ABSTRACT
Mosquitoes are important vectors for many arboviruses. It is becoming increasingly clear that various symbiotic microorganisms (including bacteria and insect-specific viruses; ISVs) in mosquitoes have the potential to modulate the ability of mosquitoes to transmit arboviruses. In this study, we compared the bacteriome and virome (both eukaryotic viruses and bacteriophages) of female adult Aedes aegypti and Culex quinquefasciatus mosquitoes fed with sucrose/water, blood, or blood spiked with Zika virus (ZIKV) or West Nile virus (WNV), respectively. Furthermore, we investigated associations between the microbiota and vector competence. We show that the influence of arboviruses on the mosquito microbiome—and vice versa—is distinct for each combination of arbovirus/mosquito species. The presence of ZIKV resulted in a temporarily increased Aedes ISV diversity. However, this effect was distinct for different ISVs: some ISVs decreased following the blood meal (Aedes aegypti totivirus), whereas other ISVs increased only when the blood contained ZIKV (Guadeloupe mosquito virus). Also, the diversity of the Aedes bacteriome depended on the diet and the presence of ZIKV, with a lower diversity observed for mosquitoes receiving blood without ZIKV. In Cx. quinquefasciatus, some ISVs increased in WNV-infected mosquitoes (Guadeloupe Culex tymo-like virus). Particularly, the presence of Wenzhou sobemo-like virus 3 (WSLV3) was associated with the absence of infectious WNV in mosquito heads, suggesting that WSLV3 might affect vector competence for WNV. Distinct profiles of bacteriophages were identified in Culex mosquitoes depending on diet, despite the lack of clear changes in the bacteriome. Overall, our data demonstrate a complex three-way interaction among arboviruses, resident microbiota, and the host, which is distinct for different arbovirus–mosquito combinations. A better understanding of these interactions may lead to the identification of microbiota able to suppress the ability of arbovirus transmission to humans, and hence improved arbovirus control measures.
KEYWORDS: single mosquito metagenomics, arbovirus, microbiome, virome, phageome, mosquito-arbovirus-microbiota interaction
INTRODUCTION
In support of the holobiont concept, increasing evidences is emerging that the interactions between the host and its complex symbiotic microbial communities have a strong influence on the host biology (e.g., evolution, immunity, and vector competence) by modifying the multipartite interaction dynamics (1, 2). Some symbiotic bacteria and insect-specific viruses (ISVs), like Wolbachia (3, 4), Chromobacterium (Csp_P) (5), Pseudomonas rhodesiae (6), Enterobacter ludwigii (6), Serratia odorifera (7), Serratia marcescens (8), Culex flavivirus (9, 10), Palm Creek virus (11) and La Crosse virus (12), have been reported to alter mosquito susceptibility to certain arboviruses. Several studies have highlighted the abundance and diversity of ISVs and bacteria in mosquito populations (13, 14); hence, the potential vector competence affecting symbiotic interactions might not result from a single virus or bacterium independently. However, no studies so far have investigated vector competence alteration from the perspective of the entire microbial community in mosquitoes.
Using single mosquito viral metagenomics, we previously described that Aedes (Ae.) aegypti and Culex (Cx.) quinquefasciatus from Guadeloupe contain distinct and stable core eukaryotic viromes (15), and some of these core eukaryotic viruses are conserved across different Aedes mosquito species from different countries and continents (16). These core eukaryotic viruses are likely to be ISVs, which live in a subtle equilibrium with the host immune system. Among the core viruses, Phasi Charoen-like phasivirus (PCLPV), together with cell fusing agent virus, could strongly inhibit the growth of Zika virus (ZIKV) and dengue virus in Aa23 cell lines (12). Therefore, the influence of those ISVs on the transmission of pathogenic arboviruses, or vice versa, is worth further investigation. Additionally, viral quantification obtained in a previous study (15) showed that the viral load of the core ISVs is variable among different individual mosquitoes of the same species. The reason for this large observed variation is currently unknown but might be immune related and could therefore be important for a better understanding of vector competence. Furthermore, the bacteriophage component (infecting bacteria) is generally neglected in mosquito virome research. Their influence on shaping the structure of bacterial microbiota in mosquitoes remains elusive.
Guadeloupe experienced a serious ZIKV outbreak in 2016, with 30,500 suspected clinical cases (17), and Ae. aegypti has been shown to be the main vector of ZIKV (18, 19). WNV seroconversions in Guadeloupe have been detected several times in horses and birds since 2002, but no human cases have been reported so far (20). Although the vector competence of Culex spp. mosquitoes from Guadeloupe has never been evaluated for WNV, Cx. quinquefasciatus is the most abundant Culex mosquito species in urban settings (21) and has the potential to transmit this virus (10).
In this study, we allowed eggs of Ae. aegypti and Cx. quinquefasciatus collected in the field from Guadeloupe in 2018 to hatch in the lab. Female Ae. aegypti and Cx. quinquefasciatus adults were fed with sucrose/water, blood, or blood spiked with ZIKV or West Nile virus (WNV), respectively. Both the bacteriome and virome (both eukaryotic viruses and bacteriophages) in individual mosquitoes of these three groups were analyzed.
After ingestion of a blood meal from a viraemic vertebrate host, arbovirus transmission to the next vertebrate hosts requires a successful viral infection and replication in the mosquito midgut and salivary glands (22 to ,24). Such a dissemination includes several critical steps: (i) invasion and establishment of infection in the midgut epithelial cells; (ii) dissemination of the virus from the midgut to the haemocoel and secondary organs, including the salivary glands, and (iii) successful replication in the salivary glands and expectoration in saliva during biting (24 to 26). Therefore, in this study, the heads of individual mosquitoes engorged with spiked blood were titrated by plaque assays to determine the presence of infectious ZIKV or WNV viral particles, as a proxy for the ability to transmit the virus during a next bite on a mammalian or avian host. The study design is summarized in Fig. S1 in the supplemental materials.
Flowchart of study design. Female adults, which hatched in the lab from eggs of Ae. aegypti and Cx. quinquefasciatus collected in the field from Guadeloupe in 2018. The 7-day-old female adults were fed with one of three meals: (a) sucrose and water; (b) regular blood; or (c) blood spiked with infectious ZIKV (Ae. aegypti) or WNV (Cx. quinquefasciatus). Individual mosquitoes from all three groups were sacrificed at 7 and 21 (ZIKV) or 14 (WNV) days postexposure (dpe). The heads of individual mosquitoes engorged with spiked blood were titrated by plaque assays to determine the presence of infectious ZIKV or WNV viral particles. Viral metagenomics and 16S rRNA sequencing were applied on the bodies of all the mosquitoes to explore the associations of the mosquito microbiome with arbovirus transmission. Note: No qRT-PCR was performed for WNV on the heads of Culex quinquefasciatus mosquitoes, due to insufficient sample. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
ZIKV and WNV, respectively, disseminated in Ae. aegypti and Cx. quinquefasciatus from Guadeloupe.
Infectious ZIKV particles in Ae. aegypti heads were only detected at 21 days postexposure (dpe) with 12.1% (5/31) dissemination efficiency (Table 1). Titers of three heads ranged from 2.7 × 102 to 3 × 104 PFU, and two heads only showed cytopathogenic effect (CPE), without clear plaques (Data Set S1). In Cx. quinquefasciatus, dissemination of WNV was observed at 7 and 14 dpe, both with 25% (5/20) dissemination efficiency. The WNV load per head ranged from 2 to 66.7 PFU for two samples at 7 dpi and two at 14 dpi. Three samples at 7 dpi and three at 14 dpi had only CPE. WNV showed a higher dissemination efficiency in Cx. quinquefasciatus compared to ZIKV in Ae. aegypti, but the viral titer of WNV was lower than ZIKV (Wilcoxon test, P value < 0.0001).
TABLE 1.
Dissemination efficiencies of ZIKV and WNV
| Mosquito species | Viral strain | Day postexposure | Dissemination efficiency | Titer (PFU per head) |
|---|---|---|---|---|
| Aedes aegypti | ZIKV Martinique strain (GenBank: KU647676) |
7 | 0% (0/33) | |
| 21 | 12.1% (5/41) | 2.7 × 102 to 3 × 104 | ||
| Culex quinquefasciatus | WNV from Camargue (France) (GenBank: AY268132) |
7 | 25% (5/20) | 2 to 66 |
| 14 | 25% (5/20) |
Eukaryotic virome in Aedes and Culex mosquitoes. Download Data Set S1, XLSX file, 0.1 MB (89KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Virome identification.
