Abstract
A fluorogenic PCR specific for ovine herpesvirus 2 (OvHV-2) DNA was developed and compared to a previously established conventional seminested PCR. Testing of a total of 152 blood samples from both positive and negative animals revealed that the results of both assays corresponded to each other in 100% of the cases. A second fluorogenic PCR for genomic sheep DNA was required to normalize the quantity of viral DNA in the sample. Separate standard curves had to be constructed for each PCR. The analytical sensitivity of the new PCRs ranged between at least 10 copies and sometimes even 1 copy of target DNA per reaction mixture. In dilution series of the target DNAs, linear decreases of the signals were observed over 7 orders of magnitude. Thus, it was possible to calculate the amounts of viral DNA in relation to the amounts of cellular DNA by normalizing the absolute quantity of OvHV-2 DNA with the amount of genomic sheep DNA. By this technique, it was possible for the first time to quantitatively characterize the course of OvHV-2 replication in naturally infected sheep.
Malignant catarrhal fever (MCF) is a sporadic but usually fatal infectious disease of cattle and other ruminant species 8, 9, 10, 12. Ovine herpesvirus 2 (OvHV-2) is believed to be the etiologic agent of the sheep-associated form of MCF (SA-MCF) that occurs almost worldwide. The agent of the wildebeest-associated form of MCF, alcelaphine herpesvirus 1 (AlcHV-1), was readily isolated and propagated in cell culture 4, 13. In contrast, the isolation of OvHV-2 in cell cultures has not been reported 5, while our own attempts remained unsuccessful (unpublished data). The establishment of a conventional PCR for the detection of OvHV-2 DNA was therefore important 2, 3. This conventional PCR was applied for detection of OvHV-2 in vivo. It turned out that the natural reservoir of this virus is sheep. Among cattle, only samples from animals suffering from MCF and some rare survivors of MCF 9, 10 react positively in this PCR (U. U. Müller-Doblies, J. Egli, H. Li, U. Braun, and M. Ackermann, in press). On the basis of these observations, PCR was established as the new “gold standard” for the diagnosis of MCF in cattle 11. Thus, without being able to grow the virus in cell culture, the technical requirements for tracing of this agent in a qualitative manner had been developed.
However, to get a better insight into the pathogenesis and replication of OvHV-2, it is essential to trace the agent in a quantitative manner along the time axis of a natural infection.
Conventional quantitative PCRs are time-consuming and carry a high risk of false-positive results due to contamination, especially when a high sample throughput is required. Therefore, a fluorogenic PCR was developed for quantitative determination of OvHV-2 DNA in samples obtained from sheep. The assay makes use of a dually labeled fluorogenic probe, which is designed to hybridize to the sequence between the primers (User's Manual, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems, Foster City, Calif., 1998). The fluorescent emission of a reporter dye, covalently attached to the 5′ end of the probe, is quenched due to the physical proximity of a quencher dye at the 3′ end. The 5′ → 3′ exonuclease activity of the DNA polymerase causes cleavage of the probe in the course of amplification, which leads to dislocation of both the reporter and the quencher. This results in an increase of the reporter dye's fluorescent emission, which is, at each time point measured, directly proportional to the amount of amplified target DNA.
An absolute quantitation of input DNA is thus accomplished by generating a standard curve with serial dilutions of the plasmid-cloned target DNA and then comparing the signals obtained from samples with unknown amounts of DNA to the standard curve (User's Manual, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems).
In order to apply this system for the quantitation of OvHV-2 DNA, a corresponding fluorogenic PCR was first established. Its sensitivity and specificity were compared to those of the conventional OvHV-2-specific PCR. In a second step, a quantitative fluorogenic PCR for genomic sheep DNA was developed in order (i) to evaluate the quality of the DNA that was extracted from samples which reacted negatively in the OvHV-2 PCR and (ii) to normalize the absolute quantity of viral DNA with the amount of genomic sheep DNA in the same sample (relative quantitation). Then, the analytical sensitivities, linearities, and efficiencies of the new tests were determined. Finally, the new methods were applied to trace and quantitate the course of a natural OvHV-2 infection in a sheep.
