ABSTRACT
Equine infectious anemia virus (EIAV) and HIV are both members of the Lentivirus genus and are similar in major virological characters. EIAV endangers the horse industry. In addition, EIAV can also be used as a model for HIV research. The maturation of the lentiviral Env protein, which is necessary for viral entry, requires Env to be folded in the endoplasmic reticulum (ER). It is currently unclear how this process is regulated. Mitochondrion-associated endoplasmic reticulum membrane (MAM) is a specialized part of the close connection between the ER and mitochondria, and one of the main functions of MAM is to promote oxidative protein production in the ER. SYNJ2BP is one of the key proteins that make up the MAM, and we found that SYNJ2BP is essential for EIAV replication. We therefore constructed a SYNJ2BP knockout HEK293T cell line in which the number of MAMs is significantly reduced. Moreover, overexpression of SYNJ2BP could increase the number of MAMs. Our study demonstrates that SYNJ2BP can improve the infectivity of the EIAV virus with elevated production of the viral Env protein through increased MAM formation. Interestingly, SYNJ2BP was able to improve the production of not only EIAV Env but also HIV. Further investigation showed that MAMs can provide more ATP and calcium ions, which are essential factors for Env production, to the ER and can also reduce ER stress induced by HIV or EIAV Envs to increase the Env production level in cells. These results may help us to understand the key production mechanisms of lentiviral Env.
IMPORTANCE Lentiviral Env proteins, which are rich in disulfide bonds, need to be fully folded in the ER; otherwise, misfolded Env proteins will induce ER stress and be degraded by ER-associated protein degradation (ERAD). To date, it is still unclear about Env production mechanism in the ER. MAM is the structure of closely connection between the ER and mitochondria. MAMs play important roles in the calcium steady state and oxidative stress, especially in the production of oxidative protein. For the first time, we found that SYNJ2BP can promote the production of lentiviral Env proteins by providing the ATP and calcium ions required for oxidative protein production in the ER and by reducing ER stress through facilitating formation of MAMs. These studies shed light on how MAMs improve lentiviral Env production, which will lay the foundation for the study of replication mechanisms in other lentiviruses from the perspective of the cellular organelle microenvironment.
KEYWORDS: EIAV, HIV, mitochondrion-associated endoplasmic reticulum membrane, SYNJ2BP, endoplasmic reticulum, envelope protein, lentivirus, mitochondria
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) and equine infectious anemia virus (EIAV) are both members of the Lentivirus genus of the Retroviridae family, which present the most serious hazards to human and animal health. HIV-1 invades CD4+ T cells and leads to the progressive loss of host immune function, eventually causing death due to various opportunistic infections (1). There is no effective vaccine for the prevention of HIV, which makes the global epidemic very serious (2). Equine infectious anemia (EIA) is a major infectious disease of equids. It is caused by infection with EIAV and is characterized by periodic episodes of fever, thrombocytopenia, anemia, rapid weight loss, and generalized lymphoid proliferation (3). Research into the replication and regulation mechanisms of lentiviruses will inform the development of antiviral drugs and vaccines and is necessary for the prevention and control of the diseases.
The lentiviral envelope (Env) glycoprotein is an indispensable viral protein that is absolutely required for viral entry into host cells. As with host cell surface and secretory proteins, HIV-1 and EIAV Env proteins are both produced through the classical secretory pathway. They are translated as the 160- and 120-kDa type I integral membrane glycoprotein precursors gp160 and gp120, respectively, on the rough endoplasmic reticulum (ER) and are then imported into the ER lumen for proper folding and modification. Both mature gp160 and gp120 proteins are exported to the cis-Golgi compartment, where they are cleaved by cellular proteases to produce surface and transmembrane proteins which are delivered to the plasma membrane for incorporation into virions (4, 5). A major procedure involved in the maturation of both HIV-1 gp160 and EIAV gp120 is the ER oxidative protein folding process. The HIV-1 gp160 protein has 20 highly conserved Cys residues, which are cross-linked into 10 disulfide bonds so that the protein can be folded into its natural conformation, and the EIAV gp120 protein has 26 highly conserved Cys residues that also need to be cross-linked into disulfide bonds (5, 6). Disulfide bond formation stabilizes the proteins and facilitates protein folding, which is controlled by ATP, calcium, and the redox state in the ER. Notably, the proteins with rich disulfide bonds are frequently misfolded (7, 8). The ER protein folding process is therefore tightly controlled by the ER-associated protein degradation (ERAD) pathway after the activation of ER stress, which retrotransports misfolded proteins to the cytosol and degrades them through the ubiquitin-proteasome system (9). It has been demonstrated that misfolded HIV Env proteins are degraded through the ERAD pathway (10). In the present study, we found that EIAV Env proteins were also degraded through the same pathway. It is known that between 20 and 90% of lentiviral Envs in infected cells are degraded due to the failure of proper folding (11, 12). However, the production mechanisms of Env proteins in the ER are still not clear.
The mitochondrion-associated endoplasmic reticulum membrane (MAM) is a specialized part of the close connection between the ER and the outer membrane of mitochondria and constitutes the physical basis of the communication between these two organelles (13). MAMs are usually composed and formed by both mitochondrial and ER proteins. MAMs engage in a wide variety of essential processes, including calcium ion (Ca2+) exchange, lipid biosynthesis, and protein translation and secretion. In addition, MAM appears to be particularly enriched in ER folding chaperones and oxidoreductases, so this domain of the ER is functionally linked to the folding of newly synthesized proteins and bridges the control of oxidative protein production to ER calcium homeostasis (14–16). Indeed, it has been demonstrated that TSPO overexpression in NKR cells can interfere with MAM, with the result that the ER cannot maintain an optimized environment to support the production of HIV Env (10). However, there is still a lack of direct evidence regarding the regulation of HIV Env folding by MAM.
Recently, a PDZ-domain-containing protein synaptojanin-2 binding protein (SYNJ2BP) was found to be highly enriched in MAM and localized in the outer mitochondrial membrane (OMM). Little is known about the function of SYNJ2BP, except that it can tether mitochondria to the ER via the ER membrane (ERM) binding partner RRBP1 to form MAM (17). In this study, we found that the number of SYNJ2BP transcripts increases as EIAV replicates in the host cell. To investigate the effect of SYNJ2BP on the virus, we analyzed EIAV replication after overexpression or knockout of SYNJ2BP (ΔSYNJ2BP) in cells. Interestingly, SYNJ2BP could increase the infectivity of the virus. Env is the key factor for virus entry into host cells, and our results showed that SYNJ2BP could increase the number of MAMs, resulting in improving the expression levels of both HIV and EIAV Envs but not those of the structural protein Gag (the precursor of capsids p24 and p26) or the nonstructural protein Rev. Furthermore, in the ΔSYNJ2BP cell, the number of MAMs was significantly reduced, and the amount of ATP and Ca2+ in ER was lower, which are essential factors for protein production, so the quantity of both HIV and EIAV Envs decreased. Simultaneously, higher ER stress was induced in cells expressing HIV or EIAV Env than in control cell, but overexpression of SYNJ2BP could reduce ER stress and increase production of HIV or EIAV Env. Our findings reveal a fundamental role for SYNJ2BP in promoting production of lentiviral Env proteins by changing the microenvironment of the ER via the MAM.
RESULTS
SYNJ2BP is a critical host factor that determines EIAV replication.
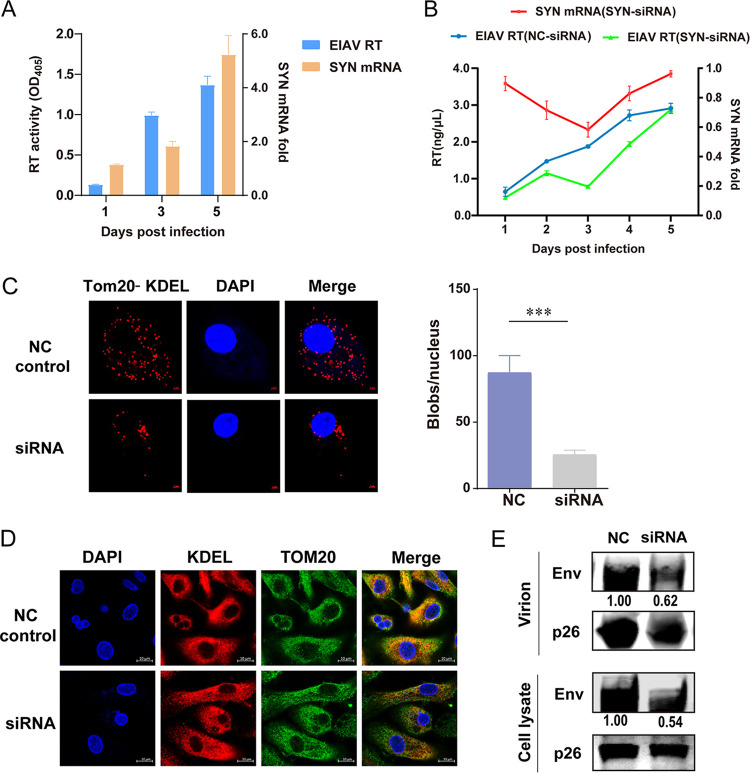
The ER contains a specialized subdomain, MAM, that physically connects the ER to mitochondria and provides a mitochondrion-ER axis that compartmentalizes both stress and metabolic signaling (18). MAM plays a central role in calcium signaling and phospholipid transport to mitochondria (14). In addition, MAM has recently been implicated in viral infection and immunity (19, 20). The PDZ-domain-containing OMM protein SYNJ2BP is one of the key proteins that make up the MAM (17). In this study, we found that the number of SYNJ2BP transcripts increases as EIAV (EIAVDLV34) replicates in the host cell (equine monocyte-derived macrophages [eMDMs]) (Fig. 1A). Therefore, we examined whether knockdown of SYNJ2BP mRNA via small interfering RNA (siRNA) in eMDMs had any effect on viral replication. After quantitative PCR (qPCR) and detection of reverse transcriptase activity, we found that, decreases in SYNJ2BP mRNA levels lead to decreases in EIAV replication efficiency (Fig. 1B). This indicated that SYNJ2BP is critical for EIAV replication. To investigate the alteration of MAMs after SYNJ2BP knockdown in eMDMs, we quantified the overlap between the mitochondria and the ER using proximity ligation assay (PLA) with the detection of translocase of outer mitochondrial membrane 20 (Tom20) and the Lys-Asp-Glu-Leu sequence (KDEL), which are common mitochondrial and ER markers, respectively. PLA permits detection of protein-protein interactions in situ (at distances <40 nm) at endogenous protein levels. In this analysis, the Tom20-KDEL pair was used to calculate the overlap between the mitochondria and the ER. When the distance between Tom20 and KDEL was <40 nm, red dots appeared in situ. The results showed that the overlap between mitochondria and the ER deceased in SYNJ2BP knockdown cells compared to those in the negative-control cells (Fig. 1C). To further verify this result, we used confocal microscopy to check for the overlap between mitochondria (green) and the ER (red) markers in these cells. Consistent with the PLA results, the overlap (yellow dots) between mitochondria and the ER deceased in the SYNJ2BP knockdown cells (Fig. 1D). These results indicate that the formation of MAM is disturbed by SYNJ2BP knockdown in eMDMs. Moreover, we found that, in SYNJ2BP knockdown eMDMs, levels of Env, but not of capsid (CA) protein p26, decreased both in the cells and in the virion-containing supernatants compared to the control (Fig. 1E). These further confirmed that SYNJ2BP is a critical host factor that determines EIAV replication. We then wanted to validate the effect of SYNJ2BP on the formation of MAM.
