rheumatol. 2013;40:866–871. doi: 10.3899/jrheum.121162. [DOI] [PubMed] [Google Scholar]
- 69.Fiocco U., Sfriso P., Oliviero F., Roux-Lombard P., Scagliori E., Cozzi L., Lunardi F., Calabrese F., Vezzù M., Dainese S., et al. Synovial effusion and synovial fluid biomarkers in psoriatic arthritis to assess intraarticular tumor necrosis factor-α blockade in the knee joint. Arthritis Res. Ther. 2010;12:R148. doi: 10.1186/ar3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Kuijk A.W.R., DeGroot J., Koeman R.C., Sakkee N., Baeten D.L., Gerlag D.M., Tak P.P. Soluble biomarkers of cartilage and bone metabolism in early proof of concept trials in psoriatic arthritis: Effects of adalimumab versus placebo. PLoS ONE. 2010;5:e12556. doi: 10.1371/journal.pone.0012556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collins E.S., Butt A.Q., Gibson D.S., Dunn M.J., Fearon U., van Kuijk A.W., Gerlag D.M., Pontifex E., Veale D.J., Tak P.P., et al. A clinically based protein discovery strategy to identify potential biomarkers of response to anti-TNF-α treatment of psoriatic arthritis. Proteom. Clin. Appl. 2016;10:645–662. doi: 10.1002/prca.201500051. [DOI] [PubMed] [Google Scholar]
- 72.Mahendran S.M., Keystone E.C., Krawetz R.J., Liang K., Diamandis E.P., Chandran V. Elucidating the endogenous synovial fluid proteome and peptidome of inflammatory arthritis using label-free mass spectrometry. Clin. Proteom. 2019;16:23. doi: 10.1186/s12014-019-9243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mulder M.L.M., van Hal T.W., Wenink M.H., Koenen H.J.P.M., van den Hoogen F.H.J., de Jong E.M.G.J., van den Reek J.M.P.A., Vriezekolk J.E. Clinical, laboratory, and genetic markers for the development or presence of psoriatic arthritis in psoriasis patients: A systematic review. Arthritis Res. Ther. 2021;23:168. doi: 10.1186/s13075-021-02545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leijten E., Tao W., Pouw J., van Kempen T., Olde Nordkamp M., Balak D., Tekstra J., Muñoz-Elías E., DePrimo S., Drylewicz J., et al. Broad proteomic screen reveals shared serum proteomic signature in patients with psoriatic arthritis and psoriasis without arthritis. Rheumatology. 2021;60:751–761. doi: 10.1093/rheumatology/keaa405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karlsson M., Zhang C., Méar L., Zhong W., Digre A., Katona B., Sjöstedt E., Butler L., Odeberg J., Dusart P., et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021;7:eabh2169. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wishart D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019;99:1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 77.López-López Á., López-Gonzálvez Á., Barker-Tejeda T.C., Barbas C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018;18:557–575. doi: 10.1080/14737159.2018.1481391. [DOI] [PubMed] [Google Scholar]
- 78.Di Y., Afifi L., Jeon C., Trivedi M., Chang H.W., Lee K., Liao W. The metabolomics of psoriatic disease. Psoriasis. 2017;7:1–15. doi: 10.2147/PTT.S118348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koussiouris J., Looby N., Anderson M., Kulasingam V., Chandran V. Metabolomics Studies in Psoriatic Disease: A Review. Metabolites. 2021;11:375. doi: 10.3390/metabo11060375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dorochow E., Köhm M., Hahnefeld L., Gurke R. Metabolic Profiling in Rheumatoid Arthritis, Psoriatic Arthritis, and Psoriasis: Elucidating Pathogenesis, Improving Diagnosis, and Monitoring Disease Activity. J. Pers. Med. 2022;12:924. doi: 10.3390/jpm12060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madsen R.K., Lundstedt T., Gabrielsson J., Sennbro C.-J., Alenius G.-M., Moritz T., Rantapää-Dahlqvist S., Trygg J. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res. Ther. 2011;13:R19. doi: 10.1186/ar3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Souto-Carneiro M., Tóth L., Behnisch R., Urbach K., Klika K.D., Carvalho R.A., Lorenz H.-M. Differences in the serum metabolome and lipidome identify potential biomarkers for seronegative rheumatoid arthritis versus psoriatic arthritis. Ann. Rheum. Dis. 2020;79:499–506. doi: 10.1136/annrheumdis-2019-216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Armstrong A.W., Wu J., Johnson M.A., Grapov D., Azizi B., Dhillon J., Fiehn O. Metabolomics in psoriatic disease: Pilot study reveals metabolite differences in psoriasis and psoriatic arthritis. F1000Research. 2014;3:248. doi: 10.12688/f1000research.4709.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coras R., Kavanaugh A., Boyd T., Huynh D., Lagerborg K.A., Xu Y.-J., Rosenthal S.B., Jain M., Guma M. Choline metabolite, trimethylamine N-oxide (TMAO), is associated with inflammation in psoriatic arthritis. Clin. Exp. Rheumatol. 2019;37:481–484. [PMC free article] [PubMed] [Google Scholar]