MATERIALS AND METHODS
Samples.
A total of 152 blood samples (EDTA-treated blood) were obtained from 109 cows, 1 moose, 40 sheep, and 2 goats. Sixty-three cows and the moose had suffered from SA-MCF and had been found to be positive for OvHV-2 DNA by conventional seminested PCR, whereas 46 healthy cows had tested negative for OvHV-2 DNA. Twenty of the sheep samples originated from known OvHV-2-free sheep. While 17 of those samples were obtained from a specific OvHV-2-free flock, 3 samples were taken from newborn lambs. Of the 20 known OvHV-2-positive sheep, 15 originated from two different conventional flocks and 5 were previously negative sheep which had been introduced into a positive flock.
Sample preparation.
A total of 40 ml of lysis buffer (0.15 M NH4Cl, 10 mM CHKO3, 0.1 mM EDTA [pH 7.2]) was added to 10 ml of EDTA-treated blood samples, and the mixture was incubated for 5 min at room temperature to lyse the erythrocytes and centrifuged at 4°C for 10 min at 868 × g. The buffy-coat cells were resuspended and washed in 50 ml of phosphate-buffered saline and centrifuged (4°C, 10 min, 868 × g), and the pellet was stored at −20°C until further use.
DNA extraction.
For the comparison of both the established seminested PCR and the new fluorogenic OvHV-2 PCR, the buffy-coat cells were thawed before being resuspended in 200 μl of extraction buffer (0.5 mM EDTA, 25 mM Tris, 0.025% Tween 20) supplemented with freshly added proteinase K (0.2 mg/ml) and RNase A (0.5 mg/ml). Then, the samples were digested overnight at 50°C. After heat inactivation of the proteinase K (95°C, 10 min), the samples were cooled on ice for 3 min and centrifuged (13,000 rpm for 30 s) in an Eppendorf microcentrifuge. The supernatant was used directly in both PCRs. For the quantitative PCR, the DNA of the buffy-coat cells was extracted with a QIAamp DNA mini kit (Qiagen, Basel, Switzerland).
Conventional seminested PCR.
The protocol for the conventional seminested OvHV-2 PCR was adapted from that of Müller-Doblies et al. 11 with the following modifications: the missing G at position 9 (indicated by the underscore in the sequence of o488) in the sequence of the original forward primer 556 (AGT CTG GGT ATA TGA ATC CAG ATG GCT CTC) was inserted, while the C at position 30 (indicated by the underscore above) was deleted to compensate for the insert. The new primer was designated o488 (AGT CTG GGG TAT ATG AAT CCA GAT GGC TCT). The reaction mixture contained 0.2 mM dTTP instead of dUTP, and only 25 cycles (instead of 39) were performed with the outer primers.
Primers and probes for fluorogenic PCRs.
Primer and probe sequences (Table 1) were designed with Perkin-Elmer Primer Express software (version 1.0, Perkin-Elmer, Foster City, Calif.) according to the manufacturer's guidelines. The design for the OvHV-2-specific primer-probe set was based on the same sequence of genomic OvHV-2 DNA which had been used for the conventional PCR (GenBank accession no. S64565) 2, 3. The sequence of an Ovis aries dispersed repeat region (GenBank accession no. AF130311) served to design oligonucleotides specific for the sheep genome (SHgenom).
TABLE 1.
OvHV-2- and SHgenom-specific primers and probes used in fluorogenic assays
| Oligonucleotide | Sequence (5′→3′) | Length of fragment (bp) |
|---|---|---|
| Forward primer (oF-OvHV-2) | TGG TAG GAG CAG GCT ACC GT | 131 |
| Rerverse primer (oR-OvHV-2) | ATC ATG CTG ACC CCT TGC AG | |
| FAMa probe (oP-OvHV-2) | TCC ACG CCG TCC GCA CTG TAA GA | |
| Forward pimer (oF-SHgenom) | GTG CAA GGT GCC CTC GAC TA | 71 |
| Reverse primer (oR-SHgenom) | AGA AGC CTC GGG AGT GTG TCT | |
| VICb probe (oP-SHgenom) | TGA CCC TAG CAT GGC ACT CAG CCC |
FAM, 6-carboxyfluorescein.
