Abstract
The extent to which horizontal gene transfer (HGT) has shaped eukaryote evolution remains an open question. Two recent studies reported four plant-like genes acquired through two HGT events by the whitefly Bemisia tabaci, a major agricultural pest (Lapadula WJ, Mascotti ML, Juri Ayub M. 2020. Whitefly genomes contain ribotoxin coding genes acquired from plants. Sci Rep. 10(1):15503; Xia J, et al. 2021. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184(7):1693–1705 e1617.). Here, we uncovered a total of 49 plant-like genes deriving from at least 24 independent HGT events in the genome of the Middle East Asia Minor 1 (MEAM1) whitefly. Orthologs of these genes are present in three cryptic B. tabaci species, they are phylogenetically nested within plant sequences, they are expressed and have evolved under purifying selection. The predicted functions of these genes suggest that most of them are involved in plant–insect interactions. Thus, substantial plant-to-insect HGT may have facilitated the evolution of B. tabaci toward adaptation to a large host spectrum. Our study shows that eukaryote-to-eukaryote HGT may be relatively common in some lineages and it provides new candidate genes that may be targeted to improve current control strategies against whiteflies.
Keywords: Bemisia tabaci, horizontal gene transfer, plants, phytophagous insects
Significance.
Horizontal gene transfer (HGT) is widespread in prokaryotes, but its extent and impact remain unclear in eukaryotes. Most cases of HGT reported so far in animals involve genes of microbial origin. Here, we show that the whitefly, Bemisia tabaci, a widespread crop pest, has acquired no less than 49 genes through at least 24 events of HGT from plant sources. The plant-like B. tabaci genes show evidence of functionality and many of them are likely involved in plant–pathogen interactions. Our study shows that plant-to-insect HGT may be more frequent than previously thought and it provides new candidate genes that may be targeted in the context of biocontrol strategies against B. tabaci.
Introduction
Horizontal gene transfer (HGT) is the passage of genetic material between organisms by means other than reproduction. The patterns, mechanisms, and vectors of HGT are well characterized in prokaryotes, in which these transfers are ubiquitous and a major source of innovation (Soucy et al. 2015). In eukaryotes, HGT has long been considered anecdotal because of multiple barriers that impede such transfers, or controversial, as resulting from phylogenetic artifacts or contaminant sequences (Martin 2017; Salzberg 2017). Yet, recent studies have reported robust HGT events in various eukaryotic organisms. In unicellular eukaryotes, HGT appears widespread, with dozens of foreign genes characterized in a large diversity of taxa, many of which are involved in adaptive functions (Lacroix and Citovsky 2016; Husnik and McCutcheon 2018; Legeret al. 2018; Sibbald et al. 2020; Van Etten and Bhattacharya 2020). An increasing number of HGT events are also reported in multicellular eukaryotes. For example, in plants, multiple cases of prokaryote-to-plant and plant-to-plant HGT have been characterized (Yang et al. 2019; Cai et al. 2021; Hibdigeet al. 2021). In metazoans, several taxa are notorious for their relatively high content in foreign genes, such as root-node nematodes (Paganini et al. 2012) and rotifers (Simion et al. 2021), and HGT from both endosymbiotic and nonendosymbiotic bacteria can be common, especially in arthropods (Moran and Jarvik 2010; Dunning Hotopp 2011; Verster et al. 2019; Cummings et al. 2022). While functional studies remain scarce, it appears that many bacterial and fungal genes independently acquired by nematodes and arthropod lineages are involved in adaptation to phytophagy (Haegeman et al. 2011; Wybouw et al. 2016). For example, a Glycosyl Hydrolase Family 32 gene acquired horizontally from rhizobial bacteria by the plant parasitic nematode Globodera pallida is expressed in the digestive system during feeding and is involved in metabolizing plant-derived sucrose (Danchin et al. 2016). In the coffee borer beetle (Hypothenemus hampei, Coleoptera), a mannanase-encoding gene acquired from Bacillus-like bacteria was shown to hydrolyze the major polysaccharide of coffee berries, thus likely facilitating adaptation of the beetle to a new food source (Acuna et al. 2012). Furthermore, in the phytophagous mite Tetranychus urticae, a cysteine synthase gene acquired from bacteria is involved in plant cyanide detoxification and thus likely enabled colonization of a new niche by this mite and other arthropods (Wybouw et al. 2014). Perhaps even more remarkable is the acquisition of four horizontally transferred genes (two BAHD acyltransferases called BtPMaT1 and BtPMaT2 and ribosome inactivating proteins [RIPs]) not from bacteria or fungi but directly from plants, by the sweet potato whitefly B. tabaci (Lapadula et al. 2020; Xia et al. 2021). Whiteflies (family Aleyrodidae) are herbivorous hemipteran insects that are important agricultural pests because of their feeding habits and the many viruses they transmit to plants. Functional assays revealed that the BtPMaT1 protein detoxifies plant phenolic glucosides that are normally used by plants to protect themselves against insect herbivores. Thus, the acquisition of a plant gene by whiteflies through HT enabled them to thwart plant defenses and may, in part, explain why these insects have become generalist phloem-feeders (Xia et al. 2021). Interestingly, whiteflies feeding on transformed tomato plants expressing BtPMaT1 gene-silencing fragments showed increased mortality and reduced fecundity (Xia et al. 2021). Therefore, identifying genes of plant origin in herbivore insects can provide targets to engineer new pest control strategies.
