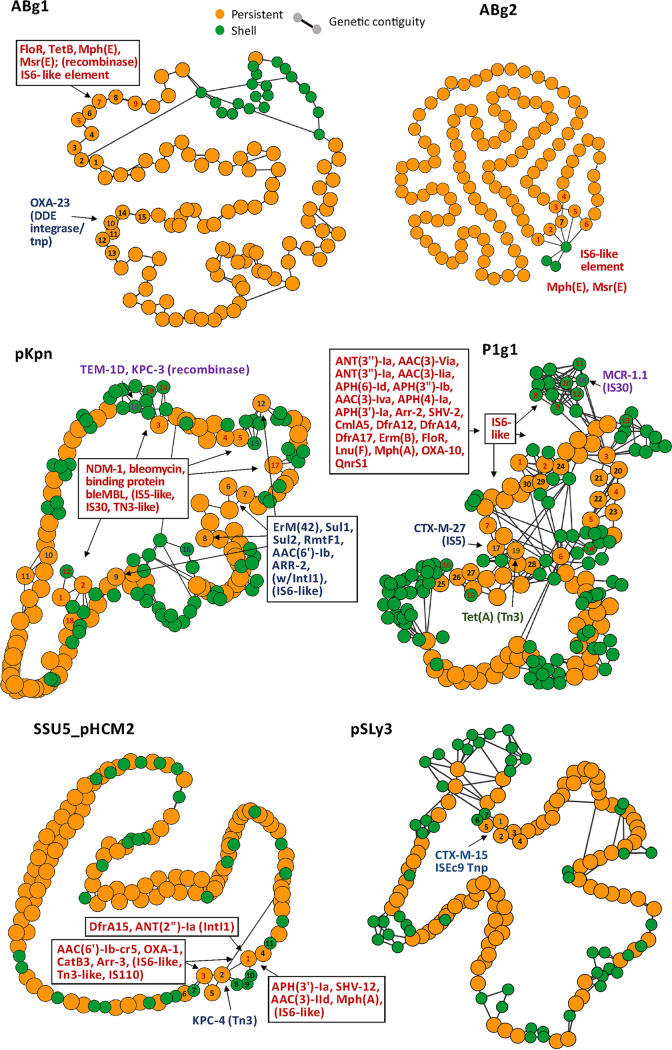
FIG 2.
Genetic environments of ARGs in P-Ps’ pangenome graphs. Nodes are gene families. Genes (including ARGs) were grouped into a gene family (default parameters of PPanGGOLiN [31]) if they had a similarity of >80% identity covering at least 80% of the sequence. Orange: persistent, green: shell. Edges are shown for adjacent genes within the gene families (genetic contiguity). Gene families with colored numbers (red, blue, violet, green) are direct neighbors of ARGs containing genetic elements (transposon, IS, integron, recombinase [separated by colors]). Black numbers are given for proximal gene families. ABg1: 1-3,6,8-9:hypothetical, 4:nucleoside, 5:ATPase AAA, 7:3′–5′ exonuclease pyrophospho-hydrolase. ABg2: 1,3:hypothetical, 2,7:ribonucleoside-diphosphate red. 4:toprim domain protein, 5:ATPase AAA, 7:3′–5′ exonuclease pyrophosphohydrolase. SSU5_pHCM2: 1:PhoH, 2-5, 7-11:hypothetical, 6:DNA ligase. P1: 1:SSB, 2-5,15,17,19-20,23-24,28: hypothetical, 6:cell division inhibitor (Icd-like), 7-11:tail fiber, 12:recombinase, 13-14:tail fiber assembly, 16:ResMod subM, 18:Ref family, 21:bleomycin hydrolase, 22:transglycosylase, 25:DNA repair, 26:Phd/YefM (T-A), 27:doc (T-A), 29:lysozyme, 30:head processing. pKpn: 1:transcriptional regulator, 2,7,9,12-15,17-19:hypothetical, 3:phoH, 4:porphyrin biosynthesis protein, 5-6: AAA family ATPase, 8: ribonucl.-diphosphate reductase subunit 10–11:tail fiber domain-containing protein, 16:HsdR. pSLy3: 1:DUF3927 family, 2:tellurite/colicin resistance, 3-7:hypothetical.