VIC, commercial fluorescent dye with maximum intensity at 555 nm.
The fluorescent reporter dye at the 5′ end of the OvHV-2 probe was 6-carboxyfluorescein (FAM; emission wavelength, 518 nm) and that at the 5′ end of the SHgenom probe was VIC (emission wavelength, 552 nm), and the quencher dyes at the 3′ ends were 6-carboxytetramethylrhodamine (TAMRA; emmission wavelength, 582 nm). The probes, which both anneal to the coding strand, were synthesized by Perkin-Elmer, Weiterstadt, Germany, and the primers were synthesized by Microsynth, Balgach, Switzerland.
Plasmid standards for quantitation. (i) Standard for OvHV-2.
To obtain a standard for quantitative OvHV-2 PCR, the 422-bp fragment, amplified with the outer primers of the seminested PCR, was cloned into a pCR 2.1-Topo plasmid vector (Invitrogen B. V., Groningen, The Netherlands) by conventional methods. The nucleotide sequence was verified (Microsynth), and the plasmid was named pOvHV-2. For use in PCR, the plasmid was purified with a Wizard Plus Midipreps kit (Catalys AG, Wallisellen, Switzerland) and its DNA content was quantitated by spectrophotometric analysis. Each dilution of the standard plasmid contained 60 μg of salmon sperm DNA (Life Technologies AG, Basel, Switzerland) per ml as a carrier. Aliquots of dilutions were frozen and used only once for fluorogenic PCR.
(ii) Standard for SHgenom.
To obtain a standard for quantitative SHgenom PCR, a 71-bp fragment was amplified from genomic sheep DNA with primers oF-SHgenom and oR-SHgenom before cloning into the pCR 2.1-Topo plasmid vector. The nucleotide sequence was verified, and the plasmid was named pSHgenom. The plasmid was purified, quantitated, and diluted in the same way as pOvHV-2.
Fluorogenic PCRs. (i) OvHV-2.
The 25-μl PCR mixture for one reaction contained 12.5 μl of TaqMan Universal Master Mix (containing AmpliTaq Gold DNA polymerase, AmpErase UNG, deoxynucleoside triphosphates with dUTP, passive reference rhodamine dye [ROX], and optimized buffer components [Perkin-Elmer Europe B. V., Rotkreuz, Switzerland]), 240 nM forward primer oF-OvHV-2, 600 nM reverse primer oR-OvHV-2, 80 nM probe oP-OvHV-2, and 10 μl of diluted standard or template DNA. The thermal cycle protocol used was the following. First, the temperature was held for 2 min at 50°C to allow the uracil-N-glycosylase (UNG) to actively digest previously amplified (contaminating) DNA. During a second hold at 95°C for 10 min, the DNA was denatured, while the AmpliTaq Gold DNA polymerase was activated and the UNG was inactivated. Then, 40 cycles followed, each of which was for 1 min at 60°C and 15 s at 95°C. The amplification was carried out in an ABI PRISM 7700 sequence detector (Perkin-Elmer, Applied Biosystems), and the data were analyzed with the appropriate sequence detector software (version 1.6). The following parameters were set: baseline, 3 to 15; threshold, 0.02.
(ii) SHgenom.
The SHgenom PCR was performed as described above for the OvHV-2 PCR, with the difference being that the PCR mixture contained 240 nM forward primer oF-SHgenom, 600 nM reverse primer oR-SHgenom, and 160 nM probe oP-SHgenom. The following parameters were set: baseline, 3 to 10; threshold, 0.16.
Relative quantitation.
Relative quantitation was performed by applying the standard curve method (User Bulletin 2, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems, 1997). Standard methods (version 5.0; Excel; Microsoft) were used to calculate mean values, standard deviations, coefficients of variation, slope values, and coefficients of determination.
OvHV-2 replication in vivo.