The 154 mosquito bodies (excluding head, legs, and wings) were individually processed for virome analyses (Table 2). The clustering of assembled contigs from all samples longer than 1 kb based on 95% nucleotide identity over 80% of the length resulted in 133,951 representative contigs (Data Set S1). After removing the viral contigs presented in both negative control samples, 127 of the remaining contigs were annotated as eukaryotic viruses by DIAMOND, BLASTn, and CAT. In total, 263 contigs were annotated as phage, of which 41 could be annotated at the family level. Besides these viral contigs, nearly half of the representative contigs belonged to mosquito genomes and 9.3% were suspected bacterial contigs. In Ae. aegypti and Cx. quinquefasciatus samples, 353 million and 163 million reads were mapped to all the nonredundant contigs, respectively. The eukaryotic viral reads and phage reads occupied 5% and 0.05% in Ae. aegypti samples, and 3.3% and 6.7% in Cx. quinquefasciatus samples, respectively.
TABLE 2.
Numbers of mosquito body samples used for NGS
| Mosquito species | Day postexposure | Virus/blooda | Blood | Sucrose/H2O | Total no. |
|---|---|---|---|---|---|
| Aedes aegypti | 7 | 33 | 5 | 5 | 43 |
| 21 | 41 | 5 | 5 | 51 | |
| Culex quinquefasciatus | 7 | 20 | 5 | 5 | 30 |
| 14 | 20 | 5 | 5 | 30 |
To maximize the statistical power of our analyses, we selected all available engorged mosquitoes, explaining the uneven distribution of included mosquitoes.
Increased eukaryotic virome richness in Ae. aegypti after receiving a ZIKV-spiked blood meal at 7 dpe.
The heatmap in Fig. 1a displays the abundance of 18 eukaryotic viral species (which have more than 500 reads in total) in each sample. The five Aedes mosquitoes with infectious ZIKV in their heads at 21 dpe displayed high levels of ZIKV nucleic acid in both their heads and bodies detected by qRT-PCR and viral metagenomics (Fig. 1a). Several mosquitoes without measurable infectious viral particles in the head, however, contained a high number of ZIKV genome copies in their bodies and heads (as measured by qRT-PCR), as well as a high number of ZIKV reads in their bodies from next-generation sequencing (NGS) analyses. The previously identified core viruses—PCLPV, Aedes aegypti totivirus (ATV), Guadeloupe mosquito virus (GMV), and Aedes aegypti anphevirus (AANV) (15), were detected in 75.5%, 78.7%, 23.4%, and 34% of Ae. aegypti mosquitoes in this study (across different conditions), respectively. GMV was not detected in mosquitoes fed on noninfectious blood (both at 7 and 21 dpe), whereas several mosquitoes in the ZIKV-spiked blood group (both at 7 and 21 dpe) showed a high abundance of GMV reads. In contrast, at 7dpe, mosquitoes fed with sucrose/water showed extremely high abundance (ranging from 366,558 to 3.5 million reads) of ATV, whereas this virus was nearly absent in regular blood-fed mosquitoes, and only present at low levels in the ZIKV-spiked blood group.
FIG 1.
Eukaryotic virome profile in Aedes aegypti and Culex quinquefasciatus. (a) Abundance of eukaryotic viral species in Aedes aegypti. The plaque assay results are indicated by “+” for positive results and “–” for negative results in the gray bars. The dot plots show the total ZIKV genome copies in Aedes aegypti heads and bodies determined by qRT-PCR. The heatmap shows the read counts on log10 scale. The viral species names shown in the heatmap are from the taxonomic annotation by DIAMOND and KronaTools. The heatmap only shows the viral species and samples that had more than 500 eukaryotic viral reads (21 viral species and two samples were removed). (b) Alpha diversity of eukaryotic viral species in Aedes aegypti at 7 and 21 dpe. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons using the Benjamini-Hochberg method. (c and d) PCoA of eukaryotic viral species in Aedes aegypti based on Bray-Curtis dissimilarity. The PERMANOVA test was performed on food sources. The “+” or “–” in the bracket after ZIKV indicates the positive or negative result of plaque assay (head), qRT-PCR detection of ZIKV in head and body, respectively. (e) Abundance of eukaryotic viral species in Culex quinquefasciatus. The plaque assay results are indicated by “+” for positive results and “–” for negative results in the gray bars. The dot plots show the total WNV genome copies in Culex quinquefasciatus heads and bodies determined by qRT-PCR. The heatmap shows the reads counts on log10 scale. The viral species names shown in the heatmap are from the taxonomic annotation by DIAMOND and KronaTools. Three samples with zero eukaryotic viral reads and 16 species containing less than 500 reads were deleted from the heatmap. Note: No qRT-PCR was performed for WNV on the heads of Culex quinquefasciatus mosquitoes, due to insufficient sample.
For further analyses, the Ae. aegypti mosquitoes that received a ZIKV-spiked blood meal were separated into groups depending on their plaque assay results (positive or negative) and ZIKV genome copy numbers in the head and body (using 1,000 copies as threshold). First, the alpha diversity (Shannon index) of eukaryotic viral species was compared between the sample collection time points and different diet groups (Fig. 1b). At 7 dpe, the Ae. aegypti group receiving a ZIKV-spiked blood meal showed a significantly higher alpha diversity compared to the blood and sucrose group. This observed high alpha diversity in the ZIKV-spiked blood group was mainly driven by the mosquitoes without ZIKV detected in their head by plaque assay or qPCR (Fig. S2). No significant difference was observed between the different sample groups collected at 21 dpe (Kruskal-Wallis, P value = 0.53). Comparison of the two time points within the same diet group showed that the alpha diversity of the ZIKV-spiked blood group decreased over time, whereas the alpha diversity in the sucrose-fed group increased over time (Fig. 1b).
The alpha diversity of eukaryotic virus in Aedes aegypti of different time points and infection status. Download FIG S2, PDF file, 0.2 MB (200.6KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bray-Curtis dissimilarities were calculated from the abundance of eukaryotic viral species (excluding ZIKV) to perform a principal coordinates analysis (PCoA) (Fig. 1c and d). The samples from mosquitoes fed with ZIKV-spiked blood and sucrose/water at 7 dpe clustered separately, whereas the samples from the blood-fed group mainly overlapped with the ZIKV-spiked blood samples with significant P value (0.001) in a permutational multivariate analysis of variance (PERMANOVA) test on food sources (R2 = 0.22) (Fig. 1c). There was no statistical difference found within the group that received ZIKV-spiked blood, based on the presence of infectious viruses in the head as determined by plaque assay. At 21 dpe, no significant difference was observed among any of the diet groups (PERMANOVA test on food sources, P value = 0.4, R2 = 0.03) (Fig. 1d).
Near complete absence of Wenzhou sobemo-like virus 3 in Culex quinquefasciatus mosquitoes disseminating WNV.
In general, Cx. quinquefasciatus samples contained fewer eukaryotic viral reads and fewer viral species compared to Ae. aegypti samples (Fig. 1a versus 1e), which is consistent with the results of our previous study (15). In 22.2% (2/9) of the mosquito bodies that were WNV plaque-assay positive, no WNV could be detected by NGS and qRT-PCR (Fig. 1e). In the samples without infectious viral particles in the head (n = 28), 11 at 7 dpe and 9 at 14 dpe harbored at least 102 WNV genome copies in their bodies (Fig. 1e). Among the previously identified core viruses of Cx. quinquefasciatus (15), PCLPV and Guadeloupe Culex tymo-like virus (GCTLV) were found in 58% and 17% of the samples, respectively (Fig. 1e), whereas GMV and Guadeloupe Culex rhabdovirus were only identified in two (with low abundance) and zero samples, respectively (Data Set S1). Notably, Wenzhou sobemo-like virus 3 (WSLV3), which was not identified in our previous study, showed high abundance in 54% of the mosquitoes fed with WNV-spiked blood without infectious WNV in their heads at 14 dpe, whereas this virus was absent in the plaque assay positive samples (except for one sample with only 80 reads). Due to the relatively low number of viral species in Cx. quinquefasciatus, no alpha and beta diversity analyses were performed.
Presence of arboviruses in blood meal modulates viral loads of specific ISVs, and vice versa.
