ABSTRACT
Laetiporus sulphureus mushroom is a complementary and alternative medicine that has anticancer, antioxidation, and analgesic effects and immunomodulatory activity; it is used as a treatment for cough and rheumatism and is a functional food that can improve physical fitness. Even though L. sulphureus has garnered considerable biotechnological and pharmacological interest due to its excellent cellulose-degrading ability and diverse biological activities, its biosynthetic potential regarding polysaccharides and secondary metabolites has not been thoroughly examined. In this study, we sequenced and assembled the whole genome of a wild L. sulphureus isolate, NWAFU-1, from the Qinling Mountains in China. Comparative genomes analysis revealed genomic differences between subspecies, and phylogenomic analysis revealed evolutionary divergence as well as genome expansion and contraction of individual Polyporaceae family species. Bioinformatics investigation identified candidate genes associated with mating type, polysaccharide synthesis, carbohydrate-active enzymes, and secondary-metabolite biosynthesis, which included multiple terpenoids, nonribosomal peptides, and polyketides. The locations of biosynthetic core genes were mapped and displayed on chromosomes and contigs. Totals of 143 proteins from 126 coding genes were identified and divided into 14 cytochrome P450 families. Furthermore, the biosynthetic network of tetracyclic triterpenoid active components was postulated by genome mining of related genes combined with the molecular network of metabolites. The genome analysis of L. sulphureus in this study improves the understanding of the biosynthesis of active compounds, which will lay a theoretical foundation for subsequent research on active-compound biosynthesis and promote the application of Laetiporus in the field of drug research and functional-food creation.
IMPORTANCE L. sulphureus is a parasitic basidiomycete fungus that causes brown rot. The fruiting bodies of L. sulphureus are used as ancient medicines in China and Europe to cure cancer, analgesia, cough, and rheumatism and are considered a functional food that regulates the body and improves health. L. sulphureus was inferred to be a tetrapolar system based on a high-quality genome, which will aid molecular breeding and artificial farming. Screening polysaccharide synthesis candidate genes and comparing carbohydrate-associated genes in brown-rot basidiomycetes help understand their growth. Identifying core genes for secondary-metabolite biosynthesis, gene cluster family analysis, and comparative cluster analysis will guide heterologous-biosynthesis investigations of these genes and help elucidate the biosynthetic pathways for L. sulphureus bioactive natural components. The biosynthesis network of tetracyclic triterpenes was mapped using metabolite profiling and genome scanning. This work explores the biosynthetic capacity of L. sulphureus-derived natural products and lays the foundation for biosynthetic studies of them.
KEYWORDS: Laetiporus sulphureus, pharmaceutical edible mushroom, near-chromosome level assembly, secondary metabolite, biosynthetic potential
INTRODUCTION
Higher fungi, mainly belonging to the phylum Basidiomycota, are commonly called mushrooms due to their ability to form fruiting bodies. Mushrooms have been extensively used in human life for thousands of years due to their unique edible and/or medicinal properties (1, 2). Mushrooms are one of the healthiest food groups in the world, and approximately 50% of edible mushrooms are considered functional foods, making them recognized as superfoods (3). The genus Laetiporus Murrill was first established by Murrill in 1904 (4), and the genus belongs to the order Polyporaceae of the class Agaricomycetes (Basidiomycota) (5). A multigene phylogenetic analysis revealed at least 17 species of the genus Laetiporus (6). The fungi of the genus Laetiporus Murrill are both highly damaging forest pathogens, parasitic on 20 species of living trees worldwide (7), including Quercus and Eucalyptus (8), and brown-rot type wood decay basidiomycetes (9), which are essential to nature’s carbon cycle due to their highly specialized biomass degradation capacity (10).
Laetiporus sulphureus (Bull.) Murrill is a model strain of the genus Laetiporus (4), which has received figurative and trivial names such as “chicken mushroom” or “wood chicken” for the shape, size, and color of its fruiting bodies (11). As wood-inhabiting macrofungi with high medicinal and edible value, the fruiting bodies of Laetiporus sulphureus have a long tradition of consumption in Europe, America, and China (12), and their medicinal uses are documented in the ancient books of traditional Chinese medicine. The bioactive compounds and extracts of L. sulphureus and their benefits for human health have been thoroughly investigated in recent years. The triterpenoids, represented by eburicoic acid, are well-known bioactive metabolites from L. sulphureus, with significant anticancer, cytotoxic, and anti-immune activities (11, 13). Several sesquiterpenoids from L. sulphureus also exhibit anticancer activity (14, 15). Antibacterial and antioxidative properties are one of the distinguishing features of Laetiporus sulphureus metabolites (16–19). Laetiporic acids derived from L. sulphureus exhibit remarkable antifungal activity against the protoplasts of Aspergillus (20) and are a group of nonisoprene polyenes containing multiple conjugated double bonds and natural dyes with food colorant potential (21, 22). In addition, the crude polysaccharides from Laetiporus sulphureus exhibited antidiabetic effects by increasing the proliferation and insulin secretion function of rat insulinoma RINm5F cells (23). In addition, the fruit body of L. sulphureus can be used as an inexpensive and safe biomass due to its high content of α-(1→3)-glucans, which have been artificially cultivated (24).