Results
Identification of Plant-to-Whitefly HGT Candidates in MEAM1
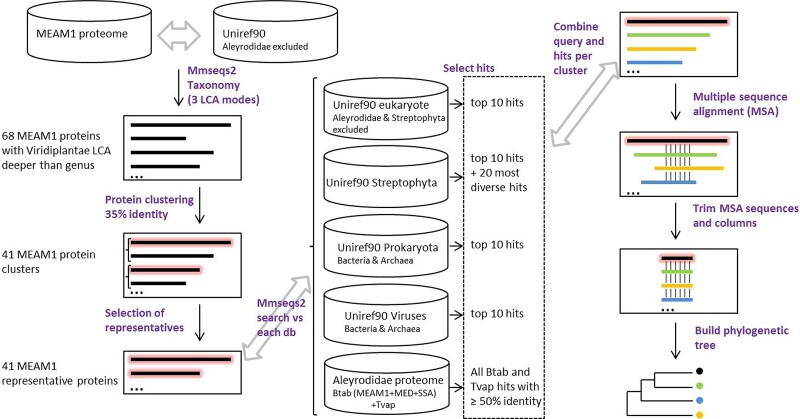
To assess the extent to which plant-to-insect HGT may have shaped interactions between whiteflies and their host plants, we conducted a systematic search for genes of plant origin in the B. tabaci Middle East Asia Minor 1 (MEAM1) genome (Chen et al. 2016). We first applied a high-throughput method to infer a last common ancestor (LCA) for each of the 15,662 predicted proteins in B. tabaci. To this end, we used MMseqs2 taxonomy (Mirdita et al. 2021) which proposes three different modes of LCA inference (i.e., single search LCA, approximate 2bLCA, and best hit) for a query protein by processing the taxonomy of proteins retrieved by similarity search. As target database, we used the Uniref90 from which we removed Aleyrodidae proteins in order to avoid confusion in LCA prediction (fig. 1). The current release of the Uniref90 database contains 119,222,328 protein sequences each representative of a group of UniprotKB proteins clusterized at 90% similarity threshold (Suzek et al. 2015). The UniprotKB database itself currently contains 230,328,648 proteins from all types of organisms, including 13,238,084 proteins from plants (Viridiplantae) and 51,638,861 proteins from non-plant eukaryotes (https://www.ebi.ac.uk/uniprot/TrEMBLstats [UniProt Consortium 2019]). In our approach, the placement of a protein LCA in plants indicates potential HGT from plants to B. tabaci after the emergence of Aleyrodidae. The deeper the LCA, the more conserved the protein across the breadth of plant taxonomy, which reduces the risk to confuse donor and recipient taxa as LCA and the risk of LCA inference based on contamination in the target database (e.g., contaminations in genome assemblies leading to erroneous taxonomic metadata). Note that the plant genes which are conserved in eukaryotes or even cellular organisms have a theoretical LCA which is deeper than plant and thus remain beyond plant-to-whitefly HGT call using this approach. We predicted an LCA for each of the MEAM1 proteins using MMseqs2 taxonomy with the three LCA inference protocols. As expected, the vast majority of the 15,662 MEAM1 proteins (e.g., 12,200 in the best hit LCA mode) were assigned to a metazoan LCA (supplementary data set 1). Interestingly, combining the LCA predictions from the different protocols, MMseqs2 predicted plant LCA with at least one of them for 365 MEAM1 proteins. Most LCA (297/365), however, were placed at tips of the plant taxonomy (genus, species, varietas and subtribe), reflecting that the target proteins used for LCA inference are poorly conserved across plants and these LCA were considered of low confidence. By contrast, we identified 68 B. tabaci MEAM1 proteins for which LCA was inferred deeper than genus level in plants and which were hence considered potentially transferred from plants to Aleyrodidae. This included the two BAHD acyltransferase genes (BtPMaT1 and BtPMaT2) reported by Xia et al. (2021) and the two RIPs identified by Lapadula et al. (2020).
Fig. 1.
Overview of the workflow used to identify plant-derived protein candidates in the MEAM1 proteome. The containers represent target databases and the different steps are indicated in purple. The MEAM1 proteins are represented by thick black lines and the representative ones are highlighted in red.
Assessment of Plant-to-whitefly HGT Candidates in MEAM1
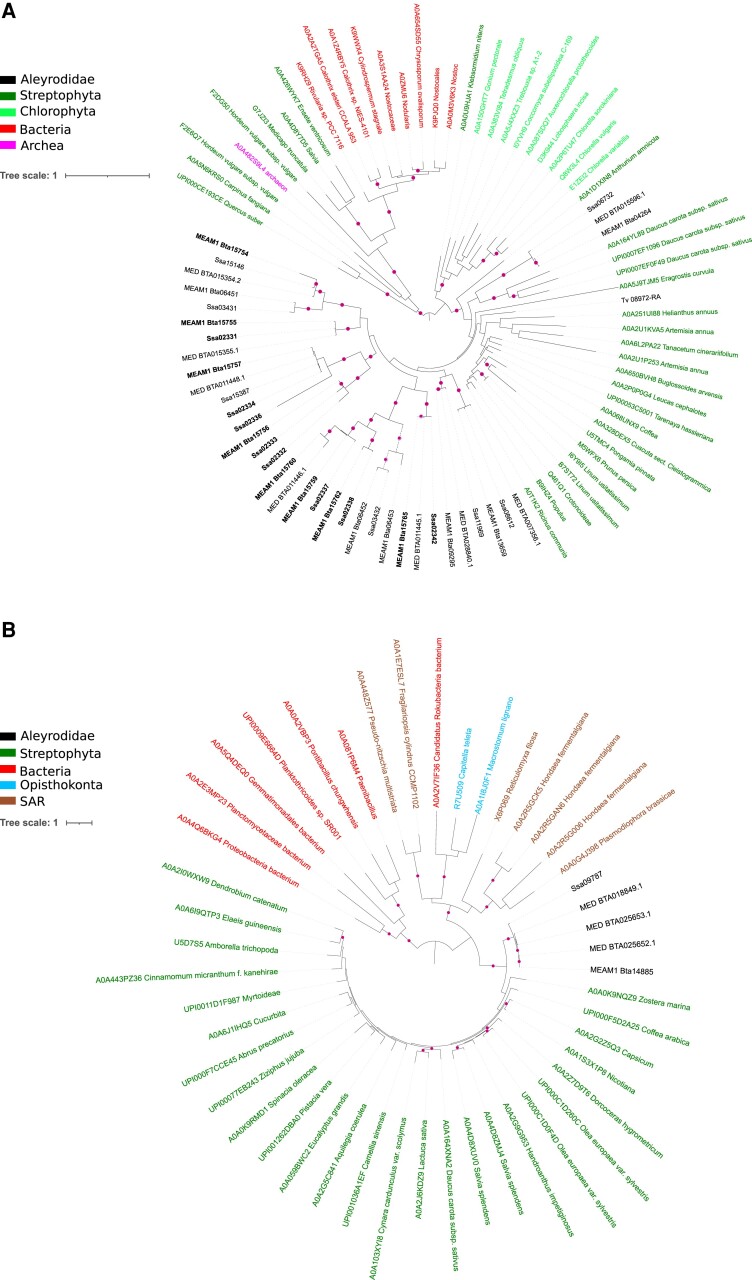
To further characterize the taxonomic origin of candidate B. tabaci plant-derived genes, we grouped them in 41 clusters of orthologous proteins and used one representative sequence per cluster to perform similarity searches against all Aleyrodidae proteomes as well as against the UniRef90 protein database of streptophytes, that of eukaryotes without streptophytes and Aleyrodidae, and those of prokaryotes and viruses (see Materials and Methods). The 10 best hits from each search supplemented with the 20 most diverse hits against streptophytes were used to build a multiple alignment which was submitted to phylogenetic analysis (fig. 1). The aim of this hit collection protocol was to address the relationships of each representative protein with regard to their best hits. Manual examination of the resulting trees and supporting alignments indicated that the signal observed in 17 of 41 data sets was not in support of plant-to-Aleyrodidae gene transfer. The reason for this was either because no or too few plant hits are present in the final alignment (which can occur because poorly aligned proteins are discarded at this stage), or the MEAM1 representative protein is absent in the final alignment because it is significantly truncated compared with selected orthologs, or the phylogenetic placement with respect to plant orthologs is in contradiction with plant-to-whitefly HGT (see supplementary fig. 1 and data set 2). For three clusters, no tree containing the representative protein was obtained as a result of applying a filter against poorly aligned proteins. Remarkably, the topology of 24 trees was consistent with the scenario of plant-to-Aleyrodidae gene transfer. In 22 of the 24 trees, the B. tabaci proteins were nested within clades of proteins of streptophyte origin (fig. 2A; supplementary fig. 2 and data set 3), as in (Lapadula et al. 2020; Xia et al. 2021). In the remaining two trees (Bta13103 and Bta14885), the B. tabaci proteins are sister to all proteins of streptophyte origin (fig. 2B and supplementary fig. 2). The topology of these two trees could be explained either by extensive sequence adaptation after transfer or by donor plant lineage being extinct or under-represented in genome databases. The alternative hypothesis of gene losses in multiple hemipterans, insects, animals and other eukaryotes may also be invoked to explain these two topologies but we deem it less likely than inferring gain of these genes in B. tabaci through HT (see the next section).