An 8-month-old ram derived from an essentially OvHV-2-free flock was introduced into a known OvHV-2-positive sheep flock. EDTA-treated blood samples were collected weekly for a total period of 39 weeks. DNA was extracted from the buffy-coat cells as described above before being quantitatively examined for their contents of both OvHV-2 DNA and genomic sheep DNA.
RESULTS
Comparison of OvHV-2 fluorogenic PCR to conventional seminested PCR.
In order to establish a fluorogenic PCR for the detection of OvHV-2 DNA, primers and probe were selected as described in Materials and Methods and a total of 152 samples with known results from the previously established conventional seminested PCR were analyzed. Buffy-coat samples from 20 OvHV-2-free sheep reacted negatively by both assays. In contrast, 20 samples obtained from sheep with known exposure to OvHV-2 reacted positively. Similarly, 63 samples from cows suffering from MCF reacted positively by both assays, whereas 46 cow samples which were negative by seminested PCR did not reveal a positive signal by the fluorogenic test. Furthermore, two goat samples and one moose sample which had been found to be positive by seminested PCR showed the same result by the newly developed test. Thus, the results of both PCRs corresponded to each other in 100% of the cases, indicating that the two methods were equally able to discriminate between OvHV-2-positive and -negative animals of different species.
Detection of genomic sheep DNA by fluorogenic PCR.
A fluorogenic PCR for the detection of genomic sheep DNA was established for two reasons. First, the DNAs of samples which had reacted negatively by the OvHV-2 PCR needed to be evaluated for their ability to be amplified (quality of DNA extraction). Second, this PCR was also thought to serve as a basis for relative quantitative analyses. For this purpose, conserved genomic sheep DNA sequences were selected and cloned after amplification. The primers and probe for this fluorogenic PCR were chosen and were found to be able to amplify either the cloned fragment (pSHgenom) or genomic sheep DNA (data not shown). In contrast, genomic bovine DNA or salmon sperm DNA was not amplified. A BLAST search 1 performed with the GenBank database revealed that under the current settings, even in theory, amplification of DNA from organisms other than sheep would be very unlikely. Therefore, this fluorogenic PCR was regarded as specific for ovine genomic DNA.
Analytical sensitivity, linearity, and efficiency.
In order to establish the analytical sensitivity of the fluorogenic OvHV-2 DNA-specific PCR, a 10-fold dilution series of pOvHV-2 (covering the range of between 108 and 10−1 plasmid molecules per reaction mixture) was tested. In four different series, the OvHV-2 fluorogenic PCR was always able to detect 10 or more molecules, and in one series it could detect even 1 molecule of standard pOvHV-2.
In parallel, the fluorogenic PCR for the detection of the sheep genome was evaluated for its sensitivity. For this purpose, three independent 10-fold dilution series of pSHgenom were analyzed. Ten or more molecules were detected in all three series, whereas in two of the series even one molecule gave a positive signal.
A dilution containing 0.1 molecule of either one of the standard plasmids always gave negative results in both systems. It was concluded that the analytical sensitivities of the two fluorogenic PCRs ranged between 1 and 10 target copies.
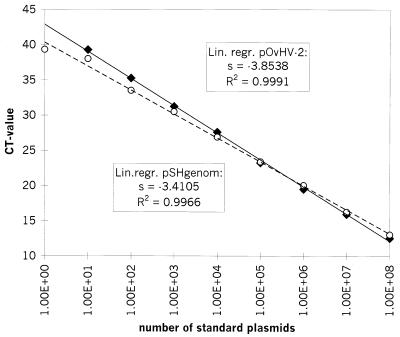
As expected, the cycle number (threshold cycle [CT] value) at which a positive signal could be discriminated from background values decreased in a linear manner with exponentially increasing numbers of DNA templates. The results, summarized in Fig. 1, indicated that in both systems the linearity persisted over the whole range of dilutions. The coefficient of determination for the linear regression of the OvHV-2 template was 0.9991, and that for the, linear regression of the genomic sheep DNA was 0.9966.
FIG. 1.