The rapid advances in genome sequencing technologies have led to investigations of the medicinal active ingredients of wood-dwelling macrofungi and their biosynthesis increasingly relying on genome sequences rather than just on the metabolites themselves. The resolution of genomic information on wood-dwelling macrofungi will help to understand their life cycle, mating types, and nutritional models. To date, the genomes of several valuable medicinal (edible) fungi, including Ganoderma lucidum (25, 26), Inonotus obliquus (27), Hericium erinaceus (28), Naematelia aurantialba (29), Antrodia camphorata (30), and Sanghuangporus sanghuang (31), have been deciphered, thereby further facilitating their medicinal utilization and industrial development. L. sulphureus is a wood-dwelling-type medical and edible fungus that has attracted much attention, although the genomes of its five subspecies (10, 32–34) have been sequenced and used to analyze the lignocellulosic biomass degradation characteristics (10, 33). However, genome mining for secondary-metabolite biosynthesis is rarely reported. This work presents the near-chromosomal-level genome assembly of a wild L. sulphureus strain from the Qinling Mountains (a famous mountain system in China). Comparative genomes reflect genomic differences among subspecies, and phylogenomic analysis reveals the timing of evolutionary divergence, genome expansion, and contraction of L. sulphureus and well-known species of Basidiomycota. Functional genes related to mating type, polysaccharide synthesis, and carbohydrate-active enzymes (CAZymes) were subjected to in silico analysis. Genome mining combined with metabolite profiling identified gene clusters involved in secondary-metabolite biosynthesis. A family classification analysis of cytochrome P450 enzymes associated with primary metabolism and postsecondary metabolic modifications was performed. The biosynthesis of tetracyclic triterpenes was systematically speculated based on the molecular network of metabolites. This investigation sheds light on the biosynthesis of active ingredients derived from L. sulphureus, which, in turn, supports their synthetic biology production and in-depth pharmacological studies.
RESULTS
Fungal strain, strain culture, and species identity.
Wild fruiting bodies (Fig. 1A) were collected from the Qinling Mountains in Ankang City, Shaanxi Province, China, and were parasitic on the trunk of a living Toxicodendron vernicifluum (Stokes) F. A. Barkl tree. The culturable mycelium (Fig. 1B) was obtained by tissue separation of the fruiting body, which was able to grow vigorously on wheat bran and wood chips (Fig. 1C). Internal transcribed spacer (ITS) sequence alignment showed that the sample was 99.83% identical to L. sulphureus var. miniatus strain NAAS04758 (see Fig. S1 in the supplemental material). This sample was identified as L. sulphureus by combining ITS analysis and morphological characteristics of the fruiting bodies; the sample was named L. sulphureus NWAFU-1. Considering its rare food and medicine value and strain resources, L. sulphureus NWAFU-1 was subjected to genome sequencing.
FIG 1.
Morphologic photographs of strain L. sulphureus NWAFU-1: wild fruiting bodies (A), mycelium growing on PDA for 3 days (B), and mycelium growing on wood chips (left) and wheat bran (right) (C).
Genome sequence assembly and annotation.
