ABSTRACT
The bacterial exometabolome consists of a vast array of specialized metabolites, many of which are only produced in response to specific environmental stimuli. For this reason, it is desirable to control the extracellular environment with a defined growth medium composed of pure ingredients. However, complex (undefined) media are expected to support the robust growth of a greater variety of microorganisms than defined media. Here, we investigate the trade-offs inherent to a range of complex and defined solid media for the growth of soil microorganisms, production of specialized metabolites, and detection of these compounds using direct infusion mass spectrometry. We find that complex media support growth of more soil microorganisms, as well as allowing for the detection of more previously discovered natural products as a fraction of total m/z features detected in each sample. However, the use of complex media often caused mass spectrometer injection failures and poor-quality mass spectra, which in some cases resulted in over a quarter of samples being removed from analysis. Defined media, while more limiting in growth, generated higher quality spectra and yielded more m/z features after background subtraction. These results inform future exometabolomic experiments requiring a medium that supports the robust growth of many soil microorganisms.
IMPORTANCE Bacteria are capable of producing and secreting a rich diversity of specialized metabolites. Yet, much of their exometabolome remains hidden due to challenges associated with eliciting specialized metabolite production, labor-intensive sample preparation, and time-consuming analysis techniques. Using our versatile three-dimensional (3D)-printed culturing platform, SubTap, we demonstrate that rapid exometabolomic data collection from a diverse set of environmental bacteria is feasible. We optimized our platform by surveying Streptomyces isolated from soil on a variety of media types to assess viability, degree of specialized metabolite production, and compatibility with downstream LESA-DIMS analysis. Ultimately, this will enable data-rich experimentation, allowing for a better understanding of bacterial exometabolomes.
KEYWORDS: specialized metabolites, natural products, DIMS, LESA-MS, cultivation conditions, exometabolome, Streptomyces
INTRODUCTION
The immense diversity of molecules that bacteria produce makes it challenging to study their secreted specialized metabolites, or exometabolome, in a holistic manner. Exometabolome composition depends on a variety of factors, including nutrient abundance, cultivation conditions, and the presence of competitors (1, 2). In particular, soil bacteria are known to secrete an array of specialized metabolites whose production often depends on environmental stimuli (3). Many studies have focused on the production of individual metabolites, such as specific antibiotic compounds (4). Yet, studying the entire breadth of molecules comprising the exometabolome poses inherent technical and practical difficulties. One of these challenges is the choice of growth medium, which affects both an organism’s ability to grow robustly and produce specialized metabolites. Furthermore, medium presents an auxiliary signal that may interfere with detection of the exometabolome (5, 6). Because it is often only practical to use a single growth medium for experiments, it is critical to choose a medium that balances multiple considerations.
Growth media optimization has largely focused on eliciting or increasing production of targeted compounds (4, 7, 8). Though it is known that increased complexity of media sometimes leads to an increased diversity of specialized metabolites (9), less is known about optimizing media toward this goal. All bacteria require sources of carbon, nitrogen, inorganic phosphate, and trace metals to grow (10). Both the ability to grow and produce specialized metabolites are dictated by how metabolic precursors are processed through the metabolic network. Complex media typically provide bacteria with a richer array of metabolic building blocks, which can support growth and elicit production of a greater abundance and diversity of specialized metabolites by reducing the energy required to synthesize metabolic precursors de novo (11).
Although complex media might be advantageous for specialized metabolite production, it can interfere with the detection of these compounds if the growth matrix is not removed prior to analysis via mass spectrometry. Direct infusion mass spectrometry (DIMS) is a common method used for untargeted metabolomics (12, 13), whereby a sample is analyzed directly by mass spectrometry without a prior separation step (e.g., liquid chromatography). The absence of a separation step may lead to the inability to detect some molecules within a sample due partly to ion competition between the different analytes and the media components present in the sample matrix (i.e., growth medium) (14). The ideal sample matrix for maximizing analyte detection would be low in salts and other nonvolatile compounds, while containing only well-characterized organic components in order to simplify background subtraction in downstream analyses. However, the minimalistic composition of defined media may constrain the growth and production of specialized metabolites by bacteria. This is particularly problematic in microbiome studies when multiple bacteria are required to grow on the same medium.
Since the number of potential growth media is vast, a heuristic procedure must be employed to narrow the possibilities. In this study, we set out to identify a solid medium that would support the robust growth of soil bacteria and allow detection of secreted specialized metabolites with DIMS. We focused on choosing a growth medium for bacteria belonging to the genus Streptomyces, which are well-known for the diversity of specialized metabolites they produce (1, 5, 15, 16). We sought a medium that would both elicit the production of specialized metabolites and facilitate DIMS detection, two potentially competing objectives. To accomplish this task in high-throughput, we relied on the SubTap growth platform we previously developed for DIMS (13). This permitted characterization of trade-offs between complex and defined media in a manner that could be applied to rationalize the selection of growth media for future studies of the exometabolome.
RESULTS AND DISCUSSION
Because the number of possible combinations of media components is endless, we began with a top-down approach. First, we studied many media for their ability to support growth of the most microorganisms in different soil samples. We then narrowed the number of organisms and media to look at the growth of unique bacteria in greater detail. This was followed by comparison of a smaller subset of media and bacteria from the genus Streptomyces to investigate their exometabolome with DIMS.
Some soil microorganisms can grow on minimalistic defined media.