- 85.Paine A., Brookes P.S., Bhattacharya S., Li D., La Garcia-Hernandez M.D.L., Tausk F., Ritchlin C. Dysregulation of bile acids, lipids, and nucleotides in psoriatic arthritis revealed by unbiased profiling of serum metabolites. Arthritis Rheumatol. 2022 doi: 10.1002/art.42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang N., Yang L., Shang L., Liang Z., Wang Y., Feng M., Yu S., Li X., Gao C., Li Z., et al. Altered Fecal Metabolomics and Potential Biomarkers of Psoriatic Arthritis Differing From Rheumatoid Arthritis. Front. Immunol. 2022;13:812996. doi: 10.3389/fimmu.2022.812996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alonso A., Julià A., Vinaixa M., Domènech E., Fernández-Nebro A., Cañete J.D., Ferrándiz C., Tornero J., Gisbert J.P., Nos P., et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016;14:133. doi: 10.1186/s12916-016-0681-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapoor S.R., Filer A., Fitzpatrick M.A., Fisher B.A., Taylor P.C., Buckley C.D., McInnes I.B., Raza K., Young S.P. Metabolic profiling predicts response to anti-tumor necrosis factor α therapy in patients with rheumatoid arthritis. Arthritis Rheum. 2013;65:1448–1456. doi: 10.1002/art.37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maceyka M., Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Serhan C.N. Discovery of specialized pro-resolving mediators marks the dawn of resolution physiology and pharmacology. Mol. Aspects Med. 2017;58:1–11. doi: 10.1016/j.mam.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang C., Wang K., Yang L., Liu R., Chu Y., Qin X., Yang P., Yu H. Lipid metabolism in inflammation-related diseases. Analyst. 2018;143:4526–4536. doi: 10.1039/C8AN01046C. [DOI] [PubMed] [Google Scholar]
- 93.Lin D.A., Boyce J.A. Lysophospholipids as Mediators of Immunity. Adv. Immunol. 2006;89:141–167. doi: 10.1016/S0065-2776(05)89004-2. [DOI] [PubMed] [Google Scholar]
- 94.Brouwers H., von Hegedus J., Toes R., Kloppenburg M., Ioan-Facsinay A. Lipid mediators of inflammation in rheumatoid arthritis and osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2015;29:741–755. doi: 10.1016/j.berh.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 95.Fan F., Mundra P.A., Fang L., Galvin A., Moore X.L., Weir J.M., Wong G., White D.A., Chin-Dusting J., Sparrow M.P., et al. Lipidomic Profiling in Inflammatory Bowel Disease: Comparison Between Ulcerative Colitis and Crohn’s Disease. Inflamm. Bowel Dis. 2015;21:1511–1518. doi: 10.1097/MIB.0000000000000394. [DOI] [PubMed] [Google Scholar]
- 96.Schwanke R.C., Marcon R., Bento A.F., Calixto J.B. EPA- and DHA-derived resolvins’ actions in inflammatory bowel disease. Eur. J. Pharmacol. 2016;785:156–164. doi: 10.1016/j.ejphar.2015.08.050. [DOI] [PubMed] [Google Scholar]
- 97.Scoville E.A., Allaman M.M., Brown C.T., Motley A.K., Horst S.N., Williams C.S., Koyama T., Zhao Z., Adams D.W., Beaulieu D.B., et al. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn’s Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics. 2018;14:17. doi: 10.1007/s11306-017-1311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grill M., Högenauer C., Blesl A., Haybaeck J., Golob-Schwarzl N., Ferreirós N., Thomas D., Gurke R., Trötzmüller M., Köfeler H.C., et al. Members of the endocannabinoid system are distinctly regulated in inflammatory bowel disease and colorectal cancer. Sci. Rep. 2019;9:2358. doi: 10.1038/s41598-019-38865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bazarganipour S., Hausmann J., Oertel S., El-Hindi K., Brachtendorf S., Blumenstein I., Kubesch A., Sprinzl K., Birod K., Hahnefeld L., et al. The Lipid Status in Patients with Ulcerative Colitis: Sphingolipids are Disease-Dependent Regulated. J. Clin. Med. 2019;8:971. doi: 10.3390/jcm8070971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zeng C., Wen B., Hou G., Lei L., Mei Z., Jia X., Chen X., Zhu W., Li J., Kuang Y., et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. Gigascience. 