FIG 1.
Alterations of SYNJ2BP in equine monocyte-derived macrophages (eMDMs) following infection with EIAV and the effects of infection on virus replication. (A) SYNJ2BP mRNA expression increased post-EIAV infection. eMDMs were infected with EIAV(EIAVDLV34) at 2 × 104 50% tissue culture infective doses (TCID50). The viral replication level was determined by the detection of reverse transcriptase (RT) activity. The transcription levels of SYNJ2BP and β-actin were quantified by qPCR. The numbers of SYNJ2BP mRNA copies were normalized to those of β-actin. The data represent the means ± the SEM from three independent experiments. The yellow and blue columns represent the SYNJ2BP mRNA fold change and the EIAV RT activity, respectively. (B) Decreases in SYNJ2BP mRNA levels lead to decreases in EIAV replication efficiency. The virus from NC and SYNJ2BP siRNA knockdown eMDMs infected with 2 × 104 TCID50 EIAV were analyzed by assessment of RT activity at 1, 2, 3, 4, and 5 days postinfection (left y axis, blue curve, NC siRNA transfection group; green curve, SYNJ2BP siRNA transfection group). Meanwhile, the fold change in levels of SYNJ2BP mRNA (right y axis, red curve) in eMDMs after siRNA knockdown was measured using qPCR at 1, 2, 3, 4, and 5 days posttransfection. The data represent the means ± the SEM from three independent experiments. (C) Proximity ligation assay (PLA) to quantify the overlap between mitochondria and ER (MAM) through detection of the endogenous marker proteins Tom20 (mitochondria) and KDEL (ER) in eMDMs after SYNJ2BP siRNA knockdown. The red dots represent the interactions between Tom20 and KDEL in situ (at distances of <40 nm); DAPI staining (blue) was performed to visualize nuclei (scale bar, 2 μm). The number of red dots (blobs/nucleus) is presented by the histogram. The data represent the means ± the SEM from 30 cells in three independent experiments (***, P < 0.001). (D) Confocal fluorescence imaging of the overlap between mitochondria and ER (MAM) detecting the endogenous marker proteins Tom20 (mitochondria) and KDEL (ER) in eMDMs after SYNJ2BP siRNA knockdown. At 2 days after siRNA transfection, the cells were fixed and stained with anti-Tom 20 and anti-KDEL antibodies to detect Tom20-KDEL (Alexa Fluor 549 [AF549] and AF488 readout); DAPI staining (blue) was performed to visualize nuclei (scale bar, 10 μm). Ten visual fields for each group were examined. (E) NC and SYNJ2BP siRNA knockdown eMDMs infected with 2 × 104 TCID50 of EIAV were analyzed by Western blotting. Cell lysate and virion-containing supernatants were analyzed using Western blotting to detect the intensity of viral Env and p26 (EIAV CA) protein bands, respectively, at 72 h postinfection. The results of the densitometry analysis to quantify the ratio of Env to p26 are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times.
SYNJ2BP is essential for the formation of MAM.
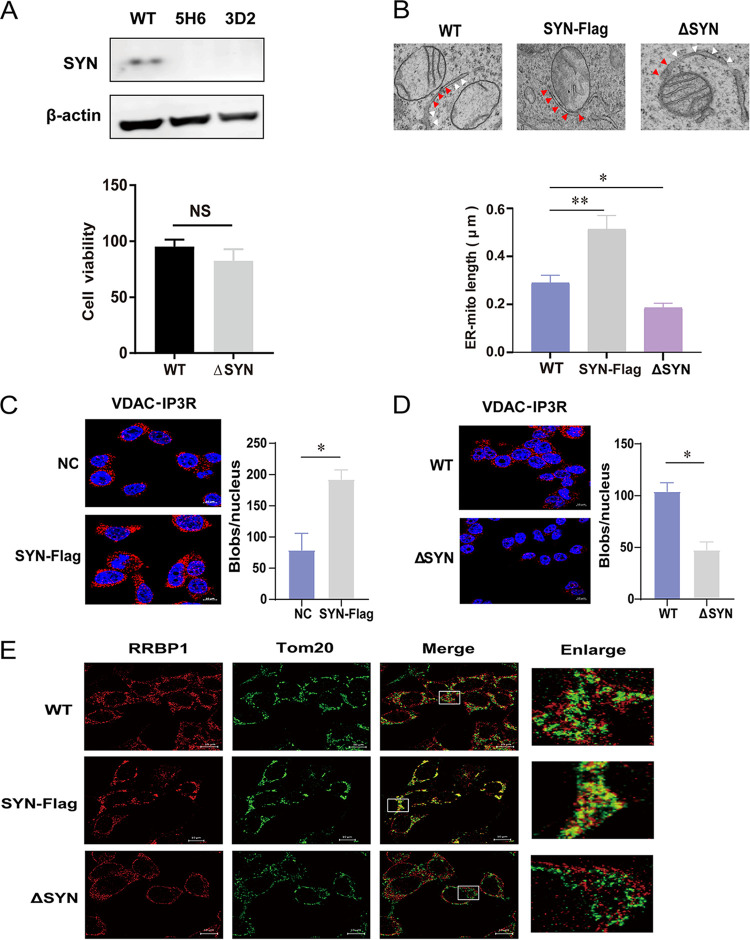
The previous study showed that SYNJ2BP dramatically increases mitochondrial contact with the rough ER when overexpressed (17). To verify this, we constructed SYNJ2BP knockout (KO) HEK293T cells using a CRISPR/Cas9 system (Fig. 2A). No difference in cell viability between wild-type (WT) and SYNJ2BP KO cells was observed. In addition, three types of imaging experiments were carried out to detect morphology and quantity of MAM in cells with either overexpression or deletion of SYNJ2BP. Transmission electron microscopy (TEM) was used to observe the detailed structure of MAM. In the SYNJ2BP-overexpressing cells, we observed more contact points between the mitochondria and the ER. However, in the SYNJ2BP KO cells, fewer contact points between the mitochondria and the ER were observed, meaning that very few MAM structures had formed (Fig. 2B). We then used PLA to quantify the overlap between the mitochondria and the ER. In this analysis, another pair of organelle markers, voltage dependent anion channel 1 (VDAC1) and inositol 1,4,5-trisphosphate receptor type 3 (IP3R), representing the interaction between mitochondria and the ER, was used to calculate the number of MAMs. The results showed that the number of MAMs increased in cells overexpressing SYNJ2BP and decreased in SYNJ2BP KO cells compared to those in control cells (Fig. 2C and D). Moreover, we then performed a direct stochastic optical reconstruction microscopy (dSTORM) analysis, which is based on high-accuracy localization of photoswitchable fluorophores providing single molecule detection up to nanometer range. dSTORM is a common tool that investigate the interactions and dynamics between organelles (21). In our results, the yellow dots indicated the overlap between OMM marker Tom20 staining (green dots) and endoplasmic reticulum membrane (ERM) marker RRBP1 (ribosome binding protein 1) staining (red dots). We found that there were fewer yellow dots in the SYNJ2BP KO cells and more in the cells overexpressing SYNJ2BP than in the control cells (Fig. 2E). All of these results indicated that the distance between the ER and mitochondria increased, followed by formation of few MAMs in cells lacking SYNJ2BP, while the number of MAMs increased in cells overexpressing SYNJ2BP.
FIG 2.
