ABSTRACT
Coastal marine macrophytes exhibit some of the highest rates of primary productivity in the world. They have been found to host a diverse set of microbes, many of which may impact the biology of their hosts through metabolisms that are unique to microbial taxa. Here, we characterized the metabolic functions of macrophyte-associated microbial communities using metagenomes collected from 2 species of kelp (Laminaria setchellii and Nereocystis luetkeana) and 3 marine angiosperms (Phyllospadix scouleri, P. serrulatus, and Zostera marina), including the rhizomes of two surfgrass species (Phyllospadix spp.), the seagrass Zostera marina, and the sediments surrounding P. scouleri and Z. marina. Using metagenomic sequencing, we describe 63 metagenome-assembled genomes (MAGs) that potentially benefit from being associated with macrophytes and may contribute to macrophyte fitness through their metabolic activity. Host-associated metagenomes contained genes for the use of dissolved organic matter from hosts and vitamin (B1, B2, B7, B12) biosynthesis in addition to a range of nitrogen and sulfur metabolisms that recycle dissolved inorganic nutrients into forms more available to the host. The rhizosphere of surfgrass and seagrass contained genes for anaerobic microbial metabolisms, including nifH genes associated with nitrogen fixation, despite residing in a well-mixed and oxygenated environment. The range of oxygen environments engineered by macrophytes likely explains the diversity of both oxidizing and reducing microbial metabolisms and contributes to the functional capabilities of microbes and their influences on carbon and nitrogen cycling in nearshore ecosystems.
IMPORTANCE Kelps, seagrasses, and surfgrasses are ecosystem engineers on rocky shorelines, where they show remarkably high levels of primary production. Through analysis of their associated microbial communities, we found a variety of microbial metabolisms that may benefit the host, including nitrogen metabolisms, sulfur oxidation, and the production of B vitamins. In turn, these microbes have the genetic capabilities to assimilate the dissolved organic compounds released by their macrophyte hosts. We describe a range of oxygen environments associated with surfgrass, including low-oxygen microhabitats in their rhizomes that host genes for nitrogen fixation. The tremendous productivity of coastal seaweeds and seagrasses is likely due in part to the activities of associated microbes, and an increased understanding of these associations is needed.
KEYWORDS: host-microbiome relationships, kelp, macrophytes, marine microbiology, oxygen, seagrass, surfgrass
INTRODUCTION
We are experiencing a paradigm shift in biology with the recognition that many species exist as a consortium with microbes (1). These microbial associations are nearly ubiquitous, spanning a diversity of hosts across ecosystems. In coastal marine environments, marine angiosperms and macroalgae show high productivity (2), are a critical component of the carbon (3) and nitrogen cycles (4, 5), and host a wide range of microbial organisms. Different macroalgal species (6, 7) and their different tissues (8, 9) host distinct microbial communities which number in the millions per cm2 of host tissue (10). Yet, we still know little about the functional role the microbiome plays in host fitness and how the host influences the microbiome. Indeed, the contributions that marine macrophytes make to global carbon and nitrogen cycling have largely ignored the roles that microbes play. Yet, bacteria can supply B vitamins (11) and affect the development of their seaweed hosts (12). Seagrasses have been shown to have microbial associates that help the host access inorganic nitrogen, including bacteria that can ammonify (13) and bacteria that can fix atmospheric nitrogen (5). Though many seaweeds and seagrasses are foundational species in the coastal ocean, our understanding of the diversity and roles of their associated microbes is nascent, even as we discover that environmental change affects these communities (14).
Marine foundational species influence their surrounding physical environment. Macrophytes alter oxygen gradients and potentially influence the associated microbial communities across a range of spatiotemporal scales. The photosynthetic and respiratory activities of the host can generate a “phycosphere” (15) in which the host influences the physical environment experienced by the microbes, sometimes over micron or mm scales. For example, the basal leaf meristem of the seagrass Zostera ranges from well-oxygenated to anoxic over a scale of 300 μm (16). At larger scales, canopy forming kelps elevate daytime dissolved oxygen (DO) levels in seawater within kelp forest stands compared to the surrounding water column (17). At night, however, the DO levels in these kelp forests may drop to between 2 to 10 mg L−1 (18). Similarly, diel fluctuations that range an order of magnitude have been measured in rocky shore tidepools that include the surfgrass Phyllospadix scouleri (19, 20). This range of oxygen concentrations likely generates temporal partitioning of oxic and anoxic metabolisms and enhances the diversity of the microbial metabolisms that may be associated with macrophytes.
In low-oxygen environments, microbial respiration with alternative terminal electron acceptors, such as nitrate or sulfate, is common. Kelp (21, 22) and surfgrasses (23, 24) release >10% of their fixed carbon as dissolved organic carbon (DOC), generating a reservoir of labile carbon in the nearshore water column. Sulfur and nitrogen reducing microbes may respire these complex carbon substrates anaerobically while in anoxic environments, altering nutrient availability in the process. The subsequent replenishment of these consumed terminal electron (SO42- and NO3-) acceptors through sulfur and nitrogen oxidizing metabolic pathways, such as sox-sulfur oxidation (25) and nitrification, (26) allows these microbial metabolisms to persist. While these biogeochemical processes are well-established, little is known about microbial nutrient cycling associated with marine macrophytes, such as kelp or seagrasses.
In coastal systems, nitrogen can limit primary production, and microbial associates that aid in accessing nitrogen might be beneficial. Oxygen fluctuations can result in nitrogen oxidation or reduction (19). In proximity to the host, this retention of nitrogen may improve host fitness. Additional microbial metabolisms that can increase the available dissolved inorganic nitrogen (DIN) for the host (13) include pathways that cleave carbon-nitrogen bonds to generate ammonium. This ammonification in biological systems can result from a diversity of hydrolases, including ureases and other enzymes that cleave C-N bonds (27). Further, microbes that fix atmospheric nitrogen have been discovered in an increasing number of taxa (28, 29), now recognized to include both heterotrophic and phototrophic taxa (30–32). Nitrogen fixation was previously assumed to be restricted to nitrogen-poor environments, but it has been quantified recently in systems thought to be nitrogen-rich (28, 33). This is an enigmatic finding, given that nitrogen fixation is a costly metabolic process that consumes 16 ATPs per N2 fixed (34). Sediments where oxygen and nutrients are low, such as the rhizosphere of seagrasses, have provided evidence of nitrogen fixation (35–39). The recent discovery that nitrogen fixation takes place on particles in the coastal ocean where nitrate is relatively abundant (28, 33) suggests that nifH genes could be abundant in other nearshore systems.
Microbial metabolisms that synthesize compounds and vitamins needed by seaweeds and seagrasses may also underlie host-microbe exchanges. The active form of Vitamin B1 (thiamine) is essential for all organisms and is involved in carbohydrate and amino acid metabolism. Vitamin B2 (riboflavin) is integrated with co-enzymes in various oxidases ais and are involved in photosynthesis and phototropism (40). Vitamin B7 (biotin) is a cofactor for acetyl coenzyme A (coA) which is essential for fatty acid synthesis. Vitamin B12 (cobalamin) is required as a coenzyme in the mitochondria, yet many algae depend upon prokaryotes to produce it (11, 41). Thus, marine macrophytes may be auxotrophic for key vitamins, and their production by host-associated bacteria may be another basis for species interactions in nature.
Here, we analyzed microbial metagenomes collected from 5 different coastal macrophytes to determine if there is functional genomic evidence of microbial metabolisms that could reciprocally benefit hosts and microbes. We examined microbial taxa and metabolisms in multiple microhabitats in which macrophytes alter the oxygen environment, including the inner bulb of the canopy kelp N. luetkeana, hypothesizing that this relatively low-oxygen environment, which is also high in carbon monoxide and nitrogen gas (42), could have unique microbial metabolisms. We also hypothesized that the rhizomes of the seagrasses P. scouleri and P. serrulatus, as well as the associated sediment around P. scouleri, would be low oxygen environments based on their ability to trap sediment in this otherwise high-energy rocky shore environment. We analyzed the microbiome of the more oxygenated surface of the blades of P. scouleri and Laminaria setchellii, comparing them with metagenome-assembled genomes (MAGs) from blades of N. luetkeana that were sampled on the same day in the same area for a separate study (43). We sampled the microbial community from the Zostera marina rhizosphere from West Falmouth Harbor, MA, USA, an environment that is well-characterized (44, 45).
We analyzed the microbial taxa present and examined their gene content to estimate functional and metabolic capacities. We hypothesized that microbial partners: (i) enhance host access to dissolved inorganic nitrogen through nitrogen recycling, ammonification, and nitrogen fixation, (ii) provide vitamins B1, B2, B7, B12, and (iii) use a diversity of abundant dissolved organic carbon exudates from the host. We tested whether microbial taxonomy and function differed across hosts and host tissue types as well as whether anaerobic metabolisms were present in the low-oxygen environments found within rhizomes and the surrounding sediment. We found that the range of oxygen environments engineered by host macrophytes likely explains the diversity of oxidizing and reducing microbial metabolisms and contributes to the diverse functional capabilities of microbes and their influences on carbon and nitrogen cycling in nearshore ecosystems.