2017;6:1–11. doi: 10.1093/gigascience/gix087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sorokin A.V., Norris P.C., English J.T., Dey A.K., Chaturvedi A., Baumer Y., Silverman J., Playford M.P., Serhan C.N., Mehta N.N. Identification of proresolving and inflammatory lipid mediators in human psoriasis. J. Clin. Lipidol. 2018;12:1047–1060. doi: 10.1016/j.jacl.2018.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coras R., Kavanaugh A., Boyd T., Huynh Q., Pedersen B., Armando A.M., Dahlberg-Wright S., Marsal S., Jain M., Paravar T., et al. Pro- and anti-inflammatory eicosanoids in psoriatic arthritis. Metabolomics. 2019;15:65. doi: 10.1007/s11306-019-1527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wójcik P., Biernacki M., Wroński A., Łuczaj W., Waeg G., Žarković N., Skrzydlewska E. Altered Lipid Metabolism in Blood Mononuclear Cells of Psoriatic Patients Indicates Differential Changes in Psoriasis Vulgaris and Psoriatic Arthritis. Int. J. Mol. Sci. 2019;20:4249. doi: 10.3390/ijms20174249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sethi S., Brietzke E. Recent advances in lipidomics: Analytical and clinical perspectives. Prostaglandins Other Lipid Mediat. 2017;128–129:8–16. doi: 10.1016/j.prostaglandins.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 105.Looby N., Roszkowska A., Reyes-Garcés N., Yu M., Bączek T., Kulasingam V., Pawliszyn J., Chandran V. Serum metabolic fingerprinting of psoriasis and psoriatic arthritis patients using solid-phase microextraction-liquid chromatography-high-resolution mass spectrometry. Metabolomics. 2021;17:59. doi: 10.1007/s11306-021-01805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ambrożewicz E., Wójcik P., Wroński A., Łuczaj W., Jastrząb A., Žarković N., Skrzydlewska E. Pathophysiological Alterations of Redox Signaling and Endocannabinoid System in Granulocytes and Plasma of Psoriatic Patients. Cells. 2018;7:159. doi: 10.3390/cells7100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kishikawa T., Arase N., Tsuji S., Maeda Y., Nii T., Hirata J., Suzuki K., Yamamoto K., Masuda T., Ogawa K., et al. Large-scale plasma-metabolome analysis identifies potential biomarkers of psoriasis and its clinical subtypes. J. Dermatol. Sci. 2021;102:78–84. doi: 10.1016/j.jdermsci.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 108.Hoch T., Schulz D., Eling N., Gómez J.M., Levesque M.P., Bodenmiller B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 2022;7:eabk1692. doi: 10.1126/sciimmunol.abk1692. [DOI] [PubMed] [Google Scholar]
- 109.Schubert W., Bonnekoh B., Pommer A.J., Philipsen L., Böckelmann R., Malykh Y., Gollnick H., Friedenberger M., Bode M., Dress A.W.M. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 110.Schriml L.M. A decade of GigaScience: 10 years of the evolving genomic and biomedical standards landscape. Gigascience. 2022;11:giac047. doi: 10.1093/gigascience/giac047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Field D., Amaral-Zettler L., Cochrane G., Cole J.R., Dawyndt P., Garrity G.M., Gilbert J., Glöckner F.O., Hirschman L., Karsch-Mizrachi I., et al. The Genomic Standards Consortium. PLoS Biol. 2011;9:e1001088. doi: 10.1371/journal.pbio.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taylor C.F., Paton N.W., Lilley K.S., Binz P.-A., Julian R.K., Jones A.R., Zhu W., Apweiler R., Aebersold R., Deutsch E.W., et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 113.Fiehn O., Kristal B., van Ommen B., Sumner L.W., Sansone S.-A., Taylor C., Hardy N., Kaddurah-Daouk R. Establishing reporting standards for metabolomic and metabonomic studies: A call for participation. Omics A J. Integr. Biol. 2006;10:158–163. doi: 10.1089/omi.2006.10.158. [DOI] [PubMed] [Google Scholar]