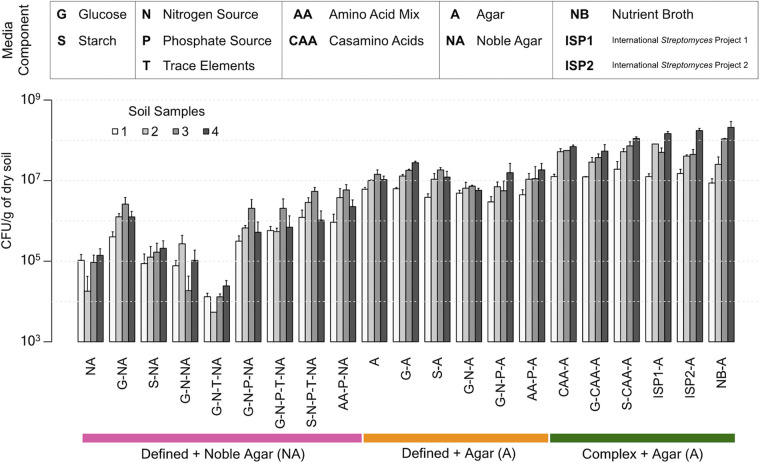
“The great plate count anomaly” is a well-known phenomenon, in which only a small fraction of soil microorganisms can be easily grown on standard laboratory growth media (17–20). Determining the concentration of colony forming units (CFU) in distinct soil samples across different media offers a simple means of comparing media for their general ability to support bacterial growth. To this end, we selected a diverse collection of 21 solid media spanning minimal defined media to complex media (Table 1; Fig. 1). Our defined media were comprised of combinations of a carbon source (glucose or starch, G or S), nitrogen source (ammonium sulfate, N), inorganic phosphate source (phosphate buffer, P), and trace elements (T). Because agar contains impurities that may support growth, we tested both agar and highly purified noble agar as our solidifying agent. We were also interested in how the inclusion of amino acids affected overall growth and included them either as a complex mixture (casamino acids, CAA) or a defined mixture of all amino acids at known proportions (AA).
TABLE 1.
Complex and defined media used in this study
| Media type | Component(s) | Product no. |
|---|---|---|
| NB | 8 g/L nutrient broth | RPI 50-488-846 |
| ISP1 | 5 g/L tryptone 3 g/L yeast extract |
BD 211705 Fisher BP1422 |
| ISP2 | 10 g/L malt extract 4 g/L yeast extract 4 g/L glucose |
BD 218630 Fisher BP1422 Fisher D16 |
| CAA | 2 g/L acid hydrolyzed casein | Fisher BP1424 |
| G-N-P-T | 55.5 mM glucose 10 mM (NH4)2SO4 25 mM potassium phosphate buffer (pH = 7.4) 1 mM MgSO4 100 nM CaCl2 100 nM NaCl 10 nM FeSO4 1 nM ZnSO4 1 nM MnSO4 1 nM CuSO4 |
Fisher D16 Fisher BP212R Fisher BP362 Fisher BP363 Fisher M65 Sigma C4901 Sigma S9888 Sigma 215422 Sigma 221376 Sigma M7634 Fisher BP346 |
| S-N-P-T | 5 g/L starch All compounds inG-N-P-T except glucose |
Sigma S9765 |
| AA-P | 5.6 g/L amino acid mix 25 mM potassium phosphate buffer (pH = 7.4) |
Teknova C0704 Fisher BP362 Fisher BP363 |
FIG 1.
Culturable bacteria from four soil samples grown on various solid media. Complex media with agar supported the growth of approximately one order of magnitude more CFU than defined media. Similarly, agar supported more growth than noble agar as a solidifying agent, presumably due to impurities that are removed during the noble agar purification process. Error bars indicate the standard deviation of three replicates.
Based on the concentration of colony-forming microorganisms per medium, defined media supported the growth of fewer CFU than complex media (Fig. 1) (two-way ANOVA, P = 5e-14). Surprisingly, agar alone only caused a roughly 10-fold reduction in growth compared to any type of complex media (i.e., those above the green horizontal bar in Fig. 1) (two-way ANOVA, P = 0.01), and this was largely unaffected by supplementation with glucose, starch, nitrogen, phosphate, trace elements, or amino acids. In contrast, noble agar resulted in about a 1,000-fold reduction in growth compared with complex media (two-way ANOVA, P = 0.004), which was partly ameliorated by supplementation with other components. Supplementation of agar or noble agar with amino acids did not support the growth of as many CFU as supplementation with CAA (two-way ANOVA, P = 0.005). Taken together, these results are consistent with the expectation that complex media supports the growth of more soil microorganisms. As this experiment was intended to assess growth of any culturable bacteria present in a given soil sample, additional data elucidating the identity of observed colonies was not collected, although it is likely that different media supported the growth of different species.
Complex media support more robust growth than defined media.
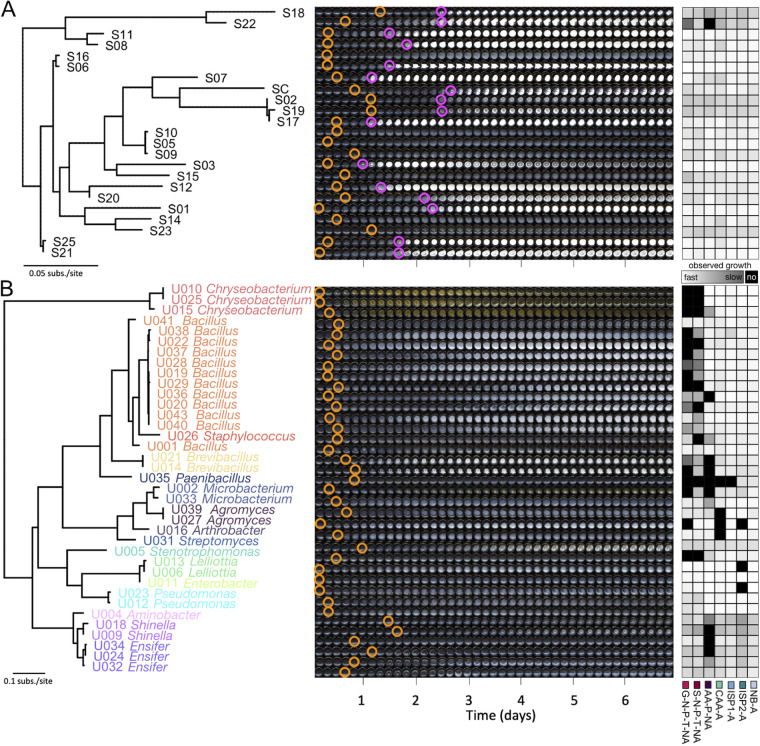
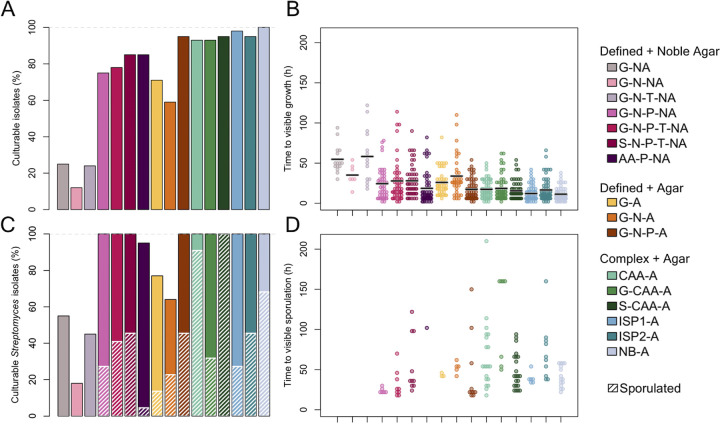
Although colony counts provide an aggregate overview of culturability, individual species of bacteria are not present at equal proportions in a single soil sample, and domination of a few key community members likely drives aggregate trends. We, therefore, isolated a group of genetically distinct bacteria from a single soil sample (Fig. 2) and compared their individual growth characteristics on six complex and 10 defined media types. As shown in Fig. 3, nearly all of the isolates grew in complex media (93% to 100%; Fig. 3A), and growth was observable within 24 h for the majority of isolates (mean time to visible growth of 15 h; Fig. 3B). Defined media with agar supported less growth (59% to 95%) and slower growth (mean time to visible growth of 25 h) than complex media. As expected, defined media with noble agar supported the least growth (7% to 50%) and slowest growth (mean time to visible growth of 29 h). Overall, these trends mirrored that of the original soil samples with the exception of inorganic phosphate addition, which greatly improved the fraction of bacterial isolates that grew and decreased the time to visible growth.