Fig. 2.
Phylogenetic relationships of potential plant-derived Bemisia tabaci genes. Phylogenetic trees showing the Bta06453 cluster, annotated as delta(12) FAD family (A) and the Bta14885 cluster, annotated as beta-(1,2)-xylosyltransferase family (B). Each tree is inferred from the multiple sequence alignment of the cluster representative sequence with its homologs found across Aleyrodidae proteins and Uniref90 proteins from eukaryotes, prokaryotes, and viruses. The labels for Uniref90 proteins are either species names or the names of the first taxonomic level comprising all the proteins of a specific Uniref90 cluster. The SSA-ECA proteins are labeled as “Ssa.” In (A), the labels of the B. tabaci proteins present in the FAD genomic hotspots found in the MEAM1 and SSA-ECA genomes are in bold and the Trialeurodes vaporariorum protein is labeled as “Tv.” The bootstrap values above 70% are indicated as magenta disks. The color legend indicating the taxonomy of proteins is indicated inset. For the ease of representation, the trees are presented in a circular mode rooted at midpoint. Unrooted trees are available in the supplementary figures 1 and 2.
Hypotheses Alternative to Horizontal Transfer
A number of earlier studies reporting cases of HGT in animals have later been dismissed and shown to be plagued by bacterial contamination (Bemm et al. 2016) or technical artifact in the similarity search or phylogenetic analyses (Salzberg 2017). In other cases, it is difficult to exclude alternative hypotheses such as multiple gene losses (Dunning Hotopp 2018). Here, in support of a plant origin, genes homologous to B. tabaci plant-like genes are absent from the proteomes of non-Aleyrodidae insects and from that of all other metazoans. Furthermore, 23 clusters are shared between MEAM1 and at least one other B. tabaci cryptic species (Mediterranean 1 [MED1] and Sub-Saharan Africa - East and Central Africa [SSA-ECA]) which were independently sequenced at a different time (2019 for SSA-ECA [Chen et al. 2019]) and even in a different laboratory for MED1 (Xie et al. 2017). Should contamination underly the presence of plant-like genes in B. tabaci, it would have had to occur at least three times independently during the sequencing process of the three genomes, and it would have had to involve three times the same plant genes, and these genes only, which is highly unlikely. While the MEAM1 assembly was produced using B. tabaci individuals collected on collard (Brassica oleracea, Brassicaceae) (Chen et al. 2015, 2016), individuals used to produce the SSA-ECA assembly were sampled from cassava (Manihot esculenta, Euphorbiaceae) (Chen et al. 2019) and those used to generate the MED1 assembly were collected on Parafilm membrane sachets containing a 25% sucrose solution (Xie et al. 2017). None of the plant-like genes we uncovered in B. tabaci were identical or nearly identical to collard or cassava genes or to genes belonging to other related Brassicaceae or Euphorbiaceae species. Thus, B. tabaci plant-like genes are bona fide B. tabaci genes, and not contaminating plant genes. We further investigated the genomic environment of MEAM1 candidate plant-derived genes by assigning a taxonomic origin to each of their nearest flanking protein-coding genes. After excluding Aleyrodidae proteins from the Uniref90 database, we were able to retrieve a hit for 61 of the 76 flanking genes investigated and found that all but two of them had a metazoan best hit (47 of 61 having a Pancrustacea best hit) (supplementary table 1). Among the 15 flanking genes that did not have a metazoan best hit, 13 of them had no hit (they are specific to Aleyrodidae), one of them had a best hit to a plant and the last one had a best hit to a bacterium. Another way to describe these results is to say that among the 49 MEAM1 plant-like genes investigated in this analysis, 45 of them had at least one nearest flanking gene showing a best hit to a metazoan gene. For the four remaining plant-like genes, we checked the best hit of the next downstream and upstream flanking genes and found that for all four genes at least one of the additional flanking genes had a metazoan best hit (supplementary table 1). These results confirm that the genes of putative plant origin stand in the context of animal contigs in the B. tabaci MEAM1 assembly.
Regarding a possible bias in terms of taxonomic representation of the target protein database, we would like to emphasize that the UniprotKB database, from which the Uniref90 database derives, is now particularly dense and diverse in terms of animal proteins. By browsing the “Proteomes” page of the “UniProt” website (https://www.uniprot.org/proteomes/), we found that it currently contains 1,093 animal proteomes comprising more than 10,000 proteins that are distributed among seven large clades (Chordata, Cnidaria, Ecdysozoa, Echinodermata, Placozoa, Porifera, Spiralia). Among Ecdysozoa, the database contains 151 insect proteomes that comprise at least 10,000 proteins and are distributed in eight orders (Coleoptera, Dictyoptera, Diptera, Hemiptera, Hymenoptera, Lepidoptera, Psocodea, and Thysanoptera). It is thus unlikely that the narrow taxonomic distribution of B. tabaci plant-like genes we observe in animal proteomes (i.e., specific to Aleyrodidae) is due to a bias in the representativeness of animals or insect taxa in the target database. Furthermore, the gene loss alternative hypothesis would posit that B. tabaci plant-like genes were inherited vertically from a common ancestor with plants (i.e., the last eukaryote common ancestor [Burki et al. 2020]) and were lost in all eukaryotes but plants and B. tabaci. A more formal way of assessing how likely hypotheses involving multiple losses are compared with those involving HT have been implemented in studies of interdomain HGT (see e.g., Cote-L’Heureux et al. 2022). Here, the density of plants, animals, and other eukaryote taxa included in the UniProt database we have used is so high that we feel hypotheses involving multiple losses may be considered very unlikely. Specifically, given the diversity of good quality eukaryote proteomes available in the UniProt database (i.e., proteomes with more than 10,000 proteins) and the phylogeny of Hemiptera (Johnson et al. 2018), Metazoa (Laumer et al. 2019) and Eukaryota (Burki et al. 2020), at least 22 independent events of losses would be required to explain the presence of the B. tabaci plant-like genes only in B. tabaci and plants. These losses would have involved all copies of the exact same gene families and would have occurred recurrently during eukaryote evolution, including after the emergence of hemipteran insects, in which at least five losses would have to be inferred based on proteomes available in UniProt (in the Aleyrodidae Trialeurodes vaporariorum, in Aphidoidea, in Psylloidea, in Fulgoroidea and in Cimicomorpha). Altogether, these lines of evidence suggest that as proposed for BAHD acyltransferases and RIPs (Lapadula et al. 2020; Xia et al. 2021), the 24 B. tabaci representative plant-like genes (supplementary data set 4) were acquired by an ancestor of B. tabaci through plant-to-insect HGT. Regarding the possibility of phylogenetic artifact, most B. tabaci plant-like genes are phylogenetically nested within plant genes, but we acknowledge that the topology of our phylogenies may not be always strongly supported. However, given the absence of B. tabaci plant-like genes in non-Aleyrodidae animals and their close phylogenetic proximity to plant genes, their acquisition through HGT seems more parsimonious to envision from a plant than a non-plant source.
