Abstract
Preserving blood samples for shipping and later DNA extraction has been performed by cooling, freezing, drying, freeze-drying, and protease treatment, among other methods. Most methods to preserve field samples for further DNA extraction do not prevent cellular and DNA damage or are useful only in preserving them for short periods. This report introduces a novel method for blood and tissue that allows preservation in freezing temperatures for a prolonged period of time. The solution reported here (20% ethylene glycol-propylene glycol) preserves cells and tissues integrity, as judged by microscopic analysis, and improves DNA yield and quality.
Storing and preserving blood and other tissue samples suitable for shipping or later DNA extraction, to be used in PCR assays and other procedures, are usually cumbersome. This often results in a poor quality of DNA and a low yield, due to the cellular lysis and further degradation of genetic material. Several methods, such as cooling, freezing (1), fixing (5), drying (6), freeze-drying (9), and the use of microbe and enzyme inhibitors (1, 2, 7), have been used to preserve the DNA from field samples. The results are mixed; the majority of these methods do not prevent cellular and DNA damage, and most of the methods are useful only in preserving samples for short periods. This report introduces an alternative method for tissue preservation with an antifreeze solution that can be kept in freezing temperatures for a prolonged period of time. Preserving the integrity of blood and tissue samples dramatically improves the quality and yield of the extracted DNA.
MATERIALS AND METHODS
Several ratios (from 0 to 30%) of autoclaved propylene glycol and ethylene glycol (Sigma Chemical Co., Gaithersburg, Md.), each alone or combined in equal parts (kept at room temperature), were mixed by gentle inversion with Na2EDTA-containing peripheral goat and human blood and directly placed in three freezers at 4, −20, and −70°C for up to 6 months. After the best concentration of antifreeze was determined, it was tested with other samples, such as bone marrow from human patients and pieces of internal organs from Brucella-infected deceased goats (spleen, brain, heart, liver, lung, kidney, stomach, uterus, lymphatic node, mammary gland, salivary gland, bladder, and pancreas). Tissues were cut into small pieces (around 2 mm per side) and kept in Eppendorf tubes containing 500 μl of a 20% ethylene glycol-propylene glycol (1:1) solution (100 μl of antifreeze mixed with 400 μl of sterile saline solution). Samples were kept at −20°C for 2 months. Samples used as controls were prepared by extracting DNA from the freshly collected blood before mixing it with antifreeze solutions. After specific periods, samples were placed at 4°C, and cellular damage or preservation was assessed by macroscopic and microscopic analysis, followed by DNA extractions.
Blood and bone marrow.
Four hundred microliters of bone marrow or blood collected in Vacutainer (Becton Dickinson) tubes containing sodium Na2EDTA (lavender stopper) was placed in 1.5-μl Eppendorf tubes, 1 ml of erythrocyte lysis solution (155 mM NH4Cl; 10 mM NaHCO3; 100 mM Na2EDTA [pH 7.4]) was added, and the contents were mixed and centrifuged. Treatment with erythrocyte lysis solution was repeated until the white cell pellets lost all reddish coloring. Pellets were treated with the universal extraction procedure described below.
Solid animal tissues.
Samples were homogenized (Kontes tissue microhomogenizers) in the presence of an equal volume (vol/wt) of sterile saline solution. Four hundred microliters of the sample was then processed to extract DNA as follows.
Universal DNA extraction procedure.
Four hundred microliters of universal lysis solution (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris-HCl [pH 8.0]) and 10 μl of proteinase K (10 mg/ml) were added to the samples, thoroughly mixed, and incubated for 30 min at 50°C. Four hundred microliters of saturated phenol (liquid phenol containing 0.1% 8-hydroxyquinoline, saturated and stabilized with 100 mM Tris-HCl [pH 8.0] and 0.2% 2-mercaptoethanol) (8) was added, mixed thoroughly, and centrifuged for 5 min at 8,000 × g. The aqueous layer was transferred to a fresh tube, and an equal volume of chloroform-isoamyl alcohol was added (24:1); the tubes were mixed thoroughly and centrifuged for 5 min at 8,000 × g. The upper layer was again transferred to a fresh tube, and 200 μl of 7.5 M ammonium acetate was added and mixed thoroughly. Samples were kept on ice for 10 min and then centrifuged for 5 min at 8,000 × g, and the aqueous content was transferred to a fresh tube. Two volumes of 95% ethanol or 1 volume of isopropanol was added, and the contents were mixed and stored overnight at −20°C. DNA was recovered by centrifuging the samples for 5 min at 8,000 × g; pellets were rinsed with 1 ml of 70% ethanol, air dried, and resuspended in 20 μl of Tris-EDTA buffer. Samples were stored at −20°C after the DNA concentrations were determined by measuring absorbance at 260 nm.