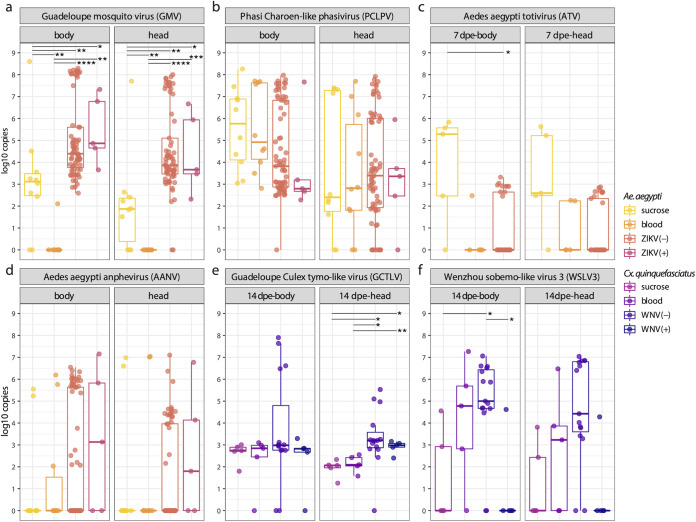
Since metagenomics is less sensitive compared to targeted PCR assays, and the abundance data from viral metagenomics are not fully quantitative, the genome copies of selected core viruses in the head and body of mosquitoes were further determined by qRT-PCR. The targeted ISVs were GMV, PCLPV, ATV, and AANV in Ae. aegypti (Fig. 2a to d), and GCTLV and WSLV3 in Cx. quinquefasciatus (Fig. 2e and f). Interestingly, the number of GMV genome copies was much higher in the body and head of the mosquito receiving ZIKV-spiked blood (with median around 104 copies) compared to blood meal without ZIKV, or the sucrose meal. The virus is almost absent in the mosquitoes that received a noninfectious blood meal (except for one sample with about 100 genome copies) (Fig. 2a). There was no significant difference of PCLPV genome copies among different groups or time points (Fig. 2b and Fig. S3). As observed in Fig. 1a, the viral genome copy number of ATV in the bodies of mosquitoes receiving a sucrose diet was higher than in mosquitoes receiving a ZIKV-spiked blood meal on 7 dpe (Fig. 2c). Although AANV appeared to show a similar pattern as GMV, this was less pronounced, and no significant difference was found for AANV among the heads or bodies of Ae. aegypti, with respect to diet (Fig. 2d and Fig. S3). GCTLV genome copy numbers were very similar among the bodies of the four groups, with a median around 103 copies, while they were significantly higher in the heads of mosquitoes receiving a WNV-spiked blood meal on 14 dpe (Fig. 2e), which was a pattern similar to GMV and AANV in Aedes (Fig. 2a and d). Interestingly, no WSLV3 genomes were detected in both body and head of mosquitoes at 14 dpe containing infectious WNV particles (except for a single sample that showed CPE in the plaque assay). In contrast, the bodies of samples without infectious WNV viral particles in the head contained significantly higher WSLV3 genome copy numbers. A similar trend was also observed in the heads, although the statistical tests were not significant (Fig. 2f). Comparable patterns were observed for the analyses of the 7 and 14 dpe analyses combined (Fig. S3).
FIG 2.
Comparison of viral genome copies in body and head mosquitoes. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons with the Benjamini–Hochberg method. The “+” or “–” in the bracket after indicates the positive or negative result of plaque assay (head).
Comparison of viral genome copies in body and head of mosquitoes. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons with the Benjamini-Hochberg method. Download FIG S3, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Presence of ZIKV in blood meal increases bacteriome diversity profile in Ae. aegypti.
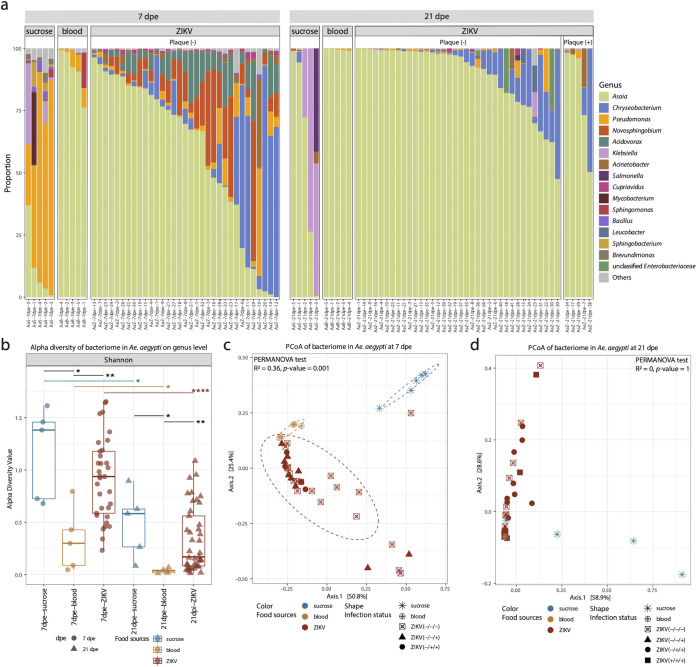
16S rRNA sequencing was performed on the same mosquito individual bodies to characterize their bacteriome. Overall, Asaia was the most prevalent genus in all Aedes samples receiving a blood meal with variable proportions (Fig. 3a). The bacteriome of the sucrose/water-fed group was mainly dominated by Pseudomonas at day 7, but Asaia and/or Klebsiella dominated after 21 days. Samples receiving a noninfectious blood meal were strongly dominated by Asaia, with very few other bacterial genera identified (i.e., Pseudomonas and Novosphingobium), both at 7 and 21 dpe. However, for the group fed with ZIKV-spiked blood at 7 dpe, a much higher bacterial diversity was observed (Fig. 3a). Asaia still dominated in most of the samples, but several samples were dominated by Chryseobacterium. Furthermore, Novosphingobium and Acidovorax were also frequently encountered in samples at 7 dpe. A similar observation could be made for samples at 21 dpe, although the dominance of Asaia was larger, followed by Chryseobacterium as the second most common bacterial genus. Novosphingobium and Acidovorax decreased dramatically at 21 dpe. In addition, for the mosquitoes receiving a ZIKV-spiked bloodmeal, the bacteriome profiles of infected (plaque-positive) and uninfected samples (no plaque) were undistinguishable (Fig. 3b).
FIG 3.
The bacteriome profile in Aedes aegypti. (a) The proportion of bacterial genera in each sample. Only the genera with total proportion above 10% are shown in the figure. (b) Alpha diversity of Aedes aegypti samples per food source group and time point on genus levels. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons using the Benjamini-Hochberg method. (c and d) PCoA of bacterial genera based on Bray-Curtis dissimilarity on 7 dpe and 21 dpe. The PERMANOVA test was performed on food sources. The “+” or “–” in the bracket after ZIKV indicates the positive or negative result of plaque assay (head), qRT-PCR detection of ZIKV in head and body, respectively.
The bacterial Shannon alpha diversity of the group fed with noninfectious blood was significantly lower than the group receiving sucrose/water at both time points. Interestingly, the alpha diversity of the ZIKV-spiked blood group was significantly higher than the group receiving noninfectious blood, and indistinguishable from that of the group receiving sucrose/water. For all three diet groups, the diversity decreased from 7 to 21 dpe. For the PCoA based on Bray-Curtis dissimilarity, the samples at 7 dpe largely clustered separately according to the food sources (PERMANOVA test: R2 = 0.36, P = 0.001) (Fig. 3c). At 21 dpe, the mosquitoes receiving a blood meal (ZIKV spiked or not) clustered together, which overlapped with two samples of the sucrose-fed group (Fig. 3d).
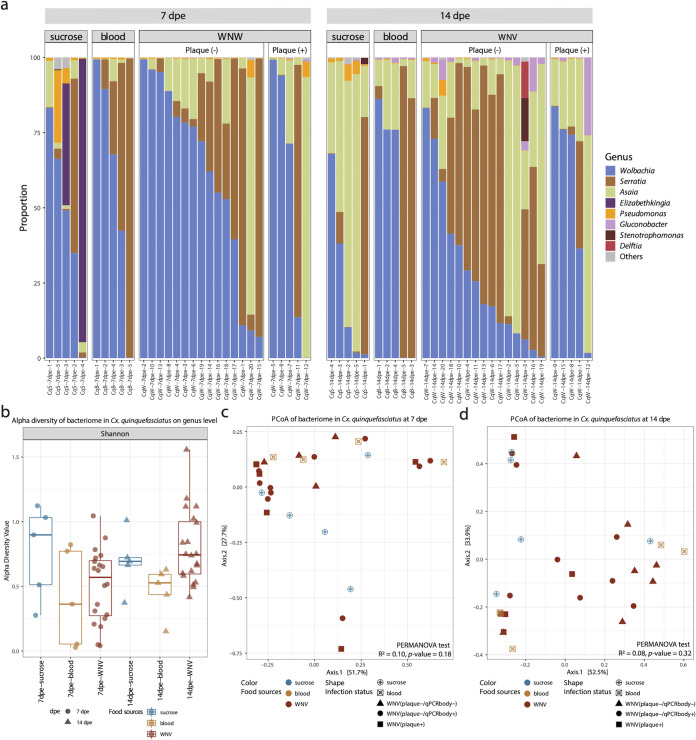
Presence of WNV in blood meal does not modify the bacteriome diversity profile in Cx. quinquefasciatus.