Amplification of DNA from standard plasmid dilutions and comparison of the amplification efficiencies of pOvHV-2 DNA (the average of four experiments is shown) and pSHgenom DNA (the average of three experiments is shown). The logarithmic value of the number of standard plasmid molecules per reaction mixture is indicated on the x axis, while the corresponding CT values are given on the y axis. The calculated standard deviation values for each point, ranging from 0.01 to 0.5, are not indicated. For both regression curves, the coefficient of determination (R2) and the slope value (s) were calculated and are indicated. Diamonds, pOvHV-2; open circles, pSHgenom. Lin. regr., linear regression.
The absolute value of the slope(s) of the standard curve (logarithm of input standard plasmid number versus CT value) is regarded as a measure of the efficiency of a fluorogenic PCR system. The smaller the absolute slope value, the more efficient the system. To compare the efficiencies of both PCRs, the absolute slope values were calculated. As a result, a slope of 3.8538 was determined for the OvHV-2 system, whereas the slope with genomic sheep DNA amounted to 3.4105 (Fig. 1). Two PCRs are considered evenly efficient only if the difference between the slope values (Δs) of their regression curves is less than 0.1 (User Bulletin 2, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems). In our case, with a Δs of 0.4433 ± 0.0076, the two PCRs were not evenly efficient: the PCR for the detection of genomic sheep DNA was more efficient than the PCR for the detection of OvHV-2 DNA. Consequently, the CT values alone could not be used for relative quantitation (User Bulletin 2, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems).
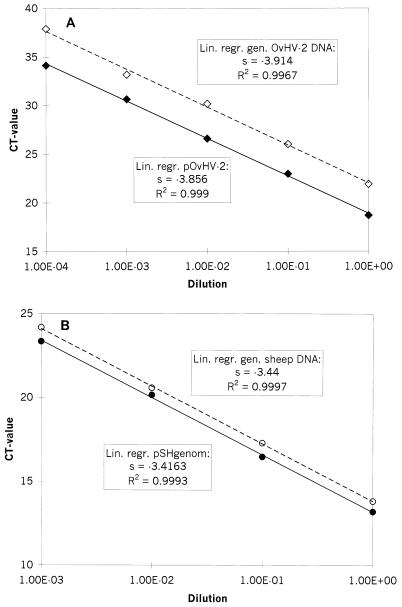
As an alternative way of relative quantitation, the amplification efficiency of each PCR system was first determined for either the plasmid or the genomic DNA template. Provided that the efficiencies with the plasmid or genomic DNA template are sufficiently similar (Δs, ≤0.1), separate standard curves with the plasmid DNA templates can be constructed for each PCR. Those standard curves are then used to calculate the absolute amount of genomic DNA templates for each experimental sample. On this basis, the relative quantitation of viral versus genomic DNA is possible (User Bulletin 2, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems). Therefore, three dilution series were made from each of the following DNA extracts: (i) buffy-coat cells of OvHV-2-positive sheep, representing genomic viral DNA as well as genomic sheep DNA, and (ii) each of the plasmids, representing the cloned counterparts of the genomic DNAs. Then, the slopes of the standard curves were determined and the Δs of cloned OvHV-2 DNA versus that of genomic OvHV-2 DNA (0.058 ± 0.009) and that of cloned OvHV-2 DNA versus that of genomic sheep DNA (0.024 ± 0.016) were calculated. In both cases the Δs was less than 0.1, which allowed these two standard curves for plasmids to be used for absolute quantitation (Fig. 2A and B).
FIG. 2.
Comparison of amplification efficiencies of genomic DNA and plasmid-cloned DNA. Average values of three experiments are shown. The calculated standard deviations for the CT values of each point, ranging from 0.03 to 0.66, are not indicated. Tenfold standard dilutions of DNA prior to amplification were used, as indicated on the x axis, whereas the corresponding CT values are presented on the y axis. The coefficients of determination (R2) and the slope values (s) of the regression curves were calculated and are indicated.(A) Open diamonds, genomic OvHV-2 DNA; black diamonds, pOvHV-2 DNA. (B) Open circles, genomic sheep DNA; black circles, pSHgenom DNA. lin. regr., linear regression; gen., genomic.
