ABSTRACT
In the human gut microbiota, Bacteroidetes break down dietary and endogenous glycosides through highly specific polysaccharide utilization loci (PULs). PULs encode a variety of sensor regulators, binding proteins, transporters, and carbohydrate-active enzymes (CAZymes). Surface glycan-binding proteins (SGBPs) are essential for the efficient capture of the glycosides present on the cell surface, providing Bacteroidetes with a competitive advantage in colonizing their habitats. Here, we present the functional and structural characterization of a SusD-like protein encoded by a xylooligosaccharide (XOS) PUL from an uncultured human gut Bacteroides strain. This locus is also conserved in Bacteroides vulgatus, thereby providing new mechanistic insights into the role of SGBPs in the metabolism of dietary fiber of importance for gut health. Various in vitro analyses, including saturation transfer difference nuclear magnetic resonance (STD-NMR) spectroscopy, revealed that the SusD-like protein cannot bind to the cognate substrate of the XOS PUL, although its presence is essential for the PUL to function. Analysis of the crystal structure of the SusD-like protein reveals an unfolded binding surface and the absence or inappropriate orientation of several key residues compared with other known SusD-like structures. These results highlight the critical role of the SusD-like protein in the transport of oligosaccharides and provide fundamental knowledge about the structure-function of SusC/D-like transporters, revealing that the binding specificity of SusD-like SGBPs does not necessarily reflect the uptake specificity of the transporter.
IMPORTANCE The metabolization of dietary fiber is a crucial function for many gut bacteria, especially Bacteroidetes, which are particularly well adapted for recognizing, binding, transporting, and degrading glycosides. In this study, we report the functional and structural characterization of a SusD-like protein involved in xylooligosaccharide utilization by an uncultured gut Bacteroides strain. We demonstrate that while this protein is structurally similar to many canonical Bacteroidetes surface glycan-binding proteins, it cannot bind the substrate taken up by the cognate SusC-like transporter. This lack of binding might be explained by the absence of several key residues known to be involved in oligosaccharide binding and/or the possible necessity of the SusC-like protein to be present to create a cooperative binding site. The term “surface glycan-binding proteins” generally used for SusD-like proteins is thus not generic. Overall, this study allowed us to revisit the concept of glycoside utilization by Bacteroidetes, in particular those strains that feed on the short fibers naturally present in some dietary compounds or on the leftovers of other microbes.
KEYWORDS: xylooligosaccharides, SusC/D, Bacteroidetes, uncultured gut bacterium
INTRODUCTION
Humans rely on their microbiota to break down dietary fiber, which is composed of oligosaccharides and polysaccharides that cannot be digested by host carbohydrate-active enzymes (CAZymes) (1). To achieve this, Bacteroidetes harbor polysaccharide utilization loci (PULs) that encode proteins involved in sensing, binding, transporting, and degrading the target glycosides into monosaccharides (2, 3). According to the starch utilization system (Sus) paradigm (4–6), polysaccharides are first degraded by enzymes attached to the cell surface. Next, SusD-like substrate-binding lipoproteins, sometimes together with the other surface glycan-binding proteins (SGBPs), SusE and/or -F, selectively recognize and capture the oligosaccharides that will be transported into the cell through the SusC-like protein.
An in-depth characterization of the first starch utilization locus of Bacteroides thetaiotaomicron to be described showed that the SusD protein has a critical role in addition to its binding ability (6). The deletion of SusD actually resulted in a loss of growth, suggesting that the presence of SusD is a prerequisite for efficient transport (7–10). SusD and SusC interact to form a complex (11). Recent findings concerning the structures of SusC/D-like complexes provided new insights into the molecular determinants of the protein-protein and glycan-protein interactions involved (12, 13). The SusC/D-like pair is well conserved among Bacteroidetes, highlighting its crucial role in glycan uptake, and the presence of genes encoding SusD-like proteins is crucial for predicting PULs (14).
Dozens of Bacteroidetes strains have been shown to use xylan as their main source of carbon (15–25). Xylan is a major constituent of hemicelluloses, an abundant component in plant cell walls, particularly in cereals, fruits, and vegetables (26). The conserved backbone of xylan is composed of a linear β-(1-4)-d-xylopyranosyl chain, which may be decorated with arabinofuranosyl, glucopyranosyl, or uronic acid derivatives (26). Xylooligosaccharides (XOSs) are hydrolyzed from xylan or are found naturally in fruits, vegetable bamboo, and honey (27). XOSs have a range of health benefits, even at a lower dose than fructooligosaccharides, and are thus considered prebiotic candidates (27–30). To date, only two PULs specific for xylan utilization by Bacteroides ovatus ATCC 8483 (PUL-XylS and PUL-XylL) have been fully biochemically characterized. Functionally, they show different specificities depending on the structural complexity of the xylan (21). In addition, in 2016, we identified an XOS-specific PUL from an uncultured gut Bacteroides strain and characterized its protein components through recombinant expression in Escherichia coli (31, 32). This locus encodes a SusC/D-like pair and a major facilitator superfamily (MFS) transport system capable of XOS uptake up to a degree of polymerization of 3 (DP3) and DP4, respectively. We also demonstrated that the deletion of the SusD-like protein abolished the growth ability of the recombinant E. coli cells on XOS, consistent with the results obtained with other SusD-like proteins. Furthermore, this PUL shares 99% DNA sequence identity with a Bacteroides vulgatus ATCC 8482 PUL (BVU_0037 to BVU_0043). We previously proved that this PUL is involved in linear XOS utilization by the Bacteroides vulgatus ATCC 8482 strain (32). On XOS, its growth is very robust, while on arabinoxylooligosaccharides (AXOSs), it is delayed and takes place at a lower rate. In addition, this strain is unable to utilize complex heavily decorated xylans or arabinoxylan, although rarely, it may utilize simpler xylans from beechwood and birchwood (32).
In the present study, we investigated how the SusD-like protein from this XOS-targeting PUL contributes to the utilization of its cognate substrate. By using various in vitro techniques to investigate the binding ability of this SusD-like protein and analyzing its crystallographic structure, we provide new mechanistic insights into XOS uptake by Bacteroidetes.
RESULTS
F5_SusD-like cannot bind to XOS. (i) Determination of the binding ability by AGE, ITC, and DSF.
The XOS locus encodes a SusD-like protein (referred to as F5_SusD-like here) sharing 99.4% identity with the SusD-like protein (BVU_0037) from Bacteroides vulgatus ATCC 8482 (with four differences at positions N249, D444, A446, and A516 corresponding to D, G, P, and V residues in BVU_0037, respectively).
