ABSTRACT
Several antibacterial compounds have recently been discovered that potentially inhibit the activity of BamA, an essential subunit of a heterooligomer (the barrel assembly machinery or BAM) that assembles outer membrane proteins (OMPs) in Gram-negative bacteria, but their mode of action is unclear. To address this issue, we examined the effect of three inhibitors on the biogenesis of a model E. coli OMP (EspP) in vivo. We found that darobactin potently inhibited the interaction of a conserved C-terminal sequence motif (the “β signal”) with BamA, but had no effect on assembly if added at a postbinding stage. In contrast, Polyphor peptide 7 and MRL-494 inhibited both binding and at least one later step of assembly. Taken together with previous studies that analyzed the binding of darobactin and Polyphor peptide 7 to BamA in vitro, our results strongly suggest that the two compounds inhibit BAM function by distinct competitive and allosteric mechanisms. In addition to providing insights into the properties of the antibacterial compounds, our results also provide direct experimental evidence that supports a model in which the binding of the β signal to BamA initiates the membrane insertion of OMPs.
KEYWORDS: antibiotics, β signal, barrel assembly machinery, Gram-negative bacteria, membrane proteins, outer membrane
INTRODUCTION
Almost all integral membrane proteins that are located in the outer membrane (OM) of Gram-negative bacteria (as well as a subset of the proteins located in the OM of mitochondria and chloroplasts) are unusual in that they lack α-helical membrane spanning segments. Instead, they are anchored to the membrane by a “β barrel,” an amphipathic β sheet that folds into a closed cylindrical structure that is held together by hydrogen bonds (1). Despite sharing a common architecture, OM proteins (OMPs) are diverse in several respects. First, β barrels vary considerably in size (from 8 to 36 β strands) and sequence (2, 3). Second, while some OMPs consist solely of an empty β barrel, other OMPs contain β barrels that are filled with peptides or plug domains as well as extracellular or periplasmic domains (4). In addition, some OMPs form homodimers or homotrimers (5). Presumably, β barrels evolved because they can be transported through the Sec machinery without being retained in the inner membrane. It is also possible that their β barrel structure facilitates insertion into the OM, an unusual membrane that is both highly rigid and asymmetrical in that it contains phospholipids in the inner leaflet and a unique glycolipid (lipopolysaccharide, or LPS) in the outer leaflet (4, 6).
It was shown over 15 years ago that the insertion of OMPs into the OM is catalyzed by a heterooligomer called the barrel assembly machinery (BAM) (7 to 9). BAM consists of BamA, a universally conserved integral OMP that contains a C-terminal β barrel and five N-terminal polypeptide transport associated (POTRA) domains that reside in the periplasm and bind a variable number of lipoproteins to the complex (10). Although the structure of the E. coli holocomplex has been solved (11 to 14), the mechanism by which it catalyzes the assembly (i.e., the folding and membrane insertion) of OMPs is unclear. A variety of models have been proposed, all of which focus on an unusual property of the BamA β barrel. Unlike most β barrels that are extremely stable, the BamA β barrel has an unstable “seam” (i.e., hydrogen bonds that hold the first and last β strands together) that has been shown to open in molecular dynamics simulations and in structural and biochemical studies (15 to 20). In “threading” models, newly synthesized OMPs enter the lumen of the BamA β barrel as unfolded polypeptides that fold into β hairpins and then bud through a “lateral gate” formed by the open BamA β barrel seam in a stepwise fashion (19, 21). At the opposite extreme, “assisted” models suggest that the dynamic BamA β barrel seam perturbs the structure of the OM to catalyze the insertion of OMP β barrels that are already partially folded in the periplasmic space (19, 21). Recently, compelling models derived from a detailed analysis of an OMP assembly intermediate bound to BAM using biochemical and structural data suggest that the open form of the BamA β barrel forms a hybrid barrel with incoming OMPs and catalyzes their insertion through a “swing” mechanism (16, 22). The results indicate that a strong interaction forms between a C-terminal peptide called the “β signal” that is conserved across the majority of OMPs and the first strand of the BamA β barrel, which likely plays a key role in the OMP assembly process.
Although the function of BAM is still poorly understood, the conservation of BamA and one of the lipoprotein subunits (BamD), the demonstration that BamA and BamD are essential for viability (9, 23), and the fact that BAM is located at the cell surface have led to the idea that it might be an excellent target for novel antibiotics. Consistent with this hypothesis, several potential BAM inhibitors that have strong antimicrobial activity have been identified. The first such compound, a β-hairpin macrocyclic peptide called JB-95, selectively disrupts the OM of E. coli and binds to a variety of OMPs, including BamA, in photolabeling experiments (24). More recently, two small molecules isolated in a screen for activators of the σE stress response have been shown to reduce the levels of OMPs in vivo and BAM activity in vitro (25). Furthermore, a naturally occurring cyclic peptide produced by Photorhabdus (darobactin), a group of structurally related synthetic cyclic peptides (Polyphor peptides 3, 4, 7, and 8), and a small molecule (MRL-494) are particularly noteworthy not only because they bind to BamA, but also because mutations that suppress their antimicrobial effects map to bamA (26 to 28). Interestingly, structural and biophysical studies have shown that darobactin mimics a β signal by binding specifically to the seam of purified BamA (primarily to β strand 1) in vitro and affects the mechanical properties of the protein (29, 30). In contrast, nuclear magnetic resonance (NMR) spectroscopy experiments have indicated that the Polyphor compounds interact with surface exposed loops of BamA (28). Available evidence also indicates that both the Polyphor compounds and MRL-494 affect the integrity of the bacterial OM (26, 28).
To gain insight into both the mechanism of action of the potential BAM inhibitors and the function of BAM, we performed a detailed analysis of the effect of darobactin, Polyphor peptide 7, and MRL-494 on OMP assembly in vivo in E. coli. We utilized a well-characterized OMP (EspP) that is a member of the autotransporter family of OMPs as a model protein in our experiments. Autotransporters contain an extracellular (“passenger”) domain attached to a β barrel by an α-helical linker that traverses the β-barrel pore (31). They are “representative” OMPs in that they contain a stable β barrel, a typical number of β strands (12), and a β motif that conforms to the consensus. We exploited the fact that two EspP mutants have been described whose assembly is either dramatically slowed or arrested at a different stage of assembly and can therefore be used as reporters of stage-specific effects (22, 32). Consistent with the notion that darobactin acts as a competitive inhibitor of BamA function, we obtained direct evidence that it completely blocks the binding of the EspP β signal to BamA in vivo and that this interaction initiates the assembly reaction. Interestingly, we found that both the Polyphor peptide 7 and MRL-494 also prevent β signal binding to BamA. The data support a model in which the compounds act as allosteric inhibitors of assembly initiation. Furthermore, we found that the Polyphor peptide 7 and MRL-494 inhibit OMP assembly at both pre- and post-BAM binding steps and strongly suggest that these compounds either block at least two steps of BamA function or inhibit OMP assembly indirectly by perturbing the structure and/or integrity of the OM. Taken together, the results imply that OMP assembly can be inhibited at multiple stages and raise the possibility that the potency of antimicrobial compounds can be enhanced by using them combinatorially.
RESULTS
Darobactin inhibits the binding of OMPs to BAM.
It was shown in a previous study that the introduction of a single arginine residue onto the lipid-facing surface of two E. coli OMPs, EspP and OmpLA, can profoundly affect their assembly (32). Interestingly, the effect is highly position specific: mutations located near the middle of the β barrel create an energy barrier that impedes membrane insertion, while mutations located near the periplasmic or extracellular side have no effect. Most of the unintegrated mutants are degraded in the periplasm, but one mutant, EspP(G1123R) (Fig. 1A), has a unique phenotype. The mutation does not affect the binding of EspP to BAM, and the mutant protein is effectively assembled, but much more slowly than wild-type EspP (t1/2 ≈ 10 min versus t1/2 < 1 min). The rate-limiting step in assembly is the exposure of the passenger domain on the cell surface. Because the passenger domain initially forms a loop that is embedded inside the EspP-BamA hybrid barrel (33), this step requires movement of the nascent β barrel into the OM. Given that EspP G1123R remains stable for >20 min (32), the results strongly suggest that the mutant β barrel docks onto BAM in a way that protects it from degradation by periplasmic proteases, but that the mutation significantly delays the membrane integration process.
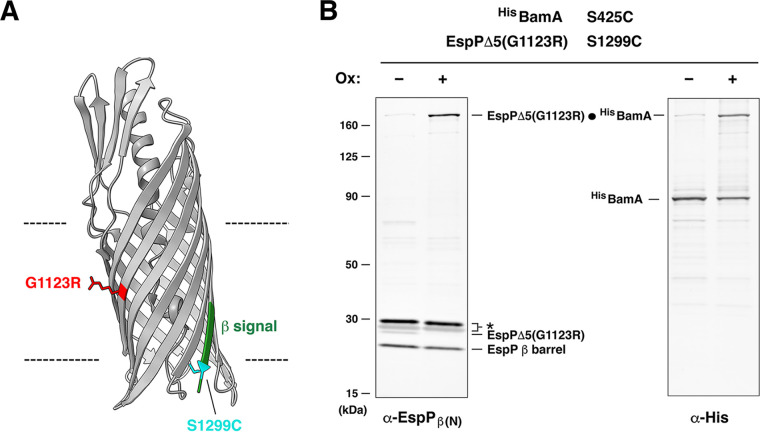
FIG 1.
