ABSTRACT
Temperate honey bees (Apis mellifera) are challenged by low temperatures and abrupt dietary shifts associated with behavioral changes during winter. Case studies have revealed drastic turnover in the gut microbiota of winter bees, highlighted by the seasonal dominance of a non-core bacterium Bartonella. However, neither biological consequence nor underlying mechanism of this microbial turnover is clear. In particular, we ask whether such changes in gut profile are related to winter dietary shift and possibly beneficial to host and associated gut microbiome? Here, we integrated evidences from genomics, metagenomics, and metabolomics in three honey bee subspecies maintained at the same locality of northern China to profile both diversity and functional variations in gut bacteria across seasons. Our results showed that winter dominance of Bartonella was shared in all tested honey bee lineages. This seasonal change was likely a consequence of winter dietary shifts characterized by greatly reduced pollen consumption and accumulation of metabolic waste due to restricted excretion. Bartonella showed expanded genomic capacity in utilizing more diverse energy substrates, such as converting metabolic wastes lactate and ethanol into pyruvate, an energy source for self-utilization and possibly also for host and other symbionts. Furthermore, Bartonella was the only bacterium capable of both producing and secreting tryptophan and phenylalanine, whose metabolic products were detected in bee guts, even though all gut bacteria lacked relevant digestion enzymes. These results thus suggested a possible mechanism where the gut bacteria might benefit the host by supplementing them with essential amino acids lacking in a protein shortage diet.
KEYWORDS: Apis mellifera, overwintering, pollen shortage, Bartonella, essential amino acids
INTRODUCTION
The honey bee (Apis mellifera) is an important pollinator that plays a critical ecosystem function in the native range, while also bearing high commercial value in producing bee products (1). As an adaptation to temperate climates, the emergence of long-lived workers (i.e., winter bees) is triggered by pollen resource dwindling (2), and the colony can survive cold winter by forming a thermoregulation cluster (“bee ball”) within the hive, generating heat via intensive vibration of flight muscles (3–6). At the same time, winter bees are confined to the hive without excretion (5), feeding mainly on stored honey (7). In addition to regulating hive temperature, winter bees will also participate in brood rearing in the coming spring (6–8). Hence, the health status of winter bees is vitally important for the whole colony, permitting successful propagation in the year round (9–12).
Honey bees harbor a relatively simple yet crucial gut microbiota, including five core gut bacterial lineages (Gilliamella, Snodgrassella, Lactobacillus Firm 4, Lactobacillus Firm 5, and Bifidobacterium) (13–15), accounting for >95% of the whole community, and ubiquitous bacteria in low quantity, such as Frischella, Commensalibacter, and Bartonella (15, 16). Increasing evidences have shown diverse beneficial effects of the core gut bacteria on honey bee host, such as immune stimulation (17), pathogenic parasites defense (18–21), detoxification (22), and growth promotion (23, 24). Contrary to extensive studies on core bacteria, the understanding of the impact of non-core bacteria (typically <5% abundance) on honey bees is limited.
The gut bacteria of honey bees are heritable and stable (13, 25), and they are transmitted via social behaviors but are also shaped by diverse factors, such as host genetics (25), antibiotics (26–28), pesticides (29), and food (30–32). In particular, food can drive the differentiation of gut bacterial strains in various animals, from Drosophila (33) to humans (34, 35). In bees, pollen diet is critical to the colonization of bee gut bacteria (36), therefore playing a vital role in shaping the gut microbiomes of the honey bees (37) and bumble bees (30). Moreover, the composition and quality of pollen may affect colony health via changing the gut community structure (31).
Given the critical role of honey bee gut bacteria and the impacts of food on both bee health and gut symbionts, an outstanding question remains to be addressed: how do honey bees and gut bacteria cope with the drastic shifts in dietary consumption during winter? During foraging seasons, honey bees consume both pollen and honey as primary food (38). Pollen is rich in nutrients, including ca. 5.9 to 11.5% fat, >20% protein, diverse fatty acids, vitamins, minerals, and antioxidant substances (39, 40), which play vital roles in bee metabolism and hormone regulation (41–43). During winter, without foraging and brood-rearing, bees mainly consume honey (5) but much less pollen (44–46). Honey constitutes primarily sugars, especially glucose and fructose (47, 48), while other nutrients are scarce (48). Therefore, winter bees can be challenged by the shortage of amino acids and lipids. In monophagous and oligophagous insects, such an unbalanced nutrition intake could be complemented by symbionts, a mechanism that effectively increases host fitness and adaptive capacities (49, 50).
Previous studies reported dramatic gut community variations in temperate honey bee colonies over winter, where a non-core bacterium, Bartonella, became dominant over core bacteria (36, 51, 52). However, it is not well known whether this microbiota variation is dependent on host lineage or geography, and the underlying cause for the increase of a non-core bacterium was unclear. Nevertheless, given the significantly decreased intake of pollen in temperate honey bees during winter (45), we hypothesize that variations in food structure may be driving the gut microbiome shift.
In this study, we sought to understand whether the gut microbiome turnover in winter bees is in concordance with pollen-reduced dietary shift at both community structure and functional levels and whether such variations are common across different honey bee lineages. Furthermore, we examined the possibility whether this seasonal variation might be beneficial to the honey bee host. Using combined evidences from subunit bacterial rRNA (16S) V4 gene fragment sequences, shotgun metagenomics and metabolomics, and multiple A. mellifera subspecies reared at the same locality in northeast China (Fig. 1; see also Table S1 in the supplemental material), we showed that seasonal bacterial community change was shared among honey bee lineages, with the non-core bacterium Bartonella becoming dominant during winter, while core bacteria remained at decreased abundances. This prominent bacterial turnover was likely due to increased fitness of Bartonella under reduced pollen diets because it is capable of utilizing alternative energy substances, e.g., lactate, acetate, and ethanol. Furthermore, comparative genomics revealed that several gut bacteria, especially Bartonella, might produce essential amino acids that could have served as a crucial supplement to the honey bee host subject to a protein deficient diet.
FIG 1.
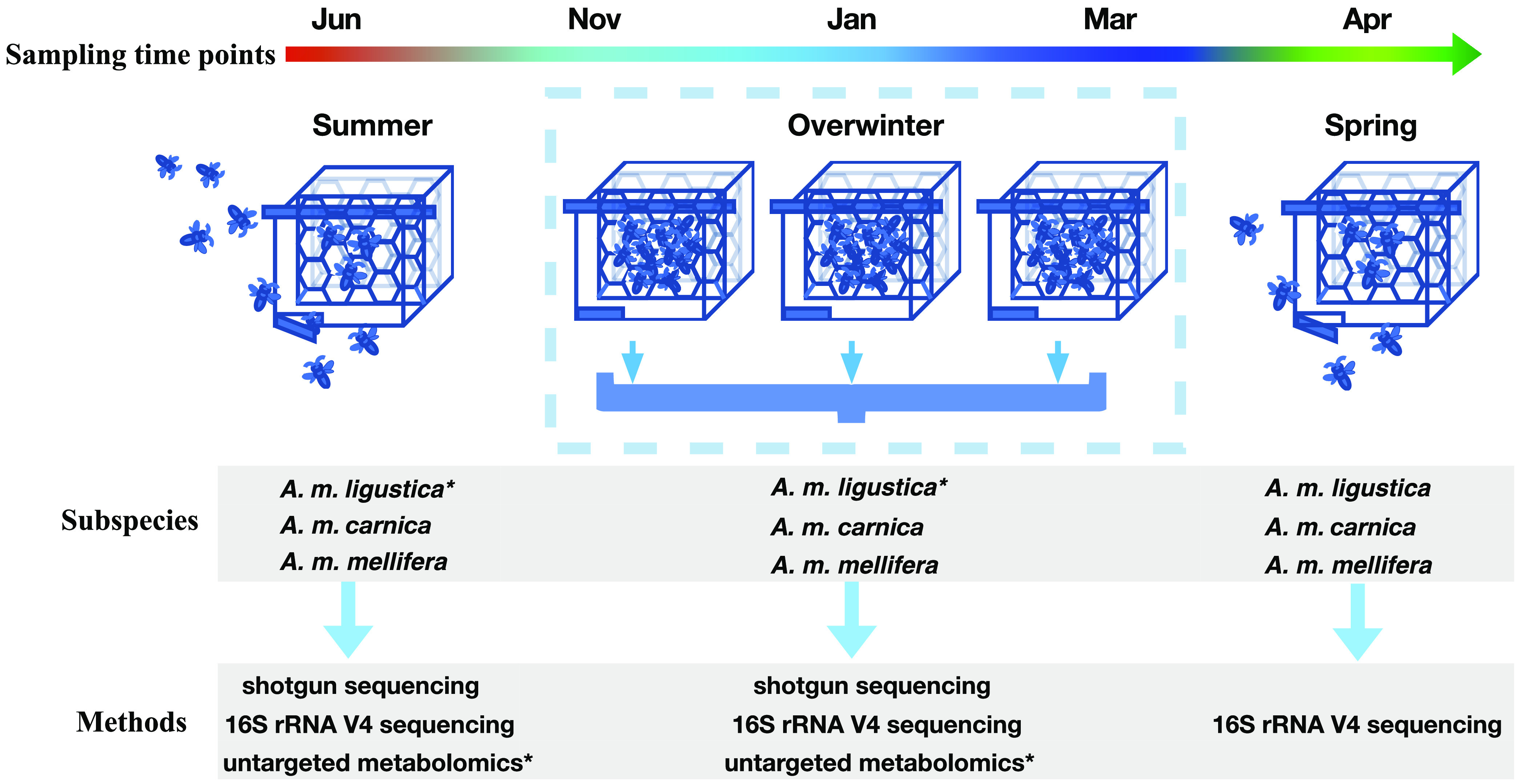
Schematic pipeline for honey bee sampling and analytical methods. Sampling time points: summer (June), early winter (November), midwinter (January), late winter (March), and spring (April). *, Samples analyzed for untargeted metabolomics.
Honey bees sampling and sequencing. Download Table S1, PDF file, 0.1 MB (122.2KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
Significant decline of pollen metabolites in winter bee guts.
Consistent with decreased pollen consumption in winter bees (44, 45, 53), metabolites derived from pollen were significantly reduced in the guts of winter bees. Untargeted metabolomic results for gut samples from four different time points (summer, June; early winter, November; midwinter, January; and late winter, March) revealed that gut metabolites varied significantly throughout the winter phase. In particular, flavonoids (kaempferol, keracyanin, and quercitrin) were all significantly reduced since the beginning of winter (Fig. 2; see also Table S2). The 9,10-dihydroxystearic acid derived from sporopollenin, spermidine, and tricoumaroyl spermidine from exosporium all decreased in the winter bees (Fig. 2; see also Table S2). These results indicated that reduced pollen consumption in the winter bees led to a decrease in relevant nutrients in honey bee guts, which would be expected to influence both the honey bees and their gut microbes.