Inference of HGT and Gene Duplication Events in Aleyrodidae
We next examined the extent to which these plant gene transfers have contributed to the genome of other Aleyrodidae available in Genbank. We clustered the 24 representative MEAM1 proteins with the whole proteomes from the B. tabaci cryptic species MEAM1, SSA-ECA and MED, and from T. vaporariorum, the only other Aleyrodidae available in Genbank at the time we performed this study. In total, the 24 corresponding Aleyrodidae clusters encompass 138 proteins, of which 131 have best hits against plant homologs including 45 from SSA-ECA, 35 from MED, and 49 from MEAM1 B. tabaci cryptic species as well as two from T. vaporariorum (table 1; supplementary table 2). Protein clustering combined to phylogenetic analysis shows that at least 24 independent HGT events are necessary to explain the presence of these genes in B. tabaci genomes. This number of plant-to-insect HGT is remarkable given that most HGT events reported so far in animals (excluding HT of transposable elements) involve genes of bacterial or fungal origins (Wybouw et al. 2016. 2018), with only few cases of gene transfers from non-fungal eukaryotes to animals (Gladyshev et al. 2008; Graham and Davies 2021). It is noteworthy that 15 out of 20 clusters are shared between the MEAM1 and the SSA-ECA cryptic species, indicating that most HGTs likely occurred before the split of these species, that is, between 5 and 40 million years ago (Santos-Garcia et al. 2015; Mugerwa et al. 2018). Chen et al. 2019 Genome of the African cassava whitefly Bemisia tabaci and distribution and genetic diversity of cassava-colonizing whiteflies in Africa, Insect Biochemistry and Molecular Biology.
Table 1.
Characteristics of Representative Sequences of Each Cluster of Plant-derived Bemisia tabaci Genes
| MEAM1 Gene Representative of Cluster at 35% ID | % ID with Best Plant Hit | Nb of Seq. in Cluster | Nb of Ka/Ks < 0.5 (Nb of sequences included in the codon alignment) | Nb of TSA Data sets | Nb of TSA Seq. (>75% cov; >95 ID4) | Predicted Functiona |
|---|---|---|---|---|---|---|
| Bta01560 | 57.8 | 9 | 35/35 (9) | 1 | 1 | BAHD acyltransferase |
| Bta01681 | 70.2 | 9 | 36/36 (9) | 7 | 8 | Methyltransferase DDB |
| Bta02746 | 49.5 | 4 | 6/6 (4) | 1 | 2 | Unknown function |
| Bta04871 | 63.2 | 3 | 3/3(3) | 5 | 8 | Nicotianamine synthase |
| Bta05675 | 63.3 | 5 | 6/6 (4) | – | – | Ornithine decarboxylase |
| Bta06115 | 72.3 | 4 | 6/6 (4) | 6 | 9 | Glucan endo-1,3-beta-glucosidase |
| Bta06453 | 58.7 | 38 | 423/435 (30) | 6 | 61 | Delta(12) FAD |
| Bta07117 | 65.9 | 2 | 0/1 (2) | 9 | 10 | Unknown function |
| Bta07485 | 66.5 | 5 | 6/6 (4) | 4 | 5 | Purple acid phosphatase |
| Bta07668 | 70.4 | 3 | 3/3 (3) | – | – | AAA-ATPase ASD |
| Bta07786 | 39.7 | 6 | 15/15 (6) | 4 | 6 | Phenolic glucoside malonyltransferase |
| Bta08104 | 57.6 | 3 | 2/3 (3) | 9 | 9 | Subtilisin-like protease/Pectinesterase |
| Bta08293 | 60 | 3 | 3/3 (3) | – | – | Chalcone_synthase |
| Bta08851 | 76.1 | 4 | 3/3 (3) | 4 | 5 | Alpha-(1,4)-fucosyltransferase |
| Bta11221 | 60.5 | 4 | 5/6 (4) | – | – | Pectinesterase |
| Bta11576 | 70.3 | 1 | NA | 8 | 15 | Delta(7)-sterol-C5(6)-desaturase |
| Bta11842 | 80.4 | 2 | 1/1 (2) | 8 | 8 | Unknown function |
| Bta13103 | 28.7 | 5 | 9/10 (5) | 7 | 13 | Ricin-like |
| Bta13961 | 64.5 | 4 | 5/6 (4) | 5 | 6 | Thaumatin |
| Bta14372 | 43.2 | 6 | 14/15 (6) | – | – | HXXXD-type acyltransferase |
| Bta14387 | 84.3 | 3 | 3/3 (3) | 8 | 13 | Pathogen-related protein |
| Bta14404 | 76.4 | 2 | 1/1 (2) | – | – | Glutelin |
| Bta14885 | 61.8 | 5 | 5/6 (4) | 6 | 7 | Beta-(1,2)-xylosyltransferase |
| Bta15659 | 74.5 | 3 | 3/3 (3) | 3 | 6 | Beta-1,4-N-acetylglucosaminyltransferase |
References supporting a potential role in host–pathogen interaction are provided in supplementary table 2.
Several transfers were apparently followed by gene duplications. The largest cluster, with predicted delta(12) fatty acid desaturase (FAD) function, comprises 38 Aleyrodidae members including 15 MEAM1 proteins. Eight of the FAD genes are organized in a genomic region spanning about 120 kb (scaffold 995: 912,024–1,039,680) which is also observed in syntenic position at least in the SSA-ECA genome (scaffold 436: 909,978–1,039,696), indicating that this hotspot evolved before the split of these species. In combination with phylogenetic analysis, the amplification of the FAD genes can be explained by a mixture of local and distal duplication events post HGT (fig. 1A).