The G1123R mutation delays EspP assembly at a step that follows the interaction of the β signal with BamA β1. (A) Crystal structure of the EspP β barrel (49) (PDB ID: 3SLJ). The G1123R mutation (red), the β signal in the final strand (green), and the S1299C mutation (cyan) that can form a disulfide bond with BamA(S425C) after β signal binding are depicted. (B) E. coli BL21(DE3) expressing HisBamAS425CBCDE and EspPΔ5(G1123R/S1299C) were mock treated (–) or treated (+) with 4-DPS oxidizing agent (Ox). Intermolecular disulfide bonds (•) were detected by double-probing with antibodies against the N termini of the EspP β barrel (α-EspPβ(N)) and HisBamA (α-His). *, nonspecific bands.
To determine if the assembly of EspP(G1123R) stalls after the protein forms an on-pathway interaction with BamA, we exploited the observation that the β signal in the final strand of the wild-type EspP β barrel (residues 1287 to 1300; see Fig. 1A) forms a tight interaction with BamA β1 during assembly (22). This interaction was previously mapped by an in vivo intermolecular cross-linking approach in which a pair of residues in EspP and BamA (residues S1299 and S425, respectively) were mutated to cysteine, and the formation of an intermolecular disulfide bond after the addition of the thiol-specific oxidant 4,4’-dipyridyl disulfide (4-DPS) was monitored. If the EspP(G1123) mutation stalls the normal assembly process after the protein interacts with BamA, then the same disulfide bond should be detected. To test this possibility, E. coli BL21(DE3) transformed with plasmids that encode a modified BAM (HisBamAS425CBCDE) and EspPΔ5(G1123R/S1299C), a mutant form of an EspP derivative that contains an extremely short passenger domain but that assembles normally in vivo (34), was grown in LB. After the expression of both constructs was induced, cells were treated with 4-DPS or mock-treated, and Western blots were conducted using antisera to simultaneously detect the N terminus of the EspP β barrel and the His-tag on BamA. Consistent with previous results, a very high fraction of EspPΔ5(G1123R/S1299C) formed a disulfide bond with HisBamAS425C (Fig. 1B). The data strongly support our hypothesis that the G1123R mutation delays a step (or steps) in assembly that occur after the EspP β signal binds to BamA.
Based on the disulfide cross-linking data and published structural data that suggest that darobactin inhibits the binding of OMP β signals to BamA, we hypothesized that darobactin would block the initial interaction of EspP(G1123R) with BamA in vivo but would not affect later stages in protein biogenesis. Because the molecular basis for darobactin toxicity has not been established, however, we first needed to determine if the compound affects OMP assembly. To this end, we transformed E. coli AD202 (MC4100 ompT::kan) with a plasmid that encodes EspP(G1123R) under the control of the trc promoter. Cells were grown in minimal medium, and the synthesis of the EspP mutant was induced by the addition of IPTG. The cells were then treated with 4 μg/mL darobactin (1× MIC) or untreated and subjected to pulse-chase radiolabeling. Radiolabeled EspP-containing polypeptides were then immunoprecipitated with an antiserum against a C-terminal peptide and resolved by SDS-PAGE. The assembly of EspP(G1123R) was then assessed by monitoring the autocatalytic release of the passenger domain of proEspP (the full-length form of the protein) from the fully folded β barrel, which has been shown to be the final step in EspP biogenesis (35, 36). It should be noted that a great advantage of the pulse-chase approach is that it facilitates the examination of the kinetics and efficiency of assembly of a cohort of newly synthesized molecules at different times both before and after the addition of molecules of interest.
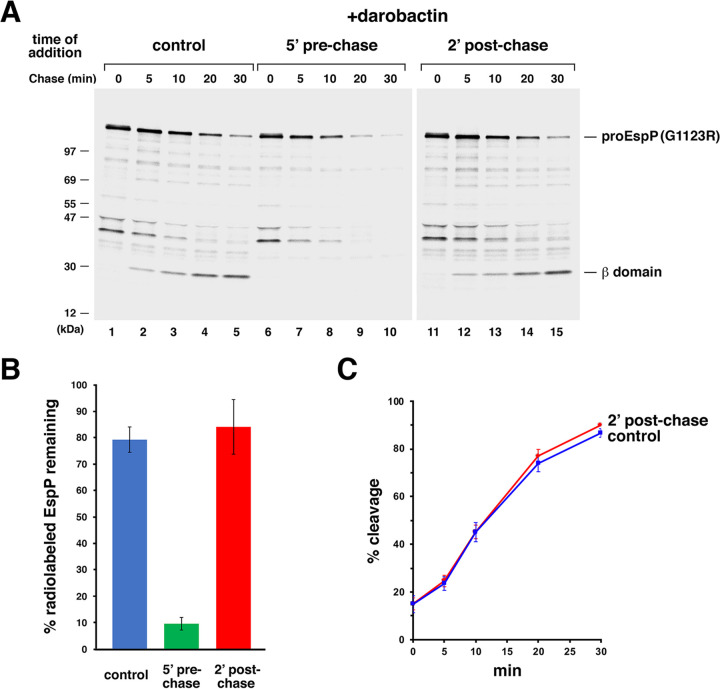
Consistent with our hypothesis, we found that darobactin completely blocked the assembly of EspP(G1123R), but only if it was added before the start of the chase. As previously observed, under normal (control) conditions, the EspP mutant underwent proteolytic maturation to form a fully folded β barrel very slowly but remained stable (Fig. 2A, lanes 1 to 5, and Fig. 2B). Treatment with proteinase K (PK) confirmed that the rate-limiting step in assembly was the exposure of the passenger domain on the cell surface (Fig. S1, top gel). In contrast, no maturation was observed when darobactin was added 5 min before the start of the chase, and the pro form of the protein was gradually degraded, presumably because its interaction with BAM was inhibited (Fig. 2A, lanes 6 to 10, and Fig. 2B). A second OMP (the porin OmpC) that was immunoprecipitated from the same samples was likewise stable in untreated cells but very rapidly degraded in the presence of darobactin presumably because it also remained in the periplasmic space (Fig. S2, lanes 1 to 10). This result shows that darobactin not only completely inhibits the assembly of EspP(G1123R), but also exerts a more general effect on OMP assembly in vivo. The same inhibitory effects were observed when darobactin was added as late as 1 min prior to the chase (Fig. S3). Remarkably, darobactin had no effect on either the assembly or stability of EspP(G11123R) (or the stability of OmpC) when it was added 2 min after the start of the chase (Fig. 2A, lanes 11 to 15, and Fig. 2B and C; Fig. S2, lanes 11 to 15). At this time point an interaction between EspP(G11123R) and BAM could be clearly detected (see below). As in untreated cells, the slow assembly correlated with a delay in the exposure of the passenger domain on the cell surface (Fig. S1, bottom gel). Taken together, these results provide strong evidence that darobactin is a direct and highly specific inhibitor of the interaction of OMPs with BAM.
FIG 2.
Darobactin blocks EspP(G1123R) assembly only if added prior to synthesis. (A) E. coli strain AD202 was transformed with pJH224 [Ptrc-EspP(G1123R)], and cultures were grown in M9 medium. At OD550 of 0.2, the expression of the EspP mutant was induced by the addition of 10 μM IPTG, and cells were subjected to pulse-chase labeling 30 min later. Culture aliquots were untreated (control) or treated with darobactin (4 μg/mL) either 5 min before or 2 min after the start of the chase (5′ prechase or 2’ postchase, respectively). Immunoprecipitations were performed using an antiserum generated against an EspP C-terminal peptide, proteins were resolved by SDS-PAGE, and radioactive proteins were imaged. A representative experiment is shown. (B) Stability of EspP(G1123R) in the presence of darobactin. The mean percentage of the total radiolabeled EspP (as defined in Materials and Methods) that was observed at the 30-min time point in three independent pulse-chase experiments is shown. Error bars represent standard error. (C) Time course of EspP(G1123R) assembly in untreated cells (control) or cells treated with darobactin 2 min after the start of the chase. The mean fraction of radiolabeled protein that was completely assembled at each time point in three independent experiments (% cleavage) is shown. Error bars represent standard error.