RESULTS
Surfgrass rhizomes have lower oxygen concentrations than surrounding seawater.
The oxygen environment in the rhizomes differed significantly from that of the surrounding seawater (Fig. 1). At near-peak photoperiod, the sediment in the rhizosphere maintained a lower dissolved oxygen (DO) concentration than did the surrounding seawater for both P. scouleri (n = 18, pairwise t-test: P < 0.001) and P. serrulatus (n = 11, pairwise t-test: P < 0.001). P. serrulatus maintained a slightly lower DO concentration in the rhizome at 2.11 mg L−1, compared to 5.61 mg L−1 for P. scouleri. However, sampling likely introduced oxygenated water from the surrounding water column to the rhizome-sediment microenvironment, suggesting that the actual DO concentrations within the sediment were lower than the values reported.
FIG 1.
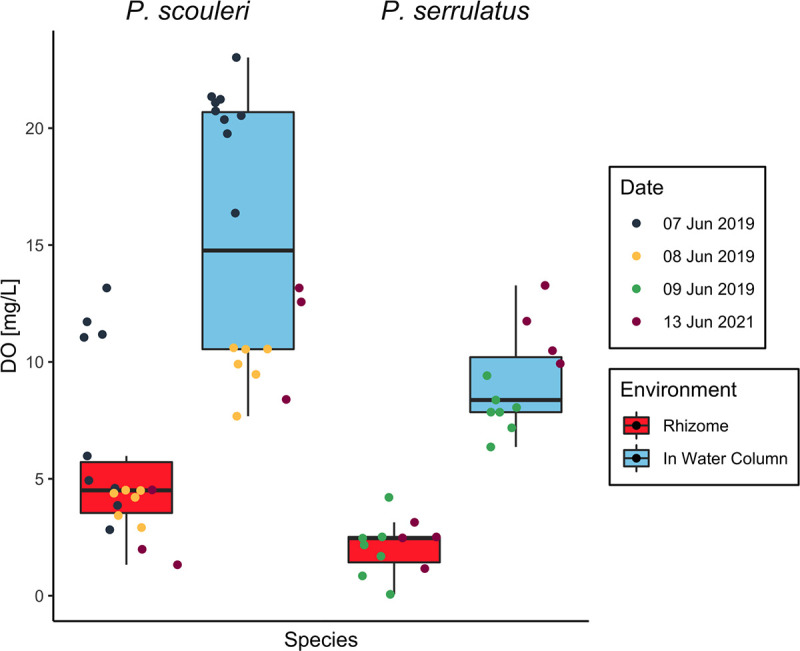
Boxplot comparing the dissolved oxygen concentrations of macrophyte tissue in the water column (blue) and in the sediment-rhizome environment (red) of P. scouleri (pairwise t-test: P < 0.001) and P. serrulatus (pairwise t-test: P < 0.001). Sampling dates are represented by different colors.
Diversity of MAGs assembled across hosts.
Following quality control and filtering, we obtained an average of 41 million sequence reads per sample (range 6.48 to 67.73 million) with 79.8% of raw reads retained on average. When these metagenomic short reads were assembled into contigs of at least 1,000 nucleotides, a mean of 42,026 contigs and a mean of 110,054 genes were present across samples (per sample summary provided in Table 1).
TABLE 1.
Summary of the features of 8 metagenomes. More information is in Data Set S1, Sheet 1, and the genome taxonomy is in Data Set S1, Sheet 2
|
Phyllospadix scouleri
|
Phyllospadix serrulatus | Laminaria setchellii | Nereocystis luetkeana |
Zostera marina
|
|||
|---|---|---|---|---|---|---|---|
| Sediment | Rhizome | Blade | Rhizome | Blade | Inner bulb | Sediment | Rhizome |
| Number of quality reads (in millions) | |||||||
| 43.68 | 67.73 | 38.41 | 37.99 | 48.58 | 6.48 | 19.37 | 65.76 |
| Bacteria | |||||||
| 63.7% | 58.3% | 63.6% | 63.0% | 63.9% | 62.2% | 33.6% | 60.6% |
| Archaea | |||||||
| 34.2% | 38.3% | 33.1% | 33.3% | 32.7% | 35.9% | 62.1% | 34.2% |
Supplementary data files containing information related to sample summaries, MAG taxonomy and binning results, genes used to generate Fig. 3, nifH genes detected, reference nifH amino acid sequences. Download Data Set S1, XLSX file, 0.1 MB (126.3KB, xlsx) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
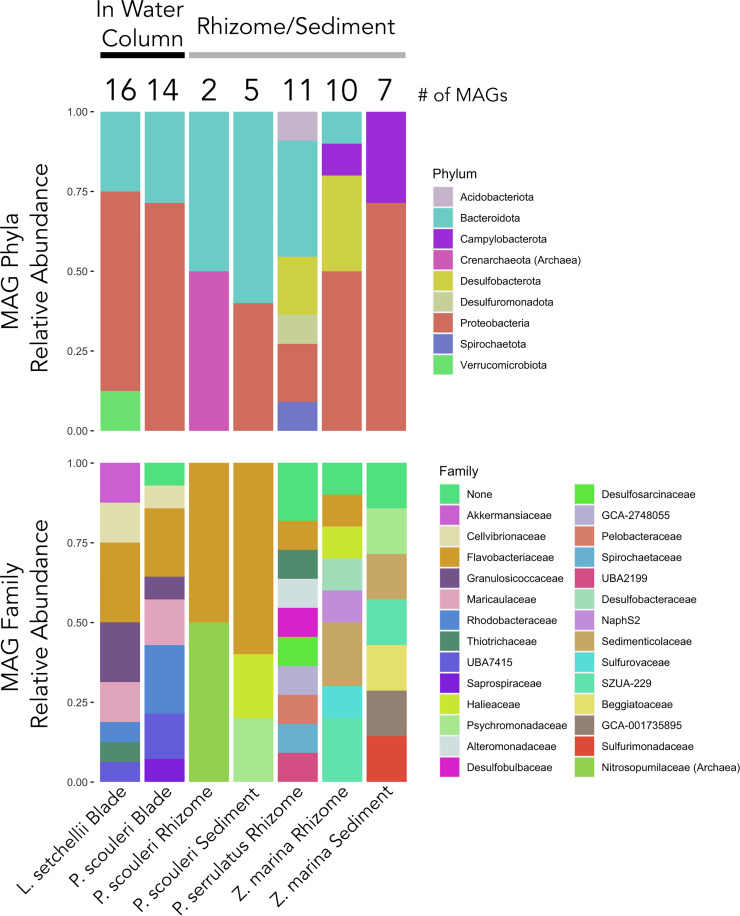
Across 8 metagenomes, we assembled 30 high quality MAGs, defined as having a completion score of >90% and a redundancy (or contamination) of < 10% (Table 2), using the criteria of Bowers et al. (46). We also identified 33 medium quality MAGs that had completion scores between 42% and 90% and redundancy scores between 0% and 11% (Data Set S1, Sheet 2). All MAGs were bacterial except for a single archaeon, Crenarchaeota, from the rhizome of P. scouleri. The bacterial MAGs spanned 8 phyla, including Proteobacteria (n = 34), Bacteroidota (n = 17), Verrucomicrobiota (n = 2), Campylobacterota (n = 3), Desulfobacterota (n = 5), and a single MAG in each of Desulfuromonadota, Acidobacteriota, and Spirochaetota. There were 43 MAGs resolved to the species level, with 14 to the genus level, 9 to the family level, 2 to the order level, and 1 to the class level. Five bins that were resolved only to the domain level (Bacteria) were classified as “Bins” and not as “MAGs” (Data Set S1, Sheet 2).
TABLE 2.