FIG 2.
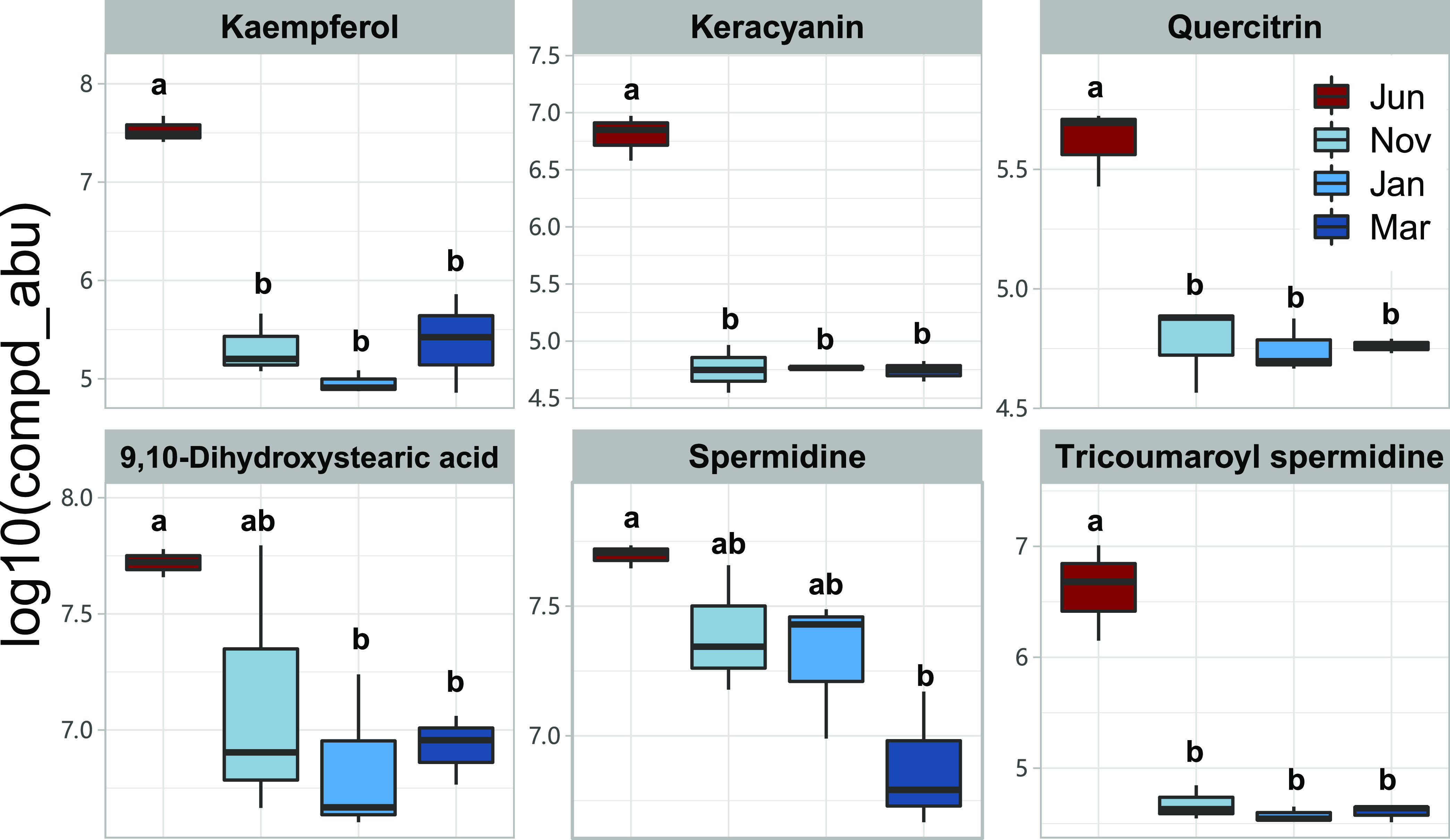
Seasonal variations in pollen-derived substances in honey bee guts. Metabolite variation in honey bee guts in summer (June), early winter (November), midwinter (January), and late winter (March). Letters above each column represent the levels of variation identified in the Wilcoxon test, where different letters indicate significant variations (P < 0.05).
Metabolites involved in metabolism of pollen and amino acids in the honey bee guts. Download Table S2, PDF file, 0.05 MB (48.9KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significant seasonal change of gut microbiota.
The gut bacterial community of the honey bee displayed seasonal variations, with the most prominent changes condensed in the transition phases of summer-winter and winter-spring (Fig. 3A to C). Throughout winter, the non-core bacterium Bartonella became dominant, while the core bacteria decreased conspicuously (Fig. 3A to C; see also Fig. S1A). A significant reduction in alpha diversity in winter bees was supported by both 16S (Kruskal-Wallis, P < 0.01) (see Fig. S1B) and shotgun metagenomics (see Fig. S1D). All the samples showed significant temporal clusters in both 16S (ANOSIM, r = 0.5597, P = 0.001; see Fig. S1C) and shotgun metagenomics results (ANOSIM, r = 0.3338, P = 0.001; see Fig. S1D). Overall, gut profiles showed clear seasonal turnover characterized by progressive changes.
FIG 3.
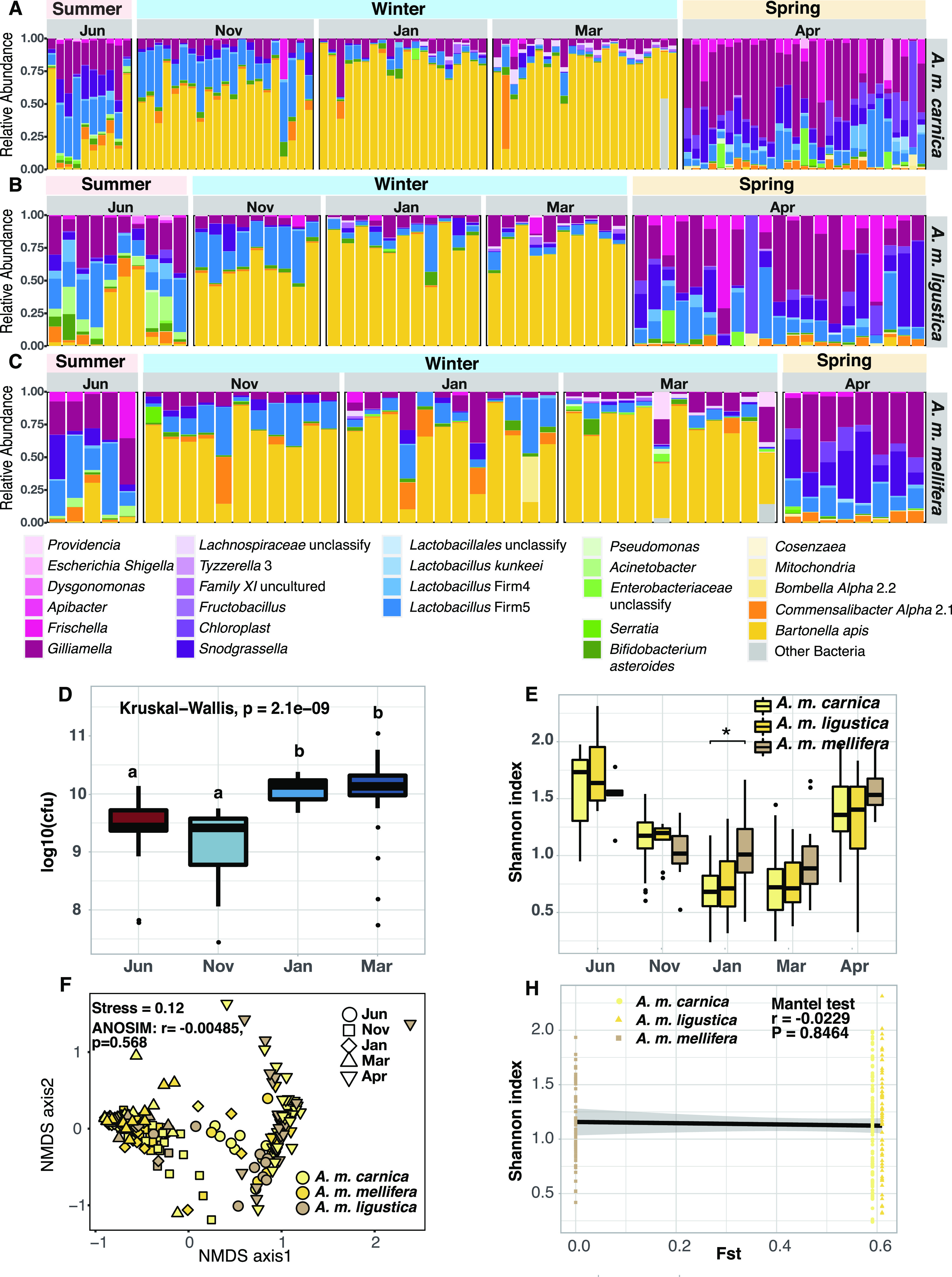
Seasonal variations in gut microbiomes in three honey bee subspecies. (A to C) Relative abundances of gut microbial phylotypes revealed by 16S rRNA V4. (A) Apis mellifera carnica; (B) Apis mellifera ligustica; (C) Apis mellifera mellifera. (D) Variations in live gut bacterial loads in honey bee gut at different times. Gut bacterial load (CFU) counting results for summer (June), early winter (November), midwinter (January), and late winter (March) are shown. (E) Alpha-diversities of gut microbiomes in three honey bee subspecies at different times. (F) NMDS (nonmetric multidimensional scaling) based on the Bray-Curtis dissimilarity determined by 16S rRNA V4. (G) Correlation between the diversity of the gut microbiome and host genetic divergence (Fst). *, P < 0.05 (Wilcoxon test).
Gut community composition and diversity differ between all times. (A) The relative community composition of the gut microbiota was determined by metagenome sequencing. (B and C) Variations in alpha-diversity (B) and NMDS based on Bray-Curtis dissimilarity (C) in microbiota across all times based on 16S rRNA. (D and E) Variations in alpha-diversity (D) and NMDS based on Bray-Curtis dissimilarity in microbiota (E) across all times based on metagenomics. The differences between groups are analyzed by Kruskal-Wallis. Letters above each column represent the levels of variation identified in the Wilcoxon test, where different letters indicate significant variations. Download FIG S1, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In addition to gut bacterial communities, distinguishable differences in the bacterial load in the bee guts were detected. Winter bees do not defecate until spring. Thus, both dead and live bacteria are included in regular metagenomics results. To exclude dead bacteria, we used CFU counts to detect live bacteria. A significant and continuous increase of live bacteria was observed in the bee guts during winter (Kruskal-Wallis, P < 0.05) (Fig. 3D). These results demonstrated that both structure and richness of gut microbiota were changed in winter.