To normalize the quantity of virus in terms of the amount of reference genomic DNA in consecutive samples from a sheep, the absolute OvHV-2 DNA copy number in each sample per average DNA copy number of the sheep genome in the different samples of this animal was calculated.
Reproducibilities of the two fluorogenic PCRs.
To determine the reproducibilities of the fluorogenic PCRs, 10-fold dilutions of the standard plasmids were repeatedly tested. The chosen amounts (102 to 105 molecules of pOvHV-2 and 105 to 108 molecules of pSHgenom per reaction mixture) corresponded to the numbers which are normally measured in experimental samples.
Ten separate runs were analyzed for each dilution of pOvHV-2 DNA, and 15 separate runs were analyzed for each dilution of pSHgenom DNA. From the resulting values, the coefficients of variation (CV) were calculated once on the basis of the CT value (CVCT) and once on the basis of the absolute values of the initial copy numbers (CVabs).
The results are shown in Table 2. In the case of pOvHV-2, the CVCT was in the range of 0.41 to 1.03% and the CVabs was in the range of 6.57 to 21.17%. The CVCT of pSHgenom was found to be between 0.43 and 0.6%, and the CVabs of pSHgenom was found to be between 5.2 and 7.12%. Thus, while some variations were recognized with very small numbers of template copies, a surprising overall constancy of the results was noted.
TABLE 2.
CVs for pOvHV-2 and pSHgenoma
| Template | Amount of template | CVCT (%) | CVabs (%) |
|---|---|---|---|
| pOvHV-2 | 102 | 1.03 | 21.17 |
| 103 | 0.63 | 10.36 | |
| 104 | 0.41 | 5.79 | |
| 105 | 0.55 | 6.57 | |
| pSHgenom | 105 | 0.43 | 6.67 |
| 106 | 0.52 | 7.12 | |
| 107 | 0.54 | 5.96 | |
| 108 | 0.60 | 5.20 |
CVs were calculated on the basis of the CVCT as well as CVabs.
Example for relative quantitation of OvHV-2.
The established quantitative PCR was then applied to observe the replication of OvHV-2 in vivo. For this purpose, a specific MCF-negative sheep was introduced into a flock of MCF-positive sheep.
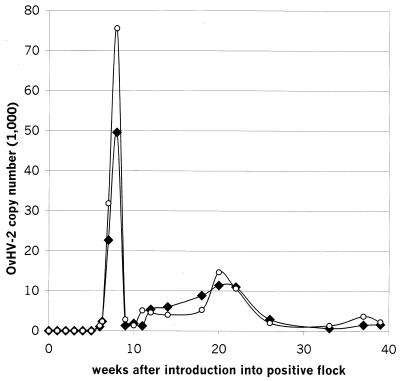
Once a week, blood samples were collected and the buffy-coat cells were examined for their absolute content of OvHV-2 DNA as well as the amount of OvHV-2 DNA in relation to the amount of genomic sheep DNA. The results are presented in Fig. 3. The first positive signal for OvHV-2 DNA was detected 6 weeks after introduction of the animal into the positive flock. At week 8, a maximum of 50,000 copies of OvHV-2 DNA per ml of blood was reached. The titer then dropped rapidly and undulated thereafter between 1,000 and 8,000 copies per ml until a second, smaller peak was observed between weeks 20 and 22 (11,000 copies). Afterward, until the end of the observation period at 39 weeks after introduction of the OvHV-2-negative sheep, the titer remained low, between 1,000 and 3,000 copies per ml.
FIG. 3.
Course of absolute and relative copy numbers of OvHV-2 DNA in the buffy-coat cells of 1 ml of EDTA-treated blood of a sheep after introduction of the sheep into an OvHV-2-positive flock. Once per week, blood samples were collected and the buffy-coat cells were examined for their absolute content of OvHV-2 DNA as well as the amount of OvHV-2 DNA in relation to the amount of genomic sheep DNA. The time (weeks after introduction into the positive flock) is indicated on the x axis. The absolute (black diamonds) or relative (open circles) number of amplifiable OvHV-2 DNA molecules per milliliter of EDTA-treated blood is given on the y axis.