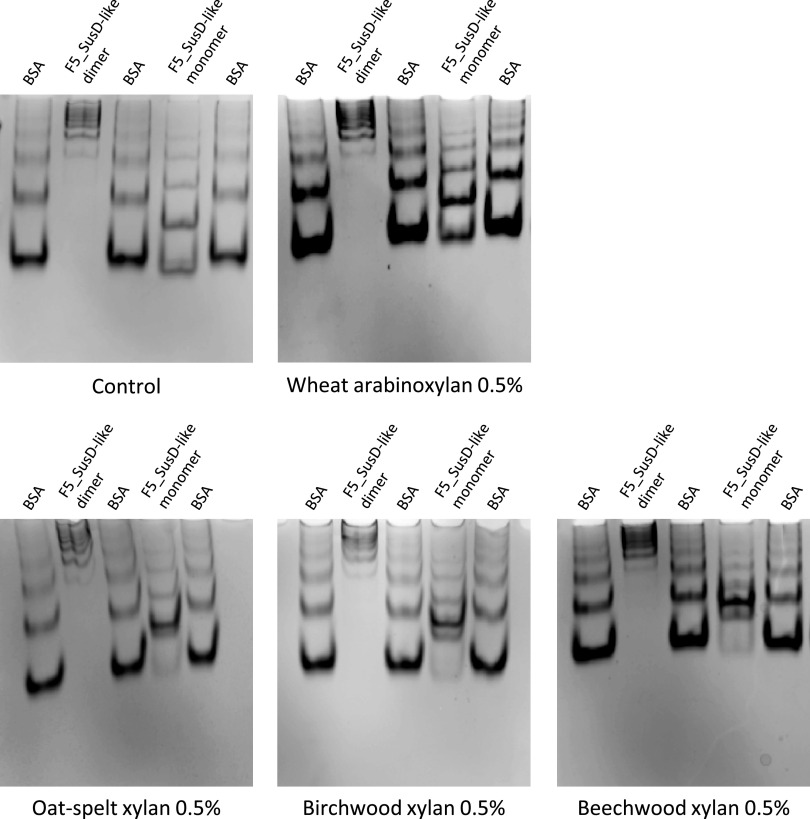
The recombinant protein was expressed in E. coli without the predicted signal peptide and N-terminal lipidation site. The ability of F5_SusD-like to bind several soluble glycans, including xylans and arabinoxylan, was assessed by affinity gel electrophoresis (AGE) (Fig. 1; see also Fig. S1 in the supplemental material). As shown by size exclusion chromatography (Fig. S2), F5_SusD-like forms monomers and dimers in solution. The dimer showed no difference in migration in the presence or absence of glycan. In contrast, monomer migration was slightly delayed by 27, 28, and 31% in the gels containing beechwood, birchwood, and oat spelt xylans, respectively. No migration delay was observed with wheat arabinoxylan. The observed smears suggest that the binding of the monomer to xylans could be unstable due to a weak affinity. F5_SusD-like is retained in the gel containing xylans to a much smaller degree than what was observed for all other characterized SusD-like proteins that efficiently bind polysaccharides (9, 10). These results indicate a weak affinity of F5_SusD-like for xylan, impaired by the presence of side chains on the xylan backbone. Isothermal titration calorimetry (ITC) was attempted using XOS as a ligand, but no binding could be detected (data not shown). Finally, differential scanning fluorimetry (DSF) was performed, but no significant change in the intrinsic fluorescence of F5_SusD-like was observed in the presence of birchwood, beechwood, or oat spelt xylans or barley β-glucan, carboxymethylcellulose, laminarin, wheat arabinoxylan, or xyloglucan (data not shown). These results indicate that the thermal stability of F5_SusD-like was not affected by significant ligand binding, contrary to what could be observed using DSF for genuine polysaccharide-binding proteins (33).
FIG 1.
Affinity gel electrophoresis of F5_SusD-like on a gel containing no polysaccharide (negative control) and on gels with 0.5% (wt/vol) different xylans.
Affinity gel electrophoresis of F5_SusD-like on a gel containing no polysaccharide (negative control) and on gels with 0.5% (wt/vol) different β-glucans. Download FIG S1, TIF file, 0.6 MB (594.5KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of F5_SusD-like oligomerization. (A) Elution profile of the protein when injected onto a Superdex 200 16/600 column in buffer containing 20 mM Tris-HCl and 150 mM NaCl (pH 7.5). (B) SDS-PAGE analysis of the fractions collected for panel A. Download FIG S2, TIF file, 0.4 MB (412.5KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Determination of the binding ability by NMR spectroscopy.
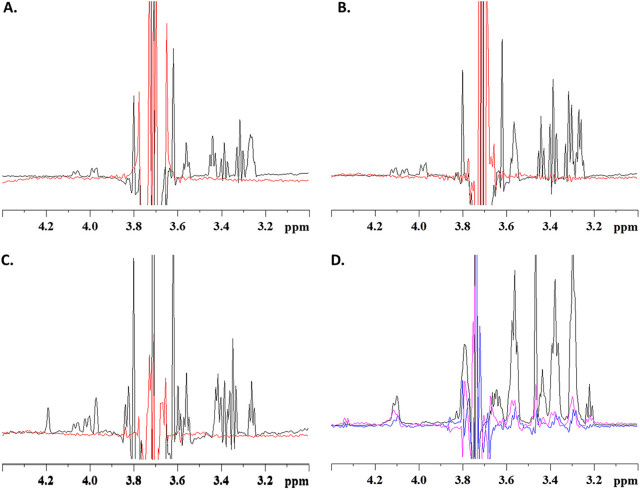
The value of saturation transfer difference nuclear magnetic resonance (STD-NMR) spectroscopy for the direct analysis of protein-ligand interactions with even weak affinities has been shown previously (34). However, to our knowledge, this approach has been used only rarely to analyze the binding ability of carbohydrate-binding proteins associated with CAZymes or with carbohydrate transporters (35, 36). Here, we used this method in order to further characterize the (in)ability of F5_SusD-like to bind to XOS. We first confirmed by one-dimensional (1D) NMR that the F5_SusD-like protein was correctly folded in solution (Fig. S3). The results of the STD experiments with xylobiose, xylotriose, or 32-α-l-arabinofuranosyl-xylobiose (AX2) on the F5_SusD-like protein were negative (Fig. 2A to C), thereby confirming that small xylooligosaccharides do not bind to the protein. A beechwood xylan sample was then prepared, but its opacity indicated that even at 500 μM, the xylan chains assemble into macroscopic structures. The NMR experiment in the presence of the F5_SusD-like protein gave a robust STD signal, but this was also the case when we probed the same xylan sample in the absence of the protein (Fig. S4). Hence, we were unable to distinguish whether binding occurred between xylan and the transporter or between the macroscopic xylan superstructures. To reduce signal saturation due to these macroscopic xylan structures, we saturated the protein indole protons (at 10.1 ppm), where xylan signals could be expected to be reduced. The STD effect with xylan alone was indeed reduced (Fig. S5) and was not only more pronounced in the presence of F5_SusD-like but also increased when we added more protein (Fig. 2D). We thus concluded that F5_SusD-like does not bind short xylooligosaccharides but binds weakly to xylan.
FIG 2.
Characterization of the ligand-binding abilities of F5_SusD-like using STD-NMR spectroscopy. (A) Zoomed-in view of the 1D xylobiose proton spectrum centered on the signals at 3.7 ppm (black) and the STD signal of the ligand upon saturation of the protein signals at 0 ppm (red). (B) Same as panel A but for the xylotriose. (C) Same as panel A but for the branched arabinoxylooligosaccharides (AX2 [32-α-l-arabinofuranosyl-xylobiose]). (D) Zoomed-in view of the 1D xylan spectrum (black), the STD signal upon saturation at 10.1 ppm of the isolated xylan (blue), or the xylan in the presence of F5_SusD-like (pink). The increased STD effect in the presence of F5_SusD-like indicates a molecular interaction.