The identified members of the bacteriome in all Culex samples were very similar regardless of collection date and food sources, mainly occupied by Wolbachia, Serratia, and Asaia (Fig. 4a). Two samples in the sucrose/water group at 7 dpe contained high proportions of Elizabethkingia. These three diet groups showed very similar Shannon indexes of alpha diversity (Fig. 4b). Furthermore, no separation among the three diet groups was observed in the PCoA (Fig. 4c and d). However, from 7 dpe to 14 dpe, the relative abundance of Asaia significantly increased in all groups except for the individuals with infectious viral particles in heads (plaque +), while the abundance of Wolbachia decreased in the group without infectious viral particles detected (plaque –) (Fig. S4).
FIG 4.
The bacteriome profile in Culex quinquefasciatus. (a) The proportion of bacterial genera in each sample. Only the genera with total proportion above 10% are shown in the figure. (b) Alpha diversity of Culex quinquefasciatus samples per food source group on genus levels. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons using the Benjamini-Hochberg method. (c) PCoA of bacterial genera based on Bray-Curtis dissimilarity of samples on 7 dpe and 14 dpe. The PERMANOVA test was performed on food sources.
Relative abundance of Asaia and Wolbachia in Culex quinquefasciatus of two time points. Download FIG S4, PDF file, 0.1 MB (84.8KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
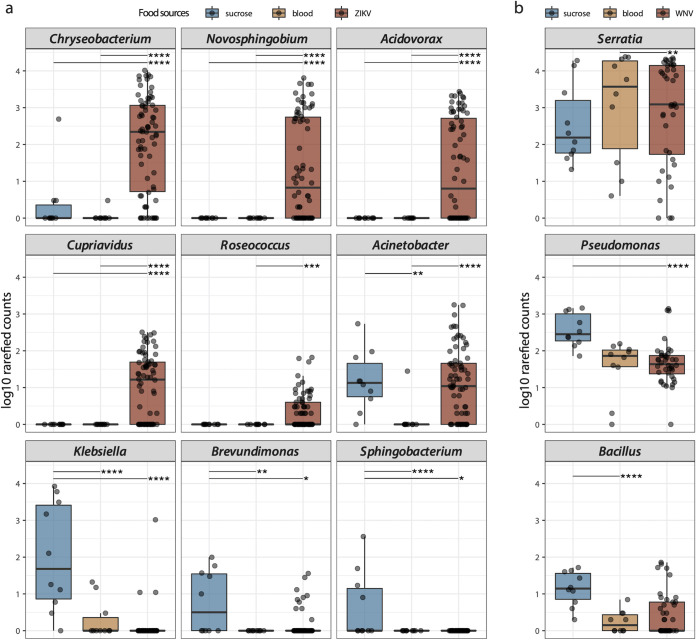
Differential effects of ZIKV on bacterial genera in Aedes mosquitoes.
The abundance of genera Chryseobacterium, Novosphingobium, Acidovorax, Cupriavidus, and Roseococcus was higher in Aedes fed with ZIKV-spiked blood compared to the noninfectious blood and sucrose/water (Fig. 5a). The noninfectious blood-fed Aedes mosquitoes showed a decreased Acinetobacter abundance. Klebsiella, Brevundimonas, and Sphingobacterium were most abundant in the Aedes fed with sucrose/water and were almost absent in mosquitoes receiving a blood meal (with and without ZIKV). The noninfectious blood-fed Culex mosquitoes showed a higher abundance of Serratia compared to WNV spiked-blood-fed mosquitoes, whereas Pseudomonas and Bacillus were more abundant in sucrose/water-fed mosquitoes (Fig. 5b).
FIG 5.
(a) The bacterial genera with differential abundance among three food source groups in Aedes aegypti. (b) The bacterial genera with differential abundance among three food source groups in Culex quinquefasciatus. These bacterial genera are determined by DESeq2 with padj <0.05 and baseMean >10 as cutoff. padj < 0.05 (*), padj < 0.01 (**), padj < 0.001 (***), padj < 0.0001 (****).
Phageome profile in mosquito samples.
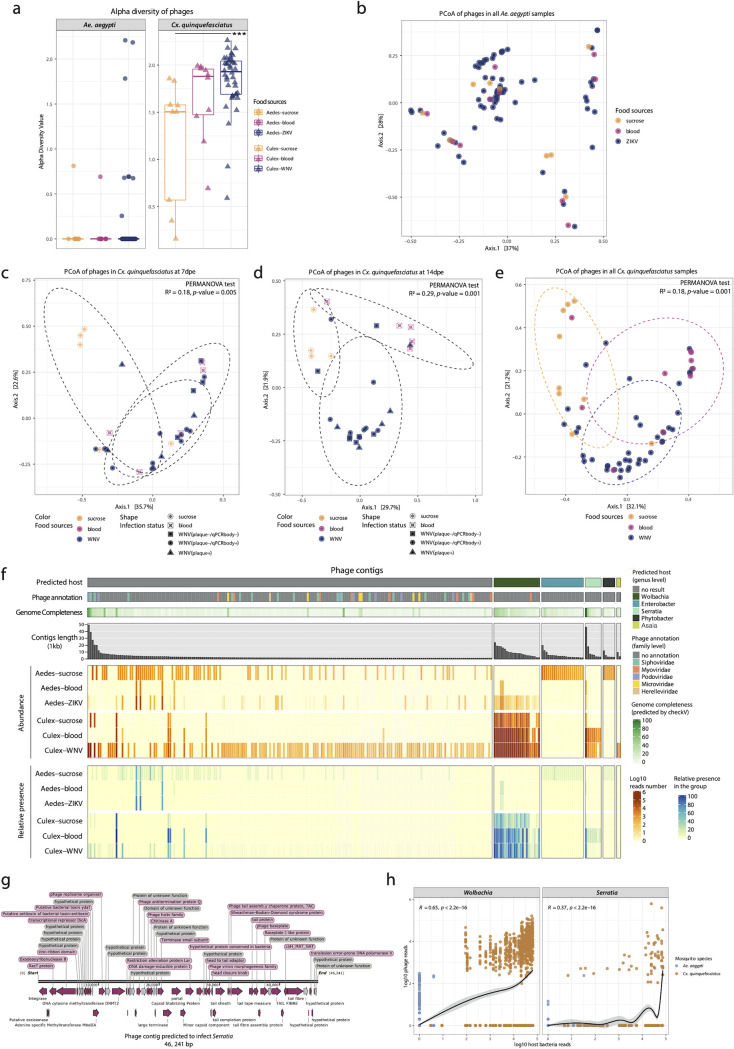
The alpha diversity (Shannon index) of the phageome in Ae. aegypti samples was indistinguishable among the three different diet groups (Fig. 6a). In contrast, the phageome alpha diversity in Cx. quinquefasciatus showed that mosquitoes receiving a WNV-spiked blood meal had a significantly higher diversity compared to the sucrose/water-fed mosquitoes. As could be expected, the PCoA analyses showed no separation among the three diet groups of Ae. aegypti (Fig. 6b). However, for the Culex samples, the sucrose/water-fed mosquitoes at 7 dpe, 14 dpe, or combined formed a separate cluster from the ones fed with blood (with or without WNV) (Fig. 6c to e). Contrarily to eukaryotic viruses, the diversity and abundance of phages in Cx. quinquefasciatus samples were much higher than Ae. aegypti (Fig. 6a and f).
FIG 6.
Phageome profile in mosquito samples. (a) Alpha diversity of the phageome in Ae. aegypti and Cx. quinquefasciatus on contigs with completeness of >20%. (b) PCoA of all Ae. aegypti samples. (c) PCoA of phageome in Cx. quinquefasciatus at 7 dpe. (d) PCoA of phageome in Cx. quinquefasciatus at 14 dpe. (e) PCoA of all Cx. quinquefasciatus samples. The PERMANOVA test was performed on food sources. The ellipses were drawn for each food type. (f) The profile of phage contigs. The columns are phage contigs split according to their predicted bacterial host (the top bar). The second bar indicates the taxonomic annotation of the phage contigs. The third bar is the genome completeness of the phage contigs evaluated by CheckV. The bar plot shows the length of each phage contig. The top heatmap (orangish) displays the abundance of phage contigs per food source in Aedes aegypti and Culex quinquefasciatus. The bottom heatmap (bluish) is the present proportion of each phage contig per food source. Samples containing more than 100 reads per contig are considered present. (g) Genome structure of Serratia phage. (h) The abundance correlation of phage contigs and their predicted host. The spearman method is used for computing correlation coefficient.