The EspP(G1123R) passenger domain is exposed on the cell surface if darobactin is added after the start of the chase. The experiment described in Fig. 2 was repeated through the pulse-chase labeling step. Culture aliquots were untreated (control) or treated with darobactin (4 μg/mL) 2 min after the start of the chase. PK was then added to half of each sample. Immunoprecipitations were performed using the anti-EspP C-terminal antiserum, proteins were resolved by SDS-PAGE, and radioactive proteins were imaged. Download FIG S1, JPG file, 1.7 MB (1.8MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Darobactin blocks OmpC assembly only if added prior to synthesis. Immunoprecipitations were performed with an antiserum generated against OmpC using the samples generated in the experiments described in Fig. 2. A representative result is shown. Download FIG S2, JPG file, 0.4 MB (431.4KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Darobactin blocks OMP assembly within 1 min of addition. The experiment described in Fig. 2A was repeated except that darobactin (4 μg/mL) was added at 1 min (-1’) or 2 min (-2’) before the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide (top) or OmpC (bottom). Download FIG S3, JPG file, 0.4 MB (399.1KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
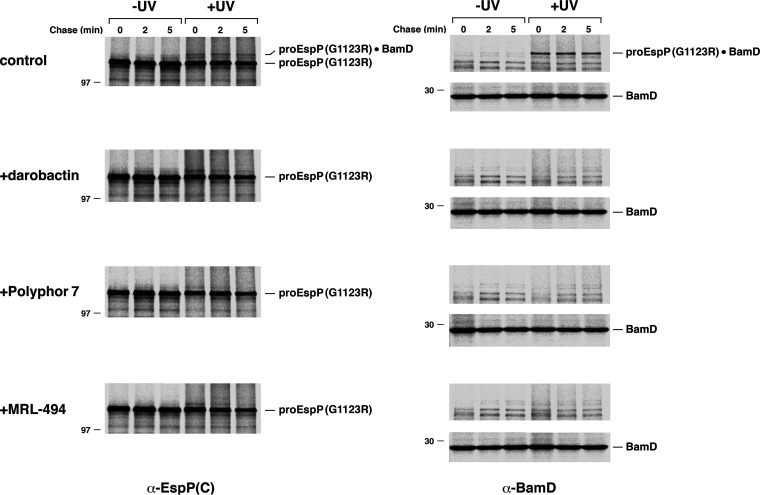
We next utilized a site-specific cross-linking approach to corroborate the conclusion that darobactin inhibits the binding of EspP(G1123R) to BAM. It has previously been shown that when the photoreactive amino acid analog pBpa is introduced into specific positions in the EspP β barrel by amber suppression, a cross-link to an individual BAM subunit can be detected after UV irradiation (37). We introduced pBpa at position 1214 because significant cross-linking between this residue and BamD is typically observed (32, 34, 37, 38) and serves as a reliable measure of the EspP-BAM interaction. AD202 was transformed with a plasmid that encodes EspP(G1123R/F1214am) and pDULE, a plasmid that encodes an amber suppressor tRNA, and an amino acyl-tRNA synthetase from Methanococcus jannaschii (39), and cells were subjected to pulse-chase labeling in either the absence or presence of darobactin and UV-irradiated (or mock-treated). Consistent with previous results (32), a high molecular weight (~160 kDa) band that could be immunoprecipitated with both anti-EspP and anti-BamD antisera was observed in UV-irradiated control cells as early as the start of the chase (Fig. 3, top-tier gels). Together with the results shown in Fig. 1B, this observation implies that in the absence of the inhibitor the assembly of the EspP(G1123R) mutant slows at a stage during which the β signal is associated with the first strand of BamA. In contrast, none of the high molecular weight band could be detected in the presence of darobactin (Fig. 3, second-tier gels). The simplest explanation of this result is that the inhibitor blocks the binding of EspP(G1123R) to BamA. Alternatively, the inhibitor could prevent the interaction of the mutant protein with BamD by altering the conformation of BAM, a notion supported by structural evidence showing that BamD moves away from the BamA substrate binding site when it is bound by darobactin (16, 29).
FIG 3.
Multiple antimicrobials block the interaction of EspP(G1123R) with BAM. AD202 transformed with pJH225 [Plac-HisEspP(G1123R/Y1214am)] and pDULE38 were grown in M9 medium. At OD550 of 0.2, Bpa (1 mM) was added to the cultures, and the expression of the EspP mutant was induced by the addition of 200 μM IPTG. Cells were subjected to pulse-chase labeling 30 min later. Culture aliquots were untreated (control) or treated with the indicated antimicrobial 5 min before the start of the chase. Samples were divided in half, and one half was UV irradiated. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide (α-EspP(C)) or BamD (α-BamD), proteins were resolved by SDS-PAGE, and radioactive proteins were imaged.
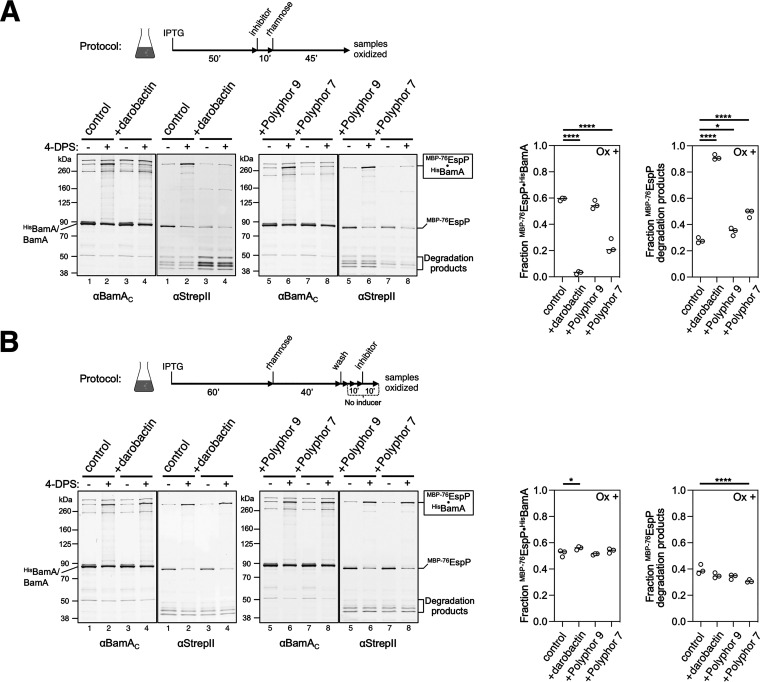
Finally, we used a second mutant, designated MBP-76EspP, to further examine the mechanism by which darobactin affects the interaction between EspP and BAM. To construct MBP-76EspP, most of the passenger domain was replaced by maltose binding protein (MBP), a polypeptide that folds rapidly in the periplasm and thereby inhibits the completion of both passenger domain secretion and β-barrel assembly by BAM (22). MBP-76EspP forms an arrested assembly intermediate in vivo in which its β barrel forms a hybrid barrel with the BamA β barrel while its passenger domain is embedded inside the BamA β barrel (22, 33). In the arrested state, the β signal forms a stable interface with BamA β1 that can be detected using the intermolecular disulfide bond formation assay described above. Consistent with previous results (22), a large fraction (~60%) of MBP-76EspPS1299C formed a high molecular weight cross-linking product with HisBamAS425C in the presence of 4-DPS that was detected by probing immunoblots with antisera directed against both a C-terminal peptide of BamA and a StrepII tag located on the N terminus of MBP-76EspP (Fig. 4A and B, lanes 1 to 2 and left graphs). Disulfide bond formation was reduced by 95%, however, if darobactin was added prior to—but not after—the induction of MBP-76EspP expression (Fig. 4A and B, lanes 3 to 4, and left graphs). Furthermore, the addition of darobactin prior to MBP-76EspP expression resulted in a considerable increase in the level of N-terminal MBP-76EspP degradation products (Fig. 4A, lanes 3 to 4 and right graphs), presumably because MBP-76EspP could not bind to BamA and was consequently targeted by periplasmic proteases. Taken together with our analysis of the EspP(G1123R) mutant, these results strongly suggest that darobactin acts as a competitive inhibitor of the interaction of the conserved OMP β signal and BamA β1 in vivo, but has no effect once a stable interaction is formed.
FIG 4.
Darobactin and the Polyphor 7 peptide inhibit the binding of the MBP-76EspP β signal to BamA β1 if added prior to synthesis. Experiment conducted as in Fig. 1B, except E. coli BL21(DE3) expressing HisBamAS425CBCDE was treated with darobactin (4 μg/mL), Polyphor peptide 7 (1.25 μg/mL), or Polyphor peptide 9 (1.25 μg/mL) either before (A) or after (B) the expression of MBP-76EspPS1299C was induced by the addition of rhamnose. Intermolecular disulfide bonds were detected by double-probing with antibodies against a C-terminal peptide of BamA (α-BamAC) and a StrepII-tag located on the N terminus of MBP-76EspP (α-StrepII). Quantitation of the fraction of MBP-76EspPS1299C cross-linked to HisBamAS425C and detected N-terminal MBP-76EspP degraded fragments from oxidized lanes are plotted in the graphs (N = 3, lines at median). *, P < 0.05; ****, P < 0.0001.