1D proton spectrum at 298 K of 500 μL of the F5_SusD-like protein at 3.8 mg/mL (54 μM) in Tris buffer. Download FIG S3, TIFF file, 0.01 MB (100KB, tiff) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Left) 1D proton spectrum at 298 K of 500 μL of xylan at 500 μM with F5_SusD-like at 0 μM (blue), 15 μM (red), or 27 μM (magenta) in Tris buffer. (Right) STD spectra of the different samples with saturation at 0 ppm for the isolated xylan (blue) in the presence of 15 μM (red) or 27 μM (magenta) F5_SusD-like. The reference xylan spectrum is shown in black. Download FIG S4, TIF file, 0.2 MB (197.1KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Left) 1D proton spectrum of 500 μL of xylan at 500 μM with F5_SusD-like at 0 μM (blue), 15 μM (red), or 27 μM (magenta) in Tris buffer at 298 K. (Right) STD spectra of the different samples with saturation at 10.1 ppm for the isolated xylan (blue) in the presence of 15 μM (red) or 27 μM (magenta) F5_SusD-like. The reference xylan spectrum is shown in black. Download FIG S5, TIF file, 0.2 MB (193.9KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Crystallographic structure of F5_SusD-like.
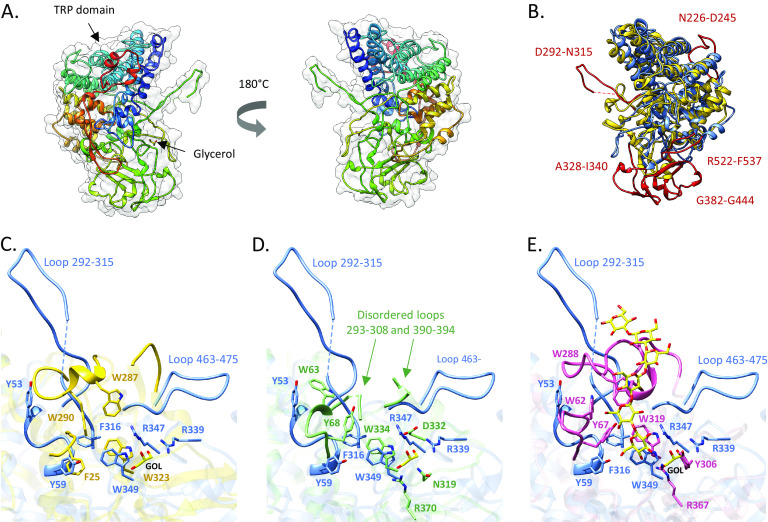
In order to better understand why F5_SusD-like does not bind the XOS substrate targeted by the PUL, we solved its three-dimensional structure by X-ray crystallography. The most similar sequence listed in the Protein Data Bank (PDB) (https://www.rcsb.org/) (37) is that of a SusD-like protein from Parabacteroides distasonis ATCC 8503 (PDB accession number 3OTN). The identity is limited, however, with only 34% identity on a short stretch of the sequence (128 amino acids), which made the structure solution by MR (molecular replacement) a challenging task. We therefore solved it using the MR-SAD (single-wavelength anomalous diffraction) technique (38) and obtained a refined structure at a 2.6-Å resolution. Data collection and refinement statistics are shown in Table 1. Despite the very low sequence similarity with structurally characterized SusD-like proteins, the F5_SusD-like protein displays a canonical “RagB/SusD” fold, which includes the conserved tetratricopeptide repeat (TPR) units. Overall, the structure of F5_SusD-like is similar to those of the 24 other SusD-like proteins that have been listed in the PDB since 2008, when the crystallographic structure of SusD was solved by Koropatkin et al. (7). F5_SusD-like presents a convex surface created by the α-helical bundle and a flat surface on the other side, where the putative sugar binding platform should be located. According to the PDB, no xylan-binding SusD-like protein has been structurally characterized so far. The closest structural homolog identified using the Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/) (39) is the laminarin-binding protein GM_SusD from Gramella sp. strain MAR_2010_102 (PDB accession number 6GCZ) (40), with a root mean square deviation (RMSD) of about 2.9 Å. The superimposition of both structures highlights the presence of a large region (A328 to I340 and G382 to G444) specific for F5_SusD-like, which is composed of random coils and some β-sheets (Fig. 3). Another interesting feature is the presence of two loops (e.g., N226 to D245 and R522 to F537) that are inserted between the conserved secondary structural elements. Furthermore, the loop from D292 to N315, which protrudes away from the folded domain, is likely to be disordered in solution, although it is partially stabilized by interactions with the neighboring SusD-like molecules in the crystal (Fig. S6). Comparison with other structurally characterized members of the SusD family shows that these features make F5_SusD-like slightly larger albeit in the same size range as those of other SusD-like proteins from xylan-targeting PULs (Table S1).
TABLE 1.
Data collection and refinement statistics for the crystal structure of the F5_SusD-like proteina
| Parameter | Value |
|
|---|---|---|
| Native | S-SAD | |
| Data collection statistics | ||
| Wavelength (Å) | 0.979 | 2.066 |
| Space group | I4122 | I4122 |
| No. of molecules per asymmetric unit | 1 | 1 |
| Cell constants a, b, c (Å) | 179.09, 179.09, 182.23 | 179.84, 179.84, 183.10 |
| Resolution (Å) (range) | 50.00–2.65 | 50.00–3.30 |
| No. of measured reflections | 307,172 | 2,114,678 |
| No. of unique reflections | 43,113 | 22,897 |
| Data completeness (%) | 100.0 | 100.0 |
| Multiplicity (%) | 7.1 | 92.4 |
| Rmerge | 0.062 (0.736) | 0.10 (0.41) |
| 〈I/σ(I)〉 | 8.4 (1.0) | 5.2 (1.5) |
| CC1/2 | 0.99 (0.86) | 1.00 (0.99) |
| Wilson B-factor (Å2) | 65.3 | 73.7 |
| Refinement statistics | ||
| Rwork/Rfree | 0.185/0.212 | |
| RMSD bond length (Å) | 0.070 | |
| RMSD bond angle (°) | 1.113 | |
| Ramachandran plot favored/allowed/ outlier regions (%) | 97/2/0 | |
| No. of atoms | ||
| Protein | 4,849 | |
| Water | 16 | |
| Other | 6 | |
| Avg B-factor (Å2) | 87.3 | |
| Cruickshank’s DPI | 0.193 | |
| MolProbity clashscore (percentile) | 4 (100th) | |
Values in parentheses are for the highest-resolution shell. DPI, diffraction precision index; S-SAD, sulfur single-wavelength anomalous diffraction phasing; CC1/2, Pearson’s correlation coefficient.
FIG 3.
F5_SusD-like crystal structure. (A) Overall structure of F5_SusD-like. (B) Secondary-structure-matching (SSM) superimposition of F5_SusD-like (blue) with GM_SusD (PDB accession number 6GCZ) (the closest structural homolog) (yellow) shown to bind to branched laminarin and linear pustulan. The loops specific for F5_SusD-like are shown in red. (C) Zoomed-in view of the superimposition of F5_SusD-like with GM_SusD. (D) Superimposition of F5_SusD-like with the nonbinding SusD-like protein BuSGBP-A (PDB accession number 7KV1) (green) present in a PUL-degrading β(1,3)-glucan and a mixed-linkage β-glucan. (E) Superimposition of F5_SusD-like with BtSGBP-A (PDB accession number 7KV3) (pink) in complex with laminarihexaose. The glycerol (GOL) present in panels C to E belongs to the F5_SusD-like structure. The figures were created with Chimera (60).