Metagenome-assembled genomes across all samples and their representation across phyla. More detailed information on the MAGs can be found in Data Set S1, Sheet 2
|
Phyllospadix scouleri
|
Phyllospadix serrulatus | Laminaria setchellii | Nereocystis luetkeana |
Zostera marina
|
|||
|---|---|---|---|---|---|---|---|
| Sediment | Rhizome | Blade | Rhizome | Blade | Inner bulb | Sediment | Rhizome |
| High Quality MAGs | |||||||
| 2 | 1 | 6 | 6 | 9 | 1 | 2 | 3 |
| Other MAGs | |||||||
| 2 | 2 | 8 | 7 | 7 | 0 | 5 | 8 |
| Proteobacteria | |||||||
| 2 | - | 10 | 2 | 10 | 1 | 5 | 5 |
| Bacteroidota | |||||||
| 3 | 1 | 4 | 4 | 4 | - | - | 1 |
| Verrucomicrobia | |||||||
| - | - | - | - | 2 | - | - | - |
| Campylobacterota | |||||||
| - | - | - | - | - | - | 2 | 1 |
| Desulfobacterota | |||||||
| - | - | - | 2 | - | - | - | 3 |
| Desulfuromonadota | |||||||
| - | - | - | 1 | - | - | - | - |
| Acidobacteriota | |||||||
| - | - | - | 1 | - | - | - | - |
| Spirochaetota | |||||||
| - | - | - | 1 | - | - | - | - |
| No ID | |||||||
| - | 1 | 0 | 2 | - | - | - | 1 |
| Crenarchaeota | |||||||
| - | 1 | - | - | - | - | - | - |
The 63 MAGs belong to diverse microbial phyla, and they were distributed across the five host species and tissue types (Fig. 2). In some cases, bacterial taxa from kelp blade tissues were most closely related to bacteria collected from the rhizome or sediment of a seagrass, suggesting that closely related bacterial taxa can associate with diverse hosts. Anaerobic sulfur cycling microbes, like Desulfobulbia, Desulfobacteria, Desulfuromonadia, and Campylobacteria (Sulfurovum sp000296775 and Sulfurimonas autotrophica), were exclusively found in the low-oxygen rhizome and sediment environments of Zostera marina and Phyllospadix spp. Conversely, Alphaproteobacteria were exclusively found on surfaces exposed to the water column, while Gammaproteobacteria was the only class found across the range of tissue types (6 out of 8 host environments) (Data Set S1, Sheet 2).
FIG 2.
Stacked bar plots showing the relative abundances of 63 metagenome-assembled genomes (MAGs) across 7 environmental samples, including 30 high quality and 33 medium quality MAGs. Only 1 MAG (UBA7415 sp002470515, a Gammaproteobacteria) was found in the bulb of N. luetkeana and is not displayed here.
Host-associated microbial genomes contain pathways to synthesize vitamins, recycle nitrogen and sulfur, and use host-generated carbon.
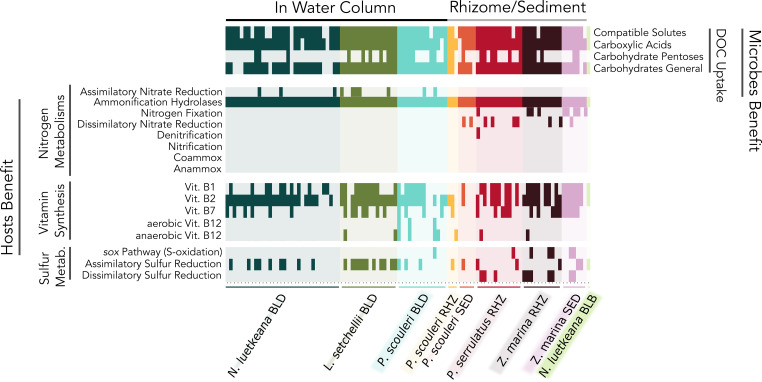
Using the Network Algorithm for Metabolism Detection (NAMeD; described in Materials and Methods), we found evidence for several metabolic pathways that are likely important for exchanges between macrophyte hosts and their microbial partners (Fig. 3). Microbes on hosts had genes for diverse carbohydrate and carboxylic acid assimilation via cell membrane transport proteins. Host-associated microbes also had genes for a diversity of nitrogen metabolisms, including ureases and hydrolases that could generate ammonium. Nitrogen metabolisms were most diverse in the rhizome and sediment samples where we identified nitrogen recycling metabolisms (dissimilatory nitrate reduction, nitrogen fixation, denitrification). Conversely, assimilatory nitrate assimilation occurred exclusively on the surface of tissues directly exposed to the water column.
FIG 3.
Boolean heatmap of microbial metabolisms in the MAGs reported in Fig. 2 and in Data Set S1, Sheet 2, across all hosts, grouped as those that might benefit the host (“hosts benefit”) and microbial metabolisms that might use host provisioned metabolites (“microbes benefit”). Each tick along the x-axis corresponds to a MAG. The N. luetkeana blade MAGs are from Weigel et al. (2022). The suite of genes for DOC Uptake (Compatible Solutes, Carboxylic Acids, Carbohydrate Pentoses, and General Carbohydrates) and Ammonification Hydrolases are shown as present if any of their genes are found in our MAGs. The presence or absence of metabolisms related to Nitrogen and Sulfur Metabolisms and Vitamin Synthesis are determined by the Network Algorithm for Metabolism Detection (NAMeD) (see supplemental code at https://github.com/kkmiranda/PNWMetagenomes/tree/main/NAMeD). The suite of genes and metabolic pathways used in this analysis can be found in Data Set S1, Sheet 3. A figure with a detailed x-axis is provided in Fig. S2.
Boolean heatmap of the microbial metabolisms described in Fig. 3 with MAG ID described on the x-axis. The MAG taxonomy corresponding to each MAG ID is found in Data Set S1, Sheet 2. Download FIG S2, TIFF file, 68.0 MB (66.5MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Every host sample had at least one gene from B-vitamin biosynthesis pathways (Fig. 3, see supplemental code at https://github.com/kkmiranda/PNWMetagenomes). We determined that all hosts had at least one MAG with the metabolic pathway to synthesize vitamins B1 (with the exception of the P. scouleri rhizome), B2, and B7 (except inside the bulb of N. luetkeana). The Vitamin B12 anaerobic biosynthesis pathway, however, was only present in MAGs found on the blades of L. setchellii (2) and P. scouleri (4) and the rhizomes of P. scouleri (1), P. serrulatus (1), and Z. marina (1). Additionally, three MAGs on the blade of P. scouleri that had this anaerobic pathway had the genes necessary to synthesize Vitamin B12 aerobically as well.
Novel detection of nifH genes in surfgrass.
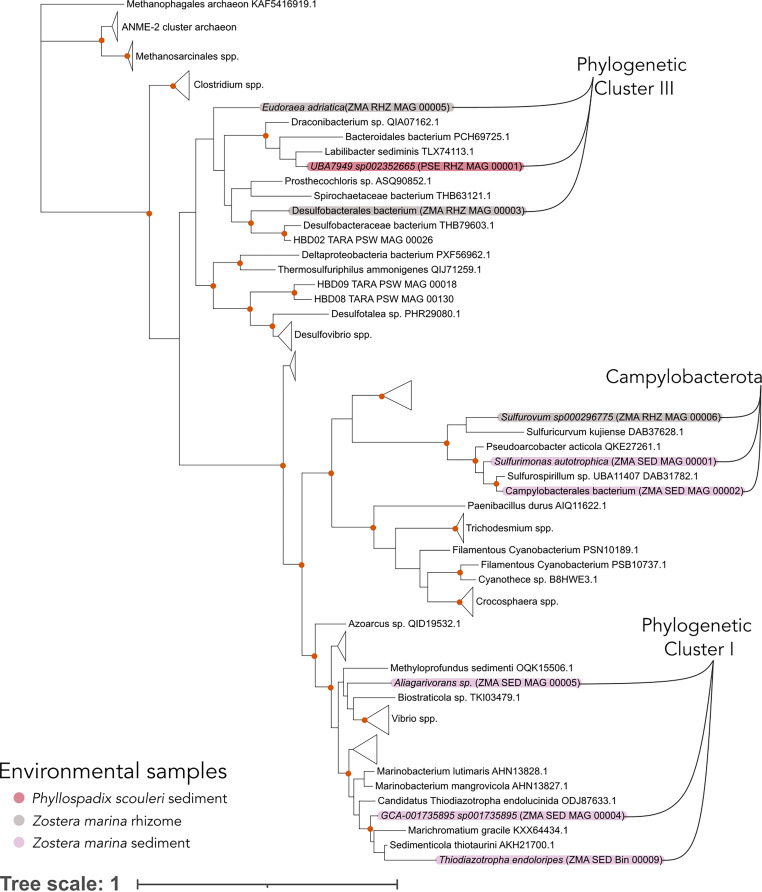
We identified the nitrogenase gene (nifH) in 9 MAGs with E value support < 1.3e-120 (KEGG) and < 1.1e-135 (COG). These 9 MAGs were assembled from the P. serrulatus rhizome (n = 1), Z. marina rhizomes (n = 3), and the surrounding sediment (n = 5). Of these 9 MAGs, 5 were resolved to the genus level (Aliagavorans, Sulfurimonas, Thiodiazotropha, Eudorea, and Sulfurovum), while others were resolved to the order and family levels, including Campylobacterales, Desulfobacterales, and 2 Flavobacteriaceae (Fig. 4, and Data Set S1, Sheet 4). The nifH genes from the rhizomes of P. serrulatus and Z. marina belonging to the class Desulfobacteria and the family Flavobacteriaceae belong to Cluster III: anaerobic nitrogen-fixers that are often coupled with sulfate-reduction (Fig. 4). The Z. marina sediment and rhizome samples also contained 3 Campylobacteria that had nifH genes that clustered together in a sister clade to the aerobic nitrogen-fixers of Cluster I.
FIG 4.