Universal pattern of bacterium shifts in winter bees regardless of host subspecies.
The same microbiota change pattern was observed in all three honey bee subspecies examined in this study: Apis mellifera carnica, Apis mellifera ligustica, and Apis mellifera mellifera (Fig. 3A to C, respectively), which were reared at the same location and were managed following the same protocol. Bartonella was dominant in winter, while core bacteria were dominated after winter among all three subspecies. Along the timescale, all subspecies maintained gut microbiota at a significantly reduced alpha-diversity during winter (Fig. 3E). No significant differentiation was detected by Shannon index among hosts, in all but one time point (between Apis mellifera carnica and Apis mellifera ligustica at midwinter, P = 0.006) across the season (Fig. 3E). Samples cannot be differentiated by subspecies (ANOSIM, r = –0.00485, P = 0.5868; Fig. 3F), and no correlation with honey bee genetics (calculated by Fst [54]) was detected (Mantel test, r = –0.0229, P = 0.8464; Fig. 3G).
Seasonal function changes of gut microbiota.
Congruent with seasonal changes in bacterial communities, the microbiome function also showed remarkable shifts. The Cluster of Orthologous Groups of protein (COG) profiles were distinguishable and clustered by seasons, within which those sampled from midwinter and late winter (January and March) were prominently distinct from others (Fig. 4A). Although all samples shared the same COG categories, the relative abundance in each COG category was different among seasons. Among the eight metabolism COG categories, the abundance of C, E, H, P, I, and Q categories increased significantly in winter, while that of categories G and F was higher in summer (see Fig. S2). Furthermore, linear discriminant effect size (LEfSe) analyses indicated apparent function transitions between summer and winter bees (Fig. 4B). For example, in midwinter, functional proteins involved in amino-acid transport and metabolism (“E”), coenzyme transport and metabolism (“H”), and inorganic ion transport and metabolism (“P”) were enriched (Fig. 4B). During late winter, the proteins participated in lipid transport and metabolism (“I”) and energy production and conservation (“C”) were featured (Fig. 4B). In contrast, carbohydrate transport and metabolism (“G”) was featured during summer when dietary pollen was available at a regular load.
FIG 4.
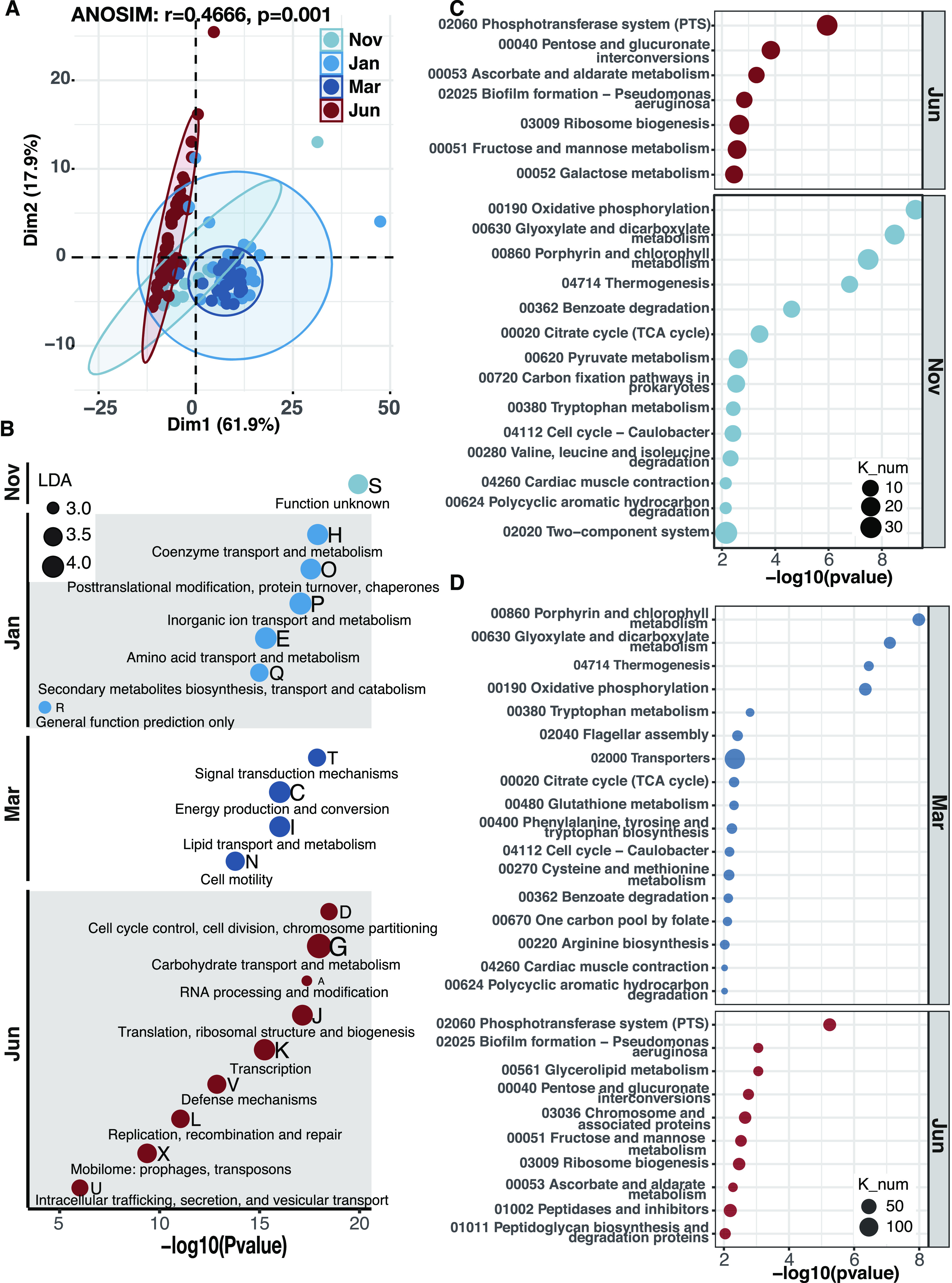
Changes in functional proteins of honey bee gut microbiome over seasons. (A) Principal coordinate analysis plots based on COG categories of different times. (B) LEfSe analysis of functional proteins of honey bee gut microbiota from different times. (C and D) Bubble plots represent enrichments of differential pathways: summer (June) versus early winter (November) (C) and summer (June) versus late winter (March) (D).
Relative abundances of COG annotations related to Metabolism between summer bees and winter bees. Labels: C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism. ****, P < 0.0001 (Wilcoxon test). Download FIG S2, EPS file, 1.5 MB (1.6MB, eps) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KEGG annotation results showed that changes in energy metabolism pathways were notable in the early winter bees compared to summer bees, showing enrichments in the oxidative phosphorylation pathway and genes involved in the utilization of pyruvate and dicarboxylic acid (Fig. 4C). The energy metabolism of gut bacteria in summer bees mainly focuses on carbohydrate transport and catabolism, including phosphotransferase system, galactose metabolism, pentose and glucuronate interconversions, and fructose and mannose metabolism (Fig. 4C). In contrast, in winter bees, carbohydrate catabolism declined significantly, while the carboxylic acid catabolism increased.
The microbiome energy functions in late winter phase were characterized by enrichments in the tricarboxylic acid (TCA) cycle pathway and dicarboxylic acid metabolism, while the pyruvate metabolism pathway was not featured (Fig. 4D). Moreover, amino acid metabolism (phenylalanine, tryptophan, and arginine synthesis pathways and cysteine, glutathione, and other metabolic pathways), as well as the metabolism of cofactors and vitamins, was significantly enriched (Fig. 4D). Also, expectedly, functional genes of the phosphotransferase system and carbohydrate metabolism pathways were significantly increased in summer, consistent with the restoration of foraged food.
Among the enrichment pathways in winter (Fig. 4C and D), Bartonella was the dominant contributor according to genome comparison among gut bacteria (see Fig. S3B and C). Moreover, the contribution from Bartonella continued to increase over winter, from ~70% (early winter; see Fig. S3B) to ~90% (late winter; see Fig. S3C), especially in amino acid metabolism and TCA cycle (see Fig. S3C). In addition, the non-core bacterium Commensalibacter was also prevalent in winter bee guts, representing larger contribution than core bacteria (see Fig. S3C). In contrast, the functional profile of summer guts, which had been restored to sugar transport and metabolism, was primarily influenced by core bacterium Gilliamella, followed by Lactobacillus and Bifidobacterium (see Fig. S3A). This pattern suggested that non-core bacteria played an important role in gut microbiome seasonal turnover and that Bartonella possessed a competitive advantage over core bacteria in winter bee guts.
Differential genes of enriched KEGG pathways and proportion of contributions from varied bacterial species. (A) Summer; (B) early winter; (C) late winter. Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bartonella was more versatile in energy production.
Based on enriched functional pathways, TCA cycle pathway was pronounced in late winter (Fig. 4) and Bartonella contributed the most (see Fig. S3C). To reveal Bartonella’s advantages, comparative analyses of whole genomes were conducted between Bartonella and core honey bee gut bacteria (see Table S4). The results showed that Bartonella had more diverse pathways in both basic energy production and the utilization of energy sources (Fig. 5A and B). All core bacteria, except Snodgrassella, could break down glucose into pyruvate through glycolysis either directly or indirectly. Snodgrassella, on the other hand, could conduct the TCA cycle (Fig. 5B), which remedies its deficiency in glycolysis pathway (Fig. 5A). In contrast, only Bartonella possessed both pathways (Fig. 5A and B).
FIG 5.