Assay of MAMs in cells overexpressing SYNJ2BP or KO HEK293T cells. (A) Wild-type HEK293T cells (WT 293T) were transfected with plenti-CRISPRv2GFP vector (a plasmid expressing GFP and Cas9) and gRNAs targeting SYNJ2BP to generate SYNJ2BP knockout cells (ΔSYN). The endogenous proteins from WT 293T cells and ΔSYN cells were identified using Western blotting with antibodies against β-actin and SYNJ2BP. 5H6 and 3D2 represent two different KO cell clones. This experiment was performed three times. A histogram representing cell viability is shown. The viability of WT and ΔSYN 293T cells was assessed using a Cell Counting Kit-8 (CCK8), in which WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5,2,(4-disulfophenyl)-2H-tetrazolium, monosodium salt] produces a water-soluble formazan dye upon reduction in the presence of an electron mediator. The data represent the means ± the SEM from three independent experiments (NS, P > 0.05). (B) Electron micrographs of changes in the connections between mitochondria and the ER in SYNJ2BP-Flag overexpression (SYN-Flag) and ΔSYN 293T cells. In the images, the red arrows indicate the connections between mitochondria and ERs (MAMs), and the white arrows indicate the part of the ER not connected to mitochondria. Compared to WT cells, more connections (MAMs) can be seen in SYN-Flag cells, and fewer connections (MAMs) can be seen in ΔSYN cells. The lengths of connections between mitochondria and the ER are presented in the histogram. The data represent the means ± the SEM from 15 cells of three independent experiments (**, P < 0.01; *, P < 0.05). (C and D) Proximity ligation assay to quantify the overlap between mitochondria and the ER (MAMs). The endogenous marker proteins VDAC (mitochondria) and IP3R (ER) were detected in NC and SYN-Flag 293T cells (C) or WT and ΔSYN 293T cells (D). The red dots represent interactions between VDAC and IP3R in situ (at distances <40 nm). DAPI staining (blue) was performed to visualize nuclei (scale bar, 10 μm). The numbers of red dots (blobs/nucleus) are presented in the histogram. The data represent the means ± the SEM from 30 cells in three independent experiments (*, P < 0.05). (E) WT and ΔSYN 293T cells transfected with empty vector or SYN-Flag were fixed and incubated together with anti-RRBP1 (ER) and anti-Tom20 (mitochondria) antibodies, followed by staining with secondary antibodies coupled to a fluorochrome. The signal was detected using a direct stochastic optical reconstruction microscopy (dSTORM) method. Single-molecule localization data from stained sections were captured and processed using high resolution light microscopy (ELYRA PS.1; Zeiss; scale bar, 10 μm). This experiment was performed three times.
SYNJ2BP increases EIAV infectivity by improving production level of Env.
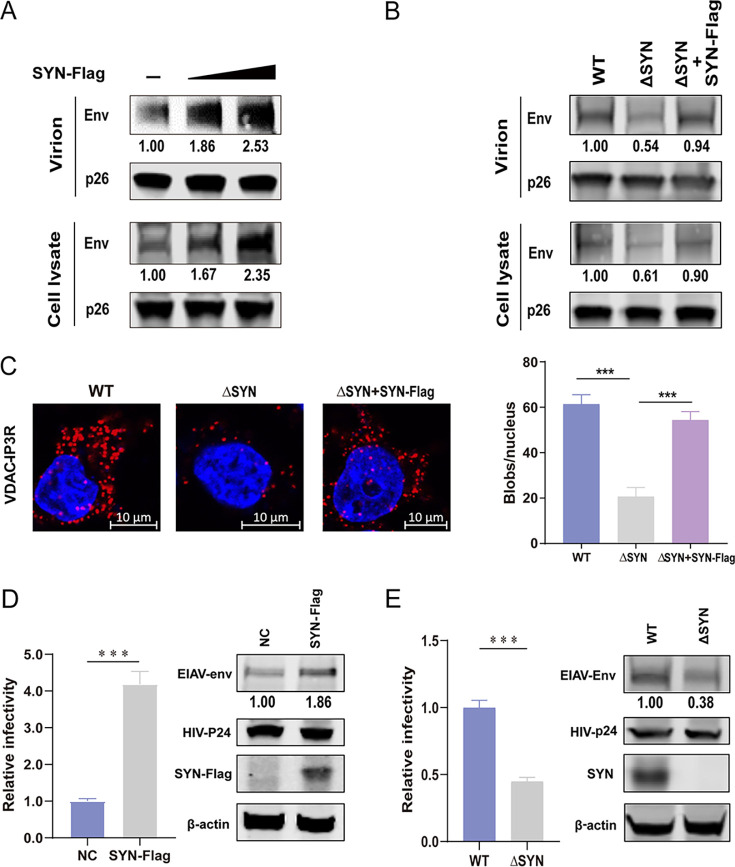
To verify whether SYNJ2BP has an effect on EIAV replication via MAM, we detected Env protein and p26 yield in virions and cells after transfection with the EIAV infectious clone (EIAVCMV3-8) into either SYNJ2BP-overexpressing or KO HEK293T cells. In cells overexpressing SYNJ2BP, the levels of Env, but not of p26 protein, increased both in the cells and in the virion-containing supernatants (Fig. 3A). However, in SYNJ2BP KO cells, the levels of Env, but not p26, decreased both in the cells and the virion-containing supernatants. When SYNJ2BP was rescued in the KO cells, levels of Env were restored (Fig. 3B). We also found that when we reconstituted SYNJ2BP in KO cells, the MAM amount was restored (Fig. 3C). The lentiviral Env is a viral protein that is absolutely required for viral entry into host cells. These findings confirm that SYNJ2BP improves Env production levels through MAM regulation and prompted us to further investigate the effect of SYNJ2BP on lentivirus infection. A luciferase-expressing HIV-EIAV pseudotyped reporter virus was therefore constructed with HIV Gag-Pol and EIAV Env. One HEK293T cell line consistently expressing equine lentiviral receptor 1 (ELR1) was used to detect the quantity of entering 293T-ELR1 cells of HIV-EIAV pseudotyped reporter virus produced by the SYNJ2BP-overexpressing or KO cells. The result showed that the infectivity of the HIV-EIAV pseudotyped reporter virus increased when produced by cells overexpressing SYNJ2BP and decreased when produced by SYNJ2BP KO cells compared to those in the control cells. Moreover, the levels of Env production were positively correlated with infectivity (Fig. 3D and E). All of these indicated that SYNJ2BP can promote EIAV infection by improving Env protein production. Further experiments were carried out to investigate whether SYNJ2BP can improve the production of other EIAV proteins and Env proteins from other lentiviruses.
FIG 3.
Analysis of EIAV replication in cells overexpressing SYNJ2BP or KO HEK293T cells. (A) 293T cells were cotransfected with EIAV infectious clone (EIAVCMV3-8) and empty vector or SYN-Flag in three doses. (B) WT and ΔSYN 293T cells were cotransfected with EIAVCMV3-8 and empty vector or SYN-Flag. For panels A and B, cell lysates and virion-containing supernatants were analyzed by Western blotting to detect the intensity of viral Env and p26 (EIAV CA) protein bands at 48 hpt. This experiment was performed three times. The results of the densitometry analysis to quantify the ratio of Env to p26 are shown at the bottom (lane 1 set as 1.0). (C) PLA to quantify the overlap between mitochondria and ER (MAM) in WT and ΔSYN 293T cells cotransfected with EIAVCMV3-8 and empty vector or SYN-Flag. The red dots represent the interactions between VDAC and IP3R in situ (at distances of <40 nm); DAPI staining (blue) was performed to visualize nuclei (scale bar, 10 μm). The number of red dots (blobs/nucleus) is presented in the histogram. The data represent the means ± the SEM from 30 cells in three independent experiments (***, P < 0.001). (D) Overexpression of SYNJ2BP increased the infectivity of a luciferase-expressing HIV-EIAV pseudotyped reporter virus. HEK293T cells were cotransfected with either SYN-Flag or an empty vector, the HIV-1 luciferase reporter proviral vector pNL4–3-lucΔVifΔEnv and pcDNA3.1-Env (EIAV). Cell lysate was analyzed by using Western blotting to detect the intensity of the EIAV Env and HIV CA (p24) protein bands at 48 hpt. Equal numbers of virions in the supernatant were used to infect a HEK293T cell line consistently expressing the equine lentiviral receptor 1 (ELR1) to determine the infectivity of the HIV-EIAV pseudotyped virus. (E) Knockout of SYNJ2BP reduced infectivity of a luciferase-expressing HIV-EIAV pseudotyped reporter virus. ΔSYN or WT HEK293T cells were transfected with HIV-1 luciferase reporter proviral vector pNL4-3-lucΔVifΔEnv and pcDNA3.1-Env (EIAV). Cell lysates and viral infectivity were analyzed as described previously (see panel D). The results of the densitometry analysis to quantify the ratio of Env to β-actin are shown at the bottom (lane 1 set as 1.0). The data in panels D and E represent the means ± the SEM from three independent experiments (***, P < 0.001).
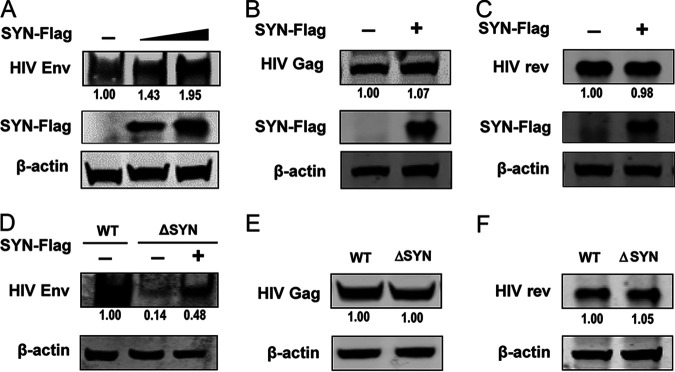
SYNJ2BP improves the production of EIAV Env but not that of Gag and Rev.
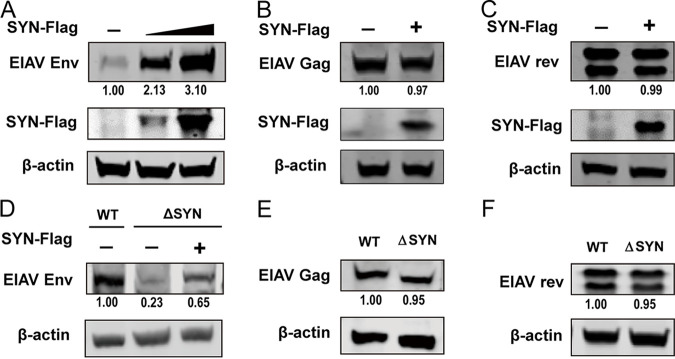
Gag and Env are the main structural proteins of lentiviruses. Unlike Env, synthesis and maturation of Gag-precursor (Pr55gag) polyprotein only requires ribosomes and not ERs. The assembly of Gag precursor proteins on the plasma membrane is essential for virus budding from the host cells. The lentivirus Gag-precursor (Pr55gag) polyprotein is cleaved by viral protease into four major internal structural proteins of the mature virion: the membrane-interacting matrix (MA), CA, and two RNA-binding nucleocapsid (NC) proteins. The regulator of expression of viral proteins (Rev) is one of the additional small nonstructural proteins that acts posttranscriptionally by allowing the essential translocation of unspliced and partially spliced RNA from the nucleus to the cytoplasm. In our study, we found that SYNJ2BP can improve the production of EIAV Env in a dose-dependent manner (Fig. 4A); however, the production levels of EIAV Gag and Rev remained constant in cells overexpressing SYNJ2BP compared to those in the control cells (Fig. 4B and C). Moreover, reconstitution of SYNJ2BP in KO cells can restore the levels of EIAV Env production (Fig. 4D), but the protein production levels of EIAV Gag and Rev were the same in SYNJ2BP KO cells as in the control cells (Fig. 4E and F). These data suggested that SYNJ2BP can improve the production of EIAV Env but not that of Gag and Rev.