A phylogenetic tree of the nifH genes found on the rhizomes of P. serrulatus (PSE, n = 1) and the rhizomes and surrounding sediment of Z. marina (ZMA, n = 3 and 5, respectively). Some nifH genes group into Cluster I, including a sulfur oxidizing taxon on the rhizome of Z. marina and other taxa in Campylobacterota, including Sulfurovum. Cluster III contains taxa associated with rhizomes, including Desulfobulbus mediterraneus on P. serrulatus and a Desulfobacterales associated with Z. marina rhizomes.A bootstrap support of >90% is indicated by an orange dot at a branch fork. A detailed figure with uncollapsed clades is available in Fig. S3.
Expanded phylogenetic tree of the nifH genes from Fig. 4 without collapsed clades. Branches may be rotated. Bootstrap support of >90% is indicated by an orange dot at a branch fork. Download FIG S3, TIFF file, 9.0 MB (9MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We note that the COG gene identified as nifH (COG1348) also includes the homologous protochlorophyllides, which are involved in photosynthetic pigment synthesis but have high sequence similarity to the nifH gene (34, 47). Instead, we used the KEGG gene (K02588) that does not match these homologs. To further confirm the presence of nitrogen fixation genes in these MAGs, we inspected genes on the same contig with nifH and found several genes related to nitrogen fixation (Data Set S1, Sheet 4). These additional genes included nifD (COG 2710) in 6 of the 9 contigs, nitrogen regulatory protein PII (COG 347), nifB (COG 535), and multiple iron containing proteins, including ferrodoxin and Fe-Mo cluster-binding proteins (Data Set S1, Sheet 4).
DISCUSSION
Macrophytes and their rhizospheres host distinct microbial taxa.
The macrophyte species we sampled in this study are foundational in coastal ecosystems (4, 48–50), yet little is known about the diversity and function of their microbiomes. All MAGs were bacterial, except for a single archaeal MAG (Crenarchaeota) in the rhizome of Phyllospadix scouleri, which was identified as Nitrosopumulis, a genus associated with nitrification (Data Set S1, Sheet 2). Together, these 5 macrophytes hosted bacteria from 9 phyla. Blades of kelp and surfgrass were a locus of microbial diversity and function, a finding that is similar to many recent studies of macroalgal and seagrass microbiomes reporting high microbial diversity (6, 8, 9, 51–53). The functional attributes of microbial taxa associated with marine macrophytes include pathogen resistance (54), the ability to provision the host with B vitamins (11), and enhanced host algal fitness (55), perhaps through some of the nitrogen or sulfur metabolisms we documented here (5, 13).
The only low-diversity sample was the interior of the bulb of Nereocystis, where we assembled only a single Gammaproteobacterial MAG (UBA7415 sp002470515), suggesting that this environment of high carbon monoxide and nitrogen gas (42) may inhibit microbial activity or pose a highly selective environment. The bulb is the oldest section of the individual and functions to keep the macrophyte elevated at the surface for access to light. Unlike the blade, which may be detached at no great expense to the individual, any structural damage to the bulb may result in changes to the drag forces experienced by whole individual, which could cause detachment and mortality (56). An area of future investigation is whether the gases within the bulb, in addition to providing buoyancy, also inhibit microbial growth.
Host-microbe interactions in a dynamic oxygen microenvironment.
Grouping MAGs by oxygen environment (Fig. 3) showed key functional differences among macrophyte hosts, likely due to the large differences in oxygen in time and space. The 3-fold differences in dissolved oxygen (DO) between the rhizosphere and the surrounding seawater were observed at near-peak photoperiod (Fig. 1). During the night, when respiration dominates, DO in the seawater of rocky shore tide pools drops to 2 mg L−1 (20), and the rhizomes are likely anoxic. While the oxygen environment may have little influence on ammonium production from ammonification hydrolases, blade tissues that were in contact with the oxygenated water column exclusively exhibited assimilatory nitrate reduction. The low oxygen rhizosphere, conversely, is an area where anaerobic nitrogen recycling metabolisms, including denitrification, dissimilatory nitrate reduction, and nitrogen fixation are possible. Ammonification, DNRA, and nitrogen fixation all may facilitate the availability of ammonium for host uptake. Interestingly, UBA7947 sp002352665 (PSE_RHZ_MAG_00001) in the rhizome of P. serrulatus had genes for both nitrogen fixation and denitrification, both of which are metabolisms that convert nitrogen gas to ammonium and vice-versa. This suggests a variable response to the nitrogen environment, as this microbe may be denitrifying during high ammonium availability and fixing nitrogen gas when ammonium is scarce. Genes for nitrification were present, though no genes in the pathway were ever complete in any MAG. The archaeon Nitrosopumulis sp. from the blades of P. scouleri surprisingly lacked nitrification genes, despite the previous demonstration that it is capable of nitrification (57).
The abundance of dissolved organic carbon from macrophytes (21–23, 48) might select for heterotrophic metabolisms. Indeed, we found an abundance of genes for dissolved organic matter assimilation and transport in all metagenomes, suggesting that heterotrophy may be common in macrophyte-associated microbial communities (43). Improved characterization of the components of dissolved organic matter and the genomes of hosts will allow us to better assess the complementarity in resource supply by hosts and resource use by microbes, with critical implications for the carbon cycle (58).
The host tissue types in this study differed in surface oxygen concentrations. Kelp and surfgrass blade tissues interact with the water column and are likely more oxygenated than rhizome tissues or sediments, though a previous study suggests that there can also be a 60% reduction in oxygen along the surface of kelp blades during respiration (59). The thick surface mucus layer, where some kelp-associated bacteria reside (10), may further create oxygen microenvironments for aerobic or anaerobic microbial processes. Over two-thirds of the bacterial taxa on the blades of N. luetkeana belonged to families associated with obligately aerobic metabolisms, demonstrating the role of oxygen in shaping macrophyte-associated microbial communities (60). In contrast, the sediment surrounding the rhizomes of Phyllospadix spp. was a low oxygen microenvironment, likely maintained by macroinvertebrate respiration (16, 61). Low rhizosphere oxygen concentrations likely structured the taxonomic composition of Z. marina to include anaerobic taxa such as Campylobacteria, Desulfatitalea, and Desulfobulbus. The presence of anaerobes like Desulfuromonadia, Desulfobacteria, Spirochaeta, and Aminicenantia in P. serrulatus rhizomes suggests that sulfate reduction also occurs, possibly coupled to the use of dissolved organic carbon as an energy source (62). Additionally, Campylobacteria and the genus Thiodiazotropha were associated with Z. marina and may remove detrimental sulfide accumulation through sulfur oxidation (63, 64).
We encountered a range of oxidizing and reducing sulfur metabolisms. As expected, assimilatory sulfate reduction was found across all samples. Dissimilatory sulfate reduction, however, was found exclusively in rhizome/sediment conditions where oxygen is less available as a terminal electron acceptor. The product of this metabolism is hydrogen sulfide, which is toxic to eukaryotic cells. However, it also contains a large amount of chemical energy which can be released through oxidation, often through the sox-oxidation pathway. Thus, microbes, like the two Gammaproteobacteria (ZMA_RHZ_MAG_00010, ZMA_SED_MAG_00004), two Sedimenticolaceae (ZMA_RHZ_MAG_00004, ZMA_RHZ_MAG_00009), and the Beggiatoales (ZMA_SED_MAG_00003) present in the Zostera sample, may have both sulfur oxidizing and reducing metabolisms (25). In the low O2 rhizome of P. serrulatus, Leucothrix sp. may couple sox-oxidation with dissimilatory nitrate reduction, where nitrate is used as the terminal electron receptor (25). Sulfur oxidation may stimulate microbial and host productivity by replenishing the available stock of sulfate, which serves as both a nutrient and a terminal electron acceptor (65).
We detected evidence for biosynthetic pathways for vitamins B1, B2, B7, and B12 that are required by the auxotrophic hosts in this study (11, 12, 66). Only the blades of P. scouleri harbored MAGs with both anaerobic and aerobic biosynthetic pathways for Vitamin B12, suggesting that the variable oxygen environment driven by host metabolism creates diverse pathways for vitamin biosynthesis. While we did not detect a complete B12 biosynthesis pathway in the N. luetkeana blade sample included in this study, Weigel et al. (2022) found all 22 genes necessary for the complete synthesis of B12 in a Granulosicoccus MAG from N. luetkeana sampled in a different year, suggesting further investigation into the extent to which microbes provide this vitamin to their macrophyte hosts.
Characteristics of previously undescribed nitrogen fixation in surfgrass.
Building on recent studies that illustrate the association of nitrogen fixing microbes with a diversity of macroalgae (67) and seagrasses (35, 36, 68, 69), we detected nifBHDK genes in a Flavobacteriaceae from P. serrulatus rhizomes. The nifH gene resolved into the Cluster III group of nifH, characterized by anaerobic nitrogen fixers. P. serrulatus, in comparison to P. scouleri, is found higher up in the intertidal zone and is often in sheltered tidepools that tend to undergo dramatic daily fluctuations in oxygen, possibly allowing for a temporal low-O2 niche during the night (Fig. 4) (20, 70). Conversely, we did not detect nitrogenase genes in the microbiome of P. scouleri, which inhabits more wave-exposed, and thus, better oxygenated, environments (Fig. 1). However, stable isotope analyses from the P. scouleri samples show a lower nitrogen isotopic signature in the rhizome compared to the rest of the plant, a possible indication of nitrogen from an atmospheric source (Fig. S1), though in situ experiments with stable isotope tracers are needed to confirm the presence of nitrogen fixation.