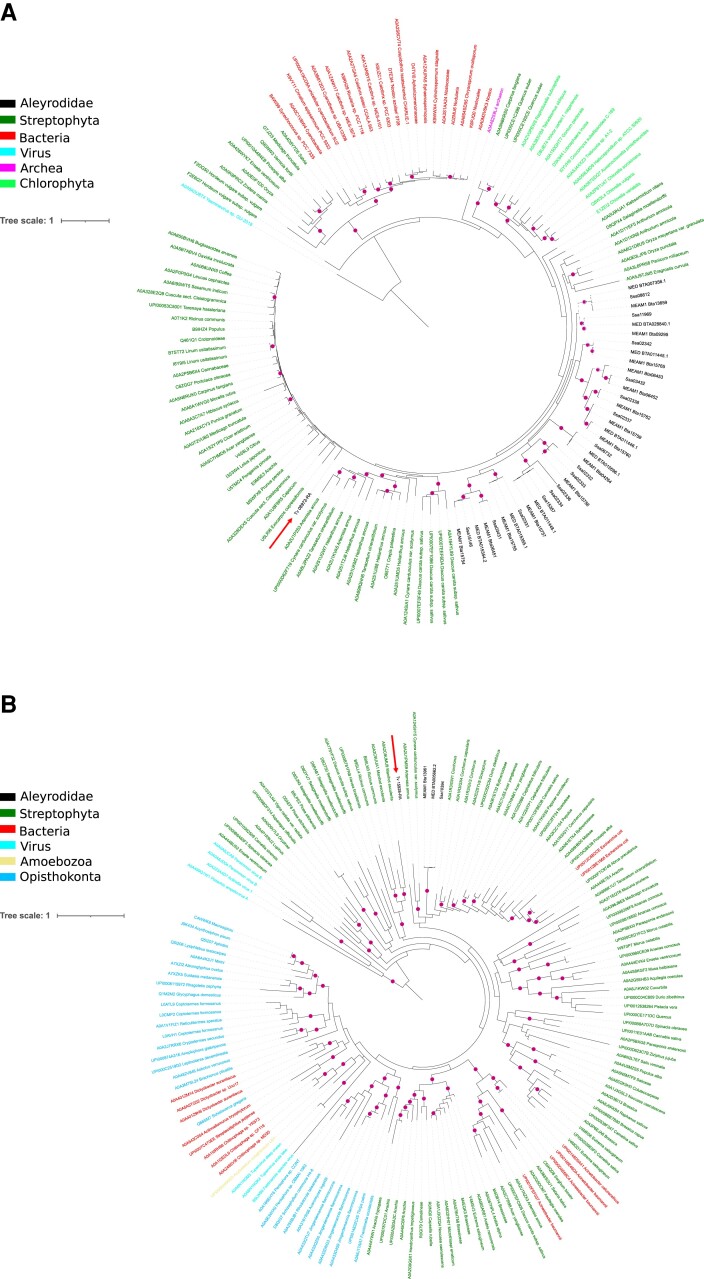
We notice, however, that in the FAD tree, the plant-related B. tabaci proteins are distributed on two distinct branches. In addition, the FAD tree is one of two clusters comprising a T. vaporariorum protein, which appears more closely related to plant than to B. tabaci proteins (fig. 2A). We reasoned that this tree topology could be biased by the hit sampling procedure which uses only the MEAM1 cluster representatives as queries. To address the monophyly of the Aleyrodidae proteins, we therefore incorporated to the initial protein alignment a collection of additional Uniref proteins collected as for the MEAM1 representative proteins above. These proteins were retrieved from similarity searches using the T. vaporariorum protein (Tv08972) and a representative of the second B. tabaci branch (Bta04264) as queries against the Uniref databases used in this study (see above and Materials and Methods). This incorporation of additional hits from the target database is meant to refine the position of Tv08972 and Bta04264 in the tree. The resulting tree confirms the monophyly of the B. tabaci proteins and suggests that the T. vaporariorum protein is more closely related to plant proteins than to B. tabaci ones (fig. 3A). However, when constraining the tree topology to place the T. vaporariorum protein as sister of the whitefly group, we could not reject the monophyly of the Aleyrodidae proteins using approximately unbiased (AU) test (Shimodaira 2002) with 95% confidence (P-value monophyly = 0.088; P-value unconstrained = 0.912).
Fig. 3.
Phylogenetic relationships of potential plant-derived genes in Aleyrodidae. Phylogenetic trees showing the Bta06453 cluster, annotated as delta(12) FAD family (A) and the Bta13961 cluster, annotated as Thaumatin (B). In (A), the tree is inferred from the multiple sequence alignment of the cluster representative sequence (MEAM1 Bta06453), MEAM1_Bta04264 and Tv_08972 with their homologs found across Aleyrodidae proteins and Uniref90 proteins from eukaryotes, prokaryotes, and viruses. In (B), the tree is inferred from the multiple sequence alignment of the cluster representative sequence (MEAM1 Bta13961) and Tv_15928 with their homologs found across Aleyrodidae proteins and Uniref90 proteins from eukaryotes, prokaryotes and viruses. The labels for Uniref90 proteins are either species names or the names of the first taxonomic level comprising all the proteins of a specific Uniref90 cluster. The SSA-ECA proteins are labeled as “Ssa.” The T. vaporariorum proteins are labeled as “Tv” and indicated by a red arrow. The bootstrap values above 70% are indicated as magenta disks. The color legend indicating the taxonomy of proteins is indicated inset. For the ease of representation, the trees are presented in a circular mode rooted at midpoint. Unrooted trees are available in the supplementary figures 1 and 2.
From the initial phylogenetic tree of the second cluster including a T. vaporariorum protein (Tv15928 in cluster Bta13961), the monophyly between the latter and the B. tabaci orthologs is also not supported. We performed the same data collection as for the FAD tree to enrich the Bta13961 protein alignment with hits of the T. vaporariorum protein against the target databases. We observe that, again, phylogenetic clustering does not support monophyly at the level of Aleyrodidae, although the T. vaporariorum protein does not group with close plant orthologs (fig. 3B). Here, the AU test did reject the monophyly of the Aleyrodidae proteins with 95% confidence (P-value monophyly = 0.0224; P-value unconstrained = 0.965).
In parallel, we compared the protein-coding genes flanking the T. vaporariorum and the B. tabaci MEAM1 homologs and we were unable to detect any synteny between the two species (data not shown). Altogether, it is difficult to conclude with confidence whether independent HGT events of homologous plant genes occurred in T. vaporariorum and in the common ancestor of the three B. tabaci cryptic species or whether these transfers instead occurred in the common ancestor of Aleyrodidae.
Number of Introns and Codon Usage Bias of Horizontally Transferred Genes
We addressed whether the structure of plant-derived genes could inform us regarding the nature of the transferred material. Indeed, genes are transferred with their introns when DNA is captured from the donor genome while they are not in the case of the capture of mRNA-derived retrogenes (Brosius 1991). Based on the gff3 annotation files of the three B. tabaci genomes, we found that plant-like genes in these genomes have an average of 1.8 introns, with 28 genes having 0 predicted intron and 2 genes having a maximum of 6 predicted introns (supplementary table 2). Therefore, we could not observe strong evidence for RNA-based transfer and the presence of introns in most plant-derived genes could be interpreted as reflecting a DNA-based transfer or an RNA-mediated mechanism followed by introns acquisitions in Aleyrodidae genomes.