- 114.Köfeler H.C., Ahrends R., Baker E.S., Ekroos K., Han X., Hoffmann N., Holčapek M., Wenk M.R., Liebisch G. Recommendations for good practice in MS-based lipidomics. J. Lipid Res. 2021;62:100138. doi: 10.1016/j.jlr.2021.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burla B., Arita M., Arita M., Bendt A.K., Cazenave-Gassiot A., Dennis E.A., Ekroos K., Han X., Ikeda K., Liebisch G., et al. MS-based lipidomics of human blood plasma: A community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018;59:2001–2017. doi: 10.1194/jlr.S087163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coman Schmid D., Crameri K., Oesterle S., Rinn B., Sengstag T., Stockinger H. SPHN—The BioMedIT Network: A Secure IT Platform for Research with Sensitive Human Data. Stud. Health Technol. Inform. 2020;270:1170–1174. doi: 10.3233/SHTI200348. [DOI] [PubMed] [Google Scholar]
- 117.Wehrens R., Hageman J.A., van Eeuwijk F., Kooke R., Flood P.J., Wijnker E., Keurentjes J.J.B., Lommen A., van Eekelen H.D.L.M., Hall R.D., et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics. 2016;12:88. doi: 10.1007/s11306-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster S.M., Sanghi A., Wu S., Snyder M.P. A Customizable Analysis Flow in Integrative Multi-Omics. Biomolecules. 2020;10:1606. doi: 10.3390/biom10121606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kohl M., Megger D.A., Trippler M., Meckel H., Ahrens M., Bracht T., Weber F., Hoffmann A.-C., Baba H.A., Sitek B., et al. A practical data processing workflow for multi-OMICS projects. Biochim. Biophys. Acta. 2014;1844:52–62. doi: 10.1016/j.bbapap.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 120.Jouan-Rimbaud Bouveresse D., Pinto R.C., Schmidtke L.M., Locquet N., Rutledge D.N. Identification of significant factors by an extension of ANOVA–PCA based on multi-block analysis. Chemom. Intell. Lab. Syst. 2011;106:173–182. doi: 10.1016/j.chemolab.2010.05.005. [DOI] [Google Scholar]
- 121.Boccard J., Rutledge D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta. 2013;769:30–39. doi: 10.1016/j.aca.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 122.Picard M., Scott-Boyer M.-P., Bodein A., Périn O., Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021;19:3735–3746. doi: 10.1016/j.csbj.2021.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kingsmore K.M., Puglisi C.E., Grammer A.C., Lipsky P.E. An introduction to machine learning and analysis of its use in rheumatic diseases. Nat. Rev. Rheumatol. 2021;17:710–730. doi: 10.1038/s41584-021-00708-w. [DOI] [PubMed] [Google Scholar]
- 124.Xing L., Yu L., Zhou N., Zhang Z., Pu Y., Wu J., Shu H. Exploration of biomarkers of psoriasis through combined multiomics analysis. Res. Sq. 2022. Preprint . [DOI] [PMC free article] [PubMed]
- 125.Stafford I.S., Kellermann M., Mossotto E., Beattie R.M., MacArthur B.D., Ennis S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit. Med. 2020;3:30. doi: 10.1038/s41746-020-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wörheide M.A., Krumsiek J., Kastenmüller G., Arnold M. Multi-omics integration in biomedical research—A metabolomics-centric review. Anal. Chim. Acta. 2021;1141:144–162. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Petricoin E.F., Ardekani A.M., Hitt B.A., Levine P.J., Fusaro V.A., Steinberg S.M., Mills G.B., Simone C., Fishman D.A., Kohn E.C., et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 128.Park Y.-J., Chung M.K., Hwang D., Kim W.-U. Proteomics in Rheumatoid Arthritis Research. Immune Netw. 2015;15:177–185. doi: 10.4110/in.2015.15.4.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsuchiya H., Ota M., Fujio K. Multiomics landscape of synovial fibroblasts in rheumatoid arthritis. Inflamm. Regen. 2021;41:7. doi: 10.1186/s41232-021-00157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tasaki S., Suzuki K., Kassai Y., Takeshita M., Murota A., Kondo Y., Ando T., Nakayama Y., Okuzono Y., Takiguchi M., et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat. Commun. 2018;9:2755. doi: 10.1038/s41467-018-05044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Karlsson A.A., Hao S., Jauhiainen A., Elfström K.M., Egevad L., Nordström T., Heintz E., Clements M.S. The cost-effectiveness of prostate cancer screening using the Stockholm3 test. PLoS ONE. 2021;16:e0246674. doi: 10.1371/journal.pone.0246674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.