FIG 4.
Assay of expression levels of EIAV viral proteins in cells overexpressing SYNJ2BP or KO HEK293T cells. (A to C) Overexpression of SYNJ2BP can promote the production of EIAV Env but not Gag or Rev. HEK293T cells were cotransfected with plasmids, either pcDNA3.1-Flag-SYNJ2BP in three doses or an empty plasmid and pcDNA3.1-Env (EIAV), or pcDNA3.1-Flag-SYNJ2BP and VR1012-Gag (EIAV) or pcDNA3.1-rev-HA (EIAV). Cell lysates were analyzed by using Western blotting to detect the intensity of the EIAV Env or Gag or Rev protein band at 48 hpt. This experiment was performed three times. (D to F) Knockout of SYNJ2BP reduced production of EIAV Env but not Gag or Rev. (D) Empty plasmid or pcDNA3.1-Flag-SYNJ2BP was cotransfected into WT or ΔSYN HEK293T cells, together with pcDNA3.1-Env (EIAV). (E and F) WT and ΔSYN HEK293T cells were transfected with VR1012-Gag (EIAV) or pcDNA3.1-rev-HA (EIAV). Cell lysates were analyzed as in panels A to C. The results of the densitometry analysis to quantify the ratio of Env, Gag, or Rev to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times.
SYNJ2BP improves the production of HIV Env but not Gag and Rev.
HIV and EIAV are known to be similar both in genome structure and in transcripts. The general organization of the EIAV env gene is similar to that described for HIV and other retroviruses. It encodes the surface (gp90) and transmembrane (gp45) glycoproteins, which are incorporated into the virus envelope (1). Similar to HIV, EIAV Rev mediates mRNA transport from the nucleus to the cytoplasm through a Rev-responsive element (RRE) located near the end of the env gene (22, 23). The production and maturation pathway of HIV Env, Gag, and Rev are similar to those of EIAV. Env is generated by ribosomes, folded in the ER, and modified by the Golgi apparatus. Neither Gag nor Rev needs to be folded in the ER. We therefore speculated that SYNJ2BP can improve HIV Env production but not that of Gag and Rev. In further experiments, we found that SYNJ2BP was able to improve the production of HIV Env in a dose-dependent manner (Fig. 5A) but that the protein production levels of HIV Gag and Rev remained constant in cells overexpressing SYNJ2BP compared to those in the control cells (Fig. 5B and C). Moreover, reconstitution of SYNJ2BP in KO cells can restore the levels of HIV Env production (Fig. 5D), but the protein production level of HIV Gag and Rev remained the same in SYNJ2BP KO cells as in the control cells (Fig. 5E and F). Taken together, these results indicated that SYNJ2BP can improve HIV Env production in the same way as that from EIAV. Further analyses were performed to investigate how SYNJ2BP improve lentiviral Env production through the MAM.
FIG 5.
Assay of expression levels of HIV proteins in cells overexpressing SYNJ2BP or KO HEK293T cells. (A to C) Overexpression of SYNJ2BP can promote the production of HIV Env but not that of Gag or Rev. HEK293T cells were cotransfected with plasmids, either pcDNA3.1-Flag-SYNJ2BP in three doses, or an empty plasmid and pcDNA3.1-Env-HA (HIV-1), or pcDNA3.1-Flag-SYNJ2BP and VR1012-Gag-HA (HIV-1), or pcDNA3.1-Rev-HA (HIV-1). Cell lysates were analyzed by using Western blotting to detect the intensity of the HIV Env or Gag or Rev protein band at 48 hpt. This experiment was performed three times. (D to F) Knockout of SYNJ2BP reduced production of HIV Env but not Gag and Rev. (D) Empty plasmid or pcDNA3.1-Flag-SYNJ2BP was cotransfected into WT or ΔSYN HEK293T cells, together with pcDNA3.1-Env-HA (HIV-1). (E and F) WT and ΔSYN HEK293T cells were transfected with VR1012-Gag-HA (HIV-1) or pcDNA3.1-Rev-HA (HIV-1). Cell lysates were analyzed as in panels A to C. The results of the densitometry analysis to quantify the ratio of Env, Gag, or Rev to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times.
SYNJ2BP can increase ATP and Ca2+ contents in the ER to promote Env production.
The production of secretory proteins in the ER depends on a ready supply of energy (ATP), as well as close monitoring of the chemical conditions (Ca2+) that favor oxidative protein folding. On the MAM, ER folding chaperones, such as calnexin and calreticulin, and oxidoreductases regulate calcium flux from the ER through reversible pathways (24). Mitochondria coalesce near the MAMs, allowing mitochondria to supply ATP to the ER, one of the major sites of ATP consumption in the cell (25). Numerous activities associated with the ER consume ATP, in particular the import of calcium and the folding of newly synthesized polypeptides (26, 27). Based on these, we determined the ATP and Ca2+ levels in the ER in SYNJ2BP KO cells. The ER was extracted, and the levels of ATP and Ca2+ in the ER were assessed by using firefly luciferase-based ATP assays and the fluorescent calcium indicator Mag-Fluo-4 AM. SYNJ2BP did not affect the amount of ER in cells with SYNJ2BP overexpression or deletion (Fig. 6A); however, ER ATP and Ca2+ levels were lower in SYNJ2BP KO cells (Fig. 6B and C). To investigate the role of ATP and Ca2+ in the ER on Env protein production, we supplemented ATP or CaCl2 into SYNJ2BP KO cells at final concentrations of either 10 or 5 mM after Env expression plasmid transfection and determined both levels of ATP or Ca2+ content in the ER and Env protein yield (Fig. 6B and C). The data demonstrated that reconstitution of ATP or Ca2+ in the ER can restore Env protein production levels in SYNJ2BP KO cells. From these results, we found that SYNJ2BP can increase ATP and Ca2+ contents in the ER by forming MAM to improve Env protein yield level.
FIG 6.
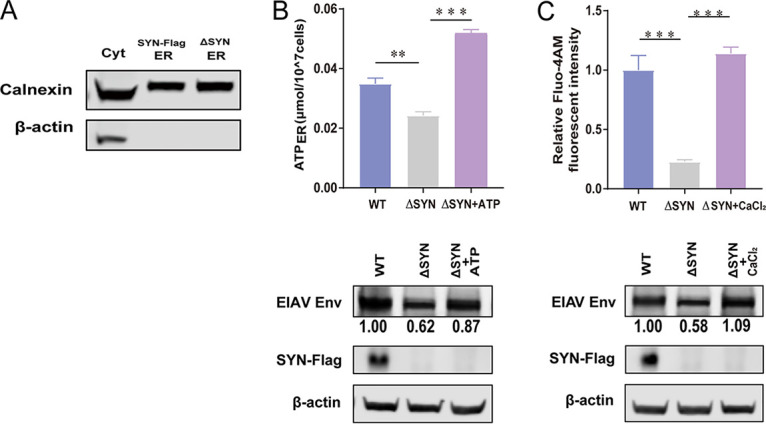
ATP and Ca2+ restore EIAV Env production in SYNJ2BP KO HEK293T cells. (A) Extraction of the same amounts of ER from cells overexpressing SYNJ2BP and KO cells. The ER extraction process is described in Materials and Methods. The lysates from the cytoplasm (Cyt) and the ER were analyzed by using Western blotting to detect the ER internal reference protein calnexin. (B or C) Supplementation of the cell culture with ATP or CaCl2 can increase the levels of ATP or Ca2+ in the ER in SYNJ2BP KO HEK293T cells, as presented by the histogram. The detection of levels of ATP or Ca2+ in the ER is described in Materials and Methods. The data represent the means ± the SEM from three independent experiments (***, P < 0.001; **, P < 0.05). EIAV Env production in WT or SYNJ2BP KO HEK293T cells with or without supplement of ATP or CaCl2 was measured by Western blotting. The results of the densitometry analysis to quantify the ratio of Env to β-actin are shown at the bottom (lane 1 set as 1.0).
SYNJ2BP can decease ER stress caused by lentiviral Env.
In the ER, the proteins rich in disulfide bonds are easily misfolded, which can cause cell responses such as ER stress. Therefore, eukaryotic cells have evolved an effective quality control system to solve this problem. When proteins are misfolded, they are targeted to the ER-associated protein degradation (ERAD) system and are then transported back to the cytoplasm and degraded through the ubiquitin-proteasome system (UPS) (9). Not only misfolded cellular proteins such as T-cell receptor and β-secretase but also misfolded viral proteins such as HIV-1 Env and influenza A virus (IAV) hemagglutinin (HA) can be degraded by ERAD (28, 29). The disruption of MAM has been implicated in increasing ER stress, and MAM is known to regulate the microenvironment in the ER needed for oxidative protein folding to reduce ER stress (30, 31). To validate this, experiments were conducted to determine ER stress level after lentiviral Env protein expression with or without SYNJ2BP coexpression. We found that EIAV Env can also induce increase mRNA and protein expression levels of glucose-regulated protein 78 (GRP78), which is a marker of ER stress, in a dose-dependent manner (Fig. 7A and B). Kifunensine (KIF) is a specific ERAD inhibitor. EIAV Env expression levels increased after treatment with KIF (100 μM) (Fig. 7C), which indicates that EIAV Env can be degraded by ERAD. To verify whether MAM can reduce ER stress induced by either EIAV or HIV Env, SYNJ2BP and EIAV or HIV Env coexpression experiments were carried out. The data showed that when EIAV or HIV Env were expressed alone in cells, the expression levels of GRP78 increased compared to those of the control group; however, when EIAV or HIV Env and SYNJ2BP were coexpressed in cells, the expression levels of GRP78 decreased (Fig. 7D and E). From these results, we concluded that SYNJ2BP is able to promote the formation of MAM to reduce ER stress and thus to increase the production level of EIAV or HIV Env.