Close-up view of the interaction between the loop spanning positions 292 to 315 and a symmetry-related molecule. Tyr307 is bound in a small amphipathic cavity and stabilized by hydrogen bonds with Asp30, Arg26, and Gln24. The 2Fo-Fc electron density map (contoured at 1.0 σ) around Tyr307 is shown. Download FIG S6, TIF file, 0.3 MB (347.1KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Molecular weight comparison of F5_SusD-like with SusD-like proteins from xylan-targeting PULs. *Experimentally shown to bind xylan. Bacova_04392 binds to relatively simple xylans, xylohexaose, but not shorter xylooligosaccharides. Bacova_03427 binds to highly decorated corn glucuronoarabinoxylan but not linear xylooligosaccharides. (1) G. V. Pereira, A. M. Abdel-Hamid, S. Dutta, C. N. D’Alessandro-Gabazza, et al., Nat Commun 12:459, 2021, https://doi.org/10.1038/s41467-020-20737-5. (2) A. Rogowski, J. A. Briggs, J. C. Mortimer, T. Tryfona, et al., Nat Commun 6:7481, 2015, https://doi.org/10.1038/ncomms8481. (3) J. Despres, E. Forano, P. Lepercq, S. Comtet-Marre, et al., BMC Genomics 17:326, 2016, https://doi.org/10.1186/s12864-016-2680-8. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We attempted to cocrystallize and soak F5_SusD-like crystals with XOSs ranging from DP2 to DP4, but no complex structure was obtained, confirming the inability of F5_SusD-like to bind to short-chain XOSs. Nevertheless, a glycerol molecule under crystallization conditions is found on the flat surface where the canonical binding site is located. The glycerol molecule is coordinated by two arginine residues (R339 and R347), and W349 makes a hydrophobic stacking platform (Fig. 3). Comparison with GM_SusD shows that two of the three tryptophan residues (W287 and W290) involved in substrate binding are missing in F5_SusD-like (Fig. 3C and Fig. S7A). Comparison to the Bacova_02651 SusD-like protein from Bacteroides ovatus (PDB accession number 5E76) (10), which displays a high glycan-binding ability toward xyloglucan (Fig. S7B), shows that an essential aromatic residue involved in substrate binding (W283) is missing in F5_SusD-like. Moreover, W75, which corresponds to W82 in Bacova_02651, displays a radically different side chain orientation.
Comparison of the oligosaccharide-binding areas of SusD-like proteins with the homologous region of F5_SusD-like. In each panel, F5_SusD-like is in blue. (A) Comparison of F5_SusD-like with the laminarin-binding GM_SusD protein (PDB accession number 6GCZ) (yellow) from Gramella sp. MAR_2010_102. (B) Comparison of F5_SusD-like with Bacova_02651 (PDB accession number 5E76) (cyan) bound to xyloglucan. The stacking platforms identified in GM_SusD and Bacova_02651 are shown with the corresponding residues in F5_SusD-like, which are noneffective in binding XOS. The glycerol (GOL) present in panels A and B belongs to the F5_SusD-like structure. The figures were created with Chimera. Download FIG S7, TIF file, 0.8 MB (861.9KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Attempting to identify the molecular determinants that make F5_SusD-like a non-glycan-binding protein is particularly difficult, especially since no xylan-binding SusD-like protein is available in the PDB. Nevertheless, a striking feature of F5_SusD-like is the long partially disordered loop (D292 to N315) located close to the canonical binding site. It is noteworthy that some of the tryptophan residues involved in substrate binding in the laminarin-binding SusD-like protein GM_SusD from Gramella sp. MAR_2010_102 (PDB accession number 6GCZ) (substrate-binding residues W287 and W290) (40) and in the xyloglucan-binding SGBP-A protein from Bacteroides ovatus (PDB accession number 5E76) (substrate-binding residue W283) (10), which are missing in F5_SusD-like, are carried by a folded loop located precisely in the area from D292 to N315. The presence of a disordered loop close to the binding site is reminiscent of the structure of the SGBP-A protein from Bacteroides uniformis (BuSGBP-A) (PDB accession number 7KV1) (RMSD, 3 Å) (41) (Fig. 3D). This SusD-like protein does not bind to β(1,3)-glucan, while it displays the key binding residues present in the SGBP-A protein from Bacteroides thetaiotaomicron (BtSGBP-A) (PDB accession number 7KV3) (41), which were shown to be involved in substrate binding (Fig. 3E). Tamura et al. correlated this lack of β(1,3)-glucan binding in BuSGBP-A with structural disorder (41). Indeed, three loops critical for shaping the binding site are structurally disordered or deformed in BuSGBP-A (positions 60 to 70, 293 to 308, and 390 to 394, corresponding to positions 54 to 58, 292 to 315, and 463 to 475 in F5_SusD-like).
Finally, the four residues N249, D444, A446, and A516 of F5_SusD-like, corresponding to D, G, P, and V residues in the nearly identical SusD-like protein BVU_0037 from Bacteroides vulgatus ATCC 8482, respectively, are not located in the binding-site area. The oligosaccharide-binding regions of canonical SusD-like proteins are thus strictly identical in F5_SusD-like and BVU_0037, indicating that BVU_0037, like F5_SusD-like, probably does not bind XOSs.
DISCUSSION
Despite the low sequence identity among SusD-like proteins, the SusC/D-like pair is used as a reference for PUL identification in genomes (14, 42). Indeed, substrate recognition and capture are key steps for glycoside utilization and for bacteria to colonize their habitat. So far, most of the SusD-like proteins that have been biochemically characterized are able to bind the polysaccharide targeted by the PUL but also, to some extent, oligosaccharides resulting from polysaccharide hydrolysis by cell-exposed enzymes. Only in rare cases have SusD-like proteins been shown to be unable to bind glycans, but their function has been investigated using only AGE or ITC (41, 43, 44). Here, we fully characterized a SusD homolog (F5_SusD-like) from an XOS PUL isolated from an uncultured bacterium, which shares 99.4% sequence identity with the B. vulgatus SusD-like protein BVU_0037. Previously, we demonstrated that the PUL containing BVU_0037 was involved in the ability of B. vulgatus to grow on XOS and, to a lesser extent, on branched arabinoxylooligosaccharides, while it utilizes xylans poorly (32). In the present study, we investigated the binding ability of the F5_SusD-like protein. Surprisingly, F5_SusD-like was not able to bind XOSs or AXOSs, while it exhibited a very low affinity for xylan. In contrast, the glycan-binding SusD-like proteins characterized to date display a strong binding affinity in the millimolar-to-micromolar range, depending on the length of the tested substrate. In most cases, the values obtained for complex polysaccharides are in the same range as the values obtained with the oligosaccharides resulting from polysaccharide hydrolysis (10, 45). Rogowski et al. characterized two SusD-like proteins involved in two distinct xylan PULs from B. ovatus (Bacova_04392 and Bacova_03427) (21). In contrast to the F5_SusD-like protein, which belongs to a PUL specific for XOS, these SusD-like proteins showed strong abilities to bind xylan, measurable by ITC, but only Bacova_04392 binds xylooligosaccharides no smaller than DP6 (KD [equilibrium dissociation constant] = 8.0 × 102 M−1 for xylohexaose) (21).
Several studies previously showed that SusD-like proteins are essential for substrate uptake via the SusC-like transporter (7, 9, 10). This is also the case for the F5_SusC/D-like system since when the F5_SusD-like gene is deleted, the F5min_SusΔSusD strain can no longer grow on XOS (32). The F5_SusC/D-like pair is thus able to transport XOS, although the F5_SusD-like protein does not bind them. If the presence of the SusD-like proteins is crucial for glycoside transport, in Bacteroidetes, their binding ability is not always required, for the reasons explained here. Both the presence and binding ability of the SusD homolog from the B. ovatus transport system are essential for growth on mixed-linkage β-glucans (9). In contrast, genetic complementation using nonbinding SusD-like mutants is sufficient to restore the functionality of the starch transport system in B. thetaiotaomicron and the xyloglucan transport system in B. ovatus (7, 10).