We also used the COUSIN index to assess whether B. tabaci plant-like genes showed any particular bias in codon usage that may further inform us on the way they have evolved after transfer (Bourret et al. 2019) (https://cousin.ird.fr/). Given the age of the transfers and the relatively poorly resolved phylogenetic relationships of B. tabaci plant-like genes among plant genes, it was not possible to identify donor plant species. Thus, we could not compare the codon preference usage (CUPref) between donor and receiving (B. tabaci) species as in (Callens et al. 2021) for example. We plotted the distribution of COUSIN indexes of plant-like B. tabaci genes and compared it to that of all B. tabaci genes. We found that a substantial fraction of the B. tabaci CDS have a codon usage preference (CUPref) that differs from the null hypothesis, that is, the distribution of COUSIN indexes is skewed toward scores of 1 or above (supplementary fig. 2). However, most B. tabaci plant-like genes fall among genes that have no CUPref, that is, the distribution of their COUSIN index is centered on zero (supplementary fig. 2). One could propose that absence of CUPref may have facilitated incorporation of these genes in the cellular environment of B. tabaci. However, it is unclear whether these genes have no CUPref in the donor species or whether they evolved toward absence of CUPref through compositional amelioration after being transferred in the B. tabaci genome (Koski et al. 2001).
Putative Function of Horizontally Transferred Genes
To search for evidence of functionality of plant-like genes in B. tabaci, we measured the evolutionary pressures acting on them by calculating pairwise ratios of nonsynonymous over synonymous substitutions (Ka/Ks) between sequences within each of the 24 clusters. Importantly, these pairwise Ka/Ks ratio are here calculated within each cluster both between paralogs and between orthologs (between B. tabaci species). We found that 593 out of the 612 ratios we obtained were lower than 0.5, indicating that most if not all plant-like genes have evolved under purifying selection in the B. tabaci species complex. Furthermore, we found transcripts supporting expression of at least one gene for 18 out of the 24 clusters, often in multiple independent transcriptome assemblies (table 1). Together with conservation across cryptic species and evidence of gene duplication, these data suggest that most if not all B. tabaci plant-like genes are functional.
Interestingly, most of these genes (21 of 24 clusters) have a predicted function based on similarity with their nearest plant relative. As for the recently described malonyltransferase (Taguchi et al. 2010; Xia et al. 2021), there is direct or indirect evidence that many of them are involved in plant–pathogen interactions (table 1). For example, the delta(12) FAD are known to produce polyunsaturated linoleic acid in plants, which are involved in response to pathogens (Dar et al. 2017). Likewise, members of subtilisin-like protease and pathogen-related protein families can both be induced following pathogen infection (van Loonet al. 2006; Figueiredo et al. 2014). Ornithine decarboxylase is also worth noting as this enzyme synthesizes putrescine in plants, a polyamine involved in pathogen response (Rhee et al. 2007) and suspected to be usurped by Hessian fly larvae to facilitate nutrient acquisition while feeding on wheat (Subramanyam et al. 2015). In the same vein, a gene resembling the nicotianamine synthase, involved in the transport of various metal ions in plants (Laffont and Arnoux 2020), may facilitate acquisition of micronutrient by whiteflies. Finally, pectinesterase is a plant cell wall degrading enzyme (PCWDE) that is also found in plant and fungal pathogens causing maceration and soft-rotting of plant tissues. In fact, horizontal acquisition of PCWDE has already been documented in insects, but the source of the gene was bacterial (Shelomi et al. 2016).
Discussion
Our study reveals that in addition to bacterial genes, which repeatedly entered arthropod genomes and fueled the evolution of herbivory (Luan et al. 2015; Wybouw et al. 2016), numerous plant genes have been acquired through HGT by B. tabaci, likely contributing to the highly polyphagous nature of this species. Our findings (supplementary data set 1) agree with earlier studies (Chen et al. 2016; Bao et al. 2021; Ren et al. 2021) and a recent large-scale analysis (Li et al. 2022) showing that numerous genes of plant but also bacterial, fungal and viral origin have been acquired through HT by B. tabaci. The significant representation of horizontally transferred genes with predicted functions potentially involved in parasitism suggests that these genes were selected from an important set of transferred genes. Using the same approach on Drosophila melanogaster, we found no gene of potential plant origin, showing that plant-to-insect HGT is not ubiquitous (see also Li et al. 2022) and suggesting that it could be facilitated by specific vectors in association with B. tabaci. It is noteworthy that viruses have been proposed to act as vectors of HT in eukaryotes (Gilbert and Cordaux 2017; Catoni et al. 2018) and that B. tabaci is known to act as a vector of dozens of plant viruses, some of which are able to replicate in insect cells (He et al. 2020). Our results call for a large-scale evaluation of plant-to-insect HGT and for a detailed functional characterization of B. tabaci plant-like genes, as is currently being done for B. tabaci genes of bacterial origin (Wang and Luan 2022), which may further contribute to control this pest.
Materials and Methods
Protein and Genome Databases
The genomes, proteomes and CDS from T. vaporariorum and the B. tabaci cryptic species MEAM1, SSA-ECA and MED were retrieved from the whitefly database (http://www.whiteflygenomics.org/ftp/). The Uniref90 (U90) database was downloaded on February 1, 2021 (https://www.uniprot.org/downloads).
Prediction of Plant-derived Genes in Aleyrodidae
We used MMseqs2 taxonomy (Mirdita et al. 2021) with three different modes of LCA inference (--lca-mode 1, 2 and 4) to infer taxonomic origin of B. tabaci MEAM1 predicted proteins against Uniref90 (U90) database from which Aleyrodidae proteins were excluded. The sequences inferred as of Viridiplantae origin were selected when ancestry in plants could be established deeper than genus level, that is, at least at the family level. The 65 MEAM1 proteins meeting this criterion were clustered using MMseqs when identity between query and target was above 35% and when target coverage by query was at least 30% (-c 0.3 --cov-mode 1 --min-seq-id 0.35) resulting in 41 clusters from which representative sequences were selected. The predicted proteins from T. vaporariorum, and B. tabaci MEAM1, SSA-ECA and MED cryptic species were clustered with the same parameters and the sequences from clusters containing MEAM1 representative plant proteins were selected. Each of these Aleyroridae proteins was compared with U90 to confirm or not their plant origin on the basis of best hit taxonomy (i.e., Viridiplantae or other).
Construction of Phylogenetic Trees
Representative MEAM1 sequences were used to search for homologs with the MMseqs2 search module against the following five databases. Four are taxonomic subsets of the U90 database corresponding to proteins from 1) Streptophyta; 2) eukaryotes without Streptophyta and without Aleyrodidae; 3) prokaryotes; and 4) viruses. The fifth database comprises Aleyrodidae proteomes from B. tabaci cryptic species MEAM1, MED and SSA-ECA and from T. vaporariorum. For each query, the fasta sequences of the top 10 hits obtained against each database supplemented with the 20 (40 in the case of Bta13961) most diverse hits obtained against Streptophyta were retrieved and aligned using MAFFT v7.475 (local alignment) (Katoh et al. 2002). Poorly aligned sequences were removed using trimal v1.4 (-resoverlap 0.7 -seqoverlap 70) (Capella-Gutierrez et al. 2009), unless this removes the MEAM1 proteins, in which case, all sequences were kept. The selected sequences were aligned again with MAFFT (local alignment) and the less informative columns of the alignments were removed using trimal (-gappyout). The resulting alignments were used for model selection and phylogenetic reconstruction using maximum likelihood (ML) in IQTREE v1.6.12 (Minh et al. 2020) with 100 bootstrap replicates. To evaluate whether topologies placing the T. vaporariorum proteins as a sister of the B. tabaci proteins would produce poorly supported trees compared with the ML trees that were obtained, we run an AU test (Shimodaira 2002) with IQTREE on the constrained and unconstrained trees using the protein models obtained for the respective ML trees (LG + R6 for Bta06453 and WAG + R6 for Bta13961).