Stable isotope analysis of δ13C and δ15N at the blade tip, meristem, and rhizome of P. scouleri. From blade tip to rhizome, water flow, and thus, elemental mixing, is reduced due to the attenuation and boundary layer effects of the surfgrass canopy. Assuming that elemental uptake occurs from the same pools of C and N, the lower the extent of mixing, the more enriched the isotopic signature should be, due to the depletion of the lighter isotope. This is observed with δ13C, which gets heavier from the blade tip to the rhizome. δ15N, like δ13C, gets heavier from the blade tip to the meristem, but it becomes lighter in the rhizome, suggesting that the rhizomal uptake of nitrogen comes from a pool of nitrogen to which the blade and meristem tissue do not have access. This different pool may be sourced from atmospheric N2 made available through N-fixation, which is only possible in the anoxic conditions of the rhizome. Download FIG S1, TIFF file, 13.0 MB (13.3MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nitrogen fixation by microbial associates provides a key means of increasing the availability of ammonium, possibly supporting primary productivity. P. scouleri biomass reaches 12.7 kg of wet mass per square meter of shore and exudes 0.93 mg C per hour, per gram of dry mass, as dissolved organic carbon (48), which may fuel microbial activity. There is evidence that nitrogen fixation can contribute to seagrass productivity (5, 71), a possible adaptation to low nitrogen environments. Our finding that nitrogen fixing microbes are associated with a rocky intertidal surfgrass is especially surprising, given that Tatoosh Island is in an area of upwelling and high DIN at the more northerly end of the California Current Large Marine Ecosystem (72). Whether nitrogen fixation forms the basis for a reciprocal host-microbe exchange is still unknown.
The metagenomic analyses we present here suggest that macrophyte-associated microbiomes may be involved in carbon, nitrogen, sulfur, and vitamin metabolisms important to their hosts, likely generating commensal or mutualistic interactions. Our findings echo those found for marine animals, in which strong gradients in oxygen in corals and sponges are associated with diverse microbial metabolisms (73–75). Fluctuating oxygen microenvironments may promote cross-feeding, where microbial taxa produce intermediate metabolites that can be further processed by other taxa, further contributing to nitrogen (76) and carbon metabolisms (77, 78) in microbial communities. Future experiments should explore the spatial extent of these metabolisms and test hypothesized interactions between the host, its microbiome, and carbon and nitrogen cycling. The importance of seaweeds and seagrasses to coastal productivity, and the demonstrated sensitivity of both the host and microbes to increasing temperatures and pH (14, 38, 52), pathogens (51), and other anthropogenic stressors, underline the importance of further studies of host-microbiome interactions in these foundational species.
MATERIALS AND METHODS
The study system.
The rocky shores of the northeast Pacific Ocean are a mosaic of seagrasses and kelp (48, 79), and they include both understory and canopy kelp species. These macrophytes interact with the water column and are also anchored directly to the substrate through attachment structures known as holdfasts and rhizomes. In the case of the surfgrasses P. scouleri and P. serrulatus, the rhizomes trap sediment, creating a unique environment around the basal parts of the surfgrasses (61). Tatoosh Island, WA, is an area of high wave energy and high macrophyte diversity (80). Although typically a well-oxygenated environment, we hypothesized that the biological activities of species in wave-influenced coastal systems can create microenvironments low in oxygen. We sampled a range of kelp and seagrass species and used metagenomic sequencing and microbial genome assembly to describe the microbial taxa and metabolisms present.
Quantifying the oxygen environment.
We quantified the oxygen microenvironment surrounding Phyllospadix spp. rhizomes by comparing the dissolved oxygen (DO) concentrations in the surrounding seawater and in the sediment around the rhizome. We used a Pyro Science Robust Oxygen Probe (OXROB10, Firesting, Pyroscience), and repeated measurements around 0900 h across 4 days (7 to 9 June 2019, 13 June 2021) within P. scouleri (n = 18) and P. serrulatus (n = 11) rhizomes. Each reading first measured the surrounding seawater, after which we gently pushed the tip of the oxygen probe into the sediment and rhizome mass to a depth of 1 to 3 mm, the typical thickness of the surrounding sediment on this rocky substrate (personal observation). We let the probe equilibrate and took a reading after 150 seconds. This allowed the rhizome oxygen environment to equilibrate after we disturbed the intact rhizome. We compared surrounding water and within-rhizome oxygen using paired t-tests in R.
Sampling and DNA extraction.
We collected metagenome samples from distinct surfaces of 5 different macrophyte species that we hypothesized differed in their oxygen environment (Data Set S1, Sheet 1). The surfaces of Phyllospadix scouleri blades and Laminaria setchellii fronds interact with a well-oxygenated water column. We swabbed their blade surfaces with a sterile swab and brushed them with an interdental brush (GUM Proxabrush Go-Betweens) to fully census all microbes adhered to the surface, including those in the mucus layer (10). We sampled the relatively low-oxygen environment of the inner bulbs of Nereocystis luetkeana in the same way.
We hypothesized that seagrass rhizomes and the associated sediment would be low-oxygen microenvironments in these high-energy rocky shores. We preserved 2 cm sections of the rhizomes of Phyllospadix scouleri, P. serrulatus, and Zostera marina, rinsing each with sterile fresh water, and enclosing it in a sterile vial. The sediment surrounding P. scouleri and Z. marina was carefully collected with an ethanol-rinsed metal spatula and was deposited in a sterile vial. All samples were collected from Tatoosh Island, WA, USA (48.393679, −124.734617), on 16 to 17 Jul 2019, except for the Z. marina samples, which were sampled from West Falmouth Bay, MA, USA (41.60708333, −70.64527778), on 19 Sept 2019. We included samples from the rhizosphere of Z. marina from the Atlantic Ocean as a known positive control for nitrogen fixation (35, 36). Swabs, tissue, and sediment were immediately frozen at 20°C and shipped to storage at −80°C. DNA was extracted from tissue pieces approximately 1 cm in length. We used a Qiagen PowerSoil Kit, following all kit methodologies. We pooled multiple individual sample extractions for each metagenome sample to increase the DNA quantity and possible discovery: P. scouleri blade, rhizome and sediment (3 pooled individuals each), P. serrulatus rhizome (3 individuals), L. setchellii blade (3 individuals), N. luetkeana interior bulb (4 individuals), and Z. marina rhizomes and sediment (2 individuals).
Shotgun metagenomic sequencing, assembly, and read recruitment.
The above 8 samples were run over 2 lanes on a HiSeq 2500 (2 × 150) with TruSeq DNA library preps at Argonne National Laboratory. For each sample, resulting DNA sequences were first quality filtered (81), then assembled with IDBA-UD v1.1.3 (82) with a minimum scaffold length of 1 kbp. Metagenomic short reads from each sample were then recruited back to their corresponding assembled contigs using Bowtie2 (83). Samtools (84) was used to generate sorted and indexed BAM files. Anvi’o v7.0 (85) was used as the command line environment for all downstream analyses. The ‘anvi-gen-contigs-database’ was used to generate anvi’o contigs databases, during which Prodigal v2.6.3 (86) identified open reading frames, and ‘anvi-run-hmms’ was used to identify genes matching to archaeal and bacterial single-copy core gene collections using HMMER (87).
Reconstructing metagenome-assembled genomes (MAGs).
To reconstruct genomes from the assembled metagenomes, we used a combination of automatic binning via CONCOCT v1.1.0 (88), followed by a manual curation of each MAG, as outlined by Shaiber et al. 2020 (89). Genome taxonomy was determined using GTDB v.1.3.0 (90) and 'anvi-run-scg-taxonomy'. We also inferred gene-level taxonomy using Centrifuge v1.0.4 (91) to aid in the manual curation.
Functional analysis of microbial communities.