As explained previously by Foley and coworkers regarding the Sus (46) and recently by Gray et al. regarding the fructooligosaccharide transport system in B. thetaiotaomicron (12), it might be because in Bacteroidetes, the cell surface glycan-binding proteins functionally complement each other in the capture of the polysaccharide. Indeed, Foley et al. (46) showed that when the SusD, -E, and -F proteins of the B. thetaiotaomicron Sus were mutated to nonbinding alleles, growth still occurred on maltooligosaccharides, even in a SusG deletion background. In that study, those authors demonstrated that SusE, independent of its starch-binding sites, can restore glycan uptake by the SusC/D system when the SusD starch-binding ability is abolished. In this context, the most probable hypothesis is that SusE interacts with the SusC/D complex, allowing SusD to open for import. Besides, in the rare examples of nonbinding SusD-like proteins that have been identified so far from loci involved in pectin and β-1,3-glucan utilization, there are additional SGBP- or SusD-like protein-encoding genes in the same locus, suggesting that they compensated the system for glycan binding (43, 44). However, two other nonbinding SusD-like proteins (BT2625 and BT3855) have been identified in the α-mannan utilization loci MAN-PUL1 and MAN-PUL3 of B. thetaiotaomicron, which do not contain any other SusD-like protein or SGBP (47). One cannot exclude that mannan-binding outer surface proteins encoded by MAN-PUL2, which is coexpressed with MAN-PUL1 and MAN-PUL3 during α-mannan utilization, could compensate for the nonbinding SusD-like proteins BT2625 and BT3855 to capture the targeted polysaccharide. Nevertheless, regarding SusD-like proteins that naturally do not bind glycans as isolated proteins, another hypothesis is that their interaction with glycans requires the presence of the cognate SusC-like protein. This hypothesis is in accordance with the data reported previously by Tauzin et al. (10), who showed by reverse genetic analysis that the presence of the SusD-like SGBP-A protein is more critical than its carbohydrate-binding ability for growth on xyloglucan, while the SGBP-B protein is not essential, although it supports the efficient capture of xyloglucooligosaccharides. In the present paper, we confirm that a SusC/D-like transport system should be functional for the uptake of the substrate targeted by the PUL without any binding ability of SusD-like or any other SGBP, supporting a model of glycan import whereby at least the SusC-like and SusD-like proteins must be associated to support glycan uptake.
In order to better understand the molecular determinants of the very weak binding ability of F5_SusD-like, we determined its crystal structure. Although F5_SusD-like could not be cocrystallized or soaked with XOS, a glycerol molecule was found at the canonical oligosaccharide-binding site, bound by R339, R347, and W349, which makes a hydrophobic stacking platform. This stacking platform might be involved in the very weak binding of xylans by F5_SusD-like. However, by comparing it with solved SusD-like structures, we hypothesized that the lack of genuine binding affinity could be due to some missing aromatic residues as well as an unfolded binding surface with the loop spanning D292 to N315 being partially disordered. Also, SusD-like affinity is driven by the surface complementation between the protein and the substrate rather than by the affinity of individual chemical groups for the substrate, as proposed in the very first description of the structure of SusD-like (7) and confirmed subsequently (9, 13, 40, 48–50). Thus, the role of “nonbinding” F5_SusD-like appears to be to effectively complement the SusC-like transporter and activate its functionality. This was first proposed by Glenwright et al. (13), who solved the crystal structure of a quaternary complex from B. thetaiotaomicron BT2261–BT2264, including the SusD-like (BT2263) and SusC-like (BT2264) transporters. Using a variety of techniques, those authors proposed a “pedal bin” mechanism in which SusD-like moves away from SusC-like in a hinge-like fashion in the absence of a ligand to expose the substrate-binding site to the extracellular milieu. According to this model, which was further confirmed by Gray et al. (12), SusC-like performs its function only when in complex with SusD-like, which acts as a lid that can open and close the transporter. Because SusC-like and SusD-like are associated proteins in vivo, the most probable hypothesis for SusD-like proteins that are not able to bind glycans as isolated proteins is that binding requires the presence of the cognate SusC-like protein. Indeed, the structural analysis of SusD-like BT1762 alone and in a complex with SusC-like BT1763 shows that a disordered loop located in the same area of positions 292 to 315 in F5_SusD-like is stabilized in the complex by the long hinge 2 loop of BT1763 (see Fig. S8 in the supplemental material). The hinge 2 loop is likely to be important for lid opening and is responsible, with hinge 1, for the majority of the interactions between BT1762 and BT1763 in the open state (12). Moreover, hinge 2 carries a phenylalanine residue (F649) making a hydrophobic interaction with the substrate. The proximity of this connection area to the SusD-like canonical binding site suggests that glycan recognition requires the presence of the cognate SusC-like protein. This could also be a hypothesis to explain why a single substitution that abolishes levan binding to BT1762 (W85A) in vitro leads to no growth defect in vivo. As discussed by Gray et al. (12), the context of the intact transporter ensures that the effects of SusD-like point mutations are much less dramatic in vivo. Among the amino acids composing the partially disordered loop at positions 292 to 315 in F5_SusD-like are 1 histidine, 3 tyrosine, and 2 glutamate residues. It would be interesting to know whether these residues are involved in the interaction with F5_SusC-like and/or whether the loop is folded in a way that enables some of these residues to form an extended binding site. Obtaining the three-dimensional (3D) structure of F5_SusC/D-like in complex with XOS would allow us to answer this question.
Structural comparison of SusD-like proteins alone and interacting with cognate SusC-like proteins. (A) Comparison of F5_SusD-like (blue) with BT1762 crystallized without BT1763 (PDB accession number 6GCZ) (salmon). (B) The structure of the BT1762-BT1763 complex (PDB accession number 6Z9A) (salmon, SusD-like; green, SusC-like) shows the molecular determinants for fructooligosaccharide binding. The figures were created with Chimera. Download FIG S8, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Finally, nonbinding or very weakly binding SusD-like proteins can be found in PULs, including (i) a SusC/D-like pair and an additional SGBP (43), (ii) two SusC/D-like pairs and an SGBP (44), or (iii) only one SusC/D-like pair (47; this study). The presence of an additional SGBP or a SusC/D-like pair in the same locus as that of a nonbinding SusD-like protein suggests compensation for the binding function lacking in the related SusD-like protein. In contrast, as previously hypothesized (47), the PULs containing only a SusC/D-like pair for which SusD-like, alone, is a nonbinding protein and no other SGBP might be involved in oligosaccharide or polysaccharide utilization that relies on the ability of the SusC/D-like complex to recognize and transport its cognate substrate.
MATERIALS AND METHODS
Heterologous protein expression and purification.
The gene fragment encoding the F5_SusD-like protein was PCR amplified from the metagenomic DNA of clone F5 (GenBank accession number HE717017), previously isolated from a metagenomic library derived from a human fecal sample (31). The PCR forward primer F5_SusD_Cloning_F (see Table S2 in the supplemental material) includes the NdeI restriction site, and the reverse primer F5_SusD_Cloning_R includes the XhoI site. The first 30 amino acids of F5_SusD-like corresponding to the lipoprotein signal peptide predicted by the SignalP 4.1 and LipoP 1.0 servers were not included in the PCR amplicon (51, 52). The gene product was cloned into the NdeI and XhoI restriction sites of the pET-28a(+) vector (Novagen, Darmstadt, Germany) preceding an N-terminal 6×His tag for affinity purification and transformed into E. coli DH5α (Invitrogen). The pET-28a(+) vector expressing F5_SusD-like was fully sequenced and transformed into E. coli BL21(DE3) for protein production. A single bacterial colony was inoculated into 5 mL of LB medium containing kanamycin (50 μg/mL) at 37°C. A preculture grown overnight was used to inoculate 200 mL of LB medium containing 50 μg/mL of kanamycin at an initial optical density at 600 nm (OD600) of 0.05. The culture was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) in the mid-exponential phase (OD600 of ~0.6) and then incubated for 4 h at 37°C.