FIG 7.
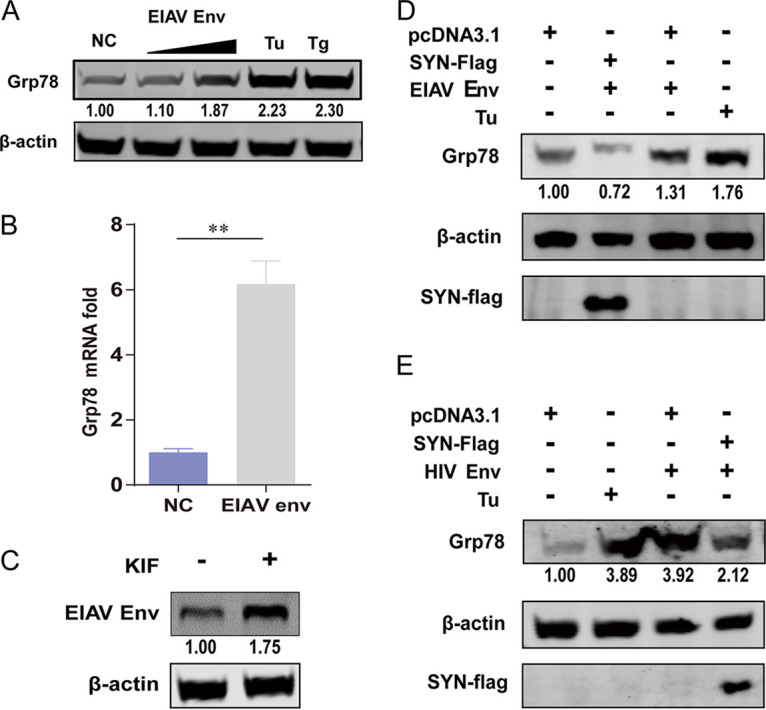
SYNJ2BP reduces ER stress induced by EIAV or HIV Env. (A) EIAV Env can induce increases in protein expression levels of the ER stress marker Grp78 in a dose-dependent manner. HEK293T cells were transfected with either pcDNA3.1-Env (EIAV) plasmids in two doses or an empty plasmid, and Tu or Tg (2 μg/mL) was used as a positive control for ER stress activation. Cell lysates were analyzed by Western blotting to detect the intensity of the Grp78 protein band at 48 hpt. The results of the densitometry analysis to quantify the ratio of Grp78 to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times. (B) EIAV Env can induce increases in mRNA levels of the ER stress marker Grp78. HEK293T cells were transfected with either pcDNA3.1-Env (EIAV) plasmids or an empty plasmid. Total RNA was isolated, and the transcriptional levels of Grp78 were measured by qPCR at 48 hpt. Fold change values were calculated according to the 2–ΔΔCT method using β-actin as an internal reference gene. The data represent the means ± the SEM from three independent experiments (**, P < 0.01). (C) EIAV Env was degraded through the ERAD pathway. KIF restores EIAV Env expression in HEK293T cells. HEK293T cells were transfected with plasmid pcDNA3.1-Env (EIAV) in treatment with KIF (100 μM) or without KIF. Cells were lysed, and EIAV Env expression levels were determined by Western blotting. The results of the densitometry analysis to quantify the ratio of Env to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times. (D) SYNJ2BP overexpression can reduce high expression levels of Grp78 induced by EIAV Env. HEK293T cells were cotransfected with either pcDNA3.1-Flag-SYNJ2BP plasmids or an empty plasmid, and pcDNA3.1-Env (EIAV) or an empty plasmid, and Tu (2 μg/mL) was used as a positive control for ER stress activation. Cell lysates were analyzed using Western blotting to detect the intensity of the Grp78 or SYNJ2BP-Flag protein bands at 48 hpt. The results of the densitometry analysis to quantify the ratio of Grp78 to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times. (E) SYNJ2BP overexpression can reduce high expression levels of Grp78 induced by HIV Env. HEK293T cells were cotransfected with either pcDNA3.1-Flag-SYNJ2BP plasmids or an empty plasmid, and pcDNA3.1-Env-HA (HIV-1) or an empty plasmid, and Tu (2 μg/mL) was used as a positive control for ER stress activation. Cell lysates were analyzed using Western blotting to detect the intensity of the Grp78 or SYNJ2BP-Flag protein bands at 48 hpt. The results of the densitometry analysis to quantify the ratio of Grp78 to β-actin are shown at the bottom (lane 1 set as 1.0). This experiment was performed three times.
DISCUSSION
Lentiviral envelope proteins are glycoproteins that are cleaved by proteases to produce surface (SU) proteins and transmembrane (TM) proteins, which are distributed on the surface of the virus. SU proteins have the function of binding to cell receptors, and TM is involved in the fusion with cell membranes during infection and the fixation on cell membranes during budding (32). However, 20 to 90% of Env in lentivirus-infected cells is known to be degraded due to incorrect folding (12) (Fig. 8). The production mechanism on lentivirus Envs, especially EIAV, is still unclear. In this study, we found that SYNJ2BP can improve production of Env from EIAV or HIV by facilitating the formation of MAM, which can in turn determine virus infection and replication (Fig. 1 and 5).
FIG 8.
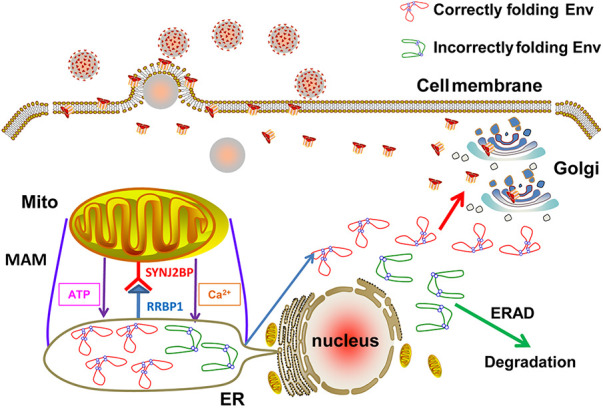
Schematic diagrams demonstrating the role of SYNJ2BP in the regulation of the ER microenvironment via MAM formation to improve production of the lentiviral Env protein. The MAMs formed by the connection of SYNJ2BP and RRBP1 can not only increase the levels of ATP and Ca2+ in the ER, which improves production of the lentiviral Env protein, but also reduce ER stress induced by the lentiviral Env protein.
Like most cellular glycoproteins, viral membrane proteins are typically translated on membrane-bound ribosomes and inserted cotranslationally into the ER in an unfolded form (33). HIV-1 Env protein has an N-terminal hydrophobic signal sequence that is essential for targeting the nascent polypeptide chains to the ER (34). This signal sequence has a number of charged residues in the N region (35), which also cause inefficient cleavage of the signal sequence and may interfere with proper folding of the targeted proteins or lead to aggregation and misfolding (36–38). In addition, the highly specialized environment in the ER plays a more important role that supports rapid and efficient protein production and assembly. In the ER, chaperones and folding enzymes, such as BiP, calnexin, calreticulin, and PDI, assist the folding of HIV-1 gp160 and isolated gp120 (39, 40). An important group of MAM proteins are the ER protein folding chaperones and oxidoreductases. Moreover, the MAM is able to support the production of secretory proteins by promoting the maintenance of millimolar concentrations of free calcium at levels that are >1,000-fold higher than the concentrations of free calcium in the cytosol (7, 41). Depletion of ER calcium stalls oxidative protein folding and prevents the export of secretory proteins from the ER (8, 42). On the other hand, mitochondria may coalesce near the MAM, allowing mitochondria to supply fuel to the ER, one of the major sites of ATP consumption in the cell (25, 43). Protein maturation and production in the ER is energy dependent (27). The inhibition of the mitochondrial electron transport chain with antimycin A led to the induction of an unfolded protein response (UPR) to cause the degradation of newly synthesized proteins, indicating that maintenance of ATP concentrations is a key factor in protein production (44). In our study, we found that SYNJ2BP can facilitate formation of MAM (Fig. 2), leading to increased ATP and Ca2+ concentrations in the ER followed by promotion of EIAV or HIV-1 Env protein production (Fig. 4, 5 and 6). This is the first time that MAM has been found to have an important role in viral membrane protein production (Fig. 8). Previous studies have shown the roles of MAM in cell signaling pathways and cellular responses after viral infection. For instance, the UL37 exon 1 of cytomegaloviruses can target to the MAM via two mitochondrial targeting signals and is able to reduce ER calcium concentrations, possibly by increasing the targeting of GRP75 to the VDAC/IP3R/GRP75 ternary complex (45, 46). The MAM also houses the cytosolic pathogen recognition receptor RIG-1 that triggers an immunity signaling cascade upon viral infection (20).