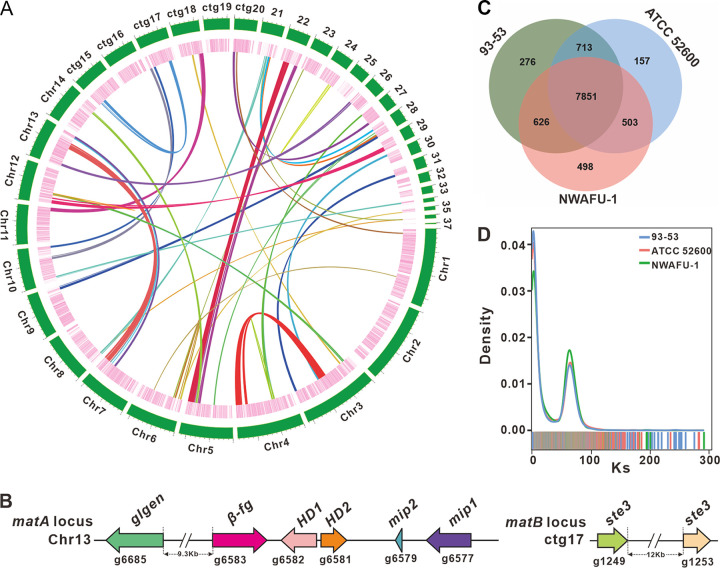
Totals of 11, 943, 393, 600 bases from 79, 622, 624 clean reads were assembled into the 48.23-Mb genome (Fig. 2A), which consisted of 14 chromosomal pseudomolecules and 23 contigs with an N50 of 1,791,120 bp and a 51.68% GC content (Tables S1 and S2). A k-mer analysis of reads showed two main peaks with similar heights, and the abscissa value of the former was half of the latter (Fig. S2). This result indicated that strain NWAFU-1 was a highly heterozygous species. Although the 99.81% coverage (Table S3) partly indicated that the genome of strain NWAFU-1 was well assembled, the BUSCO value was only 93.3% (Table S4). Sequencing, assembly parameters, and quality metrics comparisons between NWAFU-1 and reported L. sulphureus genomes further highlight the high-quality genome of strain NWAFU-1 (Table 1).
FIG 2.
Genomic characterization, mating-type loci, and comparative genomic analysis. (A) Characteristics of the de novo assembly genomic features of L. sulphureus NWAFU-1. From the outside to the inside are chromosome and contigs, gene density (the intensity of the color positively correlates with gene density), and whole-genome collinearity analysis based on protein-coding genes (sequence similarity from low to high is indicated by yellow to red). (B) Structural diagram of the genes on the matA locus and matB locus. (C) Venn schematic of comparative genomes within L. sulphureus strains. (D) Ks comparison within L. sulphureus species.
TABLE 1.
Comparison of sequencing and assembly metrics, and genome quality of L. sulphureus
| Parameter | NWAFU-1 | 93-53 | ATCC 52600 | Murrill, 1920 | MG138 |
|---|---|---|---|---|---|
| Sequencing technology | Illumina NovaSeq 6000 | Illumina HiSeq 4000 | Illumina HiSeq 2500 | PacBio Sequel II HiFi Illumina NovaSeq 6000 |
Illumina HiSeq 4000 |
| Sequencing depth | 214.04× | 85.2× | 125.0× | 106× | 90× |
| No. of scaffolds | 37 | 399 | 785 | 31 | 11,961 |
| Total assembly length (bp) | 48,323,753 | 39,909,705 | 43,372,605 | 37,410,216 | 51,778,905 |
| Largest length (bp) | 3,376,063 | 2,261,102 | 1,372,164 | 4,563,458 | 151,512 |
| Scaffold N50 (bp) | 1,791,120 | 522,580 | 211,056 | 2,582,225 | 14,997 |
| Scaffold L50 | 10 | 22 | 53 | 6 | 862 |
| GC content (%) | 51.68 | 51.4 | 51.22 | 51.7 | NAa |
| BUSCO completeness (%) | 93.3 | NA | NA | 95.9 | 85.90 |
| No. of predicted genes | 15,302 | 13,774 | 12,802 | NA | 22,479 |
| GenBank accession no. | GCA_024321985.1 | GCA_001632365 | GCA_016068325 | GCA_927399515 | GCA_003521245 |
| Isolate information | Mycelium | NA | Mycelium | Mycelium | Mycelium |
| Reference | This study | 33 | 10 | 34 | 32 |
NA, not available.
There were 15,302 protein-coding genes predicted, with an average gene length of 1,272.37 bp and a total of 111,297 exons (average length, 174.94 bp) and 95,995 introns (average length, 74.68 bp) in these coding genes (Table S5). For noncoding RNA (ncRNA), 81 tRNAs, 50 rRNAs, 19 snRNAs, and 1 sRNA were predicted (Table S6). A total of 16,663 repeats with a total length of 9,067,448 bp was predicted, accounting for 18.76% of the entire genome, with the four scattered repeats short interspersed nuclear elements (SINE), long interspersed nuclear elements (LINE), long terminal repeats (LTR), and DNA transposons accounting for 0.00% (4), 0.26% (257), 4.93% (1,835), and 1.51% (1,024), respectively (Table S7).