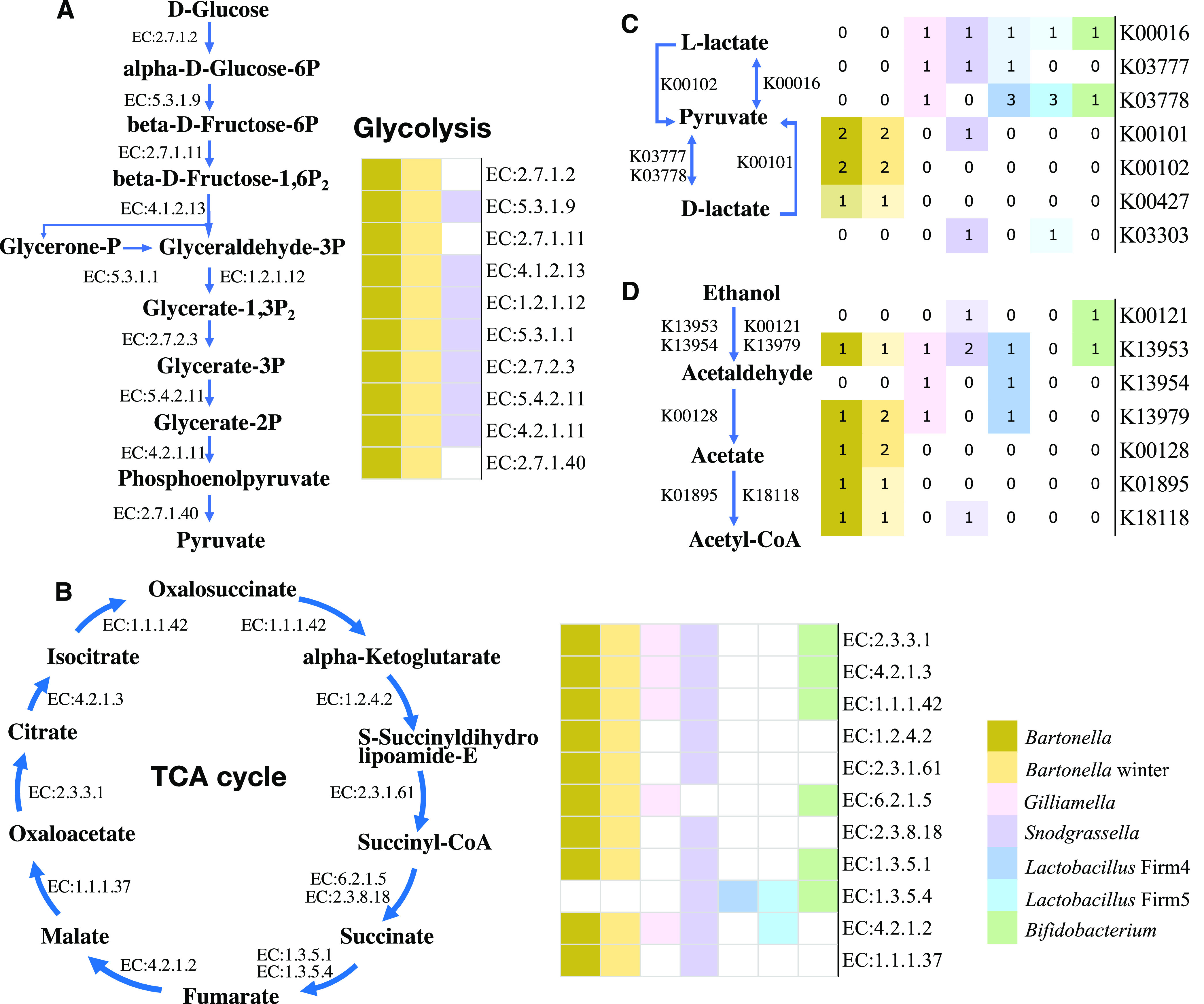
Key genes involved in basic energy pathways (glycolysis and TCA cycle) and waste degradation of Bartonella and core bacteria, as suggested by genomes. (A and B) Presence or absence of genes underlying glycolysis and TCA cycle in genomes of Bartonella (n = 6), a winter Bartonella strain (n = 1), and core bacteria (Gilliamella, n = 61; Snodgrassella, n = 9; Lactobacillus Firm 4, n = 2; Lactobacillus Firm 5, n = 13; and Bifidobacterium, n = 15) from honey bees. (A) Glycolysis; (B) TCA cycle. Colored boxes indicate presence; white boxes indicate absence. (C and D) Bartonella and core bacteria varied in copy numbers in genes involved in degradation of fermentation products (lactate, acetate, and ethanol). (C) Lactate; (D) ethanol and acetate. The numbers in the boxes represent gene copy numbers.
Genes involved in generating pyruvate and acetyl-CoA of Bartonella and core bacteria. Download Table S4, PDF file, 0.1 MB (145.8KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Congruently, Bartonella could utilize more diverse substances for energy production. In addition to glucose, Bartonella could also utilize end products of fermentation such as lactate, acetate, and ethanol produced by core bacteria, and independently produce acetyl coenzyme A (acetyl-CoA) (Fig. 5C and D), which can be introduced into TCA cycle or directly used in fatty acid synthesis. For example, Bartonella and Snodgrassella possessed lactate permease (K00427 or K03303), which could allow extracellular lactate into cells. In addition, Bartonella encoded more copies of lactate dehydrogenase (cytochrome) genes than did core bacteria (Fig. 5C), which could catalyze the oxidation of lactate to pyruvate. In addition, only Bartonella had the aldehyde dehydrogenase gene (NAD+) (K00128), which could convert ethanol into acetate and further into acetyl-CoA (Fig. 5D). Also, Bartonella possessed acetyl-CoA synthetase (K01895) for de novo synthesizing acetyl-CoA from acetate (Fig. 5D).
Bartonella was capable of supporting host overwintering with nutrients.
Metagenome function profiles showed that amino acid biosynthesis pathways were significantly enhanced in late winter (Fig. 4D), including those of arginine, phenylalanine, tryptophan, cysteine, and methionine, where Bartonella had contributed the most (see Fig. S3). Comparative genome analysis revealed that a few essential amino acids, such as phenylalanine and tryptophan, could be synthesized de novo by Bartonella, Gilliamella, and Snodgrassella (see Table S3). Interestingly, these bacteria lacked relevant genes to catabolize such amino acids. Compared to Gilliamella and Snodgrassella, only Bartonella encoded the general l-amino acid ABC transporters (AapP, AapQ, AapM, AapJ, see Table S3), which were responsible for the extracellular exportation of amino acids.
Phenylalanine, tyrosine, and tryptophan biosynthesis and general l-amino acid transport system. Download Table S3, PDF file, 0.1 MB (77.2KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The potential bacterial contribution in amino acid synthesis was further supported by our metabolomics results. With a significant reduction in pollen consumption during winter, it was expected that the essential amino acids from pollen would be decreased. However, we detected increased related metabolites in winter bees. The downstream catabolized metabolites of tryptophan and phenylalanine, e.g., tryptamine and tyramine, were significantly elevated. Other catabolites of tryptophan, such as kynurenate and 3-hydroxy-l-kynurenine, were also increased during winter. Similarly, the downstream product of tyrosine (tyramine) derived from the essential amino acid phenylalanine, was increased during winter (Fig. 6; see also Table S2). These products were not likely produced by gut bacteria since bacteria lacked complete gene sets of the catabolic pathways (Fig. 6). Furthermore, as indicated by the pathway enrichment of phenylalanine and tryptophan biosynthesis in gut bacteria, the precursors (e.g., erythrose 4-phosphate) and intermediate substrates of these amino acids (e.g., quinate, shikimate, and chorismate) were significantly increased during winter (Fig. 6; see also Table S2).
FIG 6.
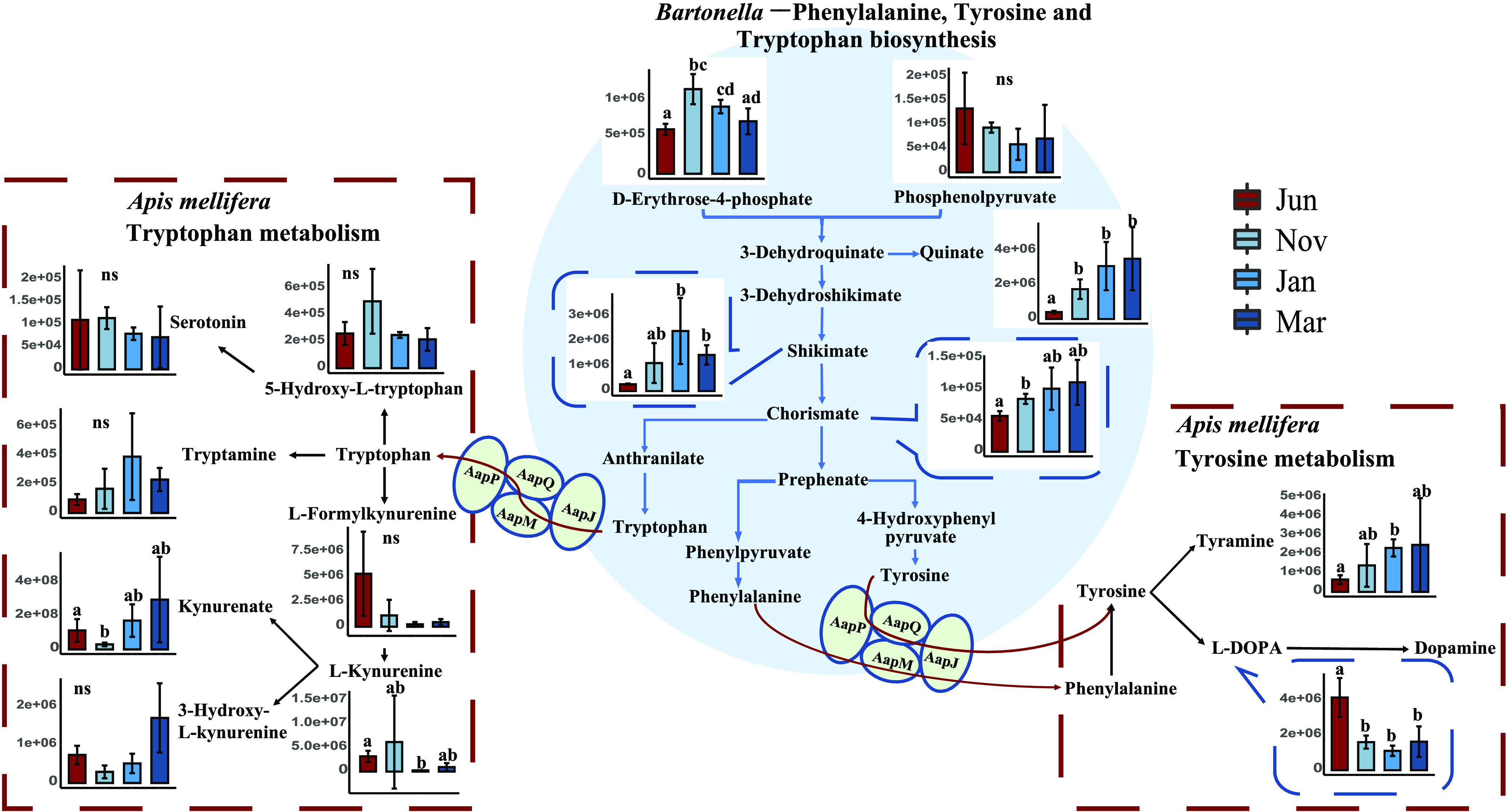
Biosynthesis and degradation of phenylalanine, tryptophan, and tyrosine. The biosynthesis of phenylalanine, tryptophan, and tyrosine by Bartonella is shown with a blue background. A simplified process for the host to decompose tryptophan, phenylalanine, and tyrosine is shown in the red box. Plots represent metabolites involved in the metabolism of tryptophan, phenylalanine, and tyrosine. Letters above each column represent the levels of variation identified in a Wilcoxon test, where different letters indicate significant variations. “ns” represents no significance.