ER stress is a cellular state in which the folding capacity of the ER is overwhelmed owing to an increase in protein load or disruption in the folding environment. The accumulation of unfolded proteins is detected by transmembrane sensors at the ER surface, which initiate a transduction cascade known as UPR (47). The activated UPR organizes a temporal decrease in protein synthesis together with the expression of a subset of genes involved not only in the folding, maturation, and stabilization of proteins but also in protein degradation via ERAD to reestablish protein homeostasis, also termed proteostasis (48). Previous reports have suggested that HIV-1 Env is targeted to the ERAD pathway for degradation in the human CD4+ T cell line CEM (10, 49). Consistent with this finding, we also found that not only HIV-1 but EIAV Env could be degraded by the ERAD pathway after induction of ER stress in HEK293T cells (Fig. 7C), indicating that the lentiviruses have the same quality control approach in the production of Env proteins. As we known, MAM can regulate the microenvironment in the ER, to allow for oxidative protein folding to reduce ER stress (30, 31). In our study, we also found that the formation of MAM could reduce ER stress induced by HIV or EIAV Env (Fig. 7D and E). This is another mechanism by which MAM promotes lentiviral Env production.
Overall, we have found a new role of MAM, which promotes lentiviral Env production, and we present our preliminarily investigations of the underlying mechanism. Further research into this mechanism is necessary and would uncover other cellular responses to lentivirus infection through the study of dynamic changes and the connection of organelles. This research is beneficial and necessary in order to understand the mechanism by which lentiviruses replicate in cells.
MATERIALS AND METHODS
Cells and viruses.
Peripheral blood was obtained from the horses using a protocol approved by the Ethics Committees of Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SYXK [Hei] 2017-009). Preparations of eMDMs were obtained from equine peripheral blood mononuclear cells (PBMCs) as described previously, with a minor modification (50). Briefly, PBMCs were isolated from 200 to 300 mL of heparinized horse peripheral blood by centrifugation through a HybriMax Histopaque cushion (density = 1.077 g/cm3; Sigma, USA). Isolated PBMCs were washed with RPMI 1640 medium (HyClone, USA) three times and resuspended in RPMI 1640 medium supplemented with 10% horse serum (HyClone). In addition, 104 U/mL penicillin, 104 μg/mL streptomycin, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 0.25 mM sodium HEPES were added into the cultures, all of which were purchased from Gibco Corporation (USA). The cells were then seeded into tissue culture flasks (Corning, USA) at 5 × 106 cells/25 cm2 and incubated at 37°C in 5% CO2 for ~12 h. Nonadherent and loosely adherent cells were removed by mildly shaking the flasks before changing the medium, and the remaining adherent cells were further incubated for 3 days to allow differentiation into eMDMs. Human embryonic kidney (HEK293T) (ATCC CRL-3216) cells were maintained in Dulbecco modified Eagle medium (HyClone) with 10% fetal bovine serum (FBS; Sigma) and 1% penicillin and streptomycin (Gibco) and kept at 37°C in 5% CO2. HEK293T cells that consistently express equine lentivirus receptor 1 (293T-ELR1) on their surfaces were constructed by and kept in our laboratory. This cell line was cultured under the same conditions as the HEK293T cells except that it required 15% FBS. Wild-type pathogenic EIAV strains isolated in China did not replicate well in cultivated cells, including primary host cells, such as donkey monocyte-derived macrophages (dMDMs). EIAVDLV34 is a dMDM-adapted EIAV strain that was derived from the pathogenic EIAV strain EIAVDV117 by 34 passages in dMDMs. This virus strain can replicate well in eMDMs.
Plasmids and chemicals.
To obtain the pcDNA3.1-Flag-SYNJ2BP plasmid, coding sequence (CDS) of human SYNJ2BP deposited in GenBank (NM_018373.3) was cloned using the primer pair 5′-ATAGTTGTAGCCACCATGAACGGAAGAGTGGATTA-3′ and 5′-GTCATCCTTGTAGTCAAGTTGTTGCCGGTATCTCA-3′ and inserted into the vector pcDNA3.1 (Invitrogen, USA) with a Flag tag at the N terminus. EIAVCMV-3-8 is an infectious EIAV clone derived from cell-adapted EIAVFDDV by replacing the U3 region of its 5′ long terminal repeat (LTR) with the cytomegalovirus (CMV) promoter. This clone was constructed as previously described and kept in our laboratory (51). The plasmid plenti-CRISPRv2GFP was constructed by replacing the puro gene plenti-CRISPRv2Puro (Addgene, USA) with the gfp gene using In-fusion HD enzyme (Clontech, USA). pcDNA3.1-Env (EIAV) and pcDNA3.1-rev-HA (EIAV) were constructed by Yin and kept in our laboratory (52). Codon-optimized plasmid VR1012-Gag (EIAV) derived from EIAVFDDV3-8 was kindly provided by Yiming Shao (Chinese Center for Disease control and Prevention). The HIV-1 luciferase reporter proviral vector pNL4-3-lucΔVifΔEnv was a gift from Yong-Hui Zheng (Michigan State University). Codon-optimized plasmid pcDNA3.1-Env-HA (HIV-1) was kindly provided by Huaxin Liao (Jinan University, China). The pcDNA3.1-Rev-HA (HIV-1) plasmid was provided by Jianhua Wang (Institute Pasteur of Shanghai, Chinese Academy of Sciences). VR1012-Gag-HA (HIV-1) was constructed by cloning the gag sequence from pNL4-3-lucΔVifΔEnv and inserting it into the VR1012 plasmid (a gift kindly provided by Xiaofang Yu at First Hospital of Jilin University).
The ER stress-specific inducers thapsigargin (Tg) and tunicamycin (Tu), and the ERAD-specific inhibitor KIF were purchased from Sigma. Tg, Tu, and KIF were dissolved in dimethyl sulfoxide to concentrations of 10 mM, 10 mM, and 2 mg/mL, respectively.
Construction of the SYNJ2BP knockout cell line.
To generate a knockout cell line for the host protein SYNJ2BP, the gRNA design tool (http://crispr.mit.edu/) was used to guide RNA (gRNA) design and off-target prediction according to the whole DNA sequence deposited in GenBank (NM_018373.3) (53). Two guide RNA sequences were selected for SYNJ2BP knockout, which were cloned into the BsmHI site of the vector plenti-CRISPRv2GFP to obtain the recombinant plasmid plenti-CRISPRv2GFP-SYNJ2BP-sgRNA. The sequences of all constructs were confirmed by DNA sequencing. After that, HEK293T cells plated into a six-well plate were transfected with 1 μg of plenti-CRISPRv2GFP-SYNJ2BP-sgRNA and 2 μg of PolyJet (SignaGen Laboratories, USA). Green fluorescent protein (GFP)-positive cells were sorted using fluorescence-activated cell sorting at 48 h posttransfection (hpt) and seeded into 96-well plates to obtain a single-cell-derived colony. The monoclonal knockout cell lines were screened with Western blotting and DNA sequencing.
Production of HIV pseudotyped retrovirus with EIAV Env.
HIV pseudotyped viruses with a luciferase reporter were produced by transfection of viral genome constructs into HEK293T cells, as described previously (54). In brief, cells overexpressing SYNJ2BP or KO HEK293T cells were plated into six-well plates and transfected with pNL4-3-lucΔVifΔEnv and pcDNA3.1-Env (EIAV) using the PolyJet DNA reagent (SignaGen Laboratories) according to the instructions in the manual. At 48 hpt, the cells were resuspended in lysis buffer and subjected to Western blotting, and the culture supernatants were collected to infect 293T-ELR1 cells that allow EIAV infection, followed by measurement of the firefly luciferase activity using a luciferase assay kit (Promega, USA) with a Centro XS LB 960 luminometer (Berthold Technologies, Germany) at 24 h postinfection.
Immunostaining and immunofluorescence microscopy.
WT or SYNJ2BP knockdown eMDMs were prepared for the detection of MAM. The cells were washed with cold phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min at room temperature. The fixed cells were permeabilized with 0.1% Triton X-100 for 15 min before blocking in 5% fat-free dry milk for 2 h and incubated at 4°C overnight using the corresponding primary antibodies in eMDMs, including mouse anti-Tom20 (Abcam, USA) at a 1:500 dilution and rabbit anti-KDEL (Abcam) at a 1:500 dilution, respectively. After three washes, the secondary antibodies coupled to a fluorochrome were added, and samples were incubated for 1 h. These secondary antibodies included Alexa Fluor 488 dye-labeled goat anti-mouse IgG (Thermo Fisher Scientific, USA) at a 1:500 dilution and Alexa Fluor 594 dye-labeled goat anti-rabbit IgG (Thermo Fisher Scientific) at a 1:500 dilution. Samples were then washed three times in PBS. Fluorescence image acquisition was performed with confocal laser scanning microscopy (LSM880; Zeiss, Germany). In addition, MAMs from cells overexpressing SYNJ2BP or KO HEK293T cells were detected using single molecule localization microscopy (SMLM). SMLM provides access to the distribution and interactions of single molecules at the nanoscale, which is especially useful for those that participate in organelle communication. dSTORM is one of most commonly used techniques for SMLM (55). The stained HEK293T cell samples were detected by dSTORM. The sample staining process is the same as for the confocal method described above, except for the antibodies used (rabbit anti-RRBP1 [Abcam] at a 1:200 dilution and Alexa Fluor 568 dye-labeled goat anti-rabbit IgG [Abcam] at a 1:500 dilution). An additional step was also required in which the samples were covered with imaging buffer (PBS [pH 7.4], 50 mM β-mercaptoethanol) to drive the fluorophores into a reversible dark state. The individual fluorophores spontaneously reentered an excitable state. By continuing to excite the samples with a laser, flashes or blinks from individual molecules can be recorded. Single-molecule localization data were acquired by high-resolution light microscopy (ELYRA PS.1; Zeiss).
Duolink proximity ligation in situ.
The Duolink II in situ proximity ligation assay (PLA; Sigma) enables the visualization and quantification of protein interactions (<40 nm) as individual fluorescent dots by microscopy (56). The fixation and permeabilization of eMDMs and HEK293T cells were performed similarly to the immunofluorescence procedure described above. Furthermore, the pair of proteins at the MAM interface were probed with the primary antibodies rabbit anti-Tom20 (Abcam) at a 1:200 dilution and rat anti-KDEL at a 1:200 dilution (Abcam) in eMDM group, and mouse anti-VDAC1 (Proteintech Group, USA) at a 1:200 dilution and rabbit anti-IP3R1 (Abcam) at a 1:200 dilution in HEK293T cells group. The secondary antibodies anti-mouse IgG and anti-rabbit IgG, as well as anti-rat IgG solution that contained PLUS and MINUS PLA probes, were used to react with the corresponding primary antibody according to the manufacturer’s instructions. The proximity ligation and amplification procedures were performed according to the manufacturer’s protocol, as described in previous studies (57). Fluorescence image acquisition was performed with confocal laser scanning microscopy (LSM880; Zeiss). The PLA signals were quantified using BlobFinder software (Olink Biosciences, Sweden) (58).