To archive comprehensive protein-coding genes function annotation, 15,302 genes were subjected to sequence similarity analysis and motif similarity search based on nine public databases (Nr, Pfam, eggCOG, UniProt, KEGG, GO, Pathway, RefSeq, and Interproscan) (Table S8). The results of the Nr library annotation found that 94.14% of the 13,307 annotated genes matched the genome of L. sulphureus 93-53 (Fig. S3), indicating that NWAFU-1 is definitely of the L. sulphureus species. Cellular components constituted the main group among the 6,689 genes annotated by the functional classification of the GO database (Fig. S4). Functional annotation based on the COG database identified 1,350 genes, of which 154 belonged to the J group (translation, ribosomal structure, and biogenesis) (Fig. S5). According to the KEGG database, 5,010 genes were identified as being involved in 5 types of pathways, with the largest number of genes involved in metabolic pathways (Fig. S6). A domain-based motif search using the Pfam database identified 9,523 genes, and the top 20 types with the largest number are shown in Fig. S7. These various perspectives and levels of annotation demonstrate the functional diversity of protein-coding genes from strain NWAFU-1.
Identification of the mating genes.
In the sexual development of mushroom-forming fungi, mating is an essential step. Genes associated with mating are in charge of directing the process. Mating-type (MAT) loci are located in distinct genomic regions that control mating (35). Heterozygous cooperation accounts for 90% of the sexual reproduction of basidiomycetes (36), which can be divided into bipolar and tetrapolar mating types. Among them, the tetrapolar mating-type system is by far the most extensive and complex sexual reproduction control system in Basidiomycetes, and its proportion is also the highest (37). Although L. sulphureus is a higher fungus of great interest, its mating type has not been identified.
In this study, the MAT-A locus was located on chromosome 14 (Chr13) by homology search with the mitochondrial intermediate peptidase (mip) codon gene and the glycosyltransferase (GT) family 8 protein codon gene (glgen) of four famous mushrooms, including Taiwanofungus camphoratus (Fig. S8A), and the MAT-B locus was located on contig 17 (ctg17) by scanning with the pheromone receptor genes (ste3) as a probe (Fig. S8B). The MAT-A locus contains two mip genes (g6577 and g6579), two homeodomain transcription factor codon genes (HD1 and HD2 [g6582 and g6581]), an unknown conserved fungal protein codon gene (β-fg [g6583]), and a glgen gene (g6685) about 9.3 kb from the β-fg gene. The MAT-B locus contains at least two unclustered ste3 genes (g1249 and g1253) (Fig. 2B and Table S9). The analysis finding that the MAT-A locus and the MAT-B locus are not in the same contig suggests that the mating type of L. sulphureus has a tetrapolar mating system. Overall, further research is needed to better understand the genomic structure of the mating-type loci in L. sulphureus.
Comparative genomic analysis within the L. sulphureus species.
The genome size of the strain NWAFU-1 is much larger than those of strains 95-53 and ATCC 52600, and accordingly, the number of protein genes encoded in NWAFU-1 is much larger than those in these two strains (Table 1). A total of 7,851 orthologous groups were identified from the three species of Laetiporus, and NWAFU-1 contained more unique orthologous groups (498) than did 93-53 (276) and ATCC 52600 (157). NWAFU-1 shared more orthologous groups with 93-53 (626) than ATCC 52600 (503), and the number of orthologous groups shared between the last two (713) was greater than the number they shared with NWAFU-1. This finding suggests that the similarity between 93-53 and ATCC 52600 is greater than the similarity of each of them to NWAFU-1 (Fig. 2C).
To further understand the differences in the genomes of the strains of L. sulphureus, a genome-wide duplication analysis based on synonymous mutation rates was performed. The consistent trends in the synonymous substitution Ks curves of these strains revealed that they are all L. sulphureus species uniform (Fig. 2D). The obvious peaks in the Ks curves suggested that genome doubling events occurred during the genomic evolution of the species L. sulphureus (Fig. 2D), as evidenced by genome-wide collinearity analysis of NWAFU-1 (Fig. 2A). The higher Ks peaks of NWAFU-1 indicated that this strain has undergone a larger-scale genome doubling event (Fig. 2D), resulting in the larger genome of this strain (Table 1).
Phylogenomic and evolutionary analysis of L. sulphureus.