FIG 2.
Phylogenetic trees, growth tracking images and speed of observed growth of bacterial isolates used in this study. (A) Maximum likelihood tree constructed from the rpoB gene of Streptomyces strains. (B) Maximum likelihood tree constructed from the 16S rRNA gene of all other soil isolates, colored based on taxonomic classification (genus level). The center of each panel shows time lapse well images of growth on NB-A at 28°C. Brightness and contrast of all images were increased uniformly by 40% to make growth and sporulation more obvious. Orange circles indicate first visible growth; magenta circles indicate first visible sporulation (when present). The right side of each panel depicts speed of observed growth on several media types used in this study.
FIG 3.
Characteristics of 59 soil bacterial isolates (including 22 Streptomyces isolates) grown on various solid media. (A) Complex media generally supported more growth than defined media. The inclusion of an inorganic phosphate source assisted growth in defined media. Note, all isolates were required to grow on NB-A for inclusion in the experiment. (B) Of the strains that grew (points), complex media enabled faster time to visible growth than defined media. Black bars indicate median time to visible growth. (C) Of the 22 Streptomyces isolates, only a fraction that grew also sporulated within 10 days, with the exception of S-CAA-A where all isolates both grew and sporulated. Crosshatching indicates percentage of Streptomyces isolates that sporulated. (D) Time of first visible sporulation for Streptomyces isolates varied considerably from strain-to-strain.
A substantial fraction (22 of 59) of our soil isolate collection was originally isolated on media designed to select for Actinomycetes, including Streptomyces species. Specialized metabolite production among Streptomyces is often synchronized with the sporulation phase of their life cycle (5). Sporulation causes a morphological change that can be identified during growth (Fig. 2A). Overall, complex media supported faster visibly identifiable sporulation (Fig. 3C and D). While CAA alone supported a greater number of isolates with visibly identifiable sporulation (91%), amino acid defined medium did not (5%). Addition of glucose to CAA greatly suppressed sporulation (from 91% to 32%), probably because sporulation is often triggered by starvation (5, 21, 22). These results indicated an overall advantage of complex media for growth and sporulation of Streptomyces.
Media differ considerably in their utility for mass spectrometry.
Injection of extracts from complex media into mass spectrometers may lead to decreased detection of microbial compounds of interest due to interference from medium components and signal suppression by salts (23). Therefore, the “ideal” media for DIMS would be the absence of media, which could only be approximated through time-consuming separation procedures (e.g., liquid chromatography) that introduce their own analytical biases. To circumvent these downsides, we sought a medium that would minimize interference in liquid extraction surface analysis (LESA)-based DIMS experiments. We selected a subset of seven media to test based on the results of our growth experiments, including three defined and four complex media. A visual summary of observed growth for this subset of media can be found on the right side of Fig. 2. Streptomyces from our isolate collection were cultured on each medium in up to four replicates using the SubTap platform that we previously developed for characterizing the exometabolome in high-throughput (see Fig. S1) (13). Mass spectra were acquired in positive ion mode using the spectral stitching method (24) that merges overlapping selected ion monitoring (SIM) windows for improved resolution and sensitivity.
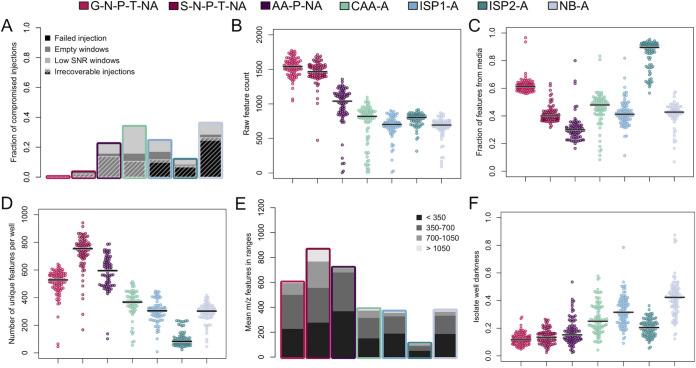
First, we assessed the overall quality of data collected on each type of medium. This included flagging injection failures and identifying spectra that contained mass-to-charge (m/z) windows with missing or poor-quality data. Electrospray destabilization prior to mass spectrometry data acquisition was the primary culprit of injection failures. Notably, the number of mass spectrometer injections that passed each of our quality filters differed considerably from medium-to-medium. Three of the four complex media had high rates of injection failure (6% to 24%), while CAA-A and all three defined media had no failed injections (Fig. 4A). For injections that were successful, we identified any spectra that contained SIM windows where no m/z features were recorded (e.g., “empty windows”), as well as windows where the total signal-to-noise ratio was less than 1% of mean window signal-to-noise ratio in a given plate (i.e., low SNR windows). Complex media suffered from a greater number of injections that contained compromised windows (5% to 34%) than defined media (0% to 22%). Of the media tested, G-N-P-T-NA and S-N-P-T-NA had the fewest injections flagged for poor quality data (0% to 3%). Finally, although many injections resulted in spectra with empty or low signal-to-noise ratio windows, a subset of these had good quality data in the rest of the spectrum. For this reason, a successful injection was removed from subsequent analysis only if it had fewer than 500 total m/z features (Fig. 4A). This quality filtering approach generally resulted in removal of a greater fraction of injections from complex media (7% to 26%) than defined media (0% to 14%). The main exception was AA-P-NA defined medium, which performed similarly to the complex media that were tested in terms of number of injections that were removed from analysis. We suspect that the relatively high total amino acid concentration in this medium (nearly 50 mM), which was prepared according to the manufacturer’s instructions, contributed to the high rate of compromised injections. It should be noted that the number of injections per isolate used in this study (four injections) surpassed the number required to have a 99% chance of observing at least one successful injection per isolate on each media type, assuming independence among injections (≥1 for G-N-T-P-NA; ≥2 for S-N-T-P-NA and ISP2-A; ≥3 for AA-P-NA, CAA-A, ISP1-A; and ≥ 4 for NB-A).