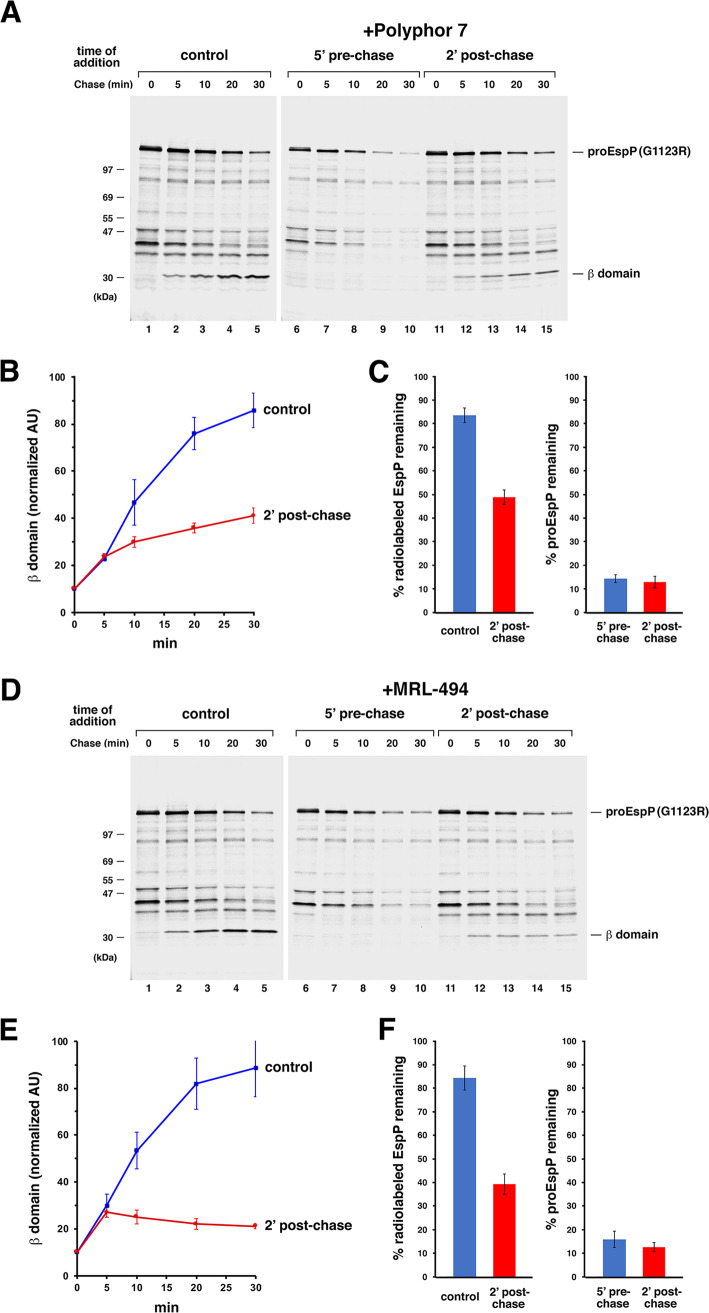
Polyphor peptide 7 and MRL-494 inhibit both early and later stages of OMP assembly.
We next wished to examine the mechanism of action of Polyphor peptide 7 and MRL-494 (26, 28), two potential BAM inhibitors that have been less well characterized than darobactin. To this end, we first repeated the experiment shown in Fig. 2, except that we added 0.5 μg/mL Polyphor peptide 7 (2× MIC) or 16.5 μg/mL MRL-494 (1× MIC) instead of darobactin at either 5 min before or 2 min after the start of the chase. Like darobactin, both compounds completely blocked the assembly of EspP(G1123R) and led to its degradation (Fig. 5A and D, lanes 1 to 10). The assembly of both EspP(G1123R) and OmpC was also completely blocked when the inhibitors were added 1 min before the start of the chase (Fig. S4). Consistent with the notion that the compounds inhibited the interaction of the EspP mutant with BAM, no cross-linking was observed between EspP(G1123R/Y1214Bpa) and BamD in the presence of either the Polyphor 7 peptide or MRL-494 (Fig. 3, third- and fourth-tier gels). Interestingly, in contrast with darobactin, both compounds significantly blocked the completion of EspP(G1123R) assembly when they were added after the start of the chase (Fig. 5A and D, lanes 11 to 15, and Fig. 5B and E). About the same level of assembly (~25%) was observed in both control cells and treated cells at the 5-min time point, but the low level of assembly that occurred between 0 min and 2 min, as well the assembly that continued briefly until the inhibitors bound to their targets, likely accounts for this similarity. While the assembly of EspP(G1123R) tapered off at later time points in the presence of the Polyphor 7 peptide, assembly appeared to stop altogether in the presence of MRL-494. Similar results were obtained even when the concentration of the inhibitors was reduced to 0.2× MIC (Fig. S5). Importantly, the total level of EspP-containing polypeptides dropped during the chase and, despite the proteolytic processing of only a fraction of the protein, the stability of proEspP was also reduced (Fig. 5C and F). These results imply that the antimicrobials either exposed proEspP to periplasmic proteases without affecting its interaction with BAM or caused its complete dissociation from BAM. Regardless, the results clearly show that the Polyphor 7 peptide and MRL-494 inhibit both the binding of EspP(G1123R) to BAM and one or more postbinding steps.
FIG 5.
The Polyphor 7 peptide and MRL-494 inhibit EspP(G1123R) assembly if added either before or after synthesis. The experiments described in Fig. 2 were repeated except that either 0.5 μg/mL Polyphor 7 peptide (A) or 16.5 μg/mL MRL-494 (D) was added in place of darobactin. Representative experiments are shown. (B and E) Time course of EspP(G1123R) assembly in untreated cells (control) or cells treated with the Polyphor 7 peptide (B) or MRL-494 (E) 2 min after the start of the chase. The mean fraction of radiolabeled protein that was completely assembled at each time point in three independent experiments (accumulated β domain in arbitrary units [AU] as defined in Materials and Methods) is shown. Error bars represent standard error. (C and F) Degradation of EspP(G1123R) upon the addition of the Polyphor 7 peptide (C) or MRL-494 (F) after synthesis. The mean percentage of the total radiolabeled EspP or proEspP that was observed at the 30-min time point in three independent experiments is shown. Error bars represent standard error.
The Polyphor 7 peptide and MRL-494 block OMP assembly within 1 min of addition. The experiment described in Fig. 5 was repeated except either the Polyphor 7 peptide or MRL-494 was added at 1 min before the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide (top) or OmpC (bottom). Download FIG S4, JPG file, 0.8 MB (797.7KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 inhibit EspP(G1123R) assembly at low concentrations when added after synthesis. The experiment described in Fig. 5 was repeated except that either the Polyphor 7 peptide was added at a concentration of 0.125 μg/mL (0.5× MIC) or 0.05 μg/mL (0.2× MIC), or MRL-494 was added at a concentration of 8.25 μg/mL (0.5× MIC) or 3.3 μg/mL (0.2× MIC) 2 min after the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide. Download FIG S5, JPG file, 1.2 MB (1.2MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
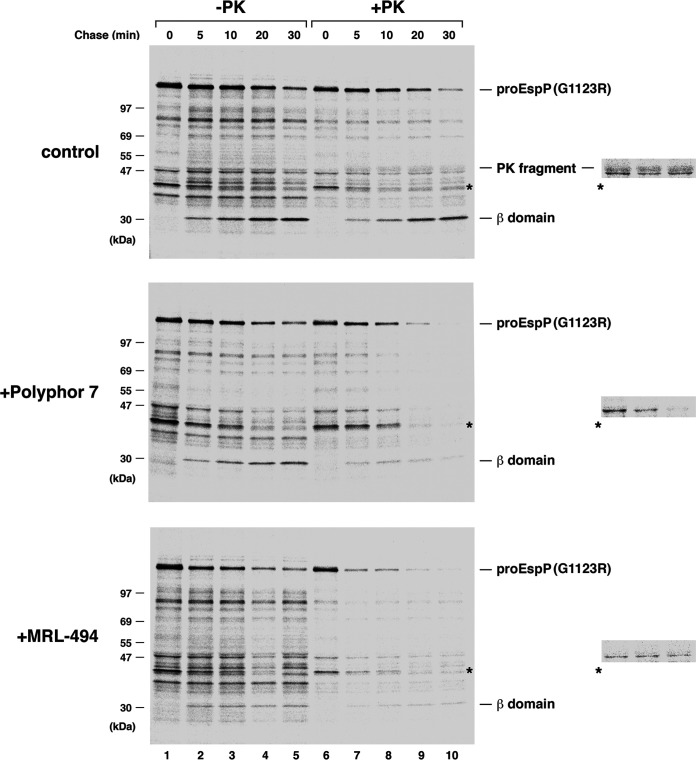
Further insights into the effect of the Polyphor 7 peptide and MRL-494 on EspP(G1123R) assembly emerged from experiments in which we added the compounds to cultures 2 min after the start of the chase and then treated cells with PK. Consistent with previous results, a small amount of an ~47 kDa C-terminal PK fragment that is derived from the pro form of the protein was detected throughout the time course in control cells (Fig. 6, top gel, lanes 6 to 10; the PK fragment migrates slightly more slowly than a background band denoted by an asterisk). The generation of this product is due to the exposure of the passenger domain on the cell surface (which indicates that translocation has been initiated) prior to its proteolytic release from the β barrel domain. In the presence of the Polyphor 7 peptide, however, none of the PK fragment could be detected at any time point even when the gel was overexposed (Fig. 6, middle gel, lanes 6 to 10). Furthermore, the observation that a significant fraction of the pro form of the protein was cleaved by the protease indicates that integrity of the OM was compromised (Fig. S6A). In the presence of MRL-494, the PK fragment was likewise undetectable, but the proteolysis was more pronounced; even some of the background bands that were resistant to PK treatment were degraded (Fig. 6, bottom gel). Interestingly, a loss of OM integrity was observed even if BamA activity was inhibited by pretreating cells with darobactin prior to the start of the chase (Fig. S7). This finding strongly suggests that the permeabilzation of the OM is not a secondary effect of the inactivation of BamA by the Polyphor peptide 7 or MRL-494. Very similar results were obtained when cells were treated with the antimicrobials at a concentration of 0.5× MIC (Fig. S6b and S8). Taken together, the results show that at growth-inhibitory concentrations, both compounds disrupt the integrity of the OM (and, in the case of MRL-494, possibly the inner membrane as well) and suggest that they inhibit the surface exposure of the EspP(G1123R) passenger domain by affecting the membrane insertion of the β barrel domain.