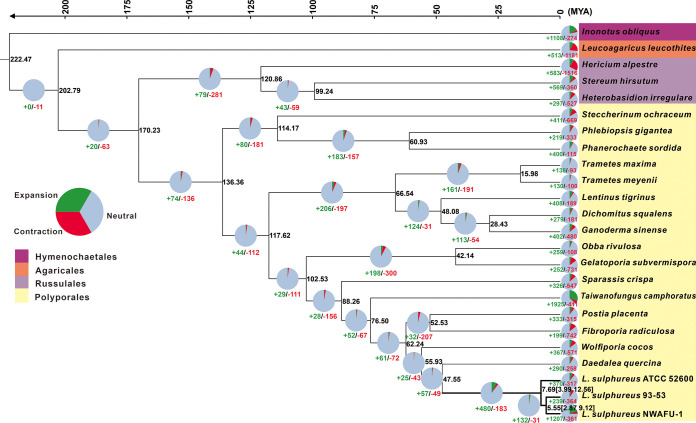
The phylogenomic tree inferred from an alignment of 579 single-copy orthologous genes from 264,508 proteins delimited phylogenetic relationships among 24 species, including three strains of L. sulphureus, with full bootstrap support (Fig. 3). The mean divergence time of the Hymenochaete outgroup represented by Inonotus obliquus is estimated to be 222.47 million years ago (MYA), while those of the Agaricales and Russulales are estimated to be 202.79 and 170.23 MYA, respectively. The divergence time between L. sulphureus and Daedalea quercina is estimated at 47.56 MYA. L. sulphureus was estimated to emerge in a mean crown age of 7.69 MYA, with a 95% highest posterior density (HPD) of 3.99 to 12.56 MYA, while the divergence time between subspecies 93-53 and NWAFU-1 was 5.55 MYA, with a 95% HPD of 2.87 to 9.12 MYA (Fig. 3). Besides three strains in L. sulphureus, other well-recognized medicinal fungal species from Polyporales, such as Ganoderma sinense, T. camphoratus, and Wolfiporia cocos, were also phylogenetically separated (Fig. 3).
FIG 3.
Maximum clade credibility tree inferred from 1,702 single-copy orthologous genes. All nodes received full bootstrap support. The divergence time is labeled as the mean crown age for each node, while the 95% highest posterior density is also given within the Laetiporus clade. The black numbers at the branches indicate the corresponding divergence times in MYA. The numbers of gene family expansion and contraction in each species are labeled with green and red symbols, respectively. The proportion of expansion, contraction, and neutrality in the genome of each species is displayed before the species name. The color of the background of each species indicates its corresponding order.
The gene family contraction occurred more frequently than the gene family expansion in the evolutionary process of the 24 fungal species studied (Fig. 3). In the case of L. sulphureus, 370, 239, and 1,207 gene families had expanded in the strains ATCC 52600, 93-53, and NWAFU-1, corresponding to 317, 364, and 361 genes being contracted, respectively. Among these gene families, 122 (50 expanded and 72 constricted), 110 (40 expanded and 70 constricted), and 181 (119 expanded and 62 contracted) in ATCC 52600, 93-53, and NWAFU-1, respectively, have undergone significant evolution (Fig. 3).
CAZyme analysis and synthesis of polysaccharides.
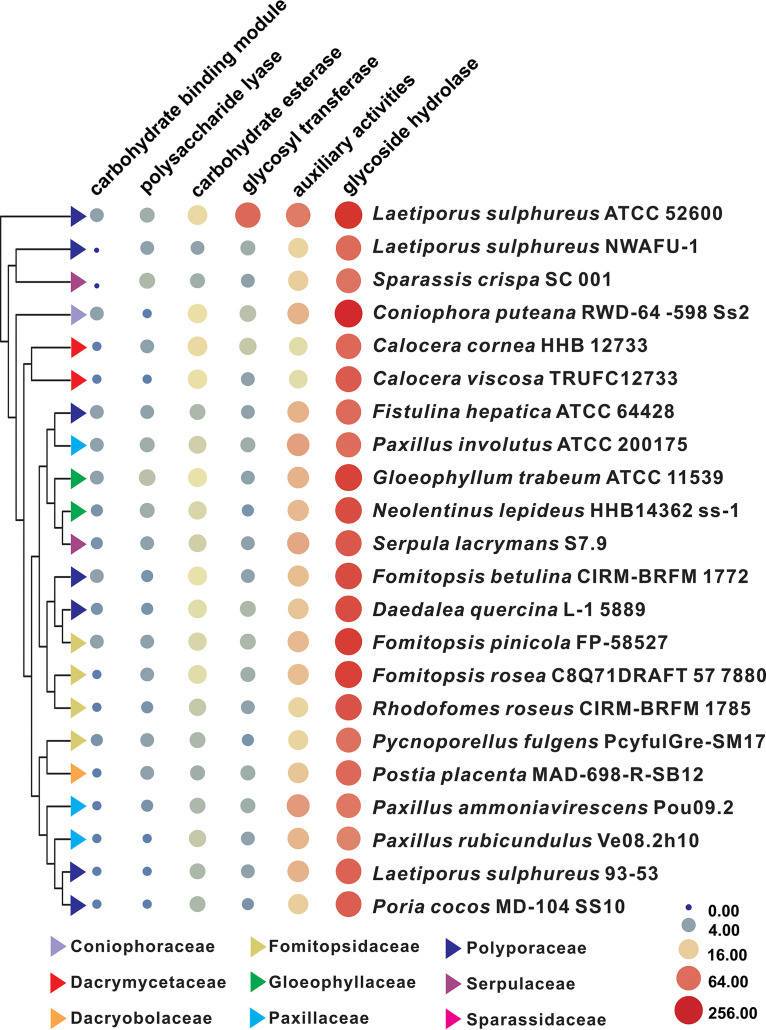