For 60 out of the 263 identified phage contigs, Wolbachia, Enterobacter, Serratia, Phytobacter, or Asaia could be predicted to be the host (Fig. 6f and Data Set S2). Six contigs were predicted to be >50% complete (Fig. 6f and Data Set S2). Among them, one Serratia infecting phage genome (46,241 bp) was predicted to be 99.5% complete with high confidence. The genome organization of this phage is visualized in Fig. 6g, which contains the essential phage proteins such as tail protein, phage baseplate, and head and tail adaptor protein. Two other contigs were estimated to be 80.4% and 57.8% complete (Data Set S2) and were annotated as Podoviridae and Siphoviridae (Fig. S5). The remaining three contigs that were predicted to be more than 50% complete did not have a taxonomic annotation or predicted bacterial host (Data Set S2). In addition, the estimated completeness of the longest predicted phage contigs infecting Wolbachia (23,868 bp), Enterobacte (19,754 bp), Phytobacter (11,746 bp), and Asaia (9,507 bp) were 46.9%, 16.1%, 12.3%, and 17.2% complete, respectively (Data Set S2).
Genome structure of phage contigs. (a) Genome structure of phage contig annotated as Podoviridae. (b) Genome structure of phage contig annotated as Siphoviridae. Download FIG S5, PDF file, 0.5 MB (560.7KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Host prediction and completeness estimation of phages. Download Data Set S2, XLSX file, 0.2 MB (171.5KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For Ae. aegypti, distinct bacteriophage profiles could be identified between the different types of diet. Many more relatively low-abundant phage contigs could be found without known host in the sucrose-fed mosquitoes (Fig. 6f). Furthermore, a clear distinction was found between phages with a known host, with Enterobacter and Phytobacter phages being more prevalent in sucrose-fed mosquitoes (and absent in blood-fed mosquitoes), and Wolbachia infecting phages in the ZIKV-spiked blood group (which were absent in the sucrose-fed mosquitoes). For Cx. quinquefasciatus, the overall phage profile was rather similar between mosquitoes receiving a different diet (blood versus sucrose) and dominated by phages predicted to infect Wolbachia. However, the number of phage reads was higher in blood-fed mosquitoes, and the weakly abundant Serratia phages were largely absent in the sucrose-fed mosquitoes.
The bacteria identified by 16S rRNA and the predicted hosts of the phage contigs were largely in agreement for Cx. quinquefasciatus, but less so for Ae. aegypti. The predicted host of the phages in Culex samples (Wolbachia, Serratia, and Asaia) were found to be highly prevalent in the Culex bacteriome (Fig. 4a, Fig. 6f). The predicted host of the phages in Aedes included Wolbachia, Enterobacter, and Phytobacter. The proportion of Wolbachia in Ae. aegypti was very low (total ranging from 0.5% to 3%, and therefore not displayed in Fig. 5a) (Data Set S3). The phage contigs predicted to infect Enterobacter and Phytobacter were derived from a single sample AaS-21dpe-3 (Data Set S2), which contained high abundance of genera Klebsiella and Salmonella (Fig. 5a). These four genera all belong the same family Enterobacteriaceae. Asaia was the most prevalent genus present in all Aedes samples, but no Asaia-infecting phage could be identified. A possible explanation could be that 203 out of the 263 phage contigs do not have a predicted bacterial host. Potentially, the phages infecting Asaia in Aedes are too divergent to be identified using currently available approaches. In addition, the abundance of Wolbachia and Serratia positively correlated with the abundance of their respective phage contigs with R = 0.65 and R = 0.37, respectively (Fig. 6h).
16S rRNA sequencing results. Download Data Set S3, XLSX file, 0.4 MB (410.4KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, adult female mosquitoes from Guadeloupe were fed with a virus-spiked blood meal (ZIKV-spiked blood for Ae. aegypti, WNV-spiked blood for Cx. quinquefasciatus), and the infection status of their heads was determined by plaque assays as a proxy for vector competence. The observed dissemination efficiencies of ZIKV and WNV are consistent with previous studies (18, 27). The observed positivity rates of the plaque assay at 7 dpe (0% for ZIKV and 25% for WNV) (Table 1) suggest that the dissemination time of the ZIKV strain used from the midgut to the head in Ae. aegypti is longer compared to that of WNV in Cx. quinquefasciatus. The ZIKV strain used in this study was isolated in Martinique, belongs to the Asian/American lineage, and has more than 99% nucleotide identity with the ZIKV strains that circulated in both humans and mosquitoes in Guadeloupe (i.e., KX673530.1, MN185332.1). However, the transmission of this strain by Ae. aegypti from Guadeloupe is less efficient compared to that of ZIKV from Senegal (African lineage) due to stronger mosquito midgut infection and escape barriers (18). The vector competence of Culex mosquitoes from Guadeloupe has never been evaluated for WNV. Although our results show a WNV dissemination efficiency of 25%, the viral titers in the heads are rather low (from 2 to 66 PFU). Viral titers up to 106 PFU per head of both insecticide-resistant and -susceptible lab mosquito lines have been reported (27). Such difference in titers suggests that Cx. quinquefasciatus from Guadeloupe is less susceptible to this particular WNV strain that was isolated from a horse in France (28, 29). In addition, in two mosquitoes with infectious WNV particles in their head, no WNV could be identified in the bodies by NGS or qRT-PCR (Fig. 3), suggesting that the virus might only transiently cause a midgut or systemic infection in some mosquitoes before entering the mosquito heads and salivary glands, or alternatively the virus could have gone directly from the gut to the head using the trachea network (30).
The ISVs and phages of the mosquitoes receiving different meals (virus-spiked blood, blood, or sucrose/water) were further explored. Generally, ISVs are more abundant and diverse than phages in Ae. aegypti, which is the opposite in Cx. quinquefasciatus (Fig. 1 and 6). This finding is consistent with the virome results from our previous study (15). The previously identified core ISVs in Ae. aegypti, such as PCLPV, GMV, ATV, and AANV, are also prevalent in samples of this study (Fig. 1a). Regarding Cx. quinquefasciatus, only two previously identified core eukaryotic viruses—GCTLV and PCLPV—are present in the majority of the samples as well as a novel virus annotated as WSLV3 (Fig. 1e). The Ae. aegypti mosquitoes receiving a blood meal showed a transiently higher ISV diversity compared to sucrose-fed animals at 7 dpe, a signal that disappeared 21 dpe (Fig. 1b). The transient nature of this change was also reflected in beta-diversity analysis (Fig. 1c and d). These observations could be explained by a major alteration or suppression of the mosquito immune response after a blood meal at 7 dpe (31), which would allow the resident ISVs to expand. Since the blood is digested 2 to 3 dpe, followed by a diet based on water and sugar, this could result in a return of the immune system to a homeostasis at 21 dpe, and a subsequent repression of ISVs. Comparable observations—correlations between changes in the host immune system and the virome—have been made in humans where immune suppressive drugs resulted in an increase in the level of anelloviruses in the blood of transplant recipients (32), or in the increase of the gut virome in nonhuman primates experimentally infected with simian immunodeficiency viruses (33). Hence, the earlier time points would be more informative to understand the effect of diet on the virome. In nature, mosquitoes undergo successive gonotrophic cycles described by getting a blood meal and laying eggs, which could result in cyclic alterations in the replication and suppression of ISV, and hence their potential vector competence for arboviruses (see above).
Since the abundance data obtained from viral metagenomics is not fully quantitative and the positive results from plaque assay need to be further confirmed, qRT-PCRs were performed to obtain the absolute load of ZIKV, WNV, and the selected core ISV genomes to better study their differential abundances among the different conditions. All selected mosquito-specific viral genomes (GMV, PCLPV, ATV, AANV, GCTLV, and WSLV3) could be detected in both head and body of most mosquitoes, sometimes reaching 108 copies (Fig. 2), supporting the hypothesis that they might significantly affect arbovirus transmission. In general, mosquitoes receiving a noninfectious blood meal showed similar loads of eukaryotic viruses (PCLPV, AANV, GCTLV, WSLV3, ATV) compared to sucrose water, except for GMV, which appeared to be suppressed due to the blood meal. Some components in the blood meal may alter the immune system and therefore specifically suppress the viral replication of GMV. Interestingly, for some viruses, the presence of an arbovirus (ZIKV/WNV) in the blood meal, compared to noninfectious blood, seemed to result in an upregulation of viral genome copies (GMV, AANV, and GCTLV), although this was only strongly significant for GMV (Fig. 2a) and weakly significant in the head for GCTLV at 14 dpe (Fig. 2e). It could be hypothesized that infection by these arboviruses suppressed a specific arm of the mosquito immune system, allowing GMV and GCTLV to grow to higher titers. The associations between these ISVs and arboviruses infection or transmission need to be further investigated for confirmation and elucidating the mechanisms driving these interactions. Notably, in Culex samples, a statistically significant difference between the number of WSLV3 genome copies in the heads of WNV-spiked blood-fed mosquitoes was found between mosquitoes able to disseminate WNV (i.e., with infectious virus detected in heads) and mosquitoes unable to disseminate WNV (Fig. 2f). A similar trend was seen in the bodies, although not significant. This could suggest that a successful migration of WNV to the head and the saliva is inhibited by the presence of WSLV3 and that only mosquitoes without, or with very low, levels of WSLV3 are efficient to transmit WNV. If this could be confirmed, WSLV3 could be a potential bio-control agent. Similar observations have been made for the most well-studied mosquito-specific virus, Culex flavivirus (CxFV). The WNV dissemination rate in Cx. pipiens was significantly lower in a CxFV-positive colony from Colorado compared to a CxFV-negative colony from Iowa (34), but the results could vary when using different types of the mosquitoes, viruses, or infection modes (10, 13).