FIG 4.
Summary of LESA-MS analysis of Streptomyces grown on complex and defined media. (A) Compromised injections occurred frequently with some media. Crosshatching indicates the fraction of injections that were removed from further analysis. (B) Prior to background subtraction, defined media have more detectable features than complex media. Each point represents a single well. (C) Media-related features comprised a large fraction of total m/z features detected in some media. (D) After removing features attributable to the media (4C) and correcting for multiple adducts, defined media contained more m/z features than complex media. (E) Most wells had few features detected above 700 m/z, with the exception of S-N-P-T-NA. (F) Complex media generally resulted in darker SubTap analysis plate wells than defined media. Horizontal black bars indicate median values.
Following our quality filtering step, the raw number of m/z features that were detected in each well were tallied. All three defined media tested produced spectra that contained a greater number of m/z features than complex media postfiltering (Fig. 4B). Processing of our data included a background subtraction step to remove any peaks present in the medium alone. For each plate, an m/z feature was subtracted if it was observed in any of the negative-control (media only) wells. The fraction of m/z features attributable to media background varied greatly from medium-to-medium and well-to-well (Fig. 4C). In particular, ISP2-A medium was responsible for the majority (90%) of m/z features in each of its wells, implying relatively few bacteria-produced compounds were detected on this medium. In contrast, S-N-P-T-NA had a large number of raw m/z features (Fig. 4B) and a relatively low fraction of features attributable to the medium (Fig. 4C).
To accurately enumerate unique specialized metabolites produced by our isolates, we collapsed common adducts (H+, Na+, K+) in each spectrum after background subtraction. The greatest number of m/z features remained in wells belonging to defined media (Fig. 4D). This was especially true for S-N-P-T-NA wells, which often had more than twice as many m/z features as complex media wells after adduct collapsing and background subtraction. Most of the detected m/z features were in the range of 70 to 700 m/z (Fig. 4E). S-N-P-T-NA had more features above 700 m/z relative to the other media types (one-sided Mann-Whitney test, p-adj = 6e-7). Frequency of detection of high m/z features has been previously used as a surrogate for specialized metabolite production in other studies because some natural products have high molecular weights (9).
Media differ considerably in selectivity for Streptomyces natural products.
We sought a medium that would permit both production and detection of secreted specialized metabolites. Although S-N-P-T-NA had the most m/z features remaining after background subtraction (Fig. 4D), it was unknown whether these features were enriched for specialized metabolites. Because many natural products are pigmented (25), measuring well darkness may provide an orthogonal metric to compare specialized metabolite production. Thus, we measured the darkness of wells in the SubTap analysis plates (Fig. S1) as a proxy for specialized metabolite production. It is worth emphasizing that the SubTap platform physically separates biomass from specialized metabolites because it contains a 0.2-μm membrane between the context plate, where bacterial growth occurs, and the analysis plate, where secreted specialized metabolites have diffused during growth. After background brightness correction for each medium by itself, complex media resulted in substantially darker substrates than defined media (Fig. 4F). If we assume well darkness is related to increased specialized metabolite production, this presents a potential trade-off between the merits of complex media for specialized metabolite production and the benefits of defined media for mass spectrometry. However, this assumption may be incorrect because well darkness could be driven by a subset of compounds that are pigmented and does not distinguish diversity of compounds from abundance of production. This is consistent with the observation that although S-N-T-P-NA does not have dark wells, it has the greatest number of unique m/z features observed via MS—highlighting an advantage of using mass spectrometry for screening production of specialized metabolites.
To further delineate this potential trade-off, we used the Natural Products Atlas (26) to obtain a list of 4,830 known compounds produced by members of the genus Streptomyces, 3,400 of which had unique masses in our mass range. We searched for the exact masses of their H+, Na+, and K+ pseudomolecular adducts (z = 1) in each well across our media. Because MS1 data are insufficient for confident identifications (27), we required the presence of multiple adducts of a compound to increase the confidence of positive matches in our search results. We reasoned that media associated with exometabolomes containing a greater fraction of known compounds better balance the trade-off between microbial growth and spectrum quality in mass spectrometry.