PCRs. (i) Brucella.
Clinical samples taken from Brucella-infected goats and patients were subjected to a PCR assay to detect this intracellular pathogen (4).
(ii) β-globin.
In human samples, the detection of β-globin was performed as an internal control for the extracted DNA, using previously described procedures (3).
RESULTS AND DISCUSSION
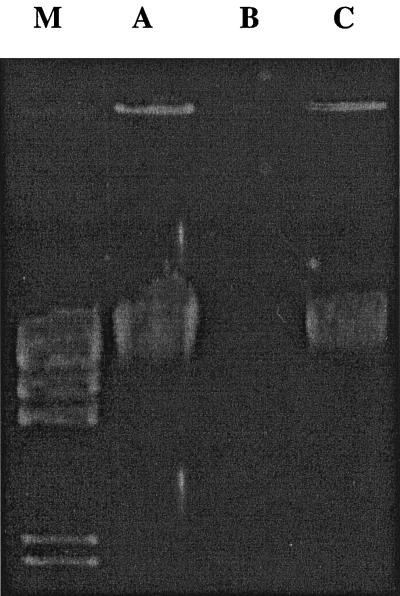
The solution that better preserved cells was 20% ethylene glycol-propylene glycol (E/P20). Samples kept at either −4 or −20°C showed excellent cellular preservation and therefore high and good yields of genomic DNA. Samples kept at −70°C and those mixed with propylene glycol alone or with concentrations of ethylene-glycol below 15% became frozen. Cells and DNA of samples stored in those solutions were greatly damaged, as judged by their microscopic and macroscopic appearance, electrophoresis results, and PCR performance. EG15 to EG30 showed complete erythrocyte lysis and 1 to 2 lymphocytes per field, and treatments E/P15, E/P20, and E/P25 showed 300 or more erythrocytes and about 2 leukocytes per field; E/P30 showed an average of fewer than 50 erythrocytes and 1 leukocyte per field (Table 1). When DNA yield and quality were evaluated, E/P20 was shown to be the best cell-preserving solution as judged by readings of optical density at 260 and 280 nm, gel appearance, and PCR performance. As shown in Fig. 1, DNA extracted from blood stored in E/P20 at −20°C for 6 months yielded 34 μg of DNA per 400 μl of blood (lane C), helping to preserve 77% of the cellular DNA contained in the preserved cells compared with the control (freshly collected blood), which yielded an average of 43.7 μg of DNA per 400 μl (lane A). The lowest DNA yield (lane B) was obtained from blood stored for six months at −20°C with no antifreeze (less than 0.06 μg of DNA per 400 μl of blood). Moreover, tissues taken from Brucella-infected goats and stored in the described antifreeze solution also conserved the histopathologic characteristics associated with this pathogen.
TABLE 1.
Blood stored at −20°C for 6 months using various concentrations of propylene glycol and/or ethylene glycola
| Antifreeze agent | Results with cells (DNA yield) for antifreeze concn (%) ofb:
|
|||||
|---|---|---|---|---|---|---|
| 5 | 10 | 15 | 20 | 25 | 30 | |
| PG | Frozen; total cell lysis (13.6 μg) | Frozen; total cell lysis (14 μg) | Frozen; total cell lysis (9.4 μg) | Frozen; total cell lysis (5.4 μg) | Frozen; total cell lysis (6.4 μg) | Frozen; total cell lysis (8.4 μg) |
| EG | Frozen; total cell lysis (1.76 μg) | Frozen; total cell lysis (2.04 μg) | Erythrocyte lysis; 1–2 leukocytes per field (16.2 μg) | Erythrocyte lysis; 1–2 leukocytes per field (8.76 μg) | Erythrocyte lysis; 1–2 leukocytes per field (8.08 μg) | Erythrocyte lysis; 1–2 leukocytes per field (7.64 μg) |
| E/P | Frozen; total cell lysis (2.88 μg) | Frozen; total cell lysis (8.96 μg) | 300 erythrocytes; 2–3 leukocytes per field (11.96 μg) | 400 erythrocytes; 2–3 leukocytes per field (34 μg) | 300 erythrocytes; 1–2 leukocytes per field (12.04 μg) | 50 erythrocytes; 0–1 leukocytes per field (5.88 μg) |
PG, propylene glycol; EG, ethylene glycol; E/P, ethylene glycol and propylene glycol combined in equal parts.