Evidence of Functionality
To assess under which selective regime plant-like genes have evolved after transfer in B. tabaci, we aligned B. tabaci sequences within each cluster at the codon level using MACSE v2 (Ranwez et al. 2018), trimmed the codon alignment using trimal v1.4 (-backtrans -ignorestopcodon -gt 0.8) (Capella-Gutierrez et al. 2009) and calculated the ratios of nonsynonymous substitutions over synonymous substitution Ka/Ks between all pairs of sequences using the seqinr R package (Charif and Lobry 2007). The number of ratios below 0.5 (indicating purifying selection) was counted and is reported in table 1 and supplementary table 2. We also searched for evidence that B. tabaci plant-like genes are transcribed by using all sequences from the MEAM1 cryptic species as queries to perform blastn (-task megablast) searches on 12 B. tabaci MEAM transcriptomes retrieved from Genbank under the following accession numbers: GAPP01.1, GAPQ01.1, GARP01.1, GARQ01.1, GAUC01.1, GBII01.1, GBIJ01.1, GCZW01.1, GEZK01.1, GFXM01.1, GIBX01.1, GICC01.1. All transcripts aligning over at least 75% of the length of the B. tabaci coding sequence with at least 95% nucleotide identity were counted and reported in supplementary table 2. Accession number and alignment features of these transcripts are provided in supplementary table 3.
Acknowledgments
This study was supported by a grant from Agence Nationale de la Recherche (ANR-18-CE02-0021-01TranspHorizon).
Contributor Information
Clément Gilbert, CNRS, IRD, UMR Evolution, Génomes, Comportement et Ecologie, Université Paris-Saclay, Gif-sur-Yvette, France.
Florian Maumus, INRAE, URGI, Université Paris-Saclay Versailles, France.
Data Availability
The data underlying this article are available in a figshare repository. It contains a supplementary table 1, which lists the best hits of the proteins coded by the genes flanking the plant-derived genes in MEAM1, a supplementary table 2, which provides features of all B. tabaci plant-derived genes included in this study and references supporting involvement in plant–pathogen interactions, a supplementary table 3, listing all transcripts covering at least 75% of the B. tabaci MEAM1 plant-derived genes with at least 95% identity, a supplementary data set 1, containing the LCA inference reports for the three taxonomy modes, a supplementary data set 2, which is an archive comprising the initial (.aln files) and trimmed (.trim files) protein alignments in fasta format as well as the phylogenetic trees in Newick format (.annot files) for each cluster of false positives, a supplementary data set 3, which is an archive comprising the initial (.aln files) and trimmed (.trim files) protein alignments in fasta format as well as the phylogenetic trees in Newick format (.annot files) for each cluster of plant-derived genes, a supplementary data set 4, which contains a fasta file combining the plant-derived MEAM1 representative sequences, a supplementary figure 1, showing the tree images combined in a single file for the false positives (.pdf), a supplementary figure 2, showing the tree images combined in a single file for plant-derived genes (.pdf), and a supplementary figure 3, showing the figure presenting the result of the COUSIN analysis. All the phylogenetic trees can be viewed interactively at https://itol.embl.de/shared/fmaumus.
Literature Cited
- UniProt Consortium . 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna R, et al. 2012. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc Natl Acad Sci U S A. 109(11):4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XY, et al. 2021. Lysine provisioning by horizontally acquired genes promotes mutual dependence between whitefly and two intracellular symbionts. PLoS Pathog. 17(11):e1010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemm F, Weiss CL, Schultz J, Forster F. 2016. Genome of a tardigrade: horizontal gene transfer or bacterial contamination? Proc Natl Acad Sci U S A. 113(22):E3054–E3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret J, Alizon S, Bravo IG. 2019. COUSIN (COdon Usage Similarity INdex): a normalized measure of codon usage preferences. Genome Biol Evol. 11(12):3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. 1991. Retroposons—seeds of evolution. Science 251(4995):753. [DOI] [PubMed] [Google Scholar]
- Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The new tree of eukaryotes. Trends Ecol Evol. 35(1):43–55. [DOI] [PubMed] [Google Scholar]
- Cai L, et al. 2021. Deeply altered genome architecture in the endoparasitic flowering plant Sapria himalayana Griff. (Rafflesiaceae). Curr Biol. 31(5):1002–1011.e9. [DOI] [PubMed] [Google Scholar]
- Callens M, Scornavacca C, Bedhomme S. 2021. Evolutionary responses to codon usage of horizontally transferred genes in Pseudomonas aeruginosa: gene retention, amelioration and compensatory evolution. Microb Genom. 7(6):000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoni M, et al. 2018. Virus-mediated export of chromosomal DNA in plants. Nat Commun. 9(1):5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D, Lobry JR. 2007. Seqinr 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. Berlin, Heidelberg: Springer. p. 207–232. [Google Scholar]
- Chen W, et al. 2015. Estimation of the whitefly Bemisia tabaci genome size based on k-mer and flow cytometric analyses. Insects 6(3):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. 2016. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. 2019. Genome of the African cassava whitefly Bemisia tabaci and distribution and genetic diversity of cassava-colonizing whiteflies in Africa. Insect Biochem Mol Biol. 110:112–120. [DOI] [PubMed] [Google Scholar]
- Cote-L’Heureux A, Maurer-Alcala XX, Katz LA. 2022. Old genes in new places: a taxon-rich analysis of interdomain lateral gene transfer events. PLoS Genet. 18(6):e1010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings TFM, et al. 2022. Citrullination was introduced into animals by horizontal gene transfer from cyanobacteria. Mol Biol Evol. 39(2):msab317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EG, Guzeeva EA, Mantelin S, Berepiki A, Jones JT. 2016. Horizontal gene transfer from bacteria has enabled the plant-parasitic nematode Globodera pallida to feed on host-derived sucrose. Mol Biol Evol. 33(6):1571–1579. [DOI] [PubMed] [Google Scholar]
- Dar AA, Choudhury AR, Kancharla PK, Arumugam N. 2017. The FAD2 gene in plants: occurrence, regulation, and role. Front Plant Sci. 8:1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC. 2011. Horizontal gene transfer between bacteria and animals. Trends Genet. 27(4):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC. 2018. Grafting or pruning in the animal tree: lateral gene transfer and gene loss? BMC Genomics 19(1):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo A, Monteiro F, Sebastiana M. 2014. Subtilisin-like proteases in plant-pathogen recognition and immune priming: a perspective. Front Plant Sci. 5:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Cordaux R. 2017. Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr Opin Virol. 25:16–22. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in bdelloid rotifers. Science 320(5880):1210–1213. [DOI] [PubMed] [Google Scholar]
- Graham LA, Davies PL. 2021. Horizontal gene transfer in vertebrates: a fishy tale. Trends Genet. 37(6):501–503. [DOI] [PubMed] [Google Scholar]
- Haegeman A, Jones JT, Danchin EG. 2011. Horizontal gene transfer in nematodes: a catalyst for plant parasitism? Mol Plant Microbe Interact. 24(8):879–887. [DOI] [PubMed] [Google Scholar]