DISCUSSION
For the honey bees, the capability to survive winter is a key adaptive mechanism during its tropic-temperate habitat expansion. However, the behavioral adaptation (forming bee balls and vibrating) and subsequent change in honey bee biology (dietary change and nonexcretion) is a double-edged sword. While it warrants a relative stable colony temperature (6, 55), it also creates a novel challenge to both the host and the gut microbiome. Our study characterized the apparent changes in honey bee gut profile, reflected in both bacterial community structure and function, to understand the underlying mechanism.
The gut profile variation is in concordance with winter dietary shifts.
It is well known that the majority of pollen-derived nutrition is engaged in honey bee brood-rearing (56) and that the reproduction rhythm of the colony is highly coordinated with the availability of pollen resource (2). For instance, pollen shortage appeared to be a direct food cue (along with environmental changes) that introduced the emergence of winter bees in the fall (2). Although few studies have examined specific changes in colony pollen consumption loads over winter, reports showed that a winter colony might only maintain a pollen hoard equivalent to what was regularly consumed by a summer colony in a single day (45, 46). In the present study, we investigated metabolic variation in winter bee guts. Considering that nutritional variations (e.g., proteins, amino acids, and lipid acids) have been reported between stored and freshly collected pollen (57, 58) and that the winter bees almost exclusively fed on stored pollen, we measured pollen wall components and bioactive constituents (flavonoids) in bee guts, instead of nutrients, as a proxy to quantify pollen consumption change over winter (Fig. 2). Pollen wall components and flavonoids are derived solely from pollen and showed no obvious reduction through storage time (59, 60). Therefore, the reduction in gut flavonoids revealed in our study provided metabolic evidence for pollen consumption decrease in winter bees. On the other hand, the nutrient variations in stored pollen may also have contributed to the reduction of pollen-derived metabolites. Although pollen consumption variation may seem to play a larger role, a systematic quantification of such changes will help to elucidate relative contributions of consumption and nutrient changes to variations in pollen-derived metabolites in winter bee gut. Nevertheless, our study supports that the winter adaptation of the temperate honeybees is associated with dietary tradeoffs between elevated honey and reduced pollen proportions, which in turn triggers a reproduction pause that lasts most of the winter. The shifts in dietary structure show a strong impact on honeybee gut microbiota on both community structure and function profiles (36). As soon as the colony stops foraging, the gut bacterial community switches rapidly to the winter configuration, with declined core bacteria and dominating Bartonella, and then returns to the “normal” phase, once food conditions become favorable in the spring. The coordinated reproduction and foraging behaviors of the colony are likely adaptive traits of the host, which is further echoed in the microbiota (Fig. 3A to C). For example, the elevation in lipid transport and metabolism in March may reflect the initiation of the spring brood.
Winter bacterial turnover is a consistent trait associated with climates but not host lineages.
The characteristic rise of Bartonella is notably associated with the temperate winter, during which the experiments were conducted, rather than taxonomic lineage of the honeybees. Previous studies conducted in temperate regions, e.g., Switzerland (36), Canada (61), and Anhui, China (51), showed a similar trend in gut change, which is congruent with findings in the present work. Conversely, seasonal surveys of honey bee gut bacteria on colonies reared in the subtropical regions, such as in Arizona (62) and Colonia, Uruguay (63), revealed much more stabilized gut microbiota throughout the seasons, where Bartonella never became dominant. On the other hand, the winter gut profile appeared not to be determined by honeybee lineages. In our study, the three subspecies we examined, reared in sympatry, exhibited a similar trend in gut community variation (Fig. 3). Similarly, multiple hives of local hybrids between Apis mellifera scutellata, Apis mellifera ligustica, and Apis mellifera mellifera shared the subtropical gut profile as described above (63). These results are very different from controlled experiments under laboratory conditions, where host genetics showed a significant indigenous effect on honey bee gut composition (25), suggesting that the seasonal change in food structure has a stronger impact on the gut microbiota in the studied system here.
Bartonella is a commensal bacterium in honey bees with high prevalence yet low abundance. However, sporadic elevation of Bartonella in summer has also been reported occasionally (52, 64). Possible reasons may include altered diet, temperature change, and behavioral variation of the workers. We expect that systematic tracking of individual biology and behavior may help to elucidate the specific cause of summer abnormality in Bartonella. On the other hand, increasing studies have pointed to its correlation to cold conditions. For example, Papp et al. (52) revealed temporal shifts in Bartonella with increased abundance were only found at cooler sites. In particular, more studies have shown that Bartonella significantly increased during winter (36, 51, 61). Our study suggests, however, that the dominance of Bartonella is probably not directly associated with temperature per se but rather that the pollen-reduced diet structure caused by seasonal change. Under this special diet structure, the diversified energy pathways may have enabled Bartonella to utilize other energy sources, showing superior capability and competitive advantages over core bacteria during winter. Such a diversified capacity may explain the abrupt and consistent increase of Bartonella under the temperate winter condition.
Bartonella might be beneficial to host and other gut bacteria.
In addition to the obvious benefit to its own survival, Bartonella may be also beneficial to the host and co-occurring gut bacteria. Our genomic inferences indicated that Bartonella could generate pyruvate. This result is in congruent with a previous study based on cross-feeding between honey bee gut symbionts, in which the increase of pyruvate was observed in bee guts mono-colonized with Bartonella (23). Pyruvate acts as an important energy source, which could be directly utilized by both bacteria and the host (65). In addition, pyruvate can promote the synthesis of trehalose, which is known for its function in improving cold hardiness in insects (66).
Furthermore, our study suggests a new possibility that Bartonella might provide essential amino acids to the host. Pollen is the major nutrient food for honey bees, containing a variety of proteins, lipids, carbohydrates, and vitamins (67, 68). The pollen-derived essential amino acids, such as tryptophan and phenylalanine, are precursors of neurotransmitters, which are involved in the regulations of physiological metabolism, nutrient intake, and labor division in honey bees (69). Thus, a shortage of pollen-derived essential amino acids and proteins may have a deleterious impact on bee social behaviors and colony health during winter or even on rear brooding potentials in the spring.
An increasing body of studies have shown that the host could obtain essential amino acids or proteins through symbionts to maintain protein balance (70, 71), such as in termites and brown planthoppers, both of which live on unbalanced diets (72–75). In the present study, our results suggest that the winter diet challenge might be remedied by corresponding changes in gut microbial community and function. Our metagenomics results show that the pathways of amino acid biosynthesis are enriched in late winter, including those of several essential amino acids of honey bees, i.e., phenylalanine, tryptophan, and methionine (Fig. 4), which is primarily contributed by Bartonella (see Fig. S3C). Consistently, metabolic intermediates (i.e., quinate, shikimate, and chorismite) produced in bacterial synthesis of phenylalanine and tryptophan have increased during winter, especially in late winter (Fig. 6), which further underlines the role of the gut microbiota as amino acids providers. Interestingly, Gilliamella, Snodgrassella, and Bartonella are all able to independently synthesize tryptophan and phenylalanine and yet are incapable of further decomposition. In principle, these amino acids could be supplied to the host. Congruently, our results reveal elevations in metabolic products derived from degradations of tryptophan and phenylalanine (Fig. 6). These products are most likely generated by the bee host, since only the honey bee possesses the complete pathways. In addition, the genes associated with l-type amino acid transports are only found in the Bartonella, allowing them to excrete amino acids out of the cell, while other core bacteria lack these genes. Hence, Bartonella may be the major contributor to provide essential amino acids for honey bees during winter, facilitating the winter bees in maintaining health and synthesizing protein for brood.
The interactive responses between host and symbionts under environmental stress have been described in many animal systems, such as in yaks (76, 77), wild mice (78), wild red squirrels (79), ground squirrels (80), stinkbugs (81), and crickets (82). Here, we report that the seasonal dynamics between host food diets and corresponding gut bacterial profiles may reflect coordinated responses of the honey bee and its bacterial symbionts under extreme food and environmental stresses. We showed multiple lines of evidence that are in accordance with the hypothesis that this seasonal variation might involve host-gut bacteria interactions, possibly reflecting adaptation. Admittedly, conclusive evidence is still lacking to provide a definite answer. We expect that further experiments designed specifically to test the impacts of Bartonella on the overall fitness of winter bees will help elucidate the evolutionary mechanism underlying the host and its gut symbionts. Finally, in addition to classic evidence where core bacteria can improve the overall honey bee fitness, our study adds yet another insight that honey bees might be able to take advantage of non-core bacteria in extreme cold conditions and are able to restore gut homeostasis when such stresses are removed.
MATERIALS AND METHODS
Honey bee sampling.
Honey bee colonies were kept in the apiary of the Jilin Bee Research Institute (located in east longitude 99.58°, northern latitude 25.55°). Sampling was carried out from November 2017 to June 2018. Nurse bees were sampled in June 2018: winter bees were sampled from November 2017 to March 2018; nurse bees transformed from over winter bees (termed “spring bees” here) were sampled in April 2018 (see Table S1). Approximately 40 workers were haphazardly sampled twice a month from each of the 8 hives (2 for Apis mellifera mellifera, 4 for Apis mellifera carnica, and 2 for Apis mellifera liguistica). All honey bee individuals were dissected with sterile forceps to obtain gut tissues, including the mid- and hindguts. Each gut sample was preserved in a 1.5-mL Eppendorf tube, frozen in liquid nitrogen, and later stored at −80°C.
Untargeted metabolomics.
Guts dissected from winter and summer bees were subject to untargeted metabolomic analyses at Novogen (Beijing, China) (Fig. 1). Each gut (ca. 30 to 70 mg) was individually ground with liquid nitrogen, and the homogenate was resuspended with prechilled 80% methanol and 0.1% formic acid and vortexed. Sample was incubated on ice for 5 min and centrifuged at 15,000 rpm and 4°C for 5 min. The supernatant was diluted to a final concentration of 60% methanol using liquid chromatography-mass spectrometry (LC-MS)-grade water. These samples were subsequently transferred to a new Eppendorf tube with a 0.22 μm filter and then centrifuged at 15,000 rpm and 4°C for 10 min. Finally, the filtrate was used for LC-MS/MS analyses.
The LC-MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Samples were injected onto a Hyperil Gold column (100 × 2.1 mm, 1.9 μm) using a 16-min linear gradient at a flow rate of 0.2 mL/min. The eluents for the positive polarity mode were eluent A (0.1% formate in water) and eluent B (methanol). The eluents for the negative polarity mode were eluent A (5 mM ammonium acetate [pH 9.0]) and eluent B (methanol). The solvent gradient was set as: 2% B for 1.5 min, an increase to 100% B until 12 min, maintenance for 2 min, and a reduction to 2% B over 0.1 min, followed by holding for 2 min. A Q Exactive HF-X mass spectrometer was operated in the positive/negative polarity mode with a spray voltage of 3.2 kV, a capillary temperature of 320°C, a sheath gas flow rate of 35 arbitrary units, and an auxiliary gas flow rate of 10 arbitrary units. The raw data files generated by UHPLC-MS/MS were processed using Compound Discoverer 3.0 (CD 3.0; Thermo Fisher).