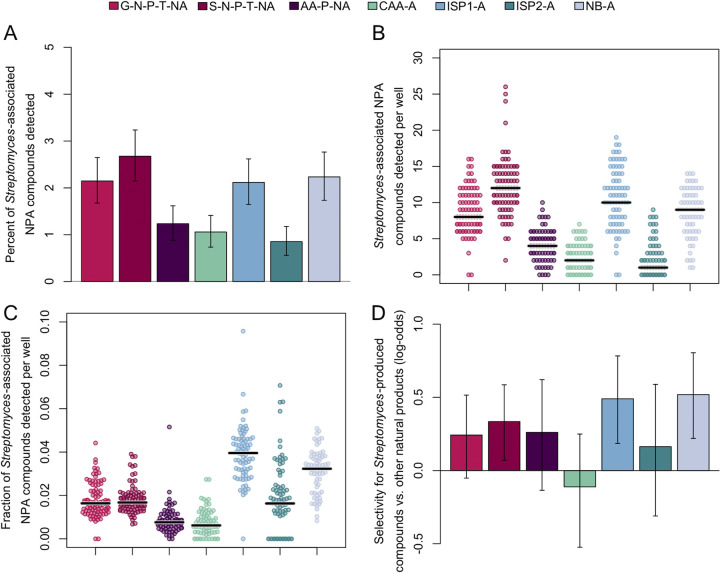
Based on fraction of known Streptomyces-produced compounds identified across all samples (Fig. 5A), S-N-P-T-NA outperformed the other media with 2.7%, followed by NB-A with 2.2%, ISP1-A and G-N-P-T-NA with 2.1%, although these differences were not statistically significant. AA-P-NA and CAA-A both performed poorly with 1.2% and 1.1% identified, respectively, while ISP2-A only matched 0.9% of the Streptomyces-produced compounds in the Natural Products Atlas. Although this measure suggests that there is a greater diversity of previously discovered specialized metabolites produced on S-N-P-T-NA, more wells were excluded from analysis on complex media (especially NB-A), inherently limiting the possible search space. On a well-by-well basis, the highest number of the Natural Products Atlas matches were detected on S-N-P-T-NA (Fig. 5B) (one-sided Mann-Whitney test, p-adj = 0.045). However, as a fraction of total observed m/z features per well, ISP1-A performed significantly better than any of the other media types (Fig. 5C) (one-sided Mann-Whitney test, p-adj = 2e-4). Finally, we quantified the selectivity for known compounds produced by Streptomyces over compounds associated with all other organisms in the Natural Products Atlas (Fig. 5D). Within each media type, only S-N-P-T-NA, ISP1-A, and NB-A displayed selectivity for Streptomyces-produced compounds that was statistically significant (p-adj = 0.049, 0.007, and 0.003, respectively). Taken together, our results suggest that some complex media elicit greater specialized metabolite production but some defined media are preferable for DIMS.
FIG 5.
Detection of compounds found in Natural Products Atlas (NPA), by media type. (A) S-N-P-T-NA, NB-A, G-N-P-T-NA, and ISP1-A had the highest coverage of Streptomyces-produced compounds found in the Natural Products Atlas detected in each plate, as well as (B) the highest number of these compounds detected per well. (C) ISP1-A and NB-A had the highest fraction of Streptomyces-produced compounds detected in each well, as a function of total number of m/z features detected per well (Fig. 4D). Horizontal black bars indicate median values. (D) ISP1-A and NB-A displayed the greatest selectivity for matching Streptomyces-produced compounds over other compounds found in the Natural Products Atlas. Error bars in (A) and (D) depict the 95% confidence interval estimated with bootstrapping.
It was previously shown that phylogenetic distance is correlated with the metabolome (28). Thus, to validate our Natural Products Atlas results, we sought to link our specialized metabolite data with the genetic relatedness of our Streptomyces isolates. Many matches to the Natural Products Atlas displayed a clustering of presence/absence patterns on a maximum likelihood phylogenetic tree constructed from the rpoB gene (Fig. S3A). We quantified the overall degree of clustering by constructing trees from the observed presence/absence pattern on each medium and quantifying the topological distance to the phylogenetic tree. Media with higher selectivity for Streptomyces-produced compounds also tended to display lower tree distances (Fig. S3B), corroborating our Natural Products Atlas search results.
Balancing trade-offs in exometabolomics.
In this study, we characterized the effect of media on variables of importance for exometabolomics. In general, we found that (i) complex media support better growth of soil microorganisms than defined media, (ii) complex media result in a greater number of failed injections and poorer quality mass spectra than defined media, (iii) complex media yield fewer m/z features after background subtraction than defined media, and (iv) complex media likely elicit greater specialized metabolite production than defined media. These findings present a trade-off between microbial growth and detection of secreted specialized metabolites.
Our results have a number of limitations. First, we did not vary the levels of media components, nor did we test many other combinations of media ingredients that are possible. Second, we only analyzed growth and sporulation over 10 days for practical reasons, although we suspect the results would not change considerably because most growth was observed long before this time point. Third, we used a single extraction solvent (70% methanol/0.1% formic acid) for our LESA-MS method, which likely would not have extracted all possible compounds present in our SubTap plates with equal probability. Alternative extraction solvents were not studied systematically. Fourth, we only analyzed the replicates that passed our quality filters, which effectively incorporated failed injections indirectly into our measures of observed m/z features because failed wells contain no features. Fifth, we used compounds in the Natural Products Atlas as a proxy for specialized metabolite detection. It is possible that these compounds have a bias associated with their inclusion in the Natural Products Atlas, such as being more likely to be produced on complex media. Similarly, it is possible that compounds in the Natural Products Atlas marked as originating from non-Streptomyces can be produced by members of the Streptomyces and should have been counted as such in the calculation of selectivity. In addition, almost all of our Streptomyces were absent from the Natural Products Atlas, so we have no prior evidence they can produce the Streptomyces compounds in the Natural Products Atlas. Lastly, we did not explore the use of negative ion mode detection in our mass spectral analyses, and it was assumed that all compounds present in the Natural Products Atlas were capable of forming positively charged pseudomolecular adducts for subsequent detection in positive ion mode.
SubTap paired with LESA-DIMS analysis was designed for efficiency and affordability, allowing for the identification of patterns of compound production by comparing across many samples. While it is true that many of the issues that were encountered may have been overcome by alternative sample preparation or pre-mass analysis chromatographic steps, our aim was to develop a workflow that would enable low-cost, high-throughput, first-pass screening with direct infusion. The number of injection failures and poor-quality spectra collected from wells with complex media partly preclude their use for high-throughput DIMS data acquisition. This resulted in over a quarter of the wells from NB-A being removed from analysis. Therefore, if an experimental design prohibits the inclusion of a high number (n ≥ 4) of replicate wells on a plate, complex media may not be a viable option. On the other hand, defined media limit the number of microorganisms that can be cultured and likely the array of compounds they can produce. For organisms that are able to grow, however, as little as a single well is necessary to ensure successful data collection, enabling high-throughput screening experiments. As it is often only practical to use a single medium type in an experiment, these tradeoffs should be kept in mind when selecting a medium. If injection failures are a minimal concern, complex media that elicit specialized metabolite production (e.g., ISP1-A and NB-A) performed well. We prefer S-N-P-T-NA to avoid injection failures and obtain the greatest diversity of m/z features, although S-N-P-T-NA did not support the growth of some soil bacteria.