- He YZ, et al. 2020. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc Natl Acad Sci U S A. 117(29):16928–16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibdige SGS, Raimondeau P, Christin PA, Dunning LT. 2021. Widespread lateral gene transfer among grasses. New Phytol. 230(6):2474–2486. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP. 2018. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol. 16(2):67–79. [DOI] [PubMed] [Google Scholar]
- Johnson KP, et al. 2018. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci U S A. 115(50):12775–12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski LB, Morton RA, Golding GB. 2001. Codon bias and base composition are poor indicators of horizontally transferred genes. Mol Biol Evol. 18:404–412. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Citovsky V. 2016. Transfer of DNA from bacteria to eukaryotes. mBio 7(4):e00863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffont C, Arnoux P. 2020. The ancient roots of nicotianamine: diversity, role, regulation and evolution of nicotianamine-like metallophores. Metallomics 12(10):1480–1493. [DOI] [PubMed] [Google Scholar]
- Lapadula WJ, Mascotti ML, Juri Ayub M. 2020. Whitefly genomes contain ribotoxin coding genes acquired from plants. Sci Rep. 10(1):15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumer CE, et al. 2019. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc Biol Sci. 286(1906):20190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger MM, Eme L, Stairs CW, Roger AJ. 2018. Demystifying eukaryote lateral gene transfer (response to Martin 2017 DOI: 10.1002/bies.201700115). Bioessays 40(5), e1700242. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2022. HGT is widespread in insects and contributes to male courtship in lepidopterans. Cell 185(16):2975–2987.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan JB, et al. 2015. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol. 7(9):2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF. 2017. Too much eukaryote LGT. Bioessays 39:12. [DOI] [PubMed] [Google Scholar]
- Minh BQ, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Steinegger M, Breitwieser F, Soding J, Levy Karin E. 2021. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics. 37(18):3029-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328(5978):624–627. [DOI] [PubMed] [Google Scholar]
- Mugerwa H, et al. 2018. African ancestry of New World, Bemisia tabaci-whitefly species. Sci Rep. 8(1):2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganini J, et al. 2012. Contribution of lateral gene transfers to the genome composition and parasitic ability of root-knot nematodes. PLoS One 7(11):e50875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE V2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35(10):2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren FR, et al. 2021. Pantothenate mediates the coordination of whitefly and symbiont fitness. ISME J. 15(6):1655–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HJ, Kim EJ, Lee JK. 2007. Physiological polyamines: simple primordial stress molecules. J Cell Mol Med. 11(4):685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL. 2017. Horizontal gene transfer is not a hallmark of the human genome. Genome Biol. 18(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Garcia D, Vargas-Chavez C, Moya A, Latorre A, Silva FJ. 2015. Genome evolution in the primary endosymbiont of whiteflies sheds light on their divergence. Genome Biol Evol. 7(3):873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelomi M, et al. 2016. Horizontal gene transfer of pectinases from bacteria preceded the diversification of stick and leaf insects. Sci Rep. 6:26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508. [DOI] [PubMed] [Google Scholar]
- Sibbald SJ, Eme L, Archibald JM, Roger AJ. 2020. Lateral gene transfer mechanisms and pan-genomes in eukaryotes. Trends Parasitol. 36(11):927–941. [DOI] [PubMed] [Google Scholar]
- Simion P, et al. 2021. Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Sci Adv. 7(41):eabg4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet. 16(8):472–482. [DOI] [PubMed] [Google Scholar]
- Subramanyam S, et al. 2015. Hessian fly larval feeding triggers enhanced polyamine levels in susceptible but not resistant wheat. BMC Plant Biol. 15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, et al. 2015. Uniref clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31(6):926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi G, et al. 2010. Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63(6):1031–1041. [DOI] [PubMed] [Google Scholar]
- Van Etten J, Bhattacharya D. 2020. Horizontal gene transfer in eukaryotes: not if, but how much? Trends Genet. 36(12):915–925. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. 2006. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 44:135–162. [DOI] [PubMed] [Google Scholar]
- Verster KI, et al. 2019. Horizontal transfer of bacterial cytolethal distending toxin B genes to insects. Mol Biol Evol. 36(10):2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Luan JB. 2022. Silencing horizontally transferred genes for the control of the whitefly Bemisia tabaci. J Pest Sci. 184(7):1693-1705.e17. [Google Scholar]
- Wybouw N, et al. 2014. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife 3:e02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw N, Pauchet Y, Heckel DG, Van Leeuwen T. 2016. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol Evol. 8(6):1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw N, Van Leeuwen T, Dermauw W. 2018. A massive incorporation of microbial genes into the genome of Tetranychus urticae, a polyphagous arthropod herbivore. Insect Mol Biol. 27(3):333–351. [DOI] [PubMed] [Google Scholar]
- Xia J, et al. 2021. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184(7):1693–1705.17. [DOI] [PubMed] [Google Scholar]
- Xie W, et al. 2017. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience 6(5):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, et al. 2019. Convergent horizontal gene transfer and cross-talk of mobile nucleic acids in parasitic plants. Nat Plants. 5(9):991–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in a figshare repository. It contains a supplementary table 1, which lists the best hits of the proteins coded by the genes flanking the plant-derived genes in MEAM1, a supplementary table 2, which provides features of all B. tabaci plant-derived genes included in this study and references supporting involvement in plant–pathogen interactions, a supplementary table 3, listing all transcripts covering at least 75% of the B. tabaci MEAM1 plant-derived genes with at least 95% identity, a supplementary data set 1, containing the LCA inference reports for the three taxonomy modes, a supplementary data set 2, which is an archive comprising the initial (.aln files) and trimmed (.trim files) protein alignments in fasta format as well as the phylogenetic trees in Newick format (.annot files) for each cluster of false positives, a supplementary data set 3, which is an archive comprising the initial (.aln files) and trimmed (.trim files) protein alignments in fasta format as well as the phylogenetic trees in Newick format (.annot files) for each cluster of plant-derived genes, a supplementary data set 4, which contains a fasta file combining the plant-derived MEAM1 representative sequences, a supplementary figure 1, showing the tree images combined in a single file for the false positives (.pdf), a supplementary figure 2, showing the tree images combined in a single file for plant-derived genes (.pdf), and a supplementary figure 3, showing the figure presenting the result of the COUSIN analysis. All the phylogenetic trees can be viewed interactively at https://itol.embl.de/shared/fmaumus.