DNA extraction.
DNA extraction of each batch of samples was conducted within a month after gut dissection. A CTAB (cetyltrimethylammonium bromide)/phenol-based extraction method (24) was used in DNA extraction with minor modification. Briefly, the whole gut was resuspended in a 2-mL tube containing 728 μL of CTAB buffer and 20 μL of 20 mg/mL proteinase K (TransGen Biotech). The mixture was then ground on ice using a TGrinder OST-Y 30, at 8000 rpm for 15 s, and this was repeated three times. Sterile zirconia beads (100 μL [diameter, 0.1 mm]; BioSpec, Bartlesville, OK) and 2 μL of mercaptoethanol were then added to the tubes. Tissues were vortexed using the MOBIO Vortex Genie for 3 min and then lysed by adding 5 μL of RNase A (TransGen Biotech), followed by incubation at 56°C overnight. Lysis was performed using centrifugation at 12,000 × g for 5 min, followed by transfer to a new 1.5-mL EP tube after the supernatant was removed. The pellet was mixed with 400 μL of phenol-chloroform-isoamyl alcohol (25:24:1), and the mixture was centrifuged at 12,000 × g for 15 min. The supernatant was transferred into a new 1.5-mL tube, and 50 μL of 3 M sodium acetate and 500 μL of isopropanol were added, followed by incubation at −20°C. After centrifugation at 17,000 × g for 30 min, the pellets were washed twice with 70% ethanol. Finally, DNA pellets were dissolved in 50 μL of Tris-EDTA buffer (pH 8.0).
High-throughput sequencing of the 16S rRNA gene fragments.
Amplification of the V4 region of the small subunit (16S) rRNA gene was performed for winter bees (November, January, and March), spring bees (April), and summer bees (June) (Fig. 1). Primers 515F and 806R and a general PCR program were used for 16S V4 amplification. The PCR master mix without DNA template was used as a negative control. Amplicons were sequenced using an Illumina Nova6000 platform with 250-bp paired-end (PE 250) reads, where >30,000 sequences were obtained for each sample. Fastp was used to control the quality of the raw data, and the default parameters were applied to remove reads with a quality value of <20 (83). The program FLASH was used for splicing (-m 15 -x 0.1) (84), and then the merged contigs were imported into QIIME 2 (v2018.8.0) (85). Quality control, denoising, and chimera elimination were performed using DADA2 (86) in QIIME 2 (v2018.8.0). Finally, the representative sequences were classified by a curated SILVA database (https://doi.org/10.5281/zenodo.6772394) for bee gut microbiota (87).
Shotgun sequencing and de novo assembly.
A total of 121 honey bee guts sampled from four time points (November, January, March, and June) were shotgun sequenced (Fig. 1) using a BGI-500 platform with 100 bp paired-end or an Illumina HiSeq X-10 platform with 150 bp paired-end. Raw Illumina reads were filtered using Fastp (v0.13.1 -q 20 -u 10 -w 16) (83), and high-quality reads were subsequently mapped onto the A. mellifera genome (GCF_000002195.4) using BWA aln (v0.7.16) (88) to filter out honey bee reads. The remaining reads were de novo assembled using Megahit (v1.1.2, k-list 51,61,71,81,91,101,111) (89) for each metagenome, where contigs longer than 500 bp were kept and blast analyzed against the NCBI nr database using DIAMOND (v0.9.22.123, blastx -f 102 -k 1 -e 1e-3) (90) for taxonomic assignment. A customized bacterial database was combined with the NCBI bacterial genomes (37). Assemblies assigned to bacteria were then blast analyzed against the customized bacterial database (blastn -outfmt 6 -e 1e-5 -max_target_seqs 5). Sequences longer than 100 bp were assigned to general bacteria or specific species, when they had a similarity of >30% or >90% to the reference, respectively. For each metagenome, clean reads were mapped onto the assemblies using SOAPaligner (v2.21, -M 4 -l 30 -r 1 -v 5 -m 200) and summarized using the SOAP.coverage script (91). Only assemblies with a sequence coverage of >90% were kept for subsequent analyses. The relative abundance of a bacterial species was defined as the number of all bases assigned to the focal species divided by the total number of bases belonging to bacteria in each sample.
Gene prediction and annotation.
Genes were predicted for bacterial assemblies using MetaGeneMark_linux_64 (92). All predicted genes were clustered using CD-HIT (v4.6.7, -c 0.95 -r 1 -G 1 -g 1 -aS 0.9 -T 24 -M 0) (93, 94) to get a nonredundant gene catalog. Finally, all amino acid sequences of clustered representative genes were annotated using COG category annotation (95) and online KEGG annotation (96). Clean reads were mapped to the nonredundant gene catalog using SOAPaligner, and the gene abundance was summarized using SOAP.coverage (91). The sequence depth of a particular gene was calculated as the total bases mapped onto the focal gene divided by the total bases mapped to any genes in each sample. The differential COG proteins and genes were using LEfSe (97). The pathway enrichment was conducted using a one-sided Fisher exact test in the R package.
Comparison of bacterial genome.
Core bacterium (100 strains) and Bartonella (6 strains) genomes (see Table S5) were downloaded from the NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/). Bartonella W7133 strain was isolated from winter bee as previously described (98). All the strains were annotated by online KEGG BlastKOALA (96).
Bacterial genomes of Bartonella and honey bee core bacteria list. Download Table S5, PDF file, 0.1 MB (85KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial quantification of gut microbes.
All gut glycerol homogenates were stored in a −80°C freezer for ca. 2 years before plate counting. They were then diluted with phosphate-buffered saline (pH 7.2) and plated on heart infusion agar with 5% sheep blood. Bacterial colonies were subsequently counted after incubation at 35°C and 5% CO2 for 48 h.
Statistical analyses.
All statistical analyses were performed in R version 3.6.0 and visualized using ggplot2 in R. Alpha- and beta-diversities were calculated using the vegan package (99). The Kruskal-Wallis test and Wilcoxon test were used for multigroups, and a P value of <0.05 indicated statistical significance.
Data availability.
Raw data for 16S rRNA and shotgun sequences of winter bees have been deposited under BioProject PRJNA797557, and raw data for 16S rRNA and shotgun sequences of summer bees have been deposited under BioProject numbers PRJNA645267 and PRJNA645015, respectively, in the NCBI database. In-house scripts are available from the corresponding authors upon request.
ACKNOWLEDGMENTS
This work was supported by National Special Support Program for High-Level Talents (10-Thousand Talents Program), Ministry of Science and Technology of the People’s Republic of China (MOST; 2018FY100403), National Natural Science Foundation of China (NSFC; 31772493), and the 2115 Talent Development Program of China Agricultural University grants to X.Z. and by NSFC (31350006) and Jilin Science and Technology Program (20200201197JC) grants to X.L.
Many colleagues and collaborators facilitated sampling or contributed samples, including Fa Zhang, Haisheng Wang, Jinzhou Wang, Xinming Wang, Kai Xu, Wenli Wu, and Zhi Wang from the Jilin Provincial Institute of Apicultural Sciences. Haoyu Lang, Qinzhi Su, Chengfeng Yang, and Jiaqiang Wu from the China Agricultural University also contributed samples to this study.
We declare there are no competing financial interests.
X.Z. and X.L. designed and coordinated the study. C.L. conducted sample collection, bacterial isolation, genome annotation, DNA extraction, and metabolic and metagenomics analyses. M.T. constructed metagenomics workflows. C.L., X.Z., M.T., and X.L. wrote the first drafts, and all authors contributed to and proofed the manuscript.
Contributor Information
Xingan Li, Email: lxingan@sina.com.
Xin Zhou, Email: xinzhou@cau.edu.cn.