When the course of OvHV-2 DNA was expressed as a function of the genomic sheep DNA signal, the same two peaks were evident. However, during peak replication of OvHV-2 about one and a half times as much viral DNA was measured relative to the amount of cellular genomic DNA measured. This observation indicated that active virus replication took place between weeks 6 and 8 as well from weeks 20 through 22.
DISCUSSION
This report describes the establishment of a fluorogenic PCR for detection and quantitation of OvHV-2 DNA. The new method was then applied to study the course of a natural OvHV-2 infection in sheep.
Upon comparison of the new method with the previously established conventional seminested OvHV-2 DNA-specific PCR, the results obtained corresponded to each other without a single exception. Thus, the new PCR was considered equally suitable for detection of OvHV-2 DNA. This interpretation is supported by previous reports, which found strong conservation between the targeted OvHV-2 sequence and the corresponding DNA sequences of other gamma herpesviruses 2, 5. Nevertheless, others have reported that there are OvHV-2-related viruses which are genetically distinct and which may not be amplified by the system described in this report 8.
Hua et al. 6 have reported on quantitative OvHV-2 DNA-specific PCRs which made use of either an endpoint dilution of the sample or the coamplification of a competitive internal standard together with the viral sequences. Both quantitative tests needed parallel reactions of standard plasmid dilutions and required time-consuming postamplification steps. Such processes might increase the risk of contamination.
To circumvent those difficulties, we wished to establish a fast, simple, and reliable fluorogenic PCR to quantitate OvHV-2 DNA. In this assay, the cycle number at which a positive signal can be discriminated from background values is termed the CT value and correlates with the amount of target templates in the initial reaction mixture. The CT values of standard plasmid dilutions allow calculation of the quantity of template molecules in a specific sample (User's Manual, ABI PRISM 7700 Sequence Detection System, Perkin-Elmer Applied Biosystems).
DNA extracted from OvHV-2-infected tissue will contain both viral DNA and cellular DNA. Amplification of cellular DNA may be used to evaluate the quality of DNA extraction from samples and provide reassurance throughout the tests that no nonspecific inhibitors have interfered with the amplification of the target DNA. Furthermore, the amount of cellular DNA in such a sample may serve as a measure of the number of cells from which DNA was extracted. Quantitation of the cellular DNA template may consequently be used to normalize the quantity of viral DNA in the sample. In order to compensate for variations in the efficiency of DNA extraction, such a normalization may be required, for example, if the course of an infection is studied in an individual animal. For this reason, a second, sheep genome-specific quantitative fluorogenic PCR was established. In a next series of experiments the analytical sensitivities, linearities, and efficiencies of the two PCRs were determined. Previously, Hua et al. 6 reported a sensitivity of 30 target copies for their assay and the linearity persisted over 4 log units. Our results indicated that the analytical sensitivity of the two fluorogenic PCRs ranged between 1 and 10 target copies. In both systems, the linearity persisted over the whole range of dilutions (at least 7 log units). To improve the analytical sensitivity (and to obtain linearity of the standard curves even at the highest plasmid dilutions), a constant amount of carrier DNA (salmon sperm DNA at 60 μg/ml) was used to prevent adsorption of the plasmids to the tube walls 7. In the case of the OvHV-2 system, the sensitivity of the pOvHV-2 dilution series with carrier DNA was 10 times greater than that of the dilution series without carrier DNA (data not shown). However, our two PCRs were not evenly efficient, and comparison of the CT values alone could not be used for relative quantitation. Therefore, separate standard curves had to be constructed for each PCR system. For each experimental sample, the absolute quantity of viral DNA as well as that of endogenous genomic reference DNA was determined from the appropriate standard curve, and then these quantities were evaluated in relation to each other. Plasmid-cloned OvHV-2 DNA was amplified with the same efficiency as genomic viral DNA. The same was true for plasmid-cloned DNA and genomic sheep DNA. Hence, plasmid-cloned templates could be used for the construction of the standard curves, which are required for quantitation. Since slight variations from run to run are possible (Table 2), it was important to construct the standard curves anew in every run.