Electron microscopy imaging.
TEM observation of MAM was carried out as previously described (59). Cells overexpressing SYNJ2BP or KO HEK293T cells were prepared. At 24 hpt, cells were fixed and embedded. Ultrathin sections were observed under a JEM2100 transmission electron microscope (JEOL, Tokyo, Japan) at 200 kV. In the cytoplasm, mitochondria, and ER, the interface between these two organelles was clearly visible.
Transfection and Western blotting.
WT or SYNJ2BP KO HEK293T cells were cultured in six-well plates and transiently transfected with the indicated plasmids using PolyJet DNA reagent (SignaGen Laboratories) according to the manufacturer’s instructions. At 48 hpt, the cells were harvested, lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM EDTA, 1% Triton X-100), and then centrifuged at 10,000 × g for 5 min to remove the cell nuclei. The proteins in the cell lysates were separated using SDS-PAGE and then transferred onto nitrocellulose membranes (Millipore, USA). Membranes were blocked with 5% fat-free dry milk in Tris-buffered saline (20 mM Tris-HCl, 150 mM NaCl) for 2 h at room temperature and then incubated with the indicated primary and secondary antibodies. The mouse anti-Flag, anti-HA, and anti-β actin monoclonal antibodies and rabbit anti-SYNJ2BP polyclonal antibody were purchased from Sigma-Aldrich (USA). Mouse anti-HIV-1-p24 monoclonal antibody and rabbit anti-Grp78 polyclonal antibodies were purchased from Sino Biological Corporation (USA) and Proteintech Group Corporation (USA), respectively. Monoclonal antibody 9H8 against p26 or Gag (EIAV) and polyclonal antibody against EIAV were prepared in our laboratory (60). DyLight 800-labeled goat anti-mouse and DyLight 680-labeled goat anti-rabbit secondary antibodies were purchased from KPL (USA). Bands were analyzed by scanning blots using an Odyssey imaging system (Li-Cor, Lincoln, NE). The relative protein levels were measured with ImageJ on the basis of at least three independent Western blots.
RNA interference.
Equine SYNJ2BP (GenBank number NM_001242552.1) siRNA and negative-control siRNA were designed and synthesized by RiboBio corporation (China). A total of 2.0 × 105 eMDMs were seeded into six-well plates, cultivated for 48 h, and then transfected with equine SYNJ2BP or negative-control siRNA using Lipofectamine RNAiMAX (Invitrogen). Briefly, 100 pmol of siRNA in 100 μL of serum-free Opti-MEM medium (Gibco) and 6 μL of Lipofectamine RNAiMAX in 100 μL of Opti-MEM were mixed, followed by incubation for 5 min at room temperature. The mixtures were then added dropwise to each well. The knockdown efficiency was determined by using real-time qPCR.
Real-time qPCR.
Total RNA was extracted from negative-control and SYNJ2BP siRNA knockdown eMDMs or negative-control and EIAV-Env-overexpressing HEK293T cells using a RNeasy minikit (Qiagen, USA) according to the manufacturer’s instructions and then subjected to reverse transcription using a Prime-Script RT reagent kit with a gDNA Eraser (TaKaRa, Japan). The expression levels of equine SYNJ2BP or human Grp78 mRNAs were quantified using an SYBR green (TaKaRa)-based real-time qPCR analysis on a StrataGene Mx3000P (Agilent, USA), according to the manufacturer’s protocols. Real-time RT-PCR was performed using the equine SYNJ2BP primers 5′-ATGTCTCCAACGACAGTGGCATCTACGT-3′ and 5′-CATCCTGGTGCAGCAGGTTCTTTAAGTC-3′ or the human Grp78 primers 5′-AAGACGGGCAAAGATGTCAGGAA-3′ and 5′-CCGAGTCAGGGTCTCAGAAAAGT-3′. ACTB was used as a housekeeping control to normalize the number of living cells. The relative fold changes in gene expression were determined by the 2–ΔΔCT threshold cycle (CT) method (61).
Viral reverse transcriptase assay.
The RT activity of EIAV in supernatant was quantified using a reverse transcriptase assay colorimetric kit (Roche, Germany) according to the manufacturer’s instructions (62). Briefly, the virus supernatant was mixed with PEG8000 and placed on ice overnight for reaction. The samples were centrifuged at 8,000 × g for 10 min, and the supernatant was discarded. The precipitate was resuspended with lysis buffer and incubated at room temperature for 30 min, followed by addition of the reaction mixture solution and incubation at 37°C for 1 h. The reactants were added to the microplate wells, followed by incubation at 37°C for 1 h. Anti-digoxigenin-peroxidase working liquid was added to each well after five washes, and samples were incubated at 37°C for 1 h. The substrate solution was then added to each well after five washes. The microplates were incubated at room temperature for 10 min. The absorbance of the samples at 405 nm was measured with a microplate reader Elx800 (Bio-Tek, USA). The reverse transcriptase activity of the sample was calculated by drawing a curve according to the optical density of the standard.
Calcium and ATP assay in ER.
Cells overexpressing SYNJ2BP and KO HEK293T cells were seeded separately into six-well plates, and WT cells were used as a control. At 24 hpt, the cells were collected and counted. The ER was extracted using a Minute ER enrichment kit (Invent, USA) according to the kit protocol. Briefly, 2 × 107 cells were collected by low-speed centrifugation (500 × g to 600 × g for 5 min). The pellet was resuspended in 550 μL of buffer A, and the cell suspension was transferred to a filter cartridge and centrifuged at 16,000 × g for 30 s. The filter was discarded, and the pellet was resuspended by vigorously vortexing for 10 s. After centrifugation at 2,000 × g for 5 min, the supernatant was transferred to a fresh 1.5-mL tube. Then, 40 μL of buffer B was added to the supernatant, and the tube was incubated at 4°C for 20 to 30 min. The tube was then centrifuged at 8,000 × g for 10 min, and the supernatant was completely removed. The pellet was resuspended in 400 μL of cold buffer A; 40 μL of buffer C was added, and the tube was vortexed briefly, followed by incubation at room temperature for 10 to 15 min. After centrifugation at 8,000 × g for 5 min, the supernatant was transferred to a fresh 1.5-mL tube. Then, 400 μL of buffer D was added, and the tube was incubated at 4°C for 20 min. The tube was centrifuged at 10,000 × g for 10 min, and the supernatant was removed and discarded. The pellet (the isolated ER fraction) was resuspended in 50 to 200 μL of PBS. To determine the quantity and quality of the extracted ER, the extracts were lysed and detected by Western blotting with the first mouse antibody against calnexin (Abcam) at 1:1,000, which is the molecular chaperone often used as a marker for ER.
ER calcium was detected as described previously (63). The extracted ERs were plated into 96-well plates, with PBS as control. After the samples had been incubated with the fluorescent calcium indicator Mag-Fluo-4 AM (AAT Bioquest, USA) for 25 min, the fluorescence signals were recorded and analyzed using an EnVision multimode plate reader (Perkin-Elmer, USA). Fluorescence signals were then monitored at 516 nm (excitation, 494 nm). The cellular ATP levels were measured using a firefly luciferase-based ATP assay kit (Beyotime Biotechnology, China) based on a fluorescence technique. Then, 40 μL of lysate was added to the ER extracted from the cells in each well of a six-well plate for subsequent assay. Next, 100 μL of ATP detection fluid was added to the detection tube. The tubes were incubated at room temperature for 3 to 5 min. The luciferase content was determined by adding 20 μL of sample or standard to the test tube, followed by rapid mixing, and then the value was read using a Berthold LB960 plate reader (Berthold, Germany) with an integration time of 5 s.
Statistical analysis.
Statistical analysis was performed using Prism version 7.0 (GraphPad, San Diego, CA). Statistical differences between groups were assessed by one-way analysis of variance, followed by a Dunnett’s post hoc test. All experiments were performed independently at least three times. Error bars represent standard deviations (SD) or standard errors of the mean (SEM) in each group, as indicated in the figure legends (NS, not significant; P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grants 31772720, 32170169, and 32172831).
We thank Zhiyuan Wen and Li Huang for helpful discussions.
Contributor Information
Xiaojun Wang, Email: wangxiaojun@caas.cn.