To address the metabolic capabilities of host-associated microbes, we annotated genes in each anvi’o contigs database using ‘anvi-run-kegg-kofams’ and ‘anvi-run-ncbi-cogs’, which used the databases of the Kyoto Encyclopedia of Genes and Genomes (KEGG) (92) and NCBI’s Clusters of Orthologous Genes (COGs) (93), respectively. We used these annotated genes to test for: (i) nitrogen cycling metabolisms, especially those within the nitrogen-fixation pathway, (ii) vitamin production, namely, vitamins B1, B2, B7, and B12, (iii) sulfur cycling metabolisms, (iv) ammonification hydrolases (EC:1.4.*, EC:3.5.*, EC:4.3.1.*), including ureases and ammonia-lyases that cleave the C-N bonds in amino acids and make ammonium available to the host, and (v) a set of dissolved organic matter (DOM) transporter genes, identified by Poretsky et al. (94), that indicate the ability of the microbial community to assimilate DOM exudates from kelps and surfgrasses. The presence or absence of these metabolisms are displayed in a Boolean heatmap in Fig. 3. For (i), (ii), and (iii), we developed and used a reproducible algorithm on KEGG definitions to detect the presence of these biosynthetic pathways, calling it the Network Algorithm for Metabolism Detection (NAMeD) (see supplementary code at https://github.com/kkmiranda/PNWMetagenomes/tree/main/NAMeD). Given that we were searching for metabolisms in novel environments where some genes might be poorly documented, we set the criteria in NAMeD for determining the presence of a metabolism at 67% of the genes present in a given pathway. For (iv) and (v), we assembled suites of genes which we searched for within each MAG (suites are described in Data Set S1, Sheet 3). To expand our functional analysis of the kelp microbiome, we included 32 MAGs from the surface of N. luetkeana blades (43), derived from metagenome samples TAT_2019a and TAT_2019b, collected from Tatoosh Island at the same time as the samples in our study.
Phylogenetic analysis of nifH genes.
To search for nifH amino acid sequences in our environmental samples, we identified 9 MAGs which contained both nifH and nifD genes using the KEGG identifiers K02588 and K02586 with E values of <1e-100. We then aligned these 9 nifH AA sequences against 89 well-characterized reference nifH AA sequences (Data Set S1, Sheet 5) using Muscle v3.8.1 (95) and refined the alignment using trimAl (gap-threshold: 0.5) (96) and ‘anvi-script-reformat-fasta’ (max-percentage-gap: 50%). A maximum-likelihood phylogeny was inferred using IQTree (97) with 1,000 bootstrap replicates, and a LG+R5 model best fit our data, found using ModelFinder (98). The nifH genes from the Zostera samples served as positive controls with which to detect nitrogen fixation genes in other samples. Fig. 2 to 4 were generated using iTol v5 (99) and R v4.0.3 (100) and were edited in Inkscape. Finally, we sampled tissues from P. scouleri rhizomes (n = 16), the basal meristematic region just distal to the sheath (n = 12), and blades 35 cm above the rhizome (n = 12) to quantify stable isotopes of δ15N and δ13C to look for signatures of nitrogen fixation (methods described in Text S1, results displayed in Fig. S1).
Additional methods to quantify carbon and nitrogen stable isotopes in P. scouleri Text S1, DOCX file, 0.08 MB (84.3KB, docx) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
In addition to the code available on github.com/kkmiranda/PNWMetagenomes, the final MAG database files generated in anvi’o are available on the FigShare repository: https://doi.org/10.6084/m9.figshare.20152949.v3. Metagenomic sequence data are available at the NCBI’s Sequence Read Archive under accession number PRJNA813168.
Supplementary Material
ACKNOWLEDGMENTS
Our gratitude is to the Makah Tribal Nation for access to Tatoosh Island. We thank The University of Chicago’s Microbiome Center for pilot award funding, Washington Department of Natural Resources, grants 93099282 and 93100399 (CAP), and NSF-DEB grant (no. 1556874) awarded to J.T. Wootton. We appreciate the work of C. Sauceda in the isotope analysis, as well as A. Wootton, A. Wood, and K. Foreman in the field sampling. S. Owens and S. Greenwald at Argonne National Lab provided expertise in sequencing. K.M. was supported by an EE Fellowship from The University of Chicago.
Contributor Information
Khashiff Miranda, Email: khashiff.miranda.1@ulaval.ca.
Christopher R. Anderton, Pacific Northwest National Laboratory
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mann KH. 1973. Seaweeds: their productivity and strategy for growth: the role of large marine algae in coastal productivity is far more important than has been suspected. Science 182:975–981. doi: 10.1126/science.182.4116.975. [DOI] [PubMed] [Google Scholar]
- 3.Duarte CM, Middelburg JJ, Caraco N. 2005. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2:1–8. doi: 10.5194/bg-2-1-2005. [DOI] [Google Scholar]
- 4.Pfister CA, Altabet MA, Weigel BL. 2019. Kelp beds and their local effects on seawater chemistry, productivity, and microbial communities. Ecology 100. doi: 10.1002/ecy.2798. [DOI] [PubMed] [Google Scholar]
- 5.Mohr W, Lehnen N, Ahmerkamp S, Marchant HK, Graf JS, Tschitschko B, Yilmaz P, Littmann S, Gruber-Vodicka H, Leisch N, Weber M, Lott C, Schubert CJ, Milucka J, Kuypers MMM. 2021. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600:105–109. doi: 10.1038/s41586-021-04063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigel BL, Pfister CA. 2019. Successional dynamics and seascape-level patterns of microbial communities on the canopy-forming kelps Nereocystis luetkeana and Macrocystis pyrifera. Front Microbiol 10:346. doi: 10.3389/fmicb.2019.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemay MA, Martone PT, Hind KR, Lindstrom SC, Wegener Parfrey L. 2018. Alternate life history phases of a common seaweed have distinct microbial surface communities. Mol Ecol 27:3555–3568. doi: 10.1111/mec.14815. [DOI] [PubMed] [Google Scholar]
- 8.Quigley CTC, Capistrant-Fossa KA, Morrison HG, Johnson LE, Morozov A, Hertzberg VS, Brawley SH. 2020. Bacterial communities show algal host (Fucus spp.)/zone differentiation across the stress gradient of the intertidal zone. Front Microbiol 11:563118. doi: 10.3389/fmicb.2020.563118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemay MA, Davis KM, Martone PT, Parfrey LW. 2021. Kelp‐associated microbiota are structured by host anatomy. J Phycol 57:1119–1130. doi: 10.1111/jpy.13169. [DOI] [PubMed] [Google Scholar]
- 10.Ramírez-Puebla ST, Weigel BL, Jack L, Schlundt C, Pfister CA, Mark Welch JL. 2022. Spatial organization of the kelp microbiome at micron scales. Microbiome 10:52. doi: 10.1186/s40168-022-01235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 12.Wichard T, Charrier B, Mineur F, Bothwell JH, Clerck OD, Coates JC. 2015. The green seaweed Ulva: a model system to study morphogenesis. Front Plant Sci 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarquinio F, Bourgoure J, Koenders A, Laverock B, Säwström C, Hyndes GA. 2018. Microorganisms facilitate uptake of dissolved organic nitrogen by seagrass leaves. ISME J 12:2796–2800. doi: 10.1038/s41396-018-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Z, Coleman MA, Provost E, Campbell AH, Kelaher BP, Dalton SJ, Thomas T, Steinberg PD, Marzinelli EM. 2019. Future climate change is predicted to affect the microbiome and condition of habitat-forming kelp. Proc Biol Sci 286:20181887. doi: 10.1098/rspb.2018.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell W, Mitchell R. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. The Biological Bull 143:265–277. doi: 10.2307/1540052. [DOI] [Google Scholar]
- 16.Brodersen KE, Siboni N, Nielsen DA, Pernice M, Ralph PJ, Seymour J, Kühl M. 2018. Seagrass rhizosphere microenvironment alters plant‐associated microbial community composition. Environ Microbiol 20:2854–2864. doi: 10.1111/1462-2920.14245. [DOI] [PubMed] [Google Scholar]
- 17.Pfister CA, Altabet MA. 2019. Enhanced microbial nitrogen transformations in association with macrobiota from the rocky intertidal. Biogeosciences 16:193–206. doi: 10.5194/bg-16-193-2019. [DOI] [Google Scholar]
- 18.Donham EM, Strope LT, Hamilton SL, Kroeker KJ. 2022. Coupled changes in pH, temperature, and dissolved oxygen impact the physiology and ecology of herbivorous kelp forest grazers. Glob Chang Biol 28:3023–3039. doi: 10.1111/gcb.16125. [DOI] [PubMed] [Google Scholar]
- 19.Pfister CA, Altabet MA, Pather S, Dwyer G. 2016. Tracer experiment and model evidence for macrofaunal shaping of microbial nitrogen functions along rocky shores. Biogeosciences 13:3519–3531. doi: 10.5194/bg-13-3519-2016. [DOI] [Google Scholar]
- 20.Fields J, Silbiger N. 2022. Foundation species loss alters multiple ecosystem functions within temperate tidepool communities. Mar Ecol Prog Ser 683:1–19. doi: 10.3354/meps13978. [DOI] [Google Scholar]