MATERIALS AND METHODS
Media composition.
Recipes for rich and defined media used throughout our experiments can be found in Table 1. To better understand the contribution of impurities present in agar, media types were combined with either agar (Fisher BP1423) or noble agar (Research Products International 50-488-478) at the following concentrations: 0.5% for SubTap experiments, 1.5% for isolate selection, and 2% for all other experiments. All media were used as-is, without adjusting pH, unless noted otherwise.
Soil communities and individual soil isolates.
Soil bacterial communities originated from four soil samples collected from locations at latitude and longitude coordinates <40.437686, −79.908732> (#1), <40.413835, −79.917433> (#2), <40.450958, −79.952528> (#3), and <40.431609, −79.963394> (#4). To create frozen soil community stocks, 1 g of each soil sample was mixed with 50 mL of ultrapure water and shaken with glass beads for 30 min. After filtering out coarse soil particulates using a paper filter, 20 mL of this solution was mixed with 30 mL of sterile glycerol for a final glycerol concentration of 60%. Aliquots were frozen at −80°C until use. Each aliquot was only thawed a single time, immediately before use, to standardize any loss in viability associated with freezing or thawing. Soil samples from each location were weighed after drying at room temperature for 7 days to determine dry weight.
Individual soil bacteria were isolated from the soil sample collected at location #4. Using a sterile pipette tip, 500 mg of soil was removed from the center bottom of the collected soil plug. The sample was serially diluted by 10-fold increments to 10−5, and 100 μL of each dilution was plated in triplicate on Luria-Bertani agar, nutrient agar with 0.5% peptone, yeast extract agar, malt extract agar, and EDM2 (29) (Table 2). We added 50 mg/L cycloheximide to all media to suppress fungal growth. All media were prepared with and without soil extract by replacing half of the water volume by an equivalent volume of soil extract, resulting in 10 total media. Soil extract was prepared by mixing 1 kg of soil from location #4 with 2 L of water. The resulting slurry was coarse filtered to remove solids, followed by passage through a 0.2-μm filter to remove fine particulates and autoclaving to ensure sterility.
TABLE 2.
Media used for initial isolation of the bacteria used in this study
| Media type | Component(s) | Product no. |
|---|---|---|
| LBA | 25 g/L Luria-Bertani broth | Fisher BP1426 |
| NA-0.5% peptone | 8 g/L nutrient broth 5 g/L peptone |
RPI 50-488-846 Fisher BP1420 |
| YEA | 3 g/L yeast extract | Fisher BP1422 |
| MEA | 30 g/L malt extract | BD 218630 |
| EDM2, pH adjusted to 7.0 using NaOH | 1 g/L starch 15 mM KH2PO4 35 mM NaCl 0.5 mM MgSO4 20 mM KNO3 |
Sigma S9765 Fisher BP362 Sigma S9888 Fisher M65 Sigma 221295 |
Soil dilutions (10−2 to 10−6 final dilution) were spread on individual agar plates of each media type in triplicate and incubated at 28°C for 7 to 14 days. Individual colonies that appeared unique based on morphological characteristics were picked with a sterile toothpick and pinned onto wells of a black 96-well plate prepared with their original growth medium. Each plate was inverted on an Epson Perfection V550 scanner, secured with packing tape to mitigate desiccation, and scanned every 4 h for 7 to 14 days at 28°C. Images were processed programmatically by clustering colonies based on time to visible growth, growth rate, pigmentation, opacity, and size. After visual inspection, a representative colony from each cluster was subsequently restreaked on its respective agar type and incubated at 28°C for 7 to 14 days. This process resulted in a set of isolates that appeared morphologically distinct, but DNA sequencing was still required to confirm uniqueness.
DNA extraction and sequencing.
To lyse the cells, individual colonies were suspended in 100 μL of Tris-EDTA buffer and sonicated for 1 min at 100% amplitude with a QSonica Model 505 Sonicator with cup horn. Each suspension was centrifuged for 1 min at 14,000 r.c.f. and supernatant containing the genomic DNA was collected. To confirm uniqueness by sequencing, quantitative PCR (qPCR) was performed using primers internal to the rpoB gene (for Streptomyces isolates grown on EDM2: rpoB-F: 5′-CTCGACATCTACCGCAAGCT and rpoB-R: 5′-GACACCATCTGGCGCG) (29) or 16S rRNA gene (27F: 5′-AGAGTTTGATYMTGGCTCA and 1492R: 5′-GGTTACCTTGTTACGACTT). A total of 0.4 μL of each (10 μM) primer, 5.0 μL iTaq Supermix, 1 μL of DNA template, and 3.2 μL of sterile water were used for each reaction. Cycling conditions on an Applied Biosystems StepOnePlus real-time PCR system began with an initial denaturation of 98°C for 2 min, followed by 40 cycles of 98°C for 15 s, 64°C for 30 s, and 80°C for 30 s, with a final melt stage at 60°C to 95°C with 10-s steps at a ramp rate of 0.3°C per step. Amplicons were Sanger sequenced and chromatograms were reviewed for quality. The isolates’ sequences were aligned to create maximum likelihood phylogenetic trees using the R package DECIPHER (30). Classification of 16S rRNA sequences was carried out with IDTAXA (31) using the SILVA training set at 60% confidence.
Growth profiling of soil communities and isolates.
To test soil community growth on different media, each frozen aliquot was diluted 10-fold six times in 60% glycerol with 30 μL of each dilution plated in triplicate. All plates contained 50 μg/mL cycloheximide to prevent fungal growth. Plates were counted after incubating for 7 days at 28°C. For each sample/media combination, we counted the dilution with the fewest countable colonies (generally, 3 < x < 30 colonies) to avoid possible growth inhibition from colonies in close proximity and interfering spreading phenotypes that were prevalent at higher concentrations.