By using the established quantitative PCR, it was possible (to our knowledge, for the first time) to witness quantitative replication of OvHV-2 in vivo. Similar results were obtained with several animals, although just one example is described in this report. In the course of 39 weeks, at least two peaks of viral replication were noticed. The first peak occurred at 8 weeks after the introduction of the OvHV-2-free animal into a conventional OvHV-2-positive flock; the second, smaller peak was observed between weeks 20 and 22. Thus, it seemed as if viral replication was curbed by an active immune response. However, the virus was not entirely eliminated from the host, and as expected for a herpesvirus, reactivation seemed to occur. Indeed, similar observations have been made with the wildebeest-associated MCF agent AlcHV-1, but that observation was described almost 30 years ago 4, 13. Thus, in the absence of appropriate cell cultures, which are able to support replication of OvHV-2 in vitro, it took such a long period of time to establish the appropriate molecular biological methods for the tracing of OvHV-2 in vivo.
ACKNOWLEDGMENT
This work was supported by the Swiss Federal Veterinary Office.
REFERENCES
- 1.Altschul S F, Boguski M S, Gish W, Wootton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 2.Baxter S I, Pow I, Bridgen A, Reid H W. PCR detection of the sheep-associated agent of malignant catarrhal fever. Arch Virol. 1993;132:145–159. doi: 10.1007/BF01309849. [DOI] [PubMed] [Google Scholar]
- 3.Bridgen A, Reid H W. Derivation of a DNA clone corresponding to the viral agent of sheep-associated malignant catarrhal fever. Res Vet Sci. 1991;50:38–44. doi: 10.1016/0034-5288(91)90051-o. [DOI] [PubMed] [Google Scholar]
- 4.Castro A E, Ramsay E C, Dotson J F, Schramke M L, Kocan A A, Whitenack D L. Characteristics of the herpesvirus of malignant catarrhal fever isolated from captive wildebeest calves. Am J Vet Res. 1984;45:409–415. [PubMed] [Google Scholar]
- 5.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hua Y, Li H, Crawford T B. Quantitation of sheep-associated malignant catarrhal fever viral DNA by competitive polymerase chain reaction. J Vet Diagn Investig. 1999;11:117–121. doi: 10.1177/104063879901100202. [DOI] [PubMed] [Google Scholar]
- 7.Leutenegger C M, Klein D, Hofmann-Lehmann R, Mislin C, Hummel U, Böni J, Boretti F, Guenzburg W H, Lutz H. Rapid feline immunodeficiency virus provirus quantitation by polymerase chain reaction using the TaqMan® fluorogenic real-time detection system. J Virol Methods. 1999;78:105–116. doi: 10.1016/s0166-0934(98)00166-9. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Dyer N, Keller J, Crawford T B. Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer. J Clin Microbiol. 2000;38:1313–1318. doi: 10.1128/jcm.38.4.1313-1318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel A L, Aspeling I A. Evidence of persistent malignant catarrhal fever infection in a cow obtained by nucleic acid hybridisation. J S Afr Vet Assoc. 1994;65:26–27. [PubMed] [Google Scholar]
- 10.Milne E M, Reid H W. Recovery of a cow from malignant catarrhal fever. Vet Rec. 1990;126:140–141. [PubMed] [Google Scholar]
- 11.Müller-Doblies U U, Li H, Hauser B, Adler H, Ackermann M. Field validation of laboratory tests for the clinical diagnosis of sheep-associated malignant catarrhal fever. J Clin Microbiol. 1998;36:2970–2972. doi: 10.1128/jcm.36.10.2970-2972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Toole D, Li H, Miller D, Williams W R, Crawford T B. Chronic and recovered cases of sheep-associated malignant catarrhal fever in cattle. Vet Rec. 1997;140:519–524. doi: 10.1136/vr.140.20.519. [DOI] [PubMed] [Google Scholar]
- 13.Reid H W, Rowe L. The attenuation of a herpes virus (malignant catarrhal fever virus) isolated from hartebeest. Res Vet Sci. 1973;15:144–146. [PubMed] [Google Scholar]