List of PCR primers used in this study. Download Table S2, DOCX file, 0.01 MB (12.3KB, docx) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For the purification of the recombinant F5_SusD-like protein, cells were harvested by centrifugation (15 min at 6,000 × g) and sonicated in Tris buffer (20 mM Tris-HCl, 300 mM NaCl [pH 8.0]). Bacterial debris was cleared by centrifugation at 11,000 × g for 30 min at 4°C, and the lysates were then passed through a column of 2 mL of Talon metal affinity resin (Clontech, USA). The F5_SusD-like protein was eluted using a gradient of 50 to 200 mM imidazole and then resuspended in Tris buffer (20 mM Tris-HCl, 150 mM NaCl [pH 7.5]) using an Amicon Ultra filter (Sigma-Aldrich) to eliminate the imidazole. Protein purity was confirmed via SDS-PAGE, and the concentration was determined by the absorbance at 280 nm using the Thermo Scientific Nanodrop 2000 instrument with an extinction coefficient of 153,350 M−1 cm−1. The F5_SusD-like protein was kept at 4°C for later analysis. The protein used for crystallization experiments was purified using an ÄKTAxpress system (GE Healthcare) with an affinity step (Talon crude, 1 mL; GE Healthcare) using the same loading buffer (20 mM Tris-HCl, 300 mM NaCl [pH 8.0]) and eluted with a 250 mM step of imidazole, followed by a gel filtration step (HiPrep 16/60 Sephacryl S-200 HR) in a solution containing 20 mM Tris-HCl and 0.15 M NaCl (pH 7.5). The fractions containing the monomeric F5_SusD-like protein were pooled, concentrated, and stored at 4°C.
Glycosides.
The mixture of XOSs contains chains of DP2 to -7 (Wako Chemicals and IRO TAIHE). Simple xylans, with sparsely decorated structures, were purchased from Sigma for beechwood xylan, Biochemika Fluka for oat spelt xylan, and Megazyme for birchwood xylan and wheat arabinoxylan. Xylobiose, xylotriose, and 32-α-l-arabinofuranosyl-xylobiose (AX2) were purchased from Megazyme. Barley β-glucan and xyloglucan from tamarind seed were also purchased from Megazyme. Laminarin from Laminaria digitata was purchased from Sigma.
Isothermal titration calorimetry.
Isothermal titration calorimetry (ITC) analysis of glycan binding by F5_SusD-like was performed using the ITC200 calorimeter (Malvern), calibrated to 25°C. Proteins (40 μM) were prepared in buffer containing 20 mM HEPES–100 mM NaCl (pH 7.0), and oligosaccharides were prepared using the same buffer. The F5_SusD-like protein was placed into the sample cell, and the syringe was loaded with 2 to 10 mM XOS.
Affinity gel electrophoresis.
The ability of F5_SusD-like to bind polysaccharides was assayed by affinity gel electrophoresis. Continuous native polyacrylamide gels were prepared, consisting of 10% (wt/vol) acrylamide in buffer containing 25 mM Tris and 250 mM glycine (pH 8.3). The substrates were added prior to polymerization at a final concentration of 0.5% (wt/vol). A total of 2.5 μg of purified F5_SusD-like was loaded onto the gels. Electrophoresis was carried out for 90 min on ice. Bovine serum albumin (BSA) was used as a noninteracting negative-control protein. The percentage of retention of F5_SusD-like in the gel containing glycans was calculated as (Dwog − Dwg)/(Dwog) × 100, where Dwog is equal to DSusD-like_wog/DBSA_wog (the distance of migration of the lowest and most intense F5_SusD-like band normalized to the distance of migration of BSA in the gel without glycan) and Dwg is equal DSusD-like_wg/DBSA_wg (the distance of migration of the lowest and most intense F5_SusD-like band normalized to the distance of migration of BSA in the gel with glycan).
NMR spectroscopy.
All experiments were performed on a Bruker Avance II 800-MHz NMR spectrometer equipped with a QCPI cryogenically cooled probe head. Spectra were recorded at 298 K, and all samples were prepared in Tris buffer (20 mM Tris and 150 mM NaCl at pH 7.5). 1D proton spectra were acquired using Watergate water suppression (53), with 16 scans for XG_SusD-like (at 327 μM) and 256 scans for F5_SusD-like (at 54 μM). STD experiments were performed with saturation of the protein resonances at 0 ppm through a train of 5-ms Gaussian 180° pulses (34, 54). Typical experiments were run with 256 scans, 2,048 acquisition points, and a 5-s relaxation delay (including the 3-s presaturation train). STD experiments were run with 256 or 512 scans. Spectra were transformed after one level of zero filling and apodization with a π/3-shifted square sine bell.
Crystallization and data collection.
The purified F5_SusD-like protein was concentrated to 20 mg/mL, and initial crystallization conditions were screened using a Mosquito robot (TPP Labtech) and JCSG I to IV commercial screens (Qiagen), from which one starting condition was identified (1.26 M sodium citrate, 0.09 M HEPES-HCl, 10% glycerol [pH 7.5]). After manual condition optimization, F5_SusD-like crystals were grown using the hanging-drop method by mixing 1 μL of the protein solution with 1 μL of the precipitant solution (1.7 M Na3-citrate, 10% glycerol, 0.1 M HEPES [pH 7.5]) and incubating the solution on a 24-well crystallization plate at 12°C. After harvesting, the crystals were soaked in cryoprotectant buffer (1.7 M Na3-citrate, 15% glycerol, 0.1 M HEPES [pH 7.5]), with or without the presence of 100 mM xylotriose, and flash-frozen in liquid N2. Data were collected at the XALOC beamline of the ALBA Synchrotron Light Source (Barcelona, Spain). A long-wavelength data set was also collected at a wavelength of 2.0 Å, using a special XALOC setup with continuous helium flow around the crystal to reduce absorption from the air (helium cone).
Structure resolution and refinement.
Using the native data set only, the structure was solved using the Morda Web server (55), which identified a low-homology molecular replacement solution. This starting model was combined with the long-wavelength data set as implemented in the Phaser program (56) and used to calculate improved phases. This also enabled the correct positioning of all of the sulfur atoms in the sequence. The structure was determined via several rounds of modeling using the Arp/Warp Web server (57), alternated with manual rebuilding using Coot (58), and passed to Refmac for restrained refinement (59). The final model was validated with MolProbity (60) and WhatIF (61).
Data availability.
The sequence of F5_SusD-like is available in the GenBank database under accession number CCG34975.1. The crystal structure of F5_SusD-like has been deposited in the Protein Data Bank under accession number 7NEK.