In spite of the fact that the carbohydrate-active enzymes (CAZymes) of L. sulphureus ATCC 52600 have been identified using multiomics approaches (10), the differences in the genome piqued our interest in the CAZymes of NWAFU-1. There were 90 genes encoding CAZymes, including 66 glycoside hydrolases (GHs), 14 auxiliary activities (AAs), 4 GTs, 3 glycoesterases (CEs), and 3 polysaccharide lyases (PLs), but no carbohydrate-binding module (CBM) was found. The GH members g12742.t1 and g2925.t1 are of interest because they both contain two HG13 structural domains. Among these six classes of genes, the number of GH genes is much higher than those of the others, and the GHs encoded by most genes are mainly involved in the degradation of pectin (GH28), cellulose (GH5), chitin (GH18), hemicellulose (GH10 and GH43), and starch (GH13 and GH31) (Table S10). The CAZyme spectra of L. sulphureus and 19 typical brown-rot basidiomycetes were compared (Data Set S1), and it was discovered that the numbers and types of CAZymes in these strains were not species specific. The strain with the characteristics of CAZymes most similar to those NWAFU-1 was Sparassis crispa SC 001, rather than strain 93-53 or ATCC 52600 (Fig. 4).
FIG 4.
Bubble plot of CAZyme analysis of L. sulphureus and related brown-rot basidiomycetes. The sizes and colors (from blue through buff to red) of circles indicate the change in quantity, and the different colored triangles indicate different families.
Exopolysaccharides (EPS), intracellular polysaccharides (IPS), and other polysaccharides have been found in the fruiting bodies (38) and fermentation broth of mycelium (23, 39) of L. sulphureus. Approximately 20 different enzymes involved in mushroom polysaccharide synthesis have been identified (40–42), including 1,3-glucan synthase (GLS), glucose phosphomutase (PGM), phosphomannose isomerase (PMI), glucokinase (GK), beta-glucan synthesis-associated protein (GSAP), phosphoglucose isomerase (PGI), UDP-glucose 4-epimerase (UGE), GDP-mannose dehydratase (GMD), phosphofructokinase (FPK), and UDP-xylose synthase (UXS). With the aid of the genes involved in the polysaccharide synthesis pathway of Basidiomycetes, 23 polysaccharide biosynthetic candidate genes were identified, including 5 GSAPs, 4 GMDs, 3 UGEs, and 2 GKs, PGMs, and GLSs, as well as only 1 FPK, PGI, and UXS (Table S11). The number of genes involved in polysaccharide biosynthesis possessed by L. sulphureus is similar to that in other medicinal and edible mushrooms such as H. erinaceus (40).
Secondary-metabolite biosynthetic gene cluster analysis.