The identified bacterial genera in mosquitoes (Fig. 3 and 4) are similar to previous reports in Aedes (13, 35, 36) and Culex mosquitoes (37, 38). For Ae. aegypti, a strict sucrose diet resulted in a highly diverse bacteriome compared to mosquitoes receiving a noninfectious blood meal, in which the bacteriome was dominated by Asaia (Fig. 3a). In particular, Klebsiella, Brevundimonas, and Sphingobacterium were highly abundant in mosquitoes fed with sucrose solution (Fig. 5a). These results are similar to a previous study, which reported that the overall diversity of bacteria declined after a noninfectious blood meal (39). This decrease may result from the oxidative stress associated with the catabolism of the blood meal (40, 41). However, if ZIKV was present in the blood meal, a strong increase in the bacterial diversity was seen (Fig. 3a and 3b), which might again be linked to an alteration of the mosquito host immune system. This increase was mainly driven by the genera Chryseobacterium, Novosphingobium, Acidovorax, Cupriavidus, and Roseococcus (Fig. 5a). The enhancement of Chryseobacterium (Flavobacteriaceae) in mosquitoes receiving a ZIKV-spiked blood meal was in agreement with a previous study, which reported that members from the family Flavobacteriaceae increased early after viral exposure (3 dpe) (42). Of note, different bacteria were dominantly present in the sucrose compared to the ZIKV-spiked blood-diet mosquitoes (Fig. 3a, Fig. 5a). These observed differences became less pronounced after 21 dpe, suggesting a return to a more “normal” immune/microbiome state. In contrast, in Cx. quinquefasciatus, the type of meal and the WNV infection status seemed to have only little or no effect on the bacteriome composition (Fig. 4), with only Pseudomonas and Bacillus being increased in the sucrose/water fed mosquitoes (Fig. 5b). Furthermore, some differences were observed in the proportion of Wolbachia, which significantly decreased from 7 dpe to 14 dpe (for the Culex fed with noninfectious blood), in favor of Asaia, which significantly increased at 14 dpe (Fig. S4). Of interest are previous observations that Wolbachia has a suppressive effect on arbovirus infection, including WNV (43, 44). This might be another explanation for the low viral titer of WNV in the head of Cx. quinquefasciatus (Table 1). A few Wolbachia reads are identified in Aedes samples, which are likely derived from integrated Wolbachia sequences in the mosquito genome (45). In addition, several other bacteria are also known to interfere with host vector competence for arboviruses. For example, the Serratia spp. (Serratia odorifera [7] and Serratia marcescens [8]) have been reported to enhance the susceptibility of field mosquitoes to dengue virus. Pseudomonas rhodesiae isolated from the Ae. albopictus midgut show direct inhibition of La Crosse virus independent of the mosquito, which possibly produce antiviral molecules (6). Thus, it will be interesting to further explore the effects of highly abundant bacteria present in mosquitoes fed with viral blood (Fig. 5), and their potential effects on the mosquito immune response to pathogenic viruses.
Although there is no difference in the alpha and beta diversity of the phageome in Ae. aegypti (Fig. 6a and b), distinct bacteriophage profiles could be identified between the different types of diet (Fig. 6f). For Culex mosquitoes, the alpha diversity indices of the phageome in mosquitoes fed with WNV-spiked blood was significantly higher than those of the ones only fed with sucrose and water (Fig. 6a), which was also reflected in the separation between blood-engorged and sucrose/water-fed mosquitoes in the PCoA analyses (Fig. 6c to e). These data suggest that although the diet had little effect on the Culex bacteriome, the diet can induce shifts in the phage communities. However, these data should be interpreted with great caution as the majority of the identified phage genome contigs do not have a known host, nor do they represent complete genomes. For only 23% of the phage contigs, the host could be predicted. The phages infecting Wolbachia and Serratia were highly abundant and mainly present in Culex mosquitoes (Fig. 6f), which also showed a high abundance of their respective bacterial hosts (Fig. 4a). In correlation analysis, the abundance of phages predicted to infect Wolbachia and Serratia, and the respective bacterial host abundance, increase together, confirming the bacterial host prediction (Fig. 6h). Although it is possible that the blood is the source of bacteriophages, it is more likely that these phages are induced from prophages inside the bacterial genomes. Whether these phages are induced from dying bacteria losing the competitive battle in a changing environment, or thrive due to strong increase of their successful bacterial host, remains to be investigated.
Our single-mosquito microbiome study strategy is a promising way to further explore the interactions among the mosquito host, symbiotic microbes, and arbovirus. However, our study also had some limitations. First, the use of mosquito saliva might be a better proxy for infectivity compared to mosquito heads. Second, we cannot completely exclude that during mosquito dissection, microbes from one body part could have contaminated other body parts. Third, quantitative interpretation of metagenomics data should be done carefully, because wet lab (e.g., virus-like particle enrichment) as well as sequencing analyses could introduce biases and all data are presented as relative data.
In summary, our results reveal that different food sources and the presence of arboviruses have variable influences on the ISVs, bacteriome, and phageome interplay in Ae. aegypti and Cx. quinquefasciatus. Furthermore, our data suggest two-way interactions between specific ISVs and arboviruses. The isolation of specific ISVs and future in vivo infection experiments will be needed to confirm their implications in vector competence.
MATERIALS AND METHODS
Mosquito populations and viral strain.
Field Ae. aegypti at larval or pupal instar and Cx. quinquefasciatus imagos from Guadeloupe collected in 2018 were used for this study. Ae. aegypti and Cx. quinquefasciatus were reared by the Institut Pasteur of Guadeloupe and Institut Pasteur in Paris, respectively. The first generation (F1) of Ae. aegypti and Cx. quinquefasciatus was used in this study.
Lyophilized ZIKV strain Martinique (GenBank: KU647676) was isolated in 2015 and provided by the Emergence Virus Unit (Marseille). The WNV strain used belongs to lineage 1a and was isolated from a horse in France (Camargue) in 2000 (28). The hatched condition and virus isolation method were described in detail in Text S1.
Detailed descriptions of methods. Download Text S1, DOCX file, 0.05 MB (51.5KB, docx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mosquito oral infections.
Seven-day-old Ae. aegypti and Cx. quinquefasciatus females were randomly separated into three groups, receiving three different diets: (i) a 10% sucrose solution; (ii) noninfectious blood meal; (iii) blood containing ZIKV (Ae. aegypti) or WNV (Cx. quinquefasciatus). Ae. aegypti and Cx. quinquefasciatus from all three groups were separately sacrificed at 7 and 21 dpe, and 7 and 14 dpe, respectively, for further analysis. More details can be found in Text S1 section “Mosquito Oral Infections.”
Viral dissemination analysis.
Each mosquito was dissected, and the head, body, legs, and wings were collected. The heads of mosquitoes engorged with infectious blood were used to assess viral dissemination beyond the mosquito midgut by plaque assays as a proxy for the ability to transmit virus. The dissemination efficiency for ZIKV and WNV were calculated, which refers to the proportion of mosquitoes with infectious viral particles in the head forming plaques (see Text S1).
Sample processing and sequencing for viral metagenomics and 16S rRNA sequencing.
The bodies of individual mosquitoes from all groups were homogenized with 400 μL PBS. One hundred fifty μL supernatant of each mosquito body was processed using an optimized sample preparation protocol for viral metagenomics—NetoVIR (46), as described previously. One hundred fifty-four mosquito samples (Table 2) together with 2 negative controls were sequenced on the Illumina NovaSeq 6000 high-throughput platform. Two hundred μL supernatant of each mosquito body homogenate was applied for 16S rRNA sequencing using standard lab protocols as optimized in a previous study (47). Sequencing was performed on the Illumina MiSeq platform.
Bioinformatic analysis for viral metagenomics and 16S rRNA sequencing data.