FIG 6.
The Polyphor 7 peptide and MRL-494 permeabilize the OM. The experiment described in Fig. 2 was repeated through the pulse-chase labeling step. Culture aliquots were untreated (control) or treated with either the Polyphor 7 peptide (0.5 μg/mL) or MRL-494 (16.5 μg/mL) 2 min after the start of the chase. PK was then added to half of each sample. Immunoprecipitations were performed using an anti-EspP C-terminal antiserum, proteins were resolved by SDS-PAGE, and radioactive proteins were imaged. A small slice of lanes 7 to 9 from each gel that was overexposed is shown on the right. A significant amount of the ~47 kDa PK fragment was only observed in the control samples. A prominent cross-reactive protein is denoted with an asterisk.
Quantitative analysis of the permeability of the OM in the presence of the Polyphor 7 peptide and MRL-494. The average percentage of proEspP(G1123R) that remained after cells were treated with PK at the 5-, 10-, 20-, and 30-min time points in the experiments illustrated in Fig. 6 (A) and Fig. S8 (B) is shown. To account for the proEspP(G1123R) that was displayed on the cell surface and therefore susceptible to PK cleavage in control cells, the percentage of the protein that was converted to the 47-kDa C-terminal fragment was added to the unprocessed proEspP(G1123R) that remained inside the cells. The percentage of proEspP(G1123R) remaining was calculated using the formula [proEspP(G1123R) + PK/proEspP(G1123R) – PK] × 100. The fraction of proEspP(G1123R) that was converted to the 47 kDa C-terminal fragment was calculated using the formula [47 kDa C-terminal fragment/proEspP(G1123R) – PK] × 100 after normalizing the 47-kDa C-terminal fragment signal based on the difference in the number of methionine residues present in the fragment and the unprocessed precursor. Error bars show the standard error. Download FIG S6, JPG file, 0.7 MB (747KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 permeabilize the OM even after BamA is inactivated by darobactin. The experiment described in Fig. 6 was repeated except that aliquots were treated with the Polyphor 7 peptide (2 min after the start of the chase) or both darobactin (5 min before the start of the chase) and the Polyphor 7 peptide (2 min after the start of the chase) (A), or treated with MRL-494 (2 min after the start of the chase) or both darobactin (5 min before the start of the chase) and MRL-494 (2 min after the start of the chase) (B). Download FIG S7, JPG file, 2.4 MB (2.5MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 permeabilize the OM at low concentrations. The experiment described in Fig. 6 was repeated except that the antimicrobials were added at 0.5× MIC (i.e., 0.125 μg/mL Polyphor 7 peptide or 8.25 μg/mL MRL-494). Download FIG S8, JPG file, 2.2 MB (2.3MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine if the Polyphor 7 peptide might affect late stages of OMP assembly, we also examined the effect of the compound on the biogenesis of MBP-76EspP. We found that, like darobactin, the Polyphor 7 peptide (but not Polyphor 9, a control peptide that lacks antimicrobial activity) significantly inhibited the formation of a disulfide bond between MBP-76EspPS1299C and BamAS425C when added prior to the expression of the fusion protein (Fig. 4A, right blots and left graph). In light of NMR data that indicate that the Polyphor 7 peptide binds to surface-exposed loops of BamA that are a significant distance from BamA β1 (28), this observation suggests that the compound may act as an allosteric inhibitor of β signal binding. Surprisingly, the addition of the Polyphor 7 peptide after MBP-76EspP was synthesized did not affect the formation of a disulfide bond between either MBP-76EspPS1299C and BamAS425C or MBP-76EspPA1043C and BamAG781C, a cysteine pair located in EspP β1 and BamA β15, respectively, at the unstable interface of the EspP-BamA hybrid barrel (22) (Fig. 4B, right blots and left graph; Fig. S9A). Furthermore, in experiments in which the assembly of MBP-76EspP was restarted after the release of the MBP moiety by PK treatment, the efficiency and kinetics of assembly completion were unaffected by the addition of the Polyphor 7 peptide (Fig. S9B). Given that the assembly of MBP-76EspP is arrested at a relatively late stage (later than the stage at which the assembly of EspP(G1123R) is delayed), these results provide clear evidence that the antimicrobial peptide does not affect the final steps of EspP biogenesis.
The Polyphor 7 peptide does not inhibit the completion of MBP-76EspP assembly after it forms a hybrid barrel with BamA. (A) Experiment conducted as in Fig. 4B, except E. coli BL21(DE3) expressed HisBamAG781CBCDE and MBP-76EspPA1043C prior to incubation with Polyphor peptide 7 (1.25 μg/mL). (B) After the expression of HisBamABCDE and MBP-76EspP was induced to generate a pool of MBP-76EspP-BamA hybrid-barrel intermediates in E. coli BL21(DE3), cells were treated with Polyphor compound 7 (0.25 μg/mL) or untreated (control) for 10 min. PK was then added to remove the MBP moiety and to monitor the completion of β-barrel assembly. Aliquots at each time point were probed by immunoblotting with α-EspPβ(N) and α-StrepII. Polypeptides that result from the degradation of the PK fragment in the periplasm are indicated. Nonspecific cross-reactive proteins are denoted with an asterisk. Download FIG S9, JPG file, 1.0 MB (1MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
In this report, we examined the effect of three recently described antimicrobial compounds on the biogenesis of EspP(G1123R), an OMP mutant that assembles extremely slowly, and MBP-76EspP, an OMP fusion protein whose assembly is arrested at a well-defined late stage as a hybrid barrel with BamA. Because the assembly of these mutants is delayed or arrested at distinct stages, we were able to obtain insights not only into the mechanism of action of the antimicrobial compounds, but also into the mechanism of assembly itself. We found that one compound, a natural product called darobactin, blocks the interaction of EspP(G1123R) with BAM and prevents its assembly if added prior to the synthesis of the protein but does not affect assembly if added after its synthesis. Consistent with these results, we also found that darobactin blocks the interaction of the MBP-76EspP β signal with BamA β1, but only if it is added prior to synthesis. In contrast, two other compounds, a chimeric peptidomimetic (the Polyphor 7 peptide) and a small molecule (MRL-494), inhibited both the interaction of EspP(G1123R) with BAM and the completion of its assembly if added at a postbinding stage. The MRL-494 compound was more potent than the Polyphor 7 compound in the sense that it completely blocked further assembly when it was added at a concentration of 1× MIC. In any case, the results clearly demonstrate that the compounds can inhibit EspP assembly at different stages. Interestingly, the observation that the Polyphor 7 peptide did not affect the assembly of MBP-76EspP when it was added at a postarrest stage suggested that the late stages of assembly that follow the formation of the EspP-BamA hybrid barrel are relatively insensitive to the effects of the compound (see below). Because all of the compounds also caused the rapid degradation of the EspP mutants and another OMP, OmpC, it is likely that they broadly inhibit OMP assembly. Perhaps even more significantly, the observation that the compounds inhibit OMP assembly within 1 min of treatment and are effective even below 1× MIC confirms that their antimicrobial activity is a direct consequence of their effects on OMP biogenesis and strongly disfavors the possibility that they trap substrates on BAM in a way that indirectly blocks the binding of incoming molecules. The rapidity of the inhibitory effects implies that toxicity must reach a specific threshold before cell death occurs (typically ~1 h after exposure in the case of darobactin [27]). Indeed, the delay in cell death is consistent with the finding that strong reductions in the levels of essential BAM subunits only modestly affect cell growth (40, 41).