Jennifer B. H. Martiny, University of California, Irvine
REFERENCES
- 1.Hoover SE, Ovinge LP. 2018. Pollen collection, honey production, and pollination services: managing honey bees in an agricultural setting. J Econ Entomol 111:1509–1516. doi: 10.1093/jee/toy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattila HR, Otis GW. 2007. Dwindling pollen resources trigger the transition to broodless populations of long-lived honeybees each autumn. Ecol Entomol 32:496–505. doi: 10.1111/j.1365-2311.2007.00904.x. [DOI] [Google Scholar]
- 3.Jacques A, Laurent M, Ribiere-Chabert M, Saussac M, Bougeard S, Budge GE, Hendrikx P, Chauzat MP, de Graaf D, Meroc E, Nguyen BK, Roelandt S, Roels S, Van der Stede Y, Tonnersen T, Kryger P, Jaarma K, Kuus M, Raie A, Heinikainen S, Pelkonen S, Vahanikkila N, Andrieux C, EPILOBEE Consortium, et al. 2017. A pan-European epidemiological study reveals honey bee colony survival depends on beekeeper education and disease control. PLoS One 12:e0172591. doi: 10.1371/journal.pone.0172591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doke MA, Frazier M, Grozinger CM. 2015. Overwintering honey bees: biology and management. Curr Opin Insect Sci 10:185–193. doi: 10.1016/j.cois.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Knoll S, Pinna W, Varcasia A, Scala A, Cappai MG. 2020. The honey bee (Apis mellifera L., 1758) and the seasonal adaptation of productions: highlights on summer to winter transition and back to summer metabolic activity—a review. Livest Sci 235:104011. doi: 10.1016/j.livsci.2020.104011. [DOI] [Google Scholar]
- 6.Johansson TSK, Johansson MP. 1979. The honeybee colony in winter. Bee World 60:155–170. doi: 10.1080/0005772X.1979.11097754. [DOI] [Google Scholar]
- 7.SEELEY TD, VISSCHER PK. 1985. Survival of honeybees in cold climates: the critical timing of colony growth and reproduction. Ecol Entomol 10:81–88. doi: 10.1111/j.1365-2311.1985.tb00537.x. [DOI] [Google Scholar]
- 8.Johnson BR. 2010. Division of labor in honeybees: form, function, and proximate mechanisms. Behav Ecol Sociobiol 64:305–316. doi: 10.1007/s00265-009-0874-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schafer MO, Ritter W, Pettis JS, Neumann P. 2010. Winter losses of honeybee colonies (Hymenoptera: Apidae): the role of infestations with Aethina tumida (Coleoptera: Nitidulidae) and Varroa destructor (Parasitiformes: Varroidae). J Econ Entomol 103:10–16. doi: 10.1603/ec09233. [DOI] [PubMed] [Google Scholar]
- 10.Bahreini R, Currie RW. 2015. Influence of honey bee genotype and wintering method on wintering performance of Varroa destructor (Parasitiformes: Varroidae)-infected honey bee (Hymenoptera: Apidae) colonies in a northern climate. J Econ Entomol 108:1495–1505. doi: 10.1093/jee/tov164. [DOI] [PubMed] [Google Scholar]
- 11.Brodschneider R, Gray A, Adjlane N, Ballis A, Brusbardis V, Charrière J-D, Chlebo R, Coffey MF, Dahle B, de Graaf DC, Maja Dražić M, Evans G, Fedoriak M, Forsythe I, Gregorc A, Grzęda U, Hetzroni A, Kauko L, Kristiansen P, Martikkala M, Martín-Hernández R, Aurelio Medina-Flores C, Mutinelli F, Raudmets A, A Ryzhikov V, Simon-Delso N, Stevanovic J, Uzunov A, Vejsnæs F, Wöhl S, Zammit-Mangion M, Danihlík J. 2018. Multi-country loss rates of honey bee colonies during winter 2016/2017 from the coloss survey. J Apic Res 57:452–457. doi: 10.1080/00218839.2018.1460911. [DOI] [Google Scholar]
- 12.Steinmann N, Corona M, Neumann P, Dainat B. 2015. Overwintering is associated with reduced expression of immune genes and higher susceptibility to virus infection in honey bees. PLoS One 10:e0129956. doi: 10.1371/journal.pone.0129956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones JC, Fruciano C, Hildebrand F, Al Toufalilia H, Balfour NJ, Bork P, Engel P, Ratnieks FLW, Hughes WOH. 2018. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol Evol 8:441–451. doi: 10.1002/ece3.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson KE, Ricigliano VA. 2017. Honey bee gut dysbiosis: a novel context of disease ecology. Curr Opin Insect Sci 22:125–132. doi: 10.1016/j.cois.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Kwong WK, Medina LA, Koch H, Sing KW, Soh E, Ascher JS, Jaffé R, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci Adv 3:e1600513. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellegaard KM, Engel P. 2019. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun 10:446. doi: 10.1038/s41467-019-08303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci 4:170003. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janashia I, Alaux C. 2016. Specific immune stimulation by endogenous bacteria in honey bees (Hymenoptera: Apidae). J Econ Entomol 109:1474–1477. doi: 10.1093/jee/tow065. [DOI] [PubMed] [Google Scholar]
- 19.Forsgren E, Olofsson TC, Vásquez A, Fries I. 2010. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108. doi: 10.1051/apido/2009065. [DOI] [Google Scholar]
- 20.Li JH, Evans JD, Li WF, Zhao YZ, DeGrandi-Hoffman G, Huang SK, Li ZG, Hamilton M, Chen YP. 2017. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS One 12:e0187505. doi: 10.1371/journal.pone.0187505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonard SP, Powell JE, Perutka J, Geng P, Heckmann LC, Horak RD, Davies BW, Ellington AD, Barrick JE, Moran NA. 2020. Engineered symbionts activate honey bee immunity and limit pathogens. Science 367:573–576. doi: 10.1126/science.aax9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA, Maleszka R, McFall-Ngai MJ. 2016. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 7:e01326-16. doi: 10.1128/mBio.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesnerova L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, Engel P. 2017. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15:e2003467. doi: 10.1371/journal.pbio.2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA 114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Lang H, Mu X, Zhang Z, Su Q, Hu X, Zheng H. 2021. Honey bee genetics shape the strain-level structure of gut microbiota in social transmission. Microbiome 9:225. doi: 10.1186/s40168-021-01174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA, Kolter R. 2012. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3:e00377-12. doi: 10.1128/mBio.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludvigsen J, Porcellato D, L’Abee-Lund TM, Amdam GV, Rudi K. 2017. Geographically widespread honeybee-gut symbiont subgroups show locally distinct antibiotic-resistant patterns. Mol Ecol 26:6590–6607. doi: 10.1111/mec.14392. [DOI] [PubMed] [Google Scholar]
- 28.Raymann K, Bobay LM, Moran NA. 2018. Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol Ecol 27:2057–2066. doi: 10.1111/mec.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Ma SL, Yan ZX, Liu F, Diao QY, Dai PL. 2019. Effects of three common pesticides on survival, food consumption and midgut bacterial communities of adult workers Apis cerana and Apis mellifera. Environ Pollut 249:860–867. doi: 10.1016/j.envpol.2019.03.077. [DOI] [PubMed] [Google Scholar]
- 30.Martinson VG, Magoc T, Koch H, Salzberg SL, Moran NA. 2014. Genomic features of a bumble bee symbiont reflect its host environment. Appl Environ Microbiol 80:3793–3803. doi: 10.1128/AEM.00322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maes PW, Rodrigues PAP, Oliver R, Mott BM, Anderson KE. 2016. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol Ecol 25:5439–5450. doi: 10.1111/mec.13862. [DOI] [PubMed] [Google Scholar]
- 32.Huang SK, Ye KT, Huang WF, Ying BH, Su X, Lin LH, Li JH, Chen YP, Li JL, Bao XL, Hu JZ, Shade A. 2018. Influence of feeding type and Nosema ceranae infection on the gut microbiota of Apis cerana workers. mSystems 3:e00177-18. doi: 10.1128/mSystems.00177-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martino ME, Joncour P, Leenay R, Gervais H, Shah M, Hughes S, Gillet B, Beisel C, Leulier F. 2018. Bacterial adaptation to the host’s diet is a key evolutionary force shaping Drosophila-Lactobacillus symbiosis. Cell Host Microbe 24:109–119.e6. doi: 10.1016/j.chom.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, Batres R, Paw B, Pergament SL, Saenyakul P, Xiong M, Kim AD, Kim G, Masopust D, Martens EC, Angkurawaranon C, McGready R, Kashyap PC, Culhane-Pera KA, Knights D. 2018. US immigration westernizes the human gut microbiome. Cell 175:962–972. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Walter J, Menon R, Koecher K, Knights D, Personalized Microbiome Class Students. 2019. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25:789–802. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Kesnerova L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. 2020. Gut microbiota structure differs between honeybees in winter and summer. ISME J 14:801–814. doi: 10.1038/s41396-019-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su Q, Tang M, Hu J, Tang J, Zhang X, Li X, Niu Q, Zhou X, Luo S. 2022. Diet outweighs genetics in shaping gut microbiomes in Asian honeybee. bioRxiv 10.1101/2022.01.23.477436. [DOI]
- 38.Tsuruda JM, Chakrabarti P, Sagili RR. 2021. Honey bee nutrition. Vet Clin North Am Food Anim Pract 37:505–519. doi: 10.1016/j.cvfa.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Khalifa SAM, Elashal M, Kieliszek M, Ghazala NE, Farag MA, Saeed A, Xiao JB, Zou XB, Khatib A, Goransson U, El-Seedi HR. 2020. Recent insights into chemical and pharmacological studies of bee bread. Trends Food Sci Technol 97:300–316. doi: 10.1016/j.tifs.2019.08.021. [DOI] [Google Scholar]
- 40.Komosinska-Vassev K, Olczyk P, Kaźmierczak J, Mencner L, Olczyk K. 2015. Bee pollen: chemical composition and therapeutic application. Evid Based Complement Alternat Med 2015:297425. doi: 10.1155/2015/297425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricigliano VA, Fitz W, Copeland DC, Mott BM, Maes P, Floyd AS, Dockstader A, Anderson KE. 2017. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch Insect Biochem Physiol 96:e21406. doi: 10.1002/arch.21406. [DOI] [PubMed] [Google Scholar]
- 42.de Jong EW, de Grandi-Hoffman G, Chen YP, Graham H, Ziolkowski N. 2019. Effects of diets containing different concentrations of pollen and pollen substitutes on physiology, Nosema burden, and virus titers in the honey bee (Apis mellifera L.). Apidologie 50:845–858. doi: 10.1007/s13592-019-00695-8. [DOI] [Google Scholar]
- 43.Gage SL, Calle S, Jacobson N, Carroll M, Degrandi-Hoffman G. 2020. Pollen alters amino acid levels in the honey bee brain and this relationship changes with age and parasitic stress. Front Neurosci 14:231. doi: 10.3389/fnins.2020.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crailsheim K, Hrassnigg N, Gmeinbauer R, Szolderits MJ, Schneider LHW, Brosch U. 1993. Pollen utilization in non-breeding honeybees in winter. J Insect Physiol 39:369–373. doi: 10.1016/0022-1910(93)90024-L. [DOI] [Google Scholar]
- 45.Jeffree EP. 1956. Winter brood and pollen in honeybee colonies. Ins Soc 3:417–422. doi: 10.1007/BF02225761. [DOI] [Google Scholar]
- 46.Avni D, Hendriksma HP, Dag A, Uni Z, Shafir S. 2014. Nutritional aspects of honey bee-collected pollen and constraints on colony development in the Eastern Mediterranean. J Insect Physiol 69:65–73. doi: 10.1016/j.jinsphys.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Nascimento K, Sattler JG, Macedo LL, González CS, Louise P, Araújo ES, Granato D, Sattler A, Almeida-Muradian LD. 2018. Phenolic compounds, antioxidant capacity and physicochemical properties of Brazilian Apis mellifera honeys. LWT-Food Sci Technol 91:85–94. doi: 10.1016/j.lwt.2018.01.016. [DOI] [Google Scholar]
- 48.Escuredo O, Seijo MC. 2019. Honey: chemical composition, stability, and authenticity. Foods 8:577. doi: 10.3390/foods8110577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol 60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 51.Liu P, Zhu Y, Ye L, Shi T, Li L, Cao H, Yu L. 2021. Overwintering honeybees maintained dynamic and stable intestinal bacteria. Sci Rep 11:22233. doi: 10.1038/s41598-021-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papp M, Bekesi L, Farkas R, Makrai L, Solymosi N. 2021. Natural diversity of honey bee (Apis mellifera) gut bacteriome in various climatic and seasonal states. bioRxiv. 10.1101/2021.01.27.428438. [DOI] [PMC free article] [PubMed]