- 21.Reed DC, Carlson CA, Halewood ER, Nelson JC, Harrer SL, Rassweiler A, Miller RJ. 2015. Patterns and controls of reef-scale production of dissolved organic carbon by giant kelp M acrocystis pyrifera: DOC production by giant kelp. Limnol Oceanogr 60:1996–2008. doi: 10.1002/lno.10154. [DOI] [Google Scholar]
- 22.Weigel BL, Pfister CA. 2021. The dynamics and stoichiometry of dissolved organic carbon release by kelp. Ecology 102. doi: 10.1002/ecy.3221. [DOI] [PubMed] [Google Scholar]
- 23.Wetzel RG, Penhale PA. 1979. Transport of carbon and excretion of dissolved organic carbon by leaves and roots/rhizomes in seagrasses and their epiphytes. Aquatic Botany 6:149–158. doi: 10.1016/0304-3770(79)90058-5. [DOI] [Google Scholar]
- 24.Barrón C, Apostolaki ET, Duarte CM. 2014. Dissolved organic carbon fluxes by seagrass meadows and macroalgal beds. Front Mar Sci 1. [Google Scholar]
- 25.Jørgensen BB, Findlay AJ, Pellerin A. 2019. The biogeochemical sulfur cycle of marine sediments. Front Microbiol 10:849. doi: 10.3389/fmicb.2019.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamersley M, Howes B. 2005. Coupled nitrification-denitrification measured in situ in a Spartina alterniflora marsh with a 15NH4+ tracer. Mar Ecol Prog Ser 299:123–135. doi: 10.3354/meps299123. [DOI] [Google Scholar]
- 27.Ladd JN, Jackson RB. 2015. Biochemistry of ammonification, p 173–228. In Stevenson FJ (ed), Agronomy Monographs. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI, USA. [Google Scholar]
- 28.Mills MM, Turk-Kubo KA, van Dijken GL, Henke BA, Harding K, Wilson ST, Arrigo KR, Zehr JP. 2020. Unusual marine cyanobacteria/haptophyte symbiosis relies on N2 fixation even in N-rich environments. ISME J 14:2395–2406. doi: 10.1038/s41396-020-0691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delmont TO, Quince C, Shaiber A, Esen ÖC, Lee ST, Rappé MS, McLellan SL, Lücker S, Eren AM. 2018. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3:804–813. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohm JA, Webb EA, Capone DG. 2011. Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol 9:499–508. doi: 10.1038/nrmicro2594. [DOI] [PubMed] [Google Scholar]
- 31.Bombar D, Paerl RW, Riemann L. 2016. Marine non-cyanobacterial diazotrophs: moving beyond molecular detection. Trends Microbiol 24:916–927. doi: 10.1016/j.tim.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Harding K, Turk-Kubo KA, Sipler RE, Mills MM, Bronk DA, Zehr JP. 2018. Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. Proc Natl Acad Sci USA 115:13371–13375. doi: 10.1073/pnas.1813658115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabello AM, Turk‐Kubo KA, Hayashi K, Jacobs L, Kudela RM, Zehr JP. 2020. Unexpected presence of the nitrogen‐fixing symbiotic cyanobacterium UCYN‐A in Monterey Bay, California. J Phycol 56:1521–1533. doi: 10.1111/jpy.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol Biol Evol 21:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 35.Patriquin D, Knowles R. 1972. Nitrogen fixation in the rhizosphere of marine angiosperms. Marine Biology 16:49–58. doi: 10.1007/BF00347847. [DOI] [Google Scholar]
- 36.Capone DG, Budin JM. 1982. Nitrogen fixation associated with rinsed roots and rhizomes of the eelgrass Zostera marina. Plant Physiol 70:1601–1604. doi: 10.1104/pp.70.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole L, McGlathery K. 2012. Nitrogen fixation in restored eelgrass meadows. Mar Ecol Prog Ser 448:235–246. doi: 10.3354/meps09512. [DOI] [Google Scholar]
- 38.Agawin NSR, Gil Atorrasagasti MG, Frank Comas A, Fernández‐Juárez V, López‐Alforja X, Hendriks IE. 2021. Response of the seagrass Posidonia oceanica and its associated N2 fixers to high business‐as‐usual climate change scenario in winter. Limnol Oceanogr 66:2346–2359. doi: 10.1002/lno.11758. [DOI] [Google Scholar]
- 39.Aoki L, McGlathery K. 2019. High rates of N fixation in seagrass sediments measured via a direct 30N2 push-pull method. Mar Ecol Prog Ser 616:1–11. doi: 10.3354/meps12961. [DOI] [Google Scholar]
- 40.Massey V. 2000. The chemical and biological versatility of riboflavin. Biochem Soc Trans 28:283–296. doi: 10.1042/bst0280283. [DOI] [PubMed] [Google Scholar]
- 41.Grossman A. 2016. Nutrient acquisition: the generation of bioactive vitamin B 12 by microalgae. Curr Biol 26:R319–R321. doi: 10.1016/j.cub.2016.02.047. [DOI] [PubMed] [Google Scholar]
- 42.Liggan LM, Martone PT. 2020. Gas composition of developing pneumatocysts in bull kelp Nereocystis luetkeana (Phaeophyceae). J Phycol 56:1367–1372. doi: 10.1111/jpy.13037. [DOI] [PubMed] [Google Scholar]
- 43.Weigel BL, Miranda KK, Fogarty EC, Watson AR, Pfister CA. 2022. Functional insights into the kelp microbiome from metagenome-assembled genomes. mSystems 7:e01422-21. doi: 10.1128/msystems.01422-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenworthy WJ, Gallegos CL, Costello C, Field D, di Carlo G. 2014. Dependence of eelgrass (Zostera marina) light requirements on sediment organic matter in Massachusetts coastal bays: implications for remediation and restoration. Mar Pollut Bull 83:446–457. doi: 10.1016/j.marpolbul.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Haviland KA, Howarth RW, Marino R, Hayn M. 2022. Variation in sediment and seagrass characteristics reflect multiple stressors along a nitrogen‐enrichment gradient in a New England lagoon. Limnology & Oceanography 67:660–672. doi: 10.1002/lno.12025. [DOI] [Google Scholar]
- 46.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Murat Eren A, Schriml L, The Genome Standards Consortium, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapili BJ, Dekas AE. 2021. PPIT: an R package for inferring microbial taxonomy from nifH sequences. Bioinformatics 37:2289–2298. doi: 10.1093/bioinformatics/btab100. [DOI] [PubMed] [Google Scholar]
- 48.Miranda KK, Weigel BL, McCoy SJ, Pfister CA. 2021. Differential impacts of alternate primary producers on carbon cycling. Ecology 102. doi: 10.1002/ecy.3455. [DOI] [PubMed] [Google Scholar]
- 49.Shelton AO. 2010. Temperature and community consequences of the loss of foundation species: surfgrass (Phyllospadix spp., Hooker) in tidepools. J Experimental Marine Biology and Ecology 391:35–42. doi: 10.1016/j.jembe.2010.06.003. [DOI] [Google Scholar]
- 50.Lefcheck JS, Wilcox DJ, Murphy RR, Marion SR, Orth RJ. 2017. Multiple stressors threaten the imperiled coastal foundation species eelgrass (Zostera marina) in Chesapeake Bay, USA. Glob Chang Biol 23:3474–3483. doi: 10.1111/gcb.13623. [DOI] [PubMed] [Google Scholar]
- 51.Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. 2013. The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol Rev 37:462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 52.Minich JJ, Morris MM, Brown M, Doane M, Edwards MS, Michael TP, Dinsdale EA. 2018. Elevated temperature drives kelp microbiome dysbiosis, while elevated carbon dioxide induces water microbiome disruption. PLoS One 13:e0192772. doi: 10.1371/journal.pone.0192772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capistrant‐Fossa KA, Morrison HG, Engelen AH, Quigley CTC, Morozov A, Serrão EA, Brodie J, Gachon CMM, Badis Y, Johnson LE, Hoarau G, Abreu MH, Tester PA, Stearns LA, Brawley SH. 2021. The microbiome of the habitat‐forming brown alga Fucus vesiculosus (Phaeophyceae) has similar cross‐Atlantic structure that reflects past and present drivers. J Phycol 57:1681–1698. doi: 10.1111/jpy.13194. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Weinberger F, Saha M, Majzoub ME, Egan S. 2021. Cross-host protection of marine bacteria against macroalgal disease. Microb Ecol doi: 10.1007/s00248-021-01909-2. [DOI] [PubMed] [Google Scholar]
- 55.Burgunter-Delamare B, KleinJan H, Frioux C, Fremy E, Wagner M, Corre E, Le Salver A, Leroux C, Leblanc C, Boyen C, Siegel A, Dittami SM. 2020. Metabolic complementarity between a brown alga and associated cultivable bacteria provide indications of beneficial interactions. Front Mar Sci 7:85. doi: 10.3389/fmars.2020.00085. [DOI] [Google Scholar]
- 56.Dobkowski KA, Crofts SB. 2021. Scaling and structural properties of juvenile bull kelp (Nereocystis luetkeana). Integrative Organismal Biology 3:obab022. doi: 10.1093/iob/obab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PSG, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sogin EM, Michellod D, Gruber-Vodicka HR, Bourceau P, Geier B, Meier DV, Seidel M, Ahmerkamp S, Schorn S, D’Angelo G, Procaccini G, Dubilier N, Liebeke M. 2022. Sugars dominate the seagrass rhizosphere. Nat Ecol Evol 6:866–877. doi: 10.1038/s41559-022-01740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noisette F, Hurd C. 2018. Abiotic and biotic interactions in the diffusive boundary layer of kelp blades create a potential refuge from ocean acidification. Funct Ecol 32:1329–1342. doi: 10.1111/1365-2435.13067. [DOI] [Google Scholar]