DNA yield is the amount of DNA per 400 μl of blood.
FIG. 1.
Genomic DNA extracted from blood stored under various conditions. Lanes: M, λ HindIII as a molecular marker; A, control, DNA isolated from freshly collected blood; B, DNA obtained from blood stored at −20°C for 6 months; C, DNA isolated from blood stored as for lane B but preserved in E/P 20.
The direct correlation between cell intactness and DNA yield and quality seems logical; performance of most methods of DNA extraction from blood relies on the primary lysis of red cells to flush their debris and concentrate by centrifuging the DNA-containing white cells before their rupture. Freezing causes all cells contained within blood samples to burst, diminishing the DNA yield. Other effects of noncontrolled cell lysis are the degrading mechanisms acting on organic molecules, such as DNA, and the difficulty in separating contaminants that remain, binding DNA and causing loss of samples and inhibition of various reactions. We have found this procedure quite convenient in several ways: (i) other people can withdraw field samples and store them to be processed at a later convenient date, (ii) researchers can collect samples, store them in any house freezer, and send them in large batches to be analyzed in specialized remote laboratories, (iii) samples can be repeatedly withdrawn from a freezer, to be tested many times, without suffering any alteration, and (iv) it allows use of the same DNA extraction procedure that is applied for fresh samples. The method described here may be tested in further studies regarding quantification of cell populations, protein and RNA preservation, and other biochemical determinations.
ACKNOWLEDGMENTS
This research was supported by CONACYT Mexico (grants 476100-5-4017M and N9507-1382P).
REFERENCES
- 1.Ahmad N N, Unjieng A B, Donoso L A. Modification of standard proteinase K/phenol method for DNA isolation to improve yield and purity from frozen blood. J Med Genet. 1995;32:129–130. doi: 10.1136/jmg.32.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albarino C G, Romanowski V. Phenol extraction revisited: a rapid method for the isolation and preservation of human genomic DNA from whole blood. Mol Cell Probes. 1994;8:423–427. doi: 10.1006/mcpr.1994.1060. [DOI] [PubMed] [Google Scholar]
- 3.Bauer H M, Manos M. PCR detection of genital human papillomavirus. In: Persing D H, Smith T F, Tenover F C, White J T, editors. Diagnostic molecular microbiology. Washington, D.C.: American Society for Microbiology; 1993. pp. 407–413. [Google Scholar]
- 4.Leal-Klevezas D S, Martínez-Vázquez I O, López-Merino A, Martínez-Soriano J P. Single-step PCR for detection of Brucella spp. from blood and milk of infected animals. J Clin Microbiol. 1995;33:3087–3090. doi: 10.1128/jcm.33.12.3087-3090.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maass M, Schreiber M, Knobloch J. Detection of Bartonella bacilliformis in cultures, blood, and formalin preserved skin biopsies by use of the polymerase chain reaction. Trop Med Parasitol. 1992;43:191–194. [PubMed] [Google Scholar]
- 6.McCabe E R B. Utility of PCR for DNA analysis from dried blood spots on filter paper blotters. PCR Methods Appl. 1991;1:99–106. doi: 10.1101/gr.1.2.99. [DOI] [PubMed] [Google Scholar]
- 7.Muralidharan K, Wemmer C. Transporting and storing field-collected specimens for DNA without refrigeration for subsequent DNA extraction and analysis. BioTechniques. 1994;17:420–422. [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Takahashi R, Matsuo S, Okuyama T, Sugiyama T. Degradation of macromolecules during preservation of lyophilized pathological tissues. Pathol Res Pract. 1995;191:420–426. doi: 10.1016/S0344-0338(11)80729-6. [DOI] [PubMed] [Google Scholar]