- 53.Moritz B, Crailsheim K. 1987. Physiology of protein digestion in the midgut of the honeybee (Apis mellifera L.). J Insect Physiol 33:923–931. doi: 10.1016/0022-1910(87)90004-7. [DOI] [Google Scholar]
- 54.Wallberg A, Han F, Wellhagen G, Dahle B, Kawata M, Haddad N, Simões ZLP, Allsopp MH, Kandemir I, De la Rúa P, Pirk CW, Webster MT. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet 46:1081–1088. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- 55.Stabentheiner A, Pressl H, Papst T, Hrassnigg N, Crailsheim K. 2003. Endothermic heat production in honeybee winter clusters. J Exp Biol 206:353–358. doi: 10.1242/jeb.00082. [DOI] [PubMed] [Google Scholar]
- 56.Hrassnigg N, Crailsheim K. 1998. Adaptation of hypopharyngeal gland development to the brood status of honeybee (Apis mellifera L.) colonies. J Insect Physiol 44:929–939. doi: 10.1016/S0022-1910(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 57.Kieliszek M, Piwowarek K, Kot AM, Błażejak S, Chlebowska-Śmigiel A, Wolska I. 2018. Pollen and bee bread as new health-oriented products: a review. Trends Food Sci Technol 71:170–180. doi: 10.1016/j.tifs.2017.10.021. [DOI] [Google Scholar]
- 58.Aylanc V, Falcão SI, Ertosun S, Vilas-Boas M. 2021. From the hive to the table: nutrition value, digestibility and bioavailability of the dietary phytochemicals present in the bee pollen and bee bread. Trends Food Sci Technol 109:464–481. doi: 10.1016/j.tifs.2021.01.042. [DOI] [Google Scholar]
- 59.Mărgăoan R, Stranț M, Varadi A, Topal E, Yücel B, Cornea-Cipcigan M, Campos MG, Vodnar DC. 2019. Bee collected pollen and bee bread: bioactive constituents and health benefits. Antioxidants 8:568–600. doi: 10.3390/antiox8120568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson KE, Carroll MJ, Sheehan T, Lanan MC, Mott BM, Maes P, Corby-Harris V. 2014. Hive-stored pollen of honey bees: many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol Ecol 23:5904–5917. doi: 10.1111/mec.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bleau N, Bouslama S, Giovenazzo P, Derome N. 2020. Dynamics of the honeybee (Apis mellifera) gut microbiota throughout the overwintering period in Canada. Microorganisms 8:1146. doi: 10.3390/microorganisms8081146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maes PW, Floyd AS, Mott BM, Anderson KE. 2021. Overwintering honey bee colonies: effect of worker age and climate on the hindgut microbiota. Insects 12:224. doi: 10.3390/insects12030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castelli L, Branchiccela B, Romero H, Zunino P, Antunez K. 2022. Seasonal dynamics of the honey bee gut microbiota in colonies under subtropical climate. Microb Ecol 83:492–500. doi: 10.1007/s00248-021-01756-1. [DOI] [PubMed] [Google Scholar]
- 64.Ribiere C, Hegarty C, Stephenson H, Whelan P, O’Toole PW. 2019. Gut and whole-body microbiota of the honey bee separate thriving and non-thriving hives. Microb Ecol 78:195–205. doi: 10.1007/s00248-018-1287-9. [DOI] [PubMed] [Google Scholar]
- 65.Panic V, Pearson S, Banks J, Tippetts TS, Velasco-Silva JN, Lee S, Simcox J, Geoghegan G, Bensard CL, van Ry T, Holland WL, Summers SA, Cox J, Ducker GS, Rutter J, Villanueva CJ. 2020. Mitochondrial pyruvate carrier is required for optimal brown fat thermogenesis. Elife 9:e52558. doi: 10.7554/eLife.52558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson NS. 2003. Trehalose: the insect “blood” sugar. Adv in Insect Phys 31:205–285. doi: 10.1016/S0065-2806(03)31004-5. [DOI] [Google Scholar]
- 67.Arathi HS, Bjostad L, Bernklau E. 2018. Metabolomic analysis of pollen from honey bee hives and from canola flowers. Metabolomics 14:86. doi: 10.1007/s11306-018-1381-5. [DOI] [PubMed] [Google Scholar]
- 68.Sillman J, Uusitalo V, Tapanen T, Salonen A, Soukka R, Kahiluoto H. 2021. Contribution of honeybees towards the net environmental benefits of food. Sci Total Environ 756:143880. doi: 10.1016/j.scitotenv.2020.143880. [DOI] [PubMed] [Google Scholar]
- 69.Fengkui Z, Baohua X, Ge Z, Hongfang W. 2015. The appropriate supplementary level of tryptophan in the diet of Apis mellifera (Hymenoptera: Apidae) worker bees. J Insect Sci 15:161. doi: 10.1093/jisesa/iev142. [DOI] [Google Scholar]
- 70.Hansen AK, Moran NA. 2014. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol 23:1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- 71.Eugen B, Niclas L, Aram M, Tim K, Kiyoto M, Andreas B. 2015. Physicochemical conditions, metabolites and community structure of the bacterial microbiota in the gut of wood-feeding cockroaches (Blaberidae: Panesthiinae). FEMS Microbiol Ecol 91:1–14. [DOI] [PubMed] [Google Scholar]
- 72.Sabree ZL, Huang CY, Arakawa G, Tokuda G, Lo N, Watanabe H, Moran NA. 2012. Genome shrinkage and loss of nutrient-providing potential in the obligate symbiont of the primitive termite Mastotermes darwiniensis. Appl Environ Microbiol 78:204–210. doi: 10.1128/AEM.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inagaki T, Matsuura K. 2018. Extended mutualism between termites and gut microbes: nutritional symbionts contribute to nest hygiene. Naturwissenschaften 105:52. doi: 10.1007/s00114-018-1580-y. [DOI] [PubMed] [Google Scholar]
- 74.Wan PJ, Yang L, Wang WX, Fan JM, Fu Q, Li GQ. 2014. Constructing the major biosynthesis pathways for amino acids in the brown planthopper, Nilaparvata lugens stal (Hemiptera: Delphacidae), based on the transcriptome data. Insect Mol Biol 23:152–164. doi: 10.1111/imb.12069. [DOI] [PubMed] [Google Scholar]
- 75.Xue J, Zhou X, Zhang CX, Yu LL, Fan HW, Wang Z, Xu HJ, Xi Y, Zhu ZR, Zhou WW, Pan PL, Li BL, Colbourne JK, Noda H, Suetsugu Y, Kobayashi T, Zheng Y, Liu S, Zhang R, Liu Y, Luo YD, Fang DM, Chen Y, Zhan DL, Lv XD, Cai Y, Wang ZB, Huang HJ, Cheng RL, Zhang XC, Lou YH, Yu B, Zhuo JC, Ye YX, Zhang WQ, Shen ZC, Yang HM, Wang J, Wang J, Bao YY, Cheng JA. 2014. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol 15:521. doi: 10.1186/s13059-014-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang ZG, Xu DM, Wang L, Hao JJ, Wang JF, Zhou X, Wang WW, Qiu Q, Huang XD, Zhou JW, Long RJ, Zhao FQ, Shi P. 2016. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr Biol 26:1873–1879. doi: 10.1016/j.cub.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 77.Guo N, Wu QF, Shi FY, Niu JH, Zhang T, Degen AA, Fang QG, Ding LM, Shang ZH, Zhang ZG, Long RJ. 2021. Seasonal dynamics of diet-gut microbiota interaction in adaptation of yaks to life at high altitude. NPJ Biofilms Microbiomes 7:38. doi: 10.1038/s41522-021-00207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maurice CF, Knowles SCL, Ladau J, Pollard KS, Fenton A, Pedersen AB, Turnbaugh PJ. 2015. Marked seasonal variation in the wild mouse gut microbiota. ISME J 9:2423–2434. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ren T, Boutin S, Humphries MM, Dantzer B, Gorrell JC, Coltman DW, McAdam AG, Wu M. 2017. Seasonal, spatial, and maternal effects on gut microbiome in wild red squirrels. Microbiome 5:163. doi: 10.1186/s40168-017-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Regan MD, Chiang E, Liu Y, Tonelli M, Verdoorn KM, Gugel SR, Suen G, Carey HV, Assadi-Porter FM. 2022. Nitrogen recycling via gut symbionts increases in ground squirrels over the hibernation season. Science 375:460–463. doi: 10.1126/science.abh2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kikuchi Y, Tada A, Musolin DL, Hari N, Hosokawa T, Fujisaki K, Fukatsu T, McFall-Ngai MJ, Goffredi S, Salem H. 2016. Collapse of insect gut symbiosis under simulated climate change. mBio 7:e01578-16. doi: 10.1128/mBio.01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferguson LV, Dhakal P, Lebenzon JE, Heinrichs DE, Bucking C, Sinclair BJ. 2018. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct Ecol 32:2357–2368. doi: 10.1111/1365-2435.13153. [DOI] [Google Scholar]
- 83.Chen SF, Zhou YQ, Chen YR, Gu J. 2018. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34:i884–890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magoc T, Salzberg SL. 2011. Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Li X, Su Q, Cao Q, Li C, Niu Q, Zheng H. 2019. A curated 16s rRNA reference database for the classification of honeybee and bumblebee gut microbiota. Biodiversity Science 27:557–566. [Google Scholar]
- 88.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li DH, Liu CM, Luo RB, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 90.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 91.Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 92.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li WZ, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 94.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV. 2019. Microbial genome analysis: the COG approach. Brief Bioinform 20:1063–1070. doi: 10.1093/bib/bbx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanehisa M, Sato Y, Morishima K. 2016. Blastkoala and Ghostkoala: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 97.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kesnerova L, Moritz R, Engel P. 2016. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol 66:414–421. doi: 10.1099/ijsem.0.000736. [DOI] [PubMed] [Google Scholar]
- 99.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Honey bees sampling and sequencing. Download Table S1, PDF file, 0.1 MB (122.2KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolites involved in metabolism of pollen and amino acids in the honey bee guts. Download Table S2, PDF file, 0.05 MB (48.9KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gut community composition and diversity differ between all times. (A) The relative community composition of the gut microbiota was determined by metagenome sequencing. (B and C) Variations in alpha-diversity (B) and NMDS based on Bray-Curtis dissimilarity (C) in microbiota across all times based on 16S rRNA. (D and E) Variations in alpha-diversity (D) and NMDS based on Bray-Curtis dissimilarity in microbiota (E) across all times based on metagenomics. The differences between groups are analyzed by Kruskal-Wallis. Letters above each column represent the levels of variation identified in the Wilcoxon test, where different letters indicate significant variations. Download FIG S1, EPS file, 1.7 MB (1.7MB, eps) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundances of COG annotations related to Metabolism between summer bees and winter bees. Labels: C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism. ****, P < 0.0001 (Wilcoxon test). Download FIG S2, EPS file, 1.5 MB (1.6MB, eps) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential genes of enriched KEGG pathways and proportion of contributions from varied bacterial species. (A) Summer; (B) early winter; (C) late winter. Download FIG S3, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes involved in generating pyruvate and acetyl-CoA of Bartonella and core bacteria. Download Table S4, PDF file, 0.1 MB (145.8KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenylalanine, tyrosine, and tryptophan biosynthesis and general l-amino acid transport system. Download Table S3, PDF file, 0.1 MB (77.2KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial genomes of Bartonella and honey bee core bacteria list. Download Table S5, PDF file, 0.1 MB (85KB, pdf) .
Copyright © 2022 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw data for 16S rRNA and shotgun sequences of winter bees have been deposited under BioProject PRJNA797557, and raw data for 16S rRNA and shotgun sequences of summer bees have been deposited under BioProject numbers PRJNA645267 and PRJNA645015, respectively, in the NCBI database. In-house scripts are available from the corresponding authors upon request.