The obtained raw paired-end reads were trimmed for quality and adapters. Reads mapping to a set of contaminating contigs known to be present in reagents were removed, and the remaining reads were de novo assembled. Contigs from all pools longer than 1,000 bp were clustered for redundancy. The representative contigs were taxonomically annotated using DIAMOND (48) against the nr database. All contigs annotated as eukaryotic virus were extracted using an in-house python script. Bacteriophages were identified using a combined approach with an optimized scoring system as described in Text S1. The trimmed and decontaminated reads from individual samples were mapped against the representative contigs to obtain the abundance.
After demultiplexing with sdm without allowing for mismatches, as part of the LotuS pipeline (49), 16S rRNA sequencing reads were further analyzed per sample using DADA2 pipeline (v1.6) (50) (see Text S1). Sequences annotated to the class Chloroplast, family mitochondria, or unknown bacteria were removed prior to the analyses. The R package decontam (51) was used to remove contaminating amplicon sequencing variants. Aedes mosquito samples with more than 11,428 reads were rarefied to 11,428 reads. Culex mosquito samples with more than 24,168 reads were rarefied to 24,168 reads. The bacterial genera with differential abundance among three food source groups were determined by R package DESeq2 with padj <0.05 and baseMean >10 as cutoff.
Statistical analyses.
All statistical analyses were performed and visualized in R using the phyloseq (52), vegan (53), and ggplot2 (54) packages. To test median difference between two groups of continuous variables (alpha diversity measures, abundances, etc.), a pairwise Wilcoxon test was performed. Multiple testing correction was performed where appropriate using the Benjamini-Hochberg procedure (false-discovery rate [FDR] adjustment set at <0.05). Observed richness was calculated by using the phyloseq (52) package. A PERMANOVA test was used to compare beta diversity among groups.
qRT-PCR to determine the viral genome copies in mosquito heads and bodies.
The extractions for the NGS analyses were used to determine the viral genome copies in the mosquito bodies. Viral RNA from the heads were extracted with the Qiagen Viral RNA minikit. qRT-PCRs were performed on the extractions to determine genome copies of ZIKV, PCLPV, GMV, AATV, and AANV in both head and body of Ae. aegypti, whereas we tested Cx. quinquefasciatus for WNV, GCTLV, and WSLV3. The reagents and analysis methods of the qRT-PCR are described in Text S1. The specific primers, probes, and qPCR conditions of each virus are in Data Set S4.
qPCR primers and probes. Download Data Set S4, XLSX file, 0.01 MB (11.7KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bioinformatic analysis of phageome.
The host prediction of phage contigs combined the results of multiple methods as described in Text S1, including blastn, CRISPR spacer arrays, and tRNA sequences. CheckV (55) was used to estimate the completeness of phage contigs. Only the contigs with >20% completeness were included in alpha diversity analysis. The phage contigs with >50% completeness were performed with cenote-taker2 (56) to annotate their genome structure and visualized by SnapGene software. The abundance correlation of phage contigs and their predicted bacterial host were calculated by cor.test and visualized by ggscatter in R.
Availability of data and material.
The viral metagenomics and 16S rRNA data for this study have been deposited in the NCBI Sequence Read Archive (SRA) repository with BioProject, accession number PRJNA738559. The source data used for the statistical analysis and R script are available at https://github.com/Matthijnssenslab/MosquitoMicrobiome.
ACKNOWLEDGMENTS
We thank Chloë Verspecht and Raul Yhossef Tito Tadeo for their help during 16S rRNA sequencing and data analysis. We thank Antoine Boullis for his technical and logistical support during mosquito oral infections.
This work was supported by the fellowship of China Postdoctoral Science Foundation (2021M691238), by the French Government's Investissement d'Avenir program, Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID), by a KU Leuven C2 grant to L.D. (C22/18/007), and by the Program Opérationnel FEDER-Guadeloupe-Conseil Régional 2014–2020 (grant 2018-FED-1084).
We declare no competing interests.
The study was conceived by J.M., C.S., A.-B.F., A.V.-R., and L.D. Mosquito collection and oral infection were carried out by L.H., A.V.-R., L.M., and N.P. Mosquito dissection and plaque assay were performed by L.W. and A.L.R.R. Library preparation for viral metagenomics and 16S rRNA sequencing was conducted by C.S. and L.D.C. qRT-PCR was performed by C.S. Bioinformatics, and statistical analysis was carried out by C.S. with help from L.B. J.M., C.S., A.-B.F., A.V.-R., N.P., L.W., and J.R. drafted the manuscript. All authors revised the article and approved the final version for publication.
Contributor Information
Jelle Matthijnssens, Email: jelle.matthijnssens@kuleuven.be.
Himanshu Batra, Boston Children's Hospital.
Marthandan Mahalingam, Catholic University of America.
REFERENCES
- 1.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 2.Altinli M, Schnettler E, Sicard M. 2021. Symbiotic interactions between mosquitoes and mosquito viruses. Front Cell Infect Microbiol 11:694020. doi: 10.3389/fcimb.2021.694020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O'Neill SL, Hoffmann AA. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 4.Hussain M, Lu G, Torres S, Edmonds JH, Kay BH, Khromykh AA, Asgari S. 2013. Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. J Virol 87:851–858. doi: 10.1128/JVI.01837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, Tripathi A, Mlambo G, Dimopoulos G. 2014. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog 10:e1004398. doi: 10.1371/journal.ppat.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyce JD, Nogueira JR, Bales AA, Pittman KE, Anderson JR. 2011. Interactions between La Crosse virus and bacteria isolated from the digestive tract of Aedes albopictus (Diptera: Culicidae). J Med Entomol 48:389–394. doi: 10.1603/me09268. [DOI] [PubMed] [Google Scholar]
- 7.Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN. 2012. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e40401. doi: 10.1371/journal.pone.0040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu P, Sun P, Nie K, Zhu Y, Shi M, Xiao C, Liu H, Liu Q, Zhao T, Chen X, Zhou H, Wang P, Cheng G. 2019. A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25:101–112.e5. doi: 10.1016/j.chom.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Kuwata R, Isawa H, Hoshino K, Sasaki T, Kobayashi M, Maeda K, Sawabe K. 2015. Analysis of mosquito-borne flavivirus superinfection in Culex tritaeniorhynchus (Diptera: Culicidae) cells persistently infected with Culex Flavivirus (Flaviviridae). J Med Entomol 52:222–229. doi: 10.1093/jme/tju059. [DOI] [PubMed] [Google Scholar]
- 10.Kent RJ, Crabtree MB, Miller BR. 2010. Transmission of West Nile virus by Culex quinquefasciatus Say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis 4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson-Peters J, Yam AWY, Lu JWF, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, Melville L, Hunt N, Webb RI, Blitvich BJ, Whelan P, Hall RA. 2013. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 8:e56534. doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz MJ, Frydman HM, Connor JH. 2018. Dual insect specific virus infection limits Arbovirus replication in Aedes mosquito cells. Virology 518:406–413. doi: 10.1016/j.virol.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guegan M, Zouache K, Démichel C, Minard G, Van TV, Potier P, Mavingui P, Moro CV. 2018. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome 6:49. doi: 10.1186/s40168-018-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 15.Shi C, Beller L, Deboutte W, Yinda KC, Delang L, Vega-Rúa A, Failloux A-B, Matthijnssens J. 2019. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome 7:121. doi: 10.1186/s40168-019-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi C, Zhao L, Atoni E, Zeng W, Hu X, Matthijnssens J, Yuan Z, Xia H. 2020. Stability of the virome in lab- and field-collected Aedes albopictus mosquitoes across different developmental stages and possible core viruses in the publicly available virome data of Aedes mosquitoes. mSystems 5:e00640-20. doi: 10.1128/mSystems.00640-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agence Régionale de Santé (ARS) Guadeloupe, 2017. Les réponses face à l’épidémie de Zika en Guadeloupe, Saint Martin, et Saint Barthélémy. ARS Guadeloupe, Gourbeyre, Guadeloupe. [Google Scholar]
- 18.Hery L, Boullis A, Delannay C, Vega-Rúa A. 2019. Transmission potential of African, Asian and American Zika virus strains by Aedes aegypti and Culex quinquefasciatus from Guadeloupe (French West Indies). Emerg Microbes Infect 8:699–706. doi: 10.1080/22221751.2019.1615849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moutailler S, Yousfi L, Mousson L, Devillers E, Vazeille M, Vega-Rúa A, Perrin Y, Jourdain F, Chandre F, Cannet A, Chantilly S, Restrepo J, Guidez A, Dusfour I, Vieira Santos de Abreu F, Pereira dos Santos T, Jiolle D, Visser TM, Koenraadt CJM, Wongsokarijo M, Diallo M, Diallo D, Gaye A, Boyer S, Duong V, Piorkowski G, Paupy C, Lourenco de Oliveira R, de Lamballerie X, Failloux A-B. 2019. A new high-throughput tool to screen mosquito-borne viruses in Zika virus endemic/epidemic areas. Viruses 11:904. doi: 10.3390/v11100904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefrancois T, Blitvich BJ, Pradel J, Molia S, Vachiéry N, Pallavicini G, Marlenee NL, Zientara S, Petitclerc M, Martinez D. 2005. West Nile virus surveillance, Guadeloupe, 2003–2004. Emerg Infect Dis 11:1100–1103. doi: 10.3201/eid1107.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosine J. 1999. Résistance d’Aedes aegypti et de Culex pipiens quinquefasciatus aux insecticides organophospohorés, biologiques et aux pyréthrinoïdes en Martinique et en Guadeloupe. Univ. Pierre et Marie Curie (Paris VI), Paris, France. [Google Scholar]