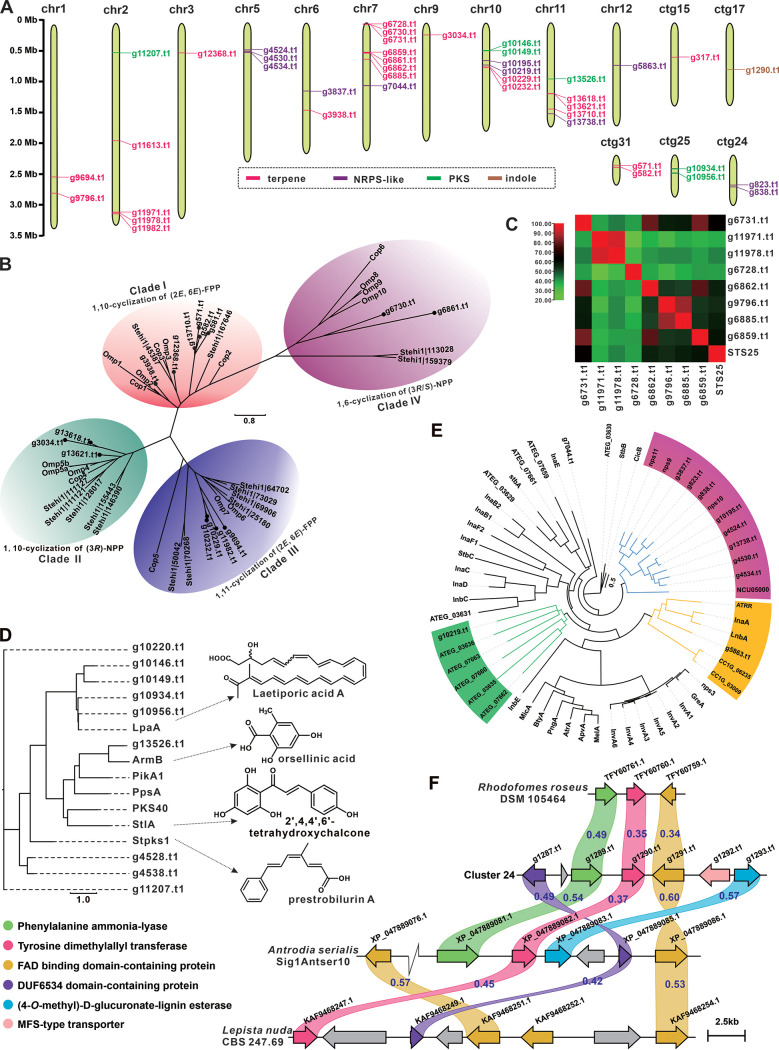
In view of the long medicinal history and significant medicinal value of L. sulphureus, we analyzed the biosynthetic potential of its secondary metabolites. The genome of NWAFU-1 was predicted using the webtool antiSMASH, and 29 secondary-metabolite biosynthetic gene clusters (BGCs) containing 46 core genes were discovered (Table 2). The core genes, consisting of genes for 25 terpene synthases, 12 nonribosomal peptide synthase enzymes (NRPS-like enzymes), 8 polyketide synthases (PKSs), and 1 indole-related enzyme (Table 2) are distributed on 10 chromosomes (Chr1, -2, -3, -5, -6, -7, -9, -10, -11, and -12) and 5 contigs (ctg15, -17, -24, -25, and -31) (Fig. 5A).
TABLE 2.
Putative BGCs responsible for secondary metabolites in strain NWAFU-1
| Cluster | Location | Start (bp) | Stop (bp) | Core gene ID | Core gene type |
|---|---|---|---|---|---|
| 1 | ctg1 | 2531802 | 2548056 | g9694.t1 | Terpene |
| 2 | ctg1 | 2813581 | 2835359 | g9796.t1 | Terpene |
| 3 | ctg2 | 453678 | 497593 | g11207.t1 | PKS |
| 4 | ctg2 | 1927623 | 1943625 | g11613.t1 | Terpene |
| 5 | ctg2 | 3109735 | 3161835 | g11971.t1 | Terpene |
| g11978.t1 | |||||
| g11982.t1 | |||||
| 6 | ctg3 | 476385 | 497617 | g12368.t1 | Terpene |
| 7 | ctg5 | 428286 | 532709 | g4524.t1 | NRPS-like |
| g4528.t1 | PKS | ||||
| g4530.t1 | NRPS-like | ||||
| g4534.t1 | NRPS-like | ||||
| g4538.t1 | PKS | ||||
| 8 | ctg6 | 1102537 | 1147198 | g3837.t1 | NRPS-like |
| 9 | ctg6 | 1441185 | 1442690 | g3938.t1 | Terpene |
| 10 | ctg7 | 6578 | 30619 | g6728.t1 | Terpene |
| g6730.t1 | |||||
| g6731.t1 | |||||
| 11 | ctg7 | 480774 | 506133 | g6859.t1 | Terpene |
| g6861.t1 | |||||
| g6862.t1 | |||||
| 12 | ctg7 | 594057 | 615281 | g6885.t1 | Terpene |
| 13 | ctg7 | 1016899 | 1064188 | g7044.t1 | NRPS-like |
| 14 | ctg9 | 183057 | 203976 | g3034.t1 | Terpene |
| 15 | ctg10 | 433898 | 478092 | g10146.t1 | PKS |
| g10149.t1 | |||||
| 16 | ctg10 | 596519 | 641283 | g10195.t1 | NRPS-like |
| 17 | ctg10 | 671144 | 742323 | g10219.t1 | NRPS-like |
| g10220.t1 | PKS | ||||
| g10229.t1 | Terpene | ||||
| g10232.t1 | |||||
| 18 | ctg11 | 907664 | 951621 | g13526.t1 | PKS |
| 19 | ctg11 | 1162331 | 1187752 | g13618.t1 | Terpene |
| g13621.t1 | |||||
| 20 | ctg11 | 1418013 | 1439351 | g13710.t1 | Terpene |
| 21 | ctg11 | 1484507 | 1528847 | g13738.t1 | NRPS-like |
| 22 | ctg12 | 682713 | 728227 | g5863.t1 | NRPS-like |
| 23 | ctg15 | 557009 | 578845 | g317.t1 | Terpene |
| 24 | ctg17 | 758001 | 779516 | g1290.t1 | Indole |
| 25 | ctg24 | 474021 | 548029 | g823.t1 | NRPS-like |
| g838.t1 | NRPS-like | ||||
| 26 | ctg25 | 204719 | 247787 | g10934.t1 | PKS |
| 27 | ctg25 | 275478 | 325614 | g10956.t1 | PKS |
| 28 | ctg31 | 161801 | 174725 | g571.t1 | Terpene |
| 29 | ctg31 | 181167 | 209940 | g581.t1 | Terpene |
| g582.t1 |
FIG 5.