- 60.Weigel BL, Pfister CA. 2021. Oxygen metabolism shapes microbial settlement on photosynthetic kelp blades compared to artificial kelp substrates. Environ Microbiol Rep 13:176–184. doi: 10.1111/1758-2229.12923. [DOI] [PubMed] [Google Scholar]
- 61.Moulton O, Hacker S. 2011. Congeneric variation in surfgrasses and ocean conditions influence macroinvertebrate community structure. Mar Ecol Prog Ser 433:53–63. doi: 10.3354/meps09180. [DOI] [Google Scholar]
- 62.Howarth RW, Hobbie JE. 1982. The regulation of decomposition and heterotrophic microbial activity in salt marsh soils: a review. p 183–207. In Estuarine Comparisons. Elsevier. [Google Scholar]
- 63.Martin BC, Middleton JA, Fraser MW, Marshall IPG, Scholz VV, Hausl B, Schmidt H. 2020. Cutting out the middle clam: lucinid endosymbiotic bacteria are also associated with seagrass roots worldwide. ISME J 14:2901–2905. doi: 10.1038/s41396-020-00771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keller AH, Schleinitz KM, Starke R, Bertilsson S, Vogt C, Kleinsteuber S. 2015. Metagenome-based metabolic reconstruction reveals the ecophysiological function of Epsilonproteobacteria in a hydrocarbon. Contaminated Sulfidic Aquifer Front Microbiol 6:1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rolando JL, Kolton M, Song T, Kostka JE. 2022. The core root microbiome of Spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes, USA. Microbiome 10:37. doi: 10.1186/s40168-021-01187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helliwell KE. 2017. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol 216:62–68. doi: 10.1111/nph.14669. [DOI] [PubMed] [Google Scholar]
- 67.Hamersley MR, Sohm JA, Burns JA, Capone DG. 2015. Nitrogen fixation associated with the decomposition of the giant kelp Macrocystis pyrifera. Aquatic Botany 125:57–63. doi: 10.1016/j.aquabot.2015.05.003. [DOI] [Google Scholar]
- 68.McGlathery K, Risgaard-Petersen N, Christensen P. 1998. Temporal and spatial variation in nitrogen fixation activity in the eelgrass Zostera marina rhizosphere. Mar Ecol Prog Ser 168:245–258. doi: 10.3354/meps168245. [DOI] [Google Scholar]
- 69.Garcias-Bonet N, Arrieta JM, Duarte CM, Marbà N. 2016. Nitrogen-fixing bacteria in Mediterranean seagrass (Posidonia oceanica) roots. Aquatic Botany 131:57–60. doi: 10.1016/j.aquabot.2016.03.002. [DOI] [Google Scholar]
- 70.Steunou A-S, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O, Bhaya D, Ward DM, Peters JW, Grossman AR, Kühl M. 2008. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J 2:364–378. doi: 10.1038/ismej.2007.117. [DOI] [PubMed] [Google Scholar]
- 71.Cardini U, van Hoytema N, Bednarz VN, Al-Rshaidat MMD, Wild C. 2018. N2 fixation and primary productivity in a red sea Halophila stipulacea meadow exposed to seasonality: N2 fixation in Halophila stipulacea. Limnol Oceanogr 63:786–798. doi: 10.1002/lno.10669. [DOI] [Google Scholar]
- 72.Pfister CA, Wootton JT, Neufeld CJ. 2007. The relative roles of coastal and oceanic processes in determining physical and chemical characteristics of an intensively sampled nearshore system. Limnol Oceanogr 52:1767–1775. doi: 10.4319/lo.2007.52.5.1767. [DOI] [Google Scholar]
- 73.Babbin AR, Tamasi T, Dumit D, Weber L, Rodríguez MVI, Schwartz SL, Armenteros M, Wankel SD, Apprill A. 2021. Discovery and quantification of anaerobic nitrogen metabolisms among oxygenated tropical Cuban stony corals. ISME J 15:1222–1235. doi: 10.1038/s41396-020-00845-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fiore CL, Jarett JK, Olson ND, Lesser MP. 2010. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol 18:455–463. doi: 10.1016/j.tim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Morris RL, Schmidt TM. 2013. Shallow breathing: bacterial life at low O2. Nat Rev Microbiol 11:205–212. doi: 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lilja EE, Johnson DR. 2019. Substrate cross-feeding affects the speed and trajectory of molecular evolution within a synthetic microbial assemblage. BMC Evol Biol 19:129. doi: 10.1186/s12862-019-1458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Jesús Astacio LM, Prabhakara KH, Li Z, Mickalide H, Kuehn S. 2021. Closed microbial communities self-organize to persistently cycle carbon. Proc Natl Acad Sci USA 118:e2013564118. doi: 10.1073/pnas.2013564118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segrè D, Mehta P, Sanchez A. 2018. Emergent simplicity in microbial community assembly. Science 361:469–474. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Menge BA, Allison GW, Blanchette CA, Farrell TM, Olson AM, Turner TA, van Tamelen P. 2005. Stasis or kinesis? Hidden dynamics of a rocky intertidal macrophyte mosaic revealed by a spatially explicit approach. J Experimental Marine Biology and Ecology 314:3–39. doi: 10.1016/j.jembe.2004.09.015. [DOI] [Google Scholar]
- 80.Leigh EG, Paine RT, Quinn JF, Suchanek TH. 1987. Wave energy and intertidal productivity. Proc Natl Acad Sci USA 84:1314–1318. doi: 10.1073/pnas.84.5.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol 12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 83.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, Trigodet F, Watson AR, Esen ÖC, Moore RM, Clayssen Q, Lee MD, Kivenson V, Graham ED, Merrill BD, Karkman A, Blankenberg D, Eppley JM, Sjödin A, Scott JJ, Vázquez-Campos X, McKay LJ, McDaniel EA, Stevens SLR, Anderson RE, Fuessel J, Fernandez-Guerra A, Maignien L, Delmont TO, Willis AD. 2021. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol 6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 89.Shaiber A, Willis AD, Delmont TO, Roux S, Chen L-X, Schmid AC, Yousef M, Watson AR, Lolans K, Esen ÖC, Lee STM, Downey N, Morrison HG, Dewhirst FE, Mark Welch JL, Eren AM. 2020. Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. Genome Biol 21:292. doi: 10.1186/s13059-020-02195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim D, Song L, Breitwieser FP, Salzberg SL. 2016. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poretsky RS, Sun S, Mou X, Moran MA. 2010. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol 12:616–627. doi: 10.1111/j.1462-2920.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 100.R Core Team. 2021. R: a Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data files containing information related to sample summaries, MAG taxonomy and binning results, genes used to generate Fig. 3, nifH genes detected, reference nifH amino acid sequences. Download Data Set S1, XLSX file, 0.1 MB (126.3KB, xlsx) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Boolean heatmap of the microbial metabolisms described in Fig. 3 with MAG ID described on the x-axis. The MAG taxonomy corresponding to each MAG ID is found in Data Set S1, Sheet 2. Download FIG S2, TIFF file, 68.0 MB (66.5MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expanded phylogenetic tree of the nifH genes from Fig. 4 without collapsed clades. Branches may be rotated. Bootstrap support of >90% is indicated by an orange dot at a branch fork. Download FIG S3, TIFF file, 9.0 MB (9MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Stable isotope analysis of δ13C and δ15N at the blade tip, meristem, and rhizome of P. scouleri. From blade tip to rhizome, water flow, and thus, elemental mixing, is reduced due to the attenuation and boundary layer effects of the surfgrass canopy. Assuming that elemental uptake occurs from the same pools of C and N, the lower the extent of mixing, the more enriched the isotopic signature should be, due to the depletion of the lighter isotope. This is observed with δ13C, which gets heavier from the blade tip to the rhizome. δ15N, like δ13C, gets heavier from the blade tip to the meristem, but it becomes lighter in the rhizome, suggesting that the rhizomal uptake of nitrogen comes from a pool of nitrogen to which the blade and meristem tissue do not have access. This different pool may be sourced from atmospheric N2 made available through N-fixation, which is only possible in the anoxic conditions of the rhizome. Download FIG S1, TIFF file, 13.0 MB (13.3MB, tiff) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional methods to quantify carbon and nitrogen stable isotopes in P. scouleri Text S1, DOCX file, 0.08 MB (84.3KB, docx) .
Copyright © 2022 Miranda et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
In addition to the code available on github.com/kkmiranda/PNWMetagenomes, the final MAG database files generated in anvi’o are available on the FigShare repository: https://doi.org/10.6084/m9.figshare.20152949.v3. Metagenomic sequence data are available at the NCBI’s Sequence Read Archive under accession number PRJNA813168.