The observation that darobactin interacts primarily with the β1 strand of purified BamA (29), together with our evidence that the compound blocks the interaction of EspP(G1123R) with BAM and the disulfide cross-linking of MBP-76EspPS1299C to BamAS425C in vivo, strongly supports the proposal that it functions as a competitive inhibitor of the β signal–BamA interaction. The fact that we could not detect any assembly defects when we added darobactin at a postbinding stage also highlights the remarkable specificity of the compound. As a corollary, the evidence that darobactin blocks the binding of the β signal to BamA β1 strongly suggests that this interaction initiates (or occurs at a very early stage of) the membrane integration of OMP β barrels. If the interaction occurred at a relatively late stage of assembly, then it seems unlikely that the compound would block the cross-linking of EspP(G1123R/Y1214am) to BamD when added prior to binding or have no effect on EspP(G1123R) assembly when added at a postbinding stage. Furthermore, the observation that the addition of darobactin at a postbinding stage did not affect the efficiency of disulfide cross-linking between MBP-76EspPS1299C and BamAS425C implies that the compound cannot displace the EspP β signal once it is bound to BamA and confirms previous results that imply that BamA β1 (as well as its mitochondrial equivalent) interacts tightly with β signals (16, 22, 42).
There are a variety of possible mechanisms by which the Polyphor 7 peptide and MRL-494 might inhibit both the interaction of OMPs with BAM and at least one postbinding step. In one possible scenario, the binding of the compounds to BamA affects conformational dynamics (or forces a conformational change) in a way that both blocks the binding of incoming substrates and alters the interaction of bound (but not yet fully integrated) substrates to prevent their movement into the membrane. Based on evidence that the Polyphor 7 peptide binds to external loops of BamA (28), it is much more likely that it functions as an allosteric inhibitor rather than as a competitive inhibitor of β signal binding. Indeed, recent structural investigations of an antibody fragment that strongly inhibits OMP assembly by binding to the surface loops of BamA and that may trap BAM in a conformation that prevents interactions with incoming OMPs provide a strong precedent for such an allosteric mechanism (43, 44). In another scenario, the Polyphor 7 peptidomimetic might exert its effects by binding to both BamA and a second molecule. Because the Polyphor peptide 7 consists of a polymyxin B1 moiety conjugated to a cyclic peptide, it very likely binds to LPS (28). LPS might serve as a critical scaffold that mediates the recruitment of the compound to BamA. Alternatively, the binding of the compound to LPS (or to another cell surface molecule) might indirectly impair BamA function or even interfere with OMP assembly by a BAM-independent mechanism in which BamA acts primarily as a receptor for the chimeric peptide. In this regard, it should be noted that recent studies suggest that LPS molecules are specifically bound to BamA in a stabilized form and that LPS might play a role in OMP assembly by maintaining the rigidity of the OM (6, 16, 43). In addition, our data raise the possibility that the Polyphor 7 peptide inhibits the binding and/or integration of OMPs by disrupting the integrity of the OM. Although MRL-494 exhibited similar properties to the peptidomimetic, it blocked postbinding steps more completely. MRL-494 likewise appears to perturb membrane integrity, but the isolation of a bamA mutant (E470K) that confers resistance to MRL-494 (26) strongly suggests that the compound does not impair OMP assembly solely by perturbing the OM. Taken together, our results favor a model in which the Polyphor 7 peptide and MRL-494 exert their effects at least partly through other OM molecules and/or the OM itself, perhaps in a synergistic manner. Regardless of which scenario is correct, both compounds promote the degradation of EspP(G1123R) molecules that are already bound to BAM, presumably by increasing their accessibility to periplasmic proteases or promoting their dissociation into the periplasm.
The finding that the addition of the Polyphor 7 peptide did not significantly affect the completion of MBP-76EspP assembly is particularly striking. Given that the assembly of MBP-76EspP is arrested after it forms a hybrid barrel with BamA (16, 22), this observation implies that the peptidomimetic does not inhibit the final stages of OMP assembly (including the closure of the β barrel) and thereby corroborates the conclusion that the compound inhibits a relatively early postbinding step. The results are surprising, however, given that the Polyphor 7 peptide impairs the integrity of the OM, and appear to be inconsistent with recent evidence that the rigidity of the OM itself plays a key role in the completion of MBP-76EspP assembly (16). The simplest interpretation of the results is that while the Polyphor 7 peptide perturbs the OM permeability barrier, it does not completely eliminate the overall tension and load-bearing features of the OM (6). Although the properties of the OM remain poorly understood, it seems likely that different biochemical, genetic, and biophysical manipulations alter the membrane in ways that differentially affect BAM function and OMP assembly, and that bacteria use a variety of mechanisms to adapt to changes in the structure and composition of the membrane. Indeed, a detailed analysis of the effects of these manipulations on the membrane in the future has the potential to yield profound insights into the OMP assembly process.
On a more general level, our results show how experiments designed to elucidate the mechanism of action of BAM inhibitors can provide insights into the OMP process itself and help to distinguish between different models of BAM function. Furthermore, our finding that compounds that inhibit multiple stages of OMP assembly by distinct mechanisms can be isolated also opens the possibility for the development of inhibitors that function in combination to reduce the emergence of drug-resistant strains of pathogenic bacteria.
MATERIALS AND METHODS
Strains, reagents, and antisera.
All experiments were conducted in the E. coli K-12 strain AD202 (MC4100 ompT::kan) (45) or the E. coli B strain BL21(DE3) (Thermo Fisher). Darobactin, Polyphor 7 and 9 peptides, and MRL-494 were obtained from Kim Lewis (Northeastern University), Polyphor, and Merck, respectively. All antimicrobials were solubilized in water and stored as stock solutions at −20°C. pBpa was purchased from Bachem. Rabbit polyclonal antisera generated against an EspP C-terminal peptide, a peptide derived from the N terminus of the EspP β barrel, a BamA C-terminal peptide, and BamD have been described previously (22, 34, 46). Mouse monoclonal anti-His and anti-Strep II antisera were obtained from Genscript and Qiagen, respectively.
Plasmids.
Plasmids pJH224 [Ptrc-EspP(G1123R)], pJH225 [Plac-HisEspP(G1123R/Y1214am)], pMTD372 [Ptrc-HisBamABCDE], pMTD607 [Prha-MBP-76EspP], pMTD710 [Ptrc-HisBamAS425CBCDE], pMTD712 [Prha-MBP-76EspPS1299C], pMTD893 [Ptrc-HisBamAG781CBCDE], pMTD951 [Prha-MBP-76EspPA1043C], and pDULE have been described previously (22, 32, 39). To construct a plasmid to express EspPΔ5(G1123R/S1299C) under the control of a rhamnose-inducible promoter, the S1299C substitution was introduced into the pJH207 derivative that harbors a G1123R mutation (32) using the QuikChange II site-directed mutagenesis kit (Agilent) and primers mtd190 and mtd191 (22).
Pulse-chase radiolabeling.
AD202 transformed with pJH224 was grown overnight at 37°C in M9 medium containing 40 μg/mL l-amino acids (except methionine and cysteine) and ampicillin (100 μg/mL). Cells were pelleted at room temperature (3,500 × g), washed, and inoculated into 50 mL fresh medium at OD550 of 0.02. IPTG (isopropyl-β-d-thiogalactopyranoside) was added (10 μM) when cultures reached OD550 of 0.2, to induce espP expression, and cells were subjected to pulse-chase labeling 30 min later as previously described (36). In all experiments, aliquots were removed and treated with an antimicrobial at a specific time either before or after the initiation of the radiolabeling (or left untreated to provide a control). In general, cells were removed at each time point and incubated with cold trichloroacetic acid (TCA; 10% final concentration) on ice. Precipitated proteins were then pelleted, and immunoprecipitations were performed as previously described (47). Proteins were resolved by SDS-PAGE on 8 to 16% minigels (Thermo Fisher), and radioactive proteins were detected using a Fujifilm FLA-9000 Phosphorimager. In some experiments, cells harvested at each time point were pipetted over ice, pelleted (3,000 × g, 4°C), and resuspended in cold M9 salts. Samples were then divided in half, and one half was treated with 200 μg/mL PK for 20 min on ice followed by 2 mM phenylmethylsulfonyl fluoride (PMSF) (10 min on ice). All samples were then incubated with TCA and processed as described above.
Quantitative analysis of pulse-chase experiments.