To profile the growth of individual soil isolates, each isolate was spotted onto an opaque (black) 96-well plate containing a single solid medium type. Each plate was inverted on an Epson Perfection V550 scanner, secured with packing tape to minimize desiccation, and full-color scans were captured every 4 h for 10 days at 28°C to monitor growth and sporulation. In total, 59 isolates were profiled (22 Streptomyces and 37 other). Though these bacteria were originally isolated from soil on different media (Table 2), at a minimum, all isolates were required to grow on NB medium to be included. Fig. 2 was generated by cropping each scanned time point and arranging them by well to create movie strips depicting how each well changed over time.
Cultivation for mass spectrometry experiments.
The 22 Streptomyces isolates were tested on a subset of media for analysis of specialized metabolite production and detection. Using the SubTap platform (13), each isolate was grown in a single well containing 120 μL media with 0.5% agar or noble agar on the culture plate, separated by a 0.2-μm polycarbonate track-etched (PCTE) membrane (GVS Group) from the analysis plate containing 30 μL of 0.5% noble agar without medium. Each isolate was grown in quadruplicate on every medium, except for S01 which was only grown in triplicate due to plate space limitations. S. coelicolor was included in the Streptomyces collection as a control strain. After 7 days of growth at 28°C, the top culture plate containing bacteria was removed and bottom analysis plate dehydrated, full-color scans were obtained using an Epson Perfection V550 scanner, and frozen until time of analysis. Analysis plate images were analyzed for average well color intensity (defined as the mean of red, blue and green channels) over a 10 pixel × 10 pixel area centered in each well.
DIMS data acquisition.
Data acquisition was performed using a Velos Pro Orbitrap mass spectrometer (Thermo Fisher Scientific) paired with a Triversa Nanomate robot (Advion) programmed using ChipSoftX to perform liquid extraction surface analysis (LESA) direct infusion experiments with the SubTap platform. With the analysis plate cooled at 8°C, each well was extracted with 70% methanol/0.1% formic acid (aspirate 20 μL solvent, dispense 15 μL on the well, hold for 4 s, aspirate 8 μL). The extract was injected into the mass spectrometer using a nanoESI chip (Advion). Mass spectra were collected in positive ion mode only. Quality control (QC) injections containing tune mix (Agilent G1969-85000) were collected every 10 injections.
Mass spectrometry data were acquired as several overlapping mass-to-charge (m/z) windows that are “stitched” together to create a complete mass spectrum (24). This method of data collection increases the dynamic range and detection sensitivity compared with collecting DIMS data as a single (wide) m/z window. Data were collected as 200 m/z SIM (selected ion monitoring) windows that overlap by 30 m/z. To allow for nanoelectrospray stabilization, data acquisition started after 45 s of sample infusion into the MS. The parameters used were 100 K mass resolution at 200 m/z, an overall mass range of 70 to 2,000 m/z with 27 total m/z windows, 15 s per m/z window acquisition time, and an AGC target of 106. The acquisition time per injection was approximately 6 min.
MS data processing and analysis.
For each plate, injection failures were manually removed and raw data files were processed using DIMSpy v1.3.0 (32) with nondefault parameters in process_scans (min_scans = 1, snr_thres = 1.0, ppm = 3.0, ringing_thresh = 0.04, min_fraction = 0.25, rsd_thres = none) and align_samples (ppm = 3.0). This resulted in a list of m/z features by signal-to-noise ratio. Further postprocessing and data analysis was performed using a custom R script available on GitHub (https://github.com/digitalwright/ExometabolomicsGrowthMedia). This included truncating the m/z range to 70 to 1,400, using QC peaks to calibrate spectra, removal of 13C isotopic peaks, removal of any features that do not occur in a plate at least once with a signal-to-noise ratio of at least 10, removal of any injections that contained low total signal-to-noise ratio windows with less than 500 m/z features, removal of features associated with polyethylene glycol, and removal of peaks associated with media or QC components. All statistical analyses were carried out in R (v3.6) (33). Where noted, P-values were adjusted for multiple comparisons using the Bonferroni correction.
Comparison with compounds in Natural Products Atlas.
The Natural Products Atlas (26) (v2020_06) was downloaded from https://www.npatlas.org/. For investigating compounds known to be produced by Streptomyces (i.e., the “Streptomyces set”), all (4,380) compounds associated with the genus Streptomyces were selected and further reduced to only unique masses within the relevant m/z range (70 to 1,400), resulting in 3,400 unique exact masses. To count as a match, all spectra were searched for the occurrence of at least two of three possible adducts (H+, Na+, K+) within a 7-ppm tolerance for each compound in the set.
To better understand selectivity for compounds produced by Streptomyces, the 8,725 compounds from the Natural Products Atlas that were not associated with and not identical to the exact masses of compounds known to be produced by Streptomyces were selected (i.e., the “non-Streptomyces set”) and searched in the same manner as the Streptomyces set. Confidence intervals (95%) were estimated through bootstrap replication by drawing 107 random (paired) samples of the Streptomyces and non-Streptomyces sets from a Poisson distribution with the expected value for each medium determined in our experiment. Selectivity (log-odds) was then defined as:
The frequency of matches to Streptomyces-produced compounds found in the Natural Products Atlas, by media type, can be found in Fig. S2. To investigate how well the Natural Products Atlas compound hits found in each media type reflect the genetic relatedness of our Streptomyces isolates, an unweighted pair group method with arithmetic mean (UPGMA) tree was constructed for each media using a binary presence/absence matrix of Natural Products Atlas hits. This tree was compared with the rpoB tree (Fig. 2A; Fig. S3A), and clustering information distance between the trees was calculated using the TreeDist R package (34) (Fig. S3B). A graphical representation of the patterns of production of four compounds in the Natural Products Atlas is also shown in Fig. S3A. Further information about the compounds from the Natural Products Atlas putatively annotated (level 3) (27) in Fig. S2 and S3 can be found in Table S1.
Data availability.
Sequences are available from GenBank under the accession numbers MN177030-MN177054, MN186620-MN186656, and OK033102-OK033103.
AKNOWLEDGMENTS
Research reported in this publication was supported by the National Institutes of Health under award number 1DP2AI145058-01 to E.S.W. T.J.D. was supported by National Library of Medicine T15 at the National Institutes of Health (grant number T15LM007059). A portion of this research was performed using the Environmental Molecular Sciences Laboratory (grid.436923.9), a DOE Office of Science User Facility sponsored by the Biological and Environmental Research program, which is located at Pacific Northwest National Laboratory.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Erik S. Wright, Email: eswright@pitt.edu.