ACKNOWLEDGMENTS
This work was carried out using the equipment at the PICT-ICEO facility and at MetaToul (Metabolomics and Fluxomics Facilities, Toulouse, France [www.metatoul.fr]). PICT-ICEO is supported by grants from the Région Midi-Pyrénées, the European Regional Development Fund, and the Institut National de la Recherche Agronomique et de l’Environnement (INRAE). MetaToul is part of the MetaboHUB-ANR-11-INBS-0010 national infrastructure (www.metabohub.fr). This research was funded by the European Union’s Horizon 2020 Framework Programme (MSCA-IF-2015_707457 [CaTSYS] and LEIT-BIO-2015-685474 [Metafluidics]). Z.W. and X.L. are supported by INSA Toulouse and the China Scholarship Council under the CSC-UT/INSA Program.
We acknowledge PICT-IPBS (Toulouse, France) and the ALBA Synchrotron Light Source (Barcelona, Spain) for access to crystallization and data collection facilities, respectively.
Contributor Information
Gabrielle Potocki-Veronese, Email: veronese@insa-toulouse.fr.
Vincent B. Young, University of Michigan—Ann Arbor
REFERENCES
- 1.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terrapon N, Lombard V, Gilbert HJ, Henrissat B. 2015. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31:647–655. doi: 10.1093/bioinformatics/btu716. [DOI] [PubMed] [Google Scholar]
- 3.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol 199:e00860-16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown HA, Koropatkin NM. 2021. Host glycan utilization within the Bacteroidetes Sus-like paradigm. Glycobiology 31:697–706. doi: 10.1093/glycob/cwaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley MH, Cockburn DW, Koropatkin NM. 2016. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K, Foley MH, Gardill BR, Dejean G, Schnizlein M, Bahr CME, Creagh AL, van Petegem F, Koropatkin NM, Brumer H. 2019. Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus. Cell Mol Life Sci 76:4319–4340. doi: 10.1007/s00018-019-03115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H, Koropatkin NM. 2016. Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. mBio 7:e02134-15. doi: 10.1128/mBio.02134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho KH, Salyers AA. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 183:7224–7230. doi: 10.1128/JB.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray DA, White JBR, Oluwole AO, Rath P, Glenwright AJ, Mazur A, Zahn M, Baslé A, Morland C, Evans SL, Cartmell A, Robinson CV, Hiller S, Ranson NA, Bolam DN, van den Berg B. 2021. Insights into SusCD-mediated glycan import by a prominent gut symbiont. Nat Commun 12:44. doi: 10.1038/s41467-020-20285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Baslé A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathöfer U, Bolam DN, Van Den Berg B. 2017. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terrapon N, Lombard V, Drula É, Lapébie P, Al-Masaudi S, Gilbert HJ, Henrissat B. 2018. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res 46:D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Chambon C, Yeoman CJ, Berg Miller ME, Fields CJ, Martens E, Terrapon N, Henrissat B, White BA, Mosoni P. 2016. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics 17:326. doi: 10.1186/s12864-016-2680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodd D, Moon Y-H, Swaminathan K, Mackie RI, Cann IKO. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J Biol Chem 285:30261–30273. doi: 10.1074/jbc.M110.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendis M, Martens EC, Simsek S. 2018. How fine structural differences of xylooligosaccharides and arabinoxylooligosaccharides regulate differential growth of Bacteroides species. J Agric Food Chem 66:8398–8405. doi: 10.1021/acs.jafc.8b01263. [DOI] [PubMed] [Google Scholar]
- 20.Mirande C, Kadlecikova E, Matulova M, Capek P, Bernalier-Donadille A, Forano E, Béra-Maillet C. 2010. Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1AT and Roseburia intestinalis XB6B4 from the human intestine. J Appl Microbiol 109:451–460. doi: 10.1111/j.1365-2672.2010.04671.x. [DOI] [PubMed] [Google Scholar]
- 21.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salyers AA, Gherardini F, O’Brien M. 1981. Utilization of xylan by two species of human colonic Bacteroides. Appl Environ Microbiol 41:1065–1068. doi: 10.1128/aem.41.4.1065-1068.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang K, Lin Y, Han Y, Jiao N. 2017. Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella flava JLT2011 using a multi-omics approach. Front Microbiol 8:220. doi: 10.3389/fmicb.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Chekan JR, Dodd D, Hong P-Y, Radlinsk L, Revindran V, Nair SK, Mackie RI, Cann I. 2014. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc Natl Acad Sci USA 111:E3708–E3717. doi: 10.1073/pnas.1406156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leth ML, Ejby M, Workman C, Ewald DA, Pedersen SS, Sternberg C, Bahl MI, Licht TR, Aachmann FL, Westereng B, Hachem MA. 2018. Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat Microbiol 3:570–580. doi: 10.1038/s41564-018-0132-8. [DOI] [PubMed] [Google Scholar]
- 26.Selvendran RR. 1984. The plant cell wall as a source of dietary fiber: chemistry and structure. Am J Clin Nutr 39:320–337. doi: 10.1093/ajcn/39.2.320. [DOI] [PubMed] [Google Scholar]
- 27.Singh RD, Banerjee J, Arora A. 2015. Prebiotic potential of oligosaccharides: a focus on xylan derived oligosaccharides. Bioact Carbohydr Diet Fibre 5:19–30. doi: 10.1016/j.bcdf.2014.11.003. [DOI] [Google Scholar]
- 28.Amorim C, Silvério SC, Prather KLJ, Rodrigues LR. 2019. From lignocellulosic residues to market: production and commercial potential of xylooligosaccharides. Biotechnol Adv 37:107397. doi: 10.1016/j.biotechadv.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 30.Goh YJ, Klaenhammer TR. 2015. Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Annu Rev Food Sci Technol 6:137–156. doi: 10.1146/annurev-food-022814-015706. [DOI] [PubMed] [Google Scholar]
- 31.Cecchini DA, Laville E, Laguerre S, Robe P, Leclerc M, Doré J, Henrissat B, Remaud-Siméon M, Monsan P, Potocki-Véronèse G. 2013. Functional metagenomics reveals novel pathways of prebiotic breakdown by human gut bacteria. PLoS One 8:e72766. doi: 10.1371/journal.pone.0072766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tauzin AS, Laville E, Xiao Y, Nouaille S, Le Bourgeois P, Heux S, Portais JC, Monsan P, Martens EC, Potocki-Veronese G, Bordes F. 2016. Functional characterization of a gene locus from an uncultured gut Bacteroides conferring xylo-oligosaccharides utilization to Escherichia coli. Mol Microbiol 102:579–592. doi: 10.1111/mmi.13480. [DOI] [PubMed] [Google Scholar]
- 33.Sun C, Liu M, Sun P, Yang M, Yates EA, Guo Z, Fernig DG. 2019. Sulfated polysaccharides interact with fibroblast growth factors and protect from denaturation. FEBS Open Bio 9:1477–1487. doi: 10.1002/2211-5463.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer M, Meyer B. 1999. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed Engl 38:1784–1788. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Bell A, Brunt J, Crost E, Vaux L, Nepravishta R, Owen CD, Latousakis D, Xiao A, Li W, Chen X, Walsh MA, Claesen J, Angulo J, Thomas GH, Juge N. 2019. Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol 4:2393–2404. doi: 10.1038/s41564-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haselhorst T, Lamerz A-C, von Itzstein M. 2009. Saturation transfer difference NMR spectroscopy as a technique to investigate protein-carbohydrate interactions in solution. Methods Mol Biol 534:375–386. doi: 10.1007/978-1-59745-022-5_26. [DOI] [PubMed] [Google Scholar]