For quantitative analysis, the signal in each band was normalized based on the relative number of methionine residues. Percent passenger domain cleavage was calculated using the formula % cleavage = (β domain/β domain + proEspP). In experiments that involved the addition of the Polyphor 7 peptide or MRL-494, however, this formula could not be used due to the continuous degradation of proEspP during the time course. Instead, the accumulation of the cleaved β domain was determined by arbitrarily assigning a value of 10 to the signal observed at the 0’ time point. (This value was chosen because approximately 10% of the passenger domain is typically cleaved at the 0’ time point in control samples). Percent radiolabeled EspP remaining (Fig. S1 and S7A) was defined as (proEspP + β domain) at 30’/(proEspP + β domain)max, where max is the maximum signal observed during the time course. In the control samples and the samples harvested from cultures that were treated with darobactin at t = 2’, the maximum signal was always observed at the t = 2’ time point because the signal inevitably increases beyond the pulse-labeling period, when a large protein is analyzed in E. coli (48). In other samples, the maximum signal was observed at the t = 0’ time point due to continuous degradation of proEspP. Percent proEspP remaining (Fig. S7B) was defined as proEspP at 30’/proEspP at 0’.
UV cross-linking.
AD202 transformed with pJH225 and pDULE was grown in M9 medium as described above except that the medium was also supplemented with 5 μg/mL tetracycline. At OD550 of 0.2, 100 μM IPTG and 1 mM pBpa were added, and pulse-chase labeling was conducted 30 min later. Antimicrobials were added to aliquots 5 min prior to the start of the chase. Samples were removed at each time point and subjected to UV irradiation essentially as described (36). All samples were ultimately TCA precipitated and processed as described above.
Disulfide bond formation assays.
BL21(DE3) transformed with plasmids containing single-cysteine substitutions in the BamA and EspP β barrels was grown overnight at 25°C with shaking at 250 rpm in 10 mL LB (Miller) medium containing ampicillin (100 μg/mL) and trimethoprim (50 μg/mL). Cultures were pelleted at (3,500 × g, 5 min, 4°C), washed with 10 mL LB, inoculated into 4 mL fresh medium at OD600 of 0.05, and grown for 4 h at 25°C (to OD600 ~0.4 to 0.6). For experiments in which the inhibitor was added prior to EspP derivative expression, IPTG (0.4 mM) was added to induce expression of BAM for 50 min, and either darobactin (4 μg/mL) or Polyphor peptide 7 (1.25 μg/mL) was then added. After 10 min, l-rhamnose (0.2%) was added to induce expression of the EspP derivative for 45 min. Culture samples were aliquoted (1 mL) into 1.5-mL tubes on ice, pelleted (10,000 × g, 2 min, 4°C), and resuspended in ice-cold phosphate-buffered saline (PBS) (1 mL). Samples were then treated with either 0.2 mM 4-DPS or an equivalent volume of ethanol (control) on ice for 30 min. For experiments in which the inhibitor was added after the synthesis of an EspP derivative, BAM expression was induced for 1 h and the EspP derivative was then expressed for 40 min. Cultures were pelleted (3,500 × g, 5 min, 4°C), washed with 10 mL LB (25°C), pelleted again, resuspended in 4 mL LB (25°C) containing ampicillin (100 μg/mL) and trimethoprim (50 μg/mL), cultured for 5 min at 25°C, and then further cultured in the presence of an inhibitor (at the concentration described above). Culture samples (1 mL) were aliquoted into 1.5-mL tubes on ice, pelleted (10,000 × g, 2 min, 4°C), and resuspended in 1 mL of PBS containing the inhibitor. Samples were then treated with either 0.2 mM 4-DPS or an equivalent volume of ethanol (control) on ice for 30 min. In all experiments, samples were subjected to immunoblotting and bands were quantitated as previously described (22).
MBP-76EspP assembly restart assays.
To monitor the progression of assembly after the formation of a BamA-EspP hybrid barrel, BL21(DE3) transformed with pMTD372 and pMTD607 were grown as in the disulfide bond formation assays. At mid-log phase, cultures were treated with IPTG (0.4 mM) for 1 h and then l-rhamnose (0.2%) for 45 min. Bacteria were pelleted (10,000 × g, 2 min, 4°C), resuspended in PBS (control) or PBS containing Polyphor compound 7 (0.25 μg/mL), and incubated for 10 min at 25°C, 350 rpm, in a Thermomixer (Eppendorf). Bacteria were then subjected to PK digestion for 0, 2, 10, and 30 min, and samples were processed as previously described (22).
ACKNOWLEDGMENTS
We thank Thushani Nilaweera for providing helpful comments on the manuscript and Kim Lewis, Polyphor Ltd., and Merck for kindly providing darobactin, Polyphor peptides 7 and 9, and MRL-494, respectively. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. M.T.D. was supported by an NIH Visiting Fellowship Award.
M.T.D. and H.D.B. designed the study, all authors performed experiments and analyzed results, and M.T.D. and H.D.B. wrote the paper.
The authors have no competing interests to declare.
Contributor Information
Harris D. Bernstein, Email: harris_bernstein@nih.gov.
M. Stephen Trent, University of Georgia
REFERENCES
- 1.Schulz GE. 2000. β-barrel membrane proteins. Curr Opin Struct Biol 10:443–447. doi: 10.1016/S0959-440X(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 2.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horne JE, Brockwell DJ, Radford SE. 2020. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J Biol Chem 295:10340–10367. doi: 10.1074/jbc.REV120.011473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng G, Fronzes R, Chandran V, Remaut H, Waksman G. 2009. Protein oligomerization in the bacterial outer membrane (review). Mol Membr Biol 26:136–145. doi: 10.1080/09687680802712422. [DOI] [PubMed] [Google Scholar]
- 6.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Webb CT, Heinz E, Lithgow T. 2012. Evolution of the β-barrel assembly machinery. Trends Microbiol 20:612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Bakelar J, Buchanan SK, Noinaj N. 2016. The structure of the β-barrel assembly machinery complex. Science 351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. 2016. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 13.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. 2016. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 14.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. 2016. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerner PA, Sousa MC. 2017. Extreme dynamics in the BamA β-barrel seam. Biochemistry 56:3142–3149. doi: 10.1021/acs.biochem.7b00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle MT, Jimah JR, Dowdy T, Ohlemacher SI, Larion M, Hinshaw JE, Bernstein HD. 2022. Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Cell 185:1143–1156.e13. doi: 10.1016/j.cell.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Gumbart JC. 2020. Membrane thinning and lateral gating are consistent features of BamA across multiple species. PLoS Comput Biol 16:e1008355. doi: 10.1371/journal.pcbi.1008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasek D, Rawson S, Lee J, Wzorek JS, Harrison SC, Li Z, Kahne D. 2020. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583:473–478. doi: 10.1038/s41586-020-2370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noinaj N, Gumbart JC, Buchanan SK. 2017. The β-barrel assembly machinery in motion. Nat Rev Microbiol 15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle MT, Bernstein HD. 2019. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat Commun 10:3358. doi: 10.1038/s41467-019-11230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 24.Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, Robinson JA. 2016. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J Biol Chem 291:1921–1932. doi: 10.1074/jbc.M115.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steenhuis M, Corona F, Ten Hagen-Jongman CM, Vollmer W, Lambin D, Selhorst P, Klaassen H, Versele M, Chaltin P, Luirink J. 2021. Combining cell envelope stress reporter assays in a screening approach to identify BAM complex inhibitors. ACS Infect Dis 7:2250–2263. doi: 10.1021/acsinfecdis.0c00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart EM, Mitchell AM, Konovalova A, Grabowicz M, Sheng J, Han X, Rodriguez-Rivera FP, Schwaid AG, Malinverni JC, Balibar CJ, Bodea S, Si Q, Wang H, Homsher MF, Painter RE, Ogawa AK, Sutterlin H, Roemer T, Black TA, Rothman DM, Walker SS, Silhavy TJ. 2019. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc Natl Acad Sci USA 116:21748–21757. doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, Honrao C, Ma X, Guo JJ, Makriyannis A, Linares-Otoya L, Böhringer N, Wuisan ZG, Kaur H, Wu R, Mateus A, Typas A, Savitski MM, Espinoza JL, O'Rourke A, Nelson KE, Hiller S, Noinaj N, Schäberle TF, D'Onofrio A, Lewis K. 2019. A new antibiotic selectively kills Gram-negative pathogens. Nature 576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luther A, Urfer M, Zahn M, Müller M, Wang S-Y, Mondal M, Vitale A, Hartmann J-B, Sharpe T, Monte FL, Kocherla H, Cline E, Pessi G, Rath P, Modaresi SM, Chiquet P, Stiegeler S, Verbree C, Remus T, Schmitt M, Kolopp C, Westwood M-A, Desjonquères N, Brabet E, Hell S, LePoupon K, Vermeulen A, Jaisson R, Rithié V, Upert G, Lederer A, Zbinden P, Wach A, Moehle K, Zerbe K, Locher HH, Bernardini F, Dale GE, Eberl L, Wollscheid B, Hiller S, Robinson JA, Obrecht D. 2019. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576:452–458. doi: 10.1038/s41586-019-1665-6. [DOI] [PubMed] [Google Scholar]