Pablo Ivan Nikel, Novo Nordisk Foundation Center for Biosustainability.
REFERENCES
- 1.Bode HB, Bethe B, Hofs R, Zeeck A. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3:619–627. . [DOI] [PubMed] [Google Scholar]
- 2.Pan R, Bai X, Chen J, Zhang H, Wang H. 2019. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: a literature Review. Front Microbiol 10:294. 10.3389/fmicb.2019.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behie SW, Bonet B, Zacharia VM, McClung DJ, Traxler MF. 2016. Molecules to ecosystems: actinomycete natural products in situ. Front Microbiol 7:2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntyre JJ, Bull AT, Bunch AW. 2000. Vancomycin production in batch and continuous culture. Biotechnol Bioeng 49:412–420. . [DOI] [PubMed] [Google Scholar]
- 5.van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333. 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 6.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. 2012. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109:E1743–E1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rateb ME, Yu Z, Yan Y, Yang D, Huang T, Vodanovic-Jankovic S, Kron MA, Shen B. 2014. Medium optimization of Streptomyces sp. 17944 for tirandamycin B production and isolation and structural elucidation of tirandamycins H, I and J. J Antibiot 67:127–132. 10.1038/ja.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim AA, El-Housseiny GS, Aboshanab KM, Yassien MA, Hassouna NA. 2019. Paromomycin production from Streptomyces rimosus NRRL 2455: statistical optimization and new synergistic antibiotic combinations against multidrug resistant pathogens. BMC Microbiol 19:18. 10.1186/s12866-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senges CHR, Al-Dilaimi A, Marchbank DH, Wibberg D, Winkler A, Haltli B, Nowrousian M, Kalinowski J, Kerr RG, Bandow JE. 2018. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc Natl Acad Sci USA 115:2490–2495. 10.1073/pnas.1715713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchman DL. 2018. Processes in microbial ecology. 2nd edition. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 11.Jenkins S, Swenson TL, Lau R, Rocha AM, Aaring A, Hazen TC, Chakraborty R, Northen TR. 2017. Construction of viable soil defined media using quantitative metabolomics analysis of soil metabolites. Front Microbiol 8:2618. 10.3389/fmicb.2017.02618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren J-L, Zhang A-H, Kong L, Wang X-J. 2018. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv 8:22335–22350. 10.1039/c8ra01574k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birer-Williams CMC, Chu RK, Anderton CR, Wright ES. 2021. SubTap, a versatile 3D printed platform for eavesdropping on extracellular interactions. mSystems 6:e0090221. 10.1128/mSystems.00902-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain CA, Rubio VY, Garrett TJ. 2019. Impact of matrix effects and ionization efficiency in non-quantitative untargeted metabolomics. Metabolomics 15:135. 10.1007/s11306-019-1597-z. [DOI] [PubMed] [Google Scholar]
- 15.Čihák M, Kameník Z, Šmídová K, Bergman N, Benada O, Kofroňová O, Petříčková K, Bobek J. 2017. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front Microbiol 8:2495. 10.3389/fmicb.2017.02495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward AC, Allenby NE. 2018. Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol Lett 365. 10.1093/femsle/fny079. [DOI] [PubMed] [Google Scholar]
- 17.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. 2016. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533:543–546. 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overmann J, Abt B, Sikorski J. 2017. Present and future of culturing bacteria. Annu Rev Microbiol 71:711–730. 10.1146/annurev-micro-090816-093449. [DOI] [PubMed] [Google Scholar]
- 19.Stewart EJ. 2012. Growing unculturable bacteria. J Bacteriol 194:4151–4160. 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of 'unculturable' bacteria. FEMS Microbiol Lett 309:1–7. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Avalos M, Guzman-Trampe S, Rodriguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 63:442–459. 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 22.van der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, van Wezel GP. 2018. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep 35:575–604. 10.1039/c8np00012c. [DOI] [PubMed] [Google Scholar]
- 23.Furey A, Moriarty M, Bane V, Kinsella B, Lehane M. 2013. Ion suppression; a critical review on causes, evaluation, prevention and applications. Talanta 115:104–122. 10.1016/j.talanta.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 24.Southam AD, Weber RJ, Engel J, Jones MR, Viant MR. 2017. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat Protoc 12:310–328. 10.1038/nprot.2016.156. [DOI] [PubMed] [Google Scholar]
- 25.Charkoudian LK, Fitzgerald JT, Khosla C, Champlin A. 2010. In living color: bacterial pigments as an untapped resource in the classroom and beyond. PLoS Biol 8:e1000510. 10.1371/journal.pbio.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castano-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee JH, Lee S, LeGrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O'Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE, Linington RG. 2019. The Natural Products Atlas: an open access knowledge base for microbial natural products discovery. ACS Cent Sci 5:1824–1833. 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3:211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, Van Treuren W, Fischer CR, Merrill BD, DeFelice BC, Sanchez JM, Higginbottom SK, Guthrie L, Fall LA, Dodd D, Fischbach MA, Sonnenburg JL. 2021. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 595:415–420. 10.1038/s41586-021-03707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright ES, Vetsigian KH. 2016. Inhibitory interactions promote frequent bistability among competing bacteria. Nat Commun 7:11274. 10.1038/ncomms11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright ES. 2015. DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics 16:322. 10.1186/s12859-015-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murali A, Bhargava A, Wright ES. 2018. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6:140. 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber RJM, Zhou J. 2020. DIMSpy: Python package for processing direct-infusion mass spectrometry-based metabolomics and lipidomics data (Version v2.0.0).
- 33.R Development Core Team. 2019. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 34.Smith MR. 2020. Information theoretic generalized Robinson–Foulds metrics for comparing phylogenetic trees. Bioinformatics 36:5007–5013. 10.1093/bioinformatics/btaa614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00922-22-s0001.pdf, PDF file, 3.6 MB (3.7MB, pdf)
Data Availability Statement
Sequences are available from GenBank under the accession numbers MN177030-MN177054, MN186620-MN186656, and OK033102-OK033103.