- 22.Chauhan C, Behura SK, Debruyn B, Lovin DD, Harker BW, Gomez-Machorro C, Mori A, Romero-Severson J, Severson DW. 2012. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. PLoS One 7:e47350. doi: 10.1371/journal.pone.0047350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett KE, Flick D, Fleming KH, Jochim R, Beaty BJ, Black WC. 2005. Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti. Genetics 170:185–194. doi: 10.1534/genetics.104.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. 2007. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol 7:9. doi: 10.1186/1471-2180-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romoser WS, Wasieloski LP, Pushko P, Kondig JP, Lerdthusnee K, Neira M, Ludwig GV. 2004. Evidence for arbovirus dissemination conduits from the mosquito (Diptera: Culicidae) midgut. J Med Entomol 41:467–475. doi: 10.1603/0022-2585-41.3.467. [DOI] [PubMed] [Google Scholar]
- 26.Girard YA, Klingler KA, Higgs S. 2004. West Nile virus dissemination and tissue tropisms in orally infected Culex pipiens quinquefasciatus. Vector Borne Zoonotic Dis 4:109–122. doi: 10.1089/1530366041210729. [DOI] [PubMed] [Google Scholar]
- 27.Atyame CM, Alout H, Mousson L, Vazeille M, Diallo M, Weill M, Failloux A-B. 2019. Insecticide resistance genes affect Culex quinquefasciatus vector competence for West Nile virus. Proc Biol Sci 286:20182273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murgue B, Murri S, Zientara S, Durand B, Durand J-P, Zeller H. 2001. West Nile outbreak in horses in southern France, 2000: the return after 35 years. Emerg Infect Dis 7:692–696. doi: 10.3201/eid0704.017417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amraoui F, Krida G, Bouattour A, Rhim A, Daaboub J, Harrat Z, Boubidi S-C, Tijane M, Sarih M, Failloux A-B. 2012. Culex pipiens, an experimental efficient vector of West Nile and Rift Valley fever viruses in the Maghreb region. PLoS One 7:e36757. doi: 10.1371/journal.pone.0036757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romoser WS, Turell MJ, Lerdthusnee K, Neira M, Dohm D, Ludwig G, Wasieloski L. 2005. Pathogenesis of Rift Valley fever virus in mosquitoes—tracheal conduits & the basal lamina as an extra-cellular barrier. Arch Virol Suppl:89–100. doi: 10.1007/3-211-29981-5_8. [DOI] [PubMed] [Google Scholar]
- 31.Sim S, Jupatanakul N, Dimopoulos G. 2014. Mosquito immunity against Arboviruses. Viruses-Basel 6:4479–4504. doi: 10.3390/v6114479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. 2013. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD. 2012. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 427:90–97. doi: 10.1016/j.virol.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David MR, Santos LMBD, Vicente ACP, Maciel-de-Freitas R. 2016. Effects of environment, dietary regime and ageing on the dengue vector microbiota: evidence of a core microbiota throughout Aedes aegypti lifespan. Mem Inst Oswaldo Cruz 111:577–587. doi: 10.1590/0074-02760160238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scolari F, Casiraghi M, Bonizzoni M. 2019. Aedes spp. and their microbiota: a review. Front Microbiol 10:2036. doi: 10.3389/fmicb.2019.02036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos-Nino ME, Fitzpatrick DM, Eckstrom KM, Tighe S, Hattaway LM, Hsueh AN, Stone DM, Dragon JA, Cheetham S. 2020. Metagenomic analysis of Aedes aegypti and Culex quinquefasciatus mosquitoes from Grenada, West Indies. PLoS One 15:e0231047. doi: 10.1371/journal.pone.0231047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minard G, Mavingui P, Moro CV. 2013. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors 6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muturi EJ, Dunlap C, Ramirez JL, Rooney AP, Kim C-H. 2018. Host blood-meal source has a strong impact on gut microbiota of Aedes aegypti. FEMS Microbiology Ecology 95:fiy213. doi: 10.1093/femsec/fiy213. [DOI] [PubMed] [Google Scholar]
- 40.Terenius O, Lindh JM, Eriksson-Gonzales K, Bussière L, Laugen AT, Bergquist H, Titanji K, Faye I. 2012. Midgut bacterial dynamics in Aedes aegypti. FEMS Microbiol Ecol 80:556–565. doi: 10.1111/j.1574-6941.2012.01317.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Gilbreath TM, Kukutla P, Yan G, Xu J. 2011. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6:e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villegas LEM, Campolina TB, Barnabe NR, Orfano AS, Chaves BA, Norris DE, Pimenta PFP, Secundino NFC. 2018. Zika virus infection modulates the bacterial diversity associated with Aedes aegypti as revealed by metagenomic analysis. PLoS One 13:e0190352. doi: 10.1371/journal.pone.0190352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goddard LB, Roth AE, Reisen WK, Scott TW. 2002. Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis 8:1385–1391. doi: 10.3201/eid0812.020536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glaser RL, Meola MA. 2010. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One 5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. 2009. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10:33. doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conceicao-Neto N, Zeller M, Lefrere H, De Bruyn P, Beller L, Deboutte W, Yinda CK, Lavigne R, Maes P, Van Ranst M, Heylen E, Matthijnssens J. 2015. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep 5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D'hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. 2016. Population-level analysis of gut microbiome variation. Science 352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 48.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 49.Hildebrand F, Tadeo R, Voigt AY, Bork P, Raes J. 2014. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan BJ, Davis NM. 2019. decontam: identify contaminants in marker-gene and metagenomics sequencing data. https://github.com/benjjneb/decontam.
- 52.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Vegetation Science 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 54.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 55.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2020. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tisza MJ, Belford AK, Domínguez-Huerta G, Bolduc B, Buck CB. 2021. Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus Evol 7:veaa100. doi: 10.1093/ve/veaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart of study design. Female adults, which hatched in the lab from eggs of Ae. aegypti and Cx. quinquefasciatus collected in the field from Guadeloupe in 2018. The 7-day-old female adults were fed with one of three meals: (a) sucrose and water; (b) regular blood; or (c) blood spiked with infectious ZIKV (Ae. aegypti) or WNV (Cx. quinquefasciatus). Individual mosquitoes from all three groups were sacrificed at 7 and 21 (ZIKV) or 14 (WNV) days postexposure (dpe). The heads of individual mosquitoes engorged with spiked blood were titrated by plaque assays to determine the presence of infectious ZIKV or WNV viral particles. Viral metagenomics and 16S rRNA sequencing were applied on the bodies of all the mosquitoes to explore the associations of the mosquito microbiome with arbovirus transmission. Note: No qRT-PCR was performed for WNV on the heads of Culex quinquefasciatus mosquitoes, due to insufficient sample. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Eukaryotic virome in Aedes and Culex mosquitoes. Download Data Set S1, XLSX file, 0.1 MB (89KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The alpha diversity of eukaryotic virus in Aedes aegypti of different time points and infection status. Download FIG S2, PDF file, 0.2 MB (200.6KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of viral genome copies in body and head of mosquitoes. Pairwise Wilcoxon test: P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). P values were corrected for multiple comparisons with the Benjamini-Hochberg method. Download FIG S3, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of Asaia and Wolbachia in Culex quinquefasciatus of two time points. Download FIG S4, PDF file, 0.1 MB (84.8KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome structure of phage contigs. (a) Genome structure of phage contig annotated as Podoviridae. (b) Genome structure of phage contig annotated as Siphoviridae. Download FIG S5, PDF file, 0.5 MB (560.7KB, pdf) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Host prediction and completeness estimation of phages. Download Data Set S2, XLSX file, 0.2 MB (171.5KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA sequencing results. Download Data Set S3, XLSX file, 0.4 MB (410.4KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed descriptions of methods. Download Text S1, DOCX file, 0.05 MB (51.5KB, docx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
qPCR primers and probes. Download Data Set S4, XLSX file, 0.01 MB (11.7KB, xlsx) .
Copyright © 2022 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.