- 37.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. 2002. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr 58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 38.Skubák P, Araç D, Bowler MW, Correia AR, Hoelz A, Larsen S, Leonard GA, McCarthy AA, McSweeney S, Mueller-Dieckmann C, Otten H, Salzman G, Pannu NS. 2018. A new MR-SAD algorithm for the automatic building of protein models from low-resolution X-ray data and a poor starting model. IUCrJ 5:166–171. doi: 10.1107/S2052252517017961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm L. 2019. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35:5326–5327. doi: 10.1093/bioinformatics/btz536. [DOI] [PubMed] [Google Scholar]
- 40.Mystkowska AA, Robb C, Vidal-Melgosa S, Vanni C, Fernandez-Guerra A, Höhne M, Hehemann J-H. 2018. Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes. FEBS J 285:4465–4481. doi: 10.1111/febs.14674. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Dejean G, Van Petegem F, Brumer H. 2021. Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota. J Biol Chem 296:100415. doi: 10.1016/j.jbc.2021.100415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Déjean G, Tamura K, Cabrera A, Jain N, Pudlo NA, Pereira G, Viborg AH, Van Petegem F, Martens EC, Brumer H. 2020. Synergy between cell surface glycosidases and glycan-binding proteins dictates the utilization of specific beta(1,3)-glucans by human gut Bacteroides. mBio 11:e00095-20. doi: 10.1128/mBio.00095-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Temple MJ, Cuskin F, Baslé A, Hickey N, Speciale G, Williams SJ, Gilbert HJ, Lowe EC. 2017. A Bacteroidetes locus dedicated to fungal 1,6-glucan degradation: unique substrate conformation drives specificity of the key endo-1,6-glucanase. J Biol Chem 292:10639–10650. doi: 10.1074/jbc.M117.787606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foley MH, Martens EC, Koropatkin NM. 2018. SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron. Mol Microbiol 108:551–566. doi: 10.1111/mmi.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron E, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsbrink J, Zhu Y, Kharade SS, Kwiatkowski KJ, Eijsink VGH, Koropatkin NM, McBride MJ, Pope PB. 2016. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol Biofuels 9:260. doi: 10.1186/s13068-016-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phansopa C, Roy S, Rafferty JB, Douglas CWI, Pandhal J, Wright PC, Kelly DJ, Stafford GP. 2014. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J 458:499–511. doi: 10.1042/BJ20131415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koropatkin N, Martens EC, Gordon JI, Smith TJ. 2009. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry 48:1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 53.Piotto M, Saudek V, Sklenár V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 54.Ley NB, Rowe ML, Williamson RA, Howard MJ. 2014. Optimising selective excitation pulses to maximise saturation transfer difference NMR spectroscopy. RSC Adv 4:7347–7351. doi: 10.1039/C3RA46246C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hungler A, Momin A, Diederichs K, Arold ST. 2016. ContaMiner and ContaBase: a webserver and database for early identification of unwantedly crystallized protein contaminants. J Appl Crystallogr 49:2252–2258. doi: 10.1107/S1600576716014965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. 1997. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr 53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB, III, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. 2018. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci 27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Affinity gel electrophoresis of F5_SusD-like on a gel containing no polysaccharide (negative control) and on gels with 0.5% (wt/vol) different β-glucans. Download FIG S1, TIF file, 0.6 MB (594.5KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of F5_SusD-like oligomerization. (A) Elution profile of the protein when injected onto a Superdex 200 16/600 column in buffer containing 20 mM Tris-HCl and 150 mM NaCl (pH 7.5). (B) SDS-PAGE analysis of the fractions collected for panel A. Download FIG S2, TIF file, 0.4 MB (412.5KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
1D proton spectrum at 298 K of 500 μL of the F5_SusD-like protein at 3.8 mg/mL (54 μM) in Tris buffer. Download FIG S3, TIFF file, 0.01 MB (100KB, tiff) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Left) 1D proton spectrum at 298 K of 500 μL of xylan at 500 μM with F5_SusD-like at 0 μM (blue), 15 μM (red), or 27 μM (magenta) in Tris buffer. (Right) STD spectra of the different samples with saturation at 0 ppm for the isolated xylan (blue) in the presence of 15 μM (red) or 27 μM (magenta) F5_SusD-like. The reference xylan spectrum is shown in black. Download FIG S4, TIF file, 0.2 MB (197.1KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Left) 1D proton spectrum of 500 μL of xylan at 500 μM with F5_SusD-like at 0 μM (blue), 15 μM (red), or 27 μM (magenta) in Tris buffer at 298 K. (Right) STD spectra of the different samples with saturation at 10.1 ppm for the isolated xylan (blue) in the presence of 15 μM (red) or 27 μM (magenta) F5_SusD-like. The reference xylan spectrum is shown in black. Download FIG S5, TIF file, 0.2 MB (193.9KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Close-up view of the interaction between the loop spanning positions 292 to 315 and a symmetry-related molecule. Tyr307 is bound in a small amphipathic cavity and stabilized by hydrogen bonds with Asp30, Arg26, and Gln24. The 2Fo-Fc electron density map (contoured at 1.0 σ) around Tyr307 is shown. Download FIG S6, TIF file, 0.3 MB (347.1KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Molecular weight comparison of F5_SusD-like with SusD-like proteins from xylan-targeting PULs. *Experimentally shown to bind xylan. Bacova_04392 binds to relatively simple xylans, xylohexaose, but not shorter xylooligosaccharides. Bacova_03427 binds to highly decorated corn glucuronoarabinoxylan but not linear xylooligosaccharides. (1) G. V. Pereira, A. M. Abdel-Hamid, S. Dutta, C. N. D’Alessandro-Gabazza, et al., Nat Commun 12:459, 2021, https://doi.org/10.1038/s41467-020-20737-5. (2) A. Rogowski, J. A. Briggs, J. C. Mortimer, T. Tryfona, et al., Nat Commun 6:7481, 2015, https://doi.org/10.1038/ncomms8481. (3) J. Despres, E. Forano, P. Lepercq, S. Comtet-Marre, et al., BMC Genomics 17:326, 2016, https://doi.org/10.1186/s12864-016-2680-8. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of the oligosaccharide-binding areas of SusD-like proteins with the homologous region of F5_SusD-like. In each panel, F5_SusD-like is in blue. (A) Comparison of F5_SusD-like with the laminarin-binding GM_SusD protein (PDB accession number 6GCZ) (yellow) from Gramella sp. MAR_2010_102. (B) Comparison of F5_SusD-like with Bacova_02651 (PDB accession number 5E76) (cyan) bound to xyloglucan. The stacking platforms identified in GM_SusD and Bacova_02651 are shown with the corresponding residues in F5_SusD-like, which are noneffective in binding XOS. The glycerol (GOL) present in panels A and B belongs to the F5_SusD-like structure. The figures were created with Chimera. Download FIG S7, TIF file, 0.8 MB (861.9KB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural comparison of SusD-like proteins alone and interacting with cognate SusC-like proteins. (A) Comparison of F5_SusD-like (blue) with BT1762 crystallized without BT1763 (PDB accession number 6GCZ) (salmon). (B) The structure of the BT1762-BT1763 complex (PDB accession number 6Z9A) (salmon, SusD-like; green, SusC-like) shows the molecular determinants for fructooligosaccharide binding. The figures were created with Chimera. Download FIG S8, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of PCR primers used in this study. Download Table S2, DOCX file, 0.01 MB (12.3KB, docx) .
Copyright © 2022 Tauzin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The sequence of F5_SusD-like is available in the GenBank database under accession number CCG34975.1. The crystal structure of F5_SusD-like has been deposited in the Protein Data Bank under accession number 7NEK.