- 29.Kaur H, Jakob RP, Marzinek JK, Green R, Imai Y, Bolla JR, Agustoni E, Robinson CV, Bond PJ, Lewis K, Maier T, Hiller S. 2021. The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature 593:125–129. doi: 10.1038/s41586-021-03455-w. [DOI] [PubMed] [Google Scholar]
- 30.Ritzmann N, Manioglu S, Hiller S, Muller DJ. 2021. Monitoring the antibiotic darobactin modulating the β-barrel assembly factor BamA. Structure 30:350–359.e3. doi: 10.1016/j.str.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein HD. 2019. Type V Secretion in Gram-negative bacteria. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson JH, Plummer AM, Fleming KG, Bernstein HD. 2017. Selective pressure for rapid membrane integration constrains the sequence of bacterial outer membrane proteins. Mol Microbiol 106:777–792. doi: 10.1111/mmi.13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle MT, Bernstein HD. 2021. BamA forms a translocation channel for polypeptide export across the bacterial outer membrane. Mol Cell 81:2000–2012.e3. doi: 10.1016/j.molcel.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova O, Peterson JH, Ieva R, Bernstein HD. 2013. Mechanistic link between β barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci USA 110:E938–E947. doi: 10.1073/pnas.1219076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dautin N, Barnard TJ, Anderson DE, Bernstein HD. 2007. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J 26:1942–1952. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ieva R, Bernstein HD. 2009. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA 106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ieva R, Tian P, Peterson JH, Bernstein HD. 2011. Sequential and spatially restricted interactions of assembly factors with an autotransporter β domain. Proc Natl Acad Sci USA 108:E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JH, Hussain S, Bernstein HD. 2018. Identification of a novel post-insertion step in the assembly of a bacterial outer membrane protein. Mol Microbiol 110:143–159. doi: 10.1111/mmi.14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. 2005. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods 2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 40.Mahoney TF, Ricci DP, Silhavy TJ. 2016. Classifying β-barrel assembly substrates by manipulating essential Bam complex members. J Bacteriol 198:1984–1992. doi: 10.1128/JB.00263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. 2012. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci USA 109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Höhr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N, Wiedemann N. 2018. Membrane protein insertion through a mitochondrial β-barrel gate. Science 359:eaah6834. doi: 10.1126/science.aah6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storek KM, Auerbach MR, Shi H, Garcia NK, Sun D, Nickerson NN, Vij R, Lin Z, Chiang N, Schneider K, Wecksler AT, Skippington E, Nakamura G, Seshasayee D, Koerber JT, Payandeh J, Smith PA, Rutherford ST. 2018. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci USA 115:3692–3697. doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White P, Haysom SF, Iadanza MG, Higgins AJ, Machin JM, Whitehouse JM, Horne JE, Schiffrin B, Carpenter-Platt C, Calabrese AN, Storek KM, Rutherford ST, Brockwell DJ, Ranson NA, Radford SE. 2021. The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding. Nat Commun 12:4174. doi: 10.1038/s41467-021-24432-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, Jacq A, Brickman E, Beckwith J, Taura T, Ueguchi C, Akiyama Y, Ito K. 1990. Characterization of cold-sensitive secY mutants of Escherichia coli. J Bacteriol 172:7005–7010. doi: 10.1128/jb.172.12.7005-7010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. 2005. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA 102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ieva R, Skillman KM, Bernstein HD. 2008. Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol 67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 48.Ulbrandt ND, Newitt JA, Bernstein HD. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187–196. doi: 10.1016/S0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 49.Barnard TJ, Gumbart J, Peterson JH, Noinaj N, Easley NC, Dautin N, Kuszak AJ, Tajkhorshid E, Bernstein HD, Buchanan SK. 2012. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J Mol Biol 415:128–142. doi: 10.1016/j.jmb.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The EspP(G1123R) passenger domain is exposed on the cell surface if darobactin is added after the start of the chase. The experiment described in Fig. 2 was repeated through the pulse-chase labeling step. Culture aliquots were untreated (control) or treated with darobactin (4 μg/mL) 2 min after the start of the chase. PK was then added to half of each sample. Immunoprecipitations were performed using the anti-EspP C-terminal antiserum, proteins were resolved by SDS-PAGE, and radioactive proteins were imaged. Download FIG S1, JPG file, 1.7 MB (1.8MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Darobactin blocks OmpC assembly only if added prior to synthesis. Immunoprecipitations were performed with an antiserum generated against OmpC using the samples generated in the experiments described in Fig. 2. A representative result is shown. Download FIG S2, JPG file, 0.4 MB (431.4KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Darobactin blocks OMP assembly within 1 min of addition. The experiment described in Fig. 2A was repeated except that darobactin (4 μg/mL) was added at 1 min (-1’) or 2 min (-2’) before the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide (top) or OmpC (bottom). Download FIG S3, JPG file, 0.4 MB (399.1KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 block OMP assembly within 1 min of addition. The experiment described in Fig. 5 was repeated except either the Polyphor 7 peptide or MRL-494 was added at 1 min before the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide (top) or OmpC (bottom). Download FIG S4, JPG file, 0.8 MB (797.7KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 inhibit EspP(G1123R) assembly at low concentrations when added after synthesis. The experiment described in Fig. 5 was repeated except that either the Polyphor 7 peptide was added at a concentration of 0.125 μg/mL (0.5× MIC) or 0.05 μg/mL (0.2× MIC), or MRL-494 was added at a concentration of 8.25 μg/mL (0.5× MIC) or 3.3 μg/mL (0.2× MIC) 2 min after the start of the chase. Immunoprecipitations were performed using antisera generated against an EspP C-terminal peptide. Download FIG S5, JPG file, 1.2 MB (1.2MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Quantitative analysis of the permeability of the OM in the presence of the Polyphor 7 peptide and MRL-494. The average percentage of proEspP(G1123R) that remained after cells were treated with PK at the 5-, 10-, 20-, and 30-min time points in the experiments illustrated in Fig. 6 (A) and Fig. S8 (B) is shown. To account for the proEspP(G1123R) that was displayed on the cell surface and therefore susceptible to PK cleavage in control cells, the percentage of the protein that was converted to the 47-kDa C-terminal fragment was added to the unprocessed proEspP(G1123R) that remained inside the cells. The percentage of proEspP(G1123R) remaining was calculated using the formula [proEspP(G1123R) + PK/proEspP(G1123R) – PK] × 100. The fraction of proEspP(G1123R) that was converted to the 47 kDa C-terminal fragment was calculated using the formula [47 kDa C-terminal fragment/proEspP(G1123R) – PK] × 100 after normalizing the 47-kDa C-terminal fragment signal based on the difference in the number of methionine residues present in the fragment and the unprocessed precursor. Error bars show the standard error. Download FIG S6, JPG file, 0.7 MB (747KB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 permeabilize the OM even after BamA is inactivated by darobactin. The experiment described in Fig. 6 was repeated except that aliquots were treated with the Polyphor 7 peptide (2 min after the start of the chase) or both darobactin (5 min before the start of the chase) and the Polyphor 7 peptide (2 min after the start of the chase) (A), or treated with MRL-494 (2 min after the start of the chase) or both darobactin (5 min before the start of the chase) and MRL-494 (2 min after the start of the chase) (B). Download FIG S7, JPG file, 2.4 MB (2.5MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide and MRL-494 permeabilize the OM at low concentrations. The experiment described in Fig. 6 was repeated except that the antimicrobials were added at 0.5× MIC (i.e., 0.125 μg/mL Polyphor 7 peptide or 8.25 μg/mL MRL-494). Download FIG S8, JPG file, 2.2 MB (2.3MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The Polyphor 7 peptide does not inhibit the completion of MBP-76EspP assembly after it forms a hybrid barrel with BamA. (A) Experiment conducted as in Fig. 4B, except E. coli BL21(DE3) expressed HisBamAG781CBCDE and MBP-76EspPA1043C prior to incubation with Polyphor peptide 7 (1.25 μg/mL). (B) After the expression of HisBamABCDE and MBP-76EspP was induced to generate a pool of MBP-76EspP-BamA hybrid-barrel intermediates in E. coli BL21(DE3), cells were treated with Polyphor compound 7 (0.25 μg/mL) or untreated (control) for 10 min. PK was then added to remove the MBP moiety and to monitor the completion of β-barrel assembly. Aliquots at each time point were probed by immunoblotting with α-EspPβ(N) and α-StrepII. Polypeptides that result from the degradation of the PK fragment in the periplasm are indicated. Nonspecific cross-reactive proteins are denoted with an asterisk. Download FIG S9, JPG file, 1.0 MB (1MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.