Cheng Du, Email: ducheng@caas.cn.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Leroux C, Cadore JL, Montelaro RC. 2004. Equine infectious anemia virus (EIAV): what has HIV’s country cousin got to tell us? Vet Res 35:485–512. 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- 2.Eisinger RW, Fauci AS. 2018. Ending the HIV/AIDS pandemic. Emerg Infect Dis 24:413–416. 10.3201/eid2403.171797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craigo JK, Montelaro RC. 2011. Equine infectious anemia virus infection and immunity: lessons for aids vaccine development. Future Virol 6:139–142. 10.2217/fvl.10.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook RF, Leroux C, Issel CJ. 2013. Equine infectious anemia and equine infectious anemia virus in 2013: a review. Vet Microbiol 167:181–204. 10.1016/j.vetmic.2013.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 265:10373–10382. 10.1016/S0021-9258(18)86956-3. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887. 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 8.Lodish HF, Kong N. 1990. Perturbation of cellular calcium blocks exit of secretory proteins from the rough endoplasmic reticulum. J Biol Chem 265:10893–10899. 10.1016/S0021-9258(19)38530-8. [DOI] [PubMed] [Google Scholar]
- 9.Meusser B, Hirsch C, Jarosch E, Sommer T. 2005. ERAD: the long road to destruction. Nat Cell Biol 7:766–772. 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 10.Zhou T, Dang Y, Zheng YH. 2014. The mitochondrial translocator protein, TSPO, inhibits HIV-1 envelope glycoprotein biosynthesis via the endoplasmic reticulum-associated protein degradation pathway. J Virol 88:3474–3484. 10.1128/JVI.03286-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA 85:9580–9584. 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fennie C, Lasky LA. 1989. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J Virol 63:639–646. 10.1128/JVI.63.2.639-646.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizzo P, Pozzan T. 2007. Mitochondrion-endoplasmic reticulum choreography: structure and signaling dynamics. Trends Cell Biol 17:511–517. 10.1016/j.tcb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. 2009. MAM: more than just a housekeeper. Trends Cell Biol 19:81–88. 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Carmine Simmen K, Cooper TJ, Thomas G, Simmen T. 2008. The subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell 19:2777–2788. 10.1091/mbc.e07-10-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilady SY, Bui M, Lynes EM, Benson MD, Watts R, Vance JE, Simmen T. 2010. Ero1alpha requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperones 15:619–629. 10.1007/s12192-010-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, Ting AY. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6:e24463. 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebiedzinska M, Szabadkai G, Jones AWE, Duszynski J, Wieckowski MR. 2009. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane, and other subcellular organelles. Int J Biochem Cell Biol 41:1805–1816. 10.1016/j.biocel.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Zhou RB, Yazdi AS, Menu P, Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469:221–225. 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 20.Horner SM, Liu HM, Park HS, Briley J, Gale M. 2011. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA 108:14590–14595. 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das A, Nag S, Mason AB, Barroso MM. 2016. Endosome-mitochondrion interactions are modulated by iron release from transferrin. J Cell Biol 214:831–845. 10.1083/jcb.201602069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belshan M, Harris ME, Shoemaker AE, Hope TJ, Carpenter S. 1998. Biological characterization of Rev variation in equine infectious anemia virus. J Virol 72:4421–4426. 10.1128/JVI.72.5.4421-4426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martarano L, Stephens R, Rice N, Derse D. 1994. Equine infectious anemia virus trans-regulatory protein Rev controls viral mRNA stability, accumulation, and alternative splicing. J Virol 68:3102–3111. 10.1128/JVI.68.5.3102-3111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmen T, Lynes EM, Gesson K, Thomas G. 2010. Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim Biophys Acta 1798:1465–1473. 10.1016/j.bbamem.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. 2003. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18:343–354. 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 26.de Meis L, Arruda AP, Carvalho DP. 2005. Role of sarco/endoplasmic reticulum Ca2+-ATPase in thermogenesis. Biosci Rep 25:181–190. 10.1007/s10540-005-2884-7. [DOI] [PubMed] [Google Scholar]
- 27.Braakman I, Helenius J, Helenius A. 1992. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature 356:260–262. 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 28.Frabutt DA, Zheng YH. 2016. Arms race between enveloped viruses and the host ERAD machinery. Viruses 8:255. 10.3390/v8090255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frabutt DA, Wang B, Riaz S, Schwartz RC, Zheng YH. 2018. Innate sensing of influenza A virus hemagglutinin glycoproteins by the host endoplasmic reticulum (ER) stress pathway triggers a potent antiviral response via ER-associated protein degradation. J Virol 92:e01690-17. 10.1128/JVI.01690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Vliet AR, Verfaillie T, Agostinis P. 2014. New functions of mitochondria associated membranes in cellular signaling. Biochim Biophys Acta 1843:2253–2262. 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Rossi A, Pizzo P, Filadi R. 2019. Calcium, mitochondria and cell metabolism: a functional triangle in bioenergetics. Biochim Biophys Acta Mol Cell Res 1866:1068–1078. 10.1016/j.bbamcr.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doms RW, Lamb RA, Rose JK, Helenius A. 1993. Folding and assembly of viral membrane proteins. Virology 193:545–562. 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 34.Johnson AE. 1997. Protein translocation at the ER membrane: a complex process becomes more so. Trends Cell Biol 7:90–95. 10.1016/S0962-8924(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 35.Pancino G, Ellerbrok H, Sitbon M, Sonigo P. 1994. Conserved framework of envelope glycoproteins among lentiviruses. Curr Top Microbiol Immunol 188:77–105. 10.1007/978-3-642-78536-8_5. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Bergeron JJ, Luo L, Ou WJ, Thomas DY, Kang CY. 1996. Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport. Proc Natl Acad Sci USA 93:9606–9611. 10.1073/pnas.93.18.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn A, Wickner W. 1985. Conserved residues of the leader peptide are essential for cleavage by leader peptidase. J Biol Chem 260:15914–15918. 10.1016/S0021-9258(17)36345-7. [DOI] [PubMed] [Google Scholar]
- 38.Christis C, Lubsen NH, Braakman I. 2008. Protein folding includes oligomerization - examples from the endoplasmic reticulum and cytosol. FEBS J 275:4700–4727. 10.1111/j.1742-4658.2008.06590.x. [DOI] [PubMed] [Google Scholar]
- 39.Otteken A, Earl PL, Moss B. 1996. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J Virol 70:3407–3415. 10.1128/JVI.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Earl PL, Moss B, Doms RW. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol 65:2047–2055. 10.1128/JVI.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert V, De Giorgi F, Massimino ML, Cantini M, Pozzan T. 1998. Direct monitoring of the calcium concentration in the sarcoplasmic and endoplasmic reticulum of skeletal muscle myotubes. J Biol Chem 273:30372–30378. 10.1074/jbc.273.46.30372. [DOI] [PubMed] [Google Scholar]
- 42.Lodish HF, Kong N, Wikstrom L. 1992. Calcium is required for folding of newly made subunits of the asialoglycoprotein receptor within the endoplasmic reticulum. J Biol Chem 267:12753–12760. 10.1016/S0021-9258(18)42340-X. [DOI] [PubMed] [Google Scholar]
- 43.Culic O, Gruwel ML, Schrader J. 1997. Energy turnover of vascular endothelial cells. Am J Physiol 273:C205–C213. 10.1152/ajpcell.1997.273.1.C205. [DOI] [PubMed] [Google Scholar]
- 44.Kuznetsov G, Bush KT, Zhang PL, Nigam SK. 1996. Perturbations in maturation of secretory proteins and their association with endoplasmic reticulum chaperones in a cell culture model for epithelial ischemia. Proc Natl Acad Sci USA 93:8584–8589. 10.1073/pnas.93.16.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bozidis P, Williamson CD, Wong DS, Colberg-Poley AM. 2010. Trafficking of UL37 proteins into mitochondrion-associated membranes during permissive human cytomegalovirus infection. J Virol 84:7898–7903. 10.1128/JVI.00885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson CD, Colberg-Poley AM. 2010. Intracellular sorting signals for sequential trafficking of human cytomegalovirus UL37 proteins to the endoplasmic reticulum and mitochondria. J Virol 84:6400–6409. 10.1128/JVI.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder M, Kaufman RJ. 2005. The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Kaufman RJ. 2016. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529:326–335. 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 49.Zhou T, Frabutt DA, Moremen KW, Zheng YH. 2015. ERManI (Endoplasmic Reticulum class I alpha-Mannosidase) is required for HIV-1 envelope glycoprotein degradation via endoplasmic reticulum-associated protein degradation pathway. J Biol Chem 290:22184–22192. 10.1074/jbc.M115.675207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim WS, Edwards JF, Boyd NK, Payne SL, Ball JM. 2003. Simultaneous quantitation of equine cytokine mRNAs using a multi-probe ribonuclease protection assay. Vet Immunol Immunopathol 91:45–51. 10.1016/s0165-2427(02)00263-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang XF, Bai B, Lin Y, Qi T, Du C, Song M, Wang X. 2019. High-efficiency rescue of equine infectious anemia virus from a CMV-driven infectious clone. Virol Sin 34:725–728. 10.1007/s12250-019-00153-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin X, Hu Z, Gu Q, Wu X, Zheng YH, Wei P, Wang X. 2014. Equine tetherin blocks retrovirus release and its activity is antagonized by equine infectious anemia virus envelope protein. J Virol 88:1259–1270. 10.1128/JVI.03148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li YQ, Fine EJ, Wu XB, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832. +. 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji S, Na L, Ren HL, Wang YJ, Wang XJ. 2018. Equine myxovirus resistance protein 2 restricts lentiviral replication by blocking nuclear uptake of capsid protein. J Virol 92:e00499-18. 10.1128/JVI.00499-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van de Linde S, Loschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc 6:991–1009. 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 56.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 20:473–477. 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 57.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 58.Allalou A, Wahlby C. 2009. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed 94:58–65. 10.1016/j.cmpb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, Zhuang H, Zhang X, Chen H, Li S, Yang Y, Lu Y, Wang J, Zhu R, Zhang L, Sui S, Tan N, Zhao B, Zhang J, Li L, Feng D. 2016. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J 35:1368–1384. 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Z, Chang H, Chu XY, Li S, Wang MY, Wang XJ. 2016. Identification and characterization of a common B-cell epitope on EIAV capsid proteins. Appl Microbiol Biotechnol 100:10531–10542. 10.1007/s00253-016-7817-9. [DOI] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Ma JA, Shi N, Jiang CG, Lin YZ, Wang XF, Wang SA, Lv XL, Zhao LP, Shao YM, Kong XG, Zhou JH, Shen RX. 2011. A proviral derivative from a reference attenuated EIAV vaccine strain failed to elicit protective immunity. Virology 410:96–106. 10.1016/j.virol.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 63.Hu H, Jiang M, Cao Y, Zhang Z, Jiang B, Tian F, Feng J, Dou Y, Gorospe M, Zheng M, Zheng L, Yang Z, Wang W. 2020. HuR regulates phospholamban expression in isoproterenol-induced cardiac remodelling. Cardiovasc Res 116:944–955. 10.1093/cvr/cvz205. [DOI] [PMC free article] [PubMed] [Google Scholar]