Analysis of genes involved in secondary-metabolite biosynthesis. (A) Distribution of biosynthetic core genes for natural products on the chromosomes (contigs). (B, D, and E) Phylogenetic tree analysis for sesquiterpene synthases (B), PKSs (D), and NRPS-like enzymes (E) from NWAFU-1 and their respective homologs. (C) Percent identity matrix of 8 STSs from NWAFU-1 and STS25. (F) Comparison of indole-type BGC and similar BGCs; similarity is marked between the corresponding two genes.
Terpenoids, particularly triterpenoids, are the main bioactive constituents of L. sulphureus. Among the 25 predicted terpenoid synthases, there are 15 sesquiterpene synthases (STSs) and 8 monoterpene synthases (mono-TPSs), 1 squalene synthase (SQS), and 1 phytoene synthase (PSY) (Table S12). Cluster analysis of NWAFU-1-derived STSs revealed 15 STSs grouped into four clades using known STSs from mushrooms such as Omphalotus olearius (43), Stereum hirsutum (44), and Coprinus cinerea (45). Clade I had the most STSs (6), followed by clade II with four, clade III with three, and clade IV with two (Fig. 5B and Table S13). The structural characteristics of the catalytic products of these STSs can be inferred from the grouping information. The most similar homologies of the eight mono-TPSs are all monoterpene synthase 25 (STS25) from Postia placenta, which catalyzes the cyclization of geranyl diphosphate (GPP) to myristene and linalool (46). STS25 and these 8 mono-TPSs show high similarity, with identifies ranging from 37.78 to 64.95 (Fig. 5C and Table S14). The mevalonate pathway (MVP) is a recognized upstream pathway of fungal terpenoid metabolism. KEGG annotation identifies key genes for MVP in NWAFU-1 (Fig. S9).
Cluster analysis of 9 PKSs and their six most similar homologs (Table S15), as well as LpaA (20), an identified PKS from L. sulphureus, showed that four (g10146.t1, g10149.t1, g10956.t1, and g10934.t1) of these PKSs were more closely related to LpaA, but their most similar hits in UniProtKB are not LpaA. The PKS g13526.t1 exhibited 48.52% identify with ArmB (47), an orsellinic acid synthase, and it was speculated that this gene was responsible for the synthesis of orsellinic acid in L. sulphureus. The other four PKSs, although showing the most similar hits to Stpks1 in UniProtKB (48), did not point to a usable reference in the current clustering analysis (Fig. 5D).
NRPS-like enzymes other than g4530.t1 all contain at least two functional structural domains (Fig. S9). Further clustering of these NRPS-like enzymes (Table S16) with a total of 46 NRPS-like enzymes (Table S17) identified so far showed that 8 formed a branch with nps9 to -11 and NCU05000. nps9 to -11 were structurally consistent, all containing three structural domains, including an adenylation (A) domain, short-chain dehydrogenase/reductase domain, and thiolation domain. NRPS-like enzymes g5863.t1 and g10129.t1 are in two different clades. In addition to both an A domain and PKS_PP-binding domain, the former contains an NADP_Rossmann domain, while the latter contains a ketide_synthase domain. The remaining NRPS-like enzyme, g7044.t1, did not form clusters, probably because of the additional NUC-45 central domain in its structure (Fig. 5E and Fig. S10).
Cluster 24, containing g1290, was predicted by antiSMASH to be an indole-type BGC, and its core gene, 1290.t1, was annotated as tyrosine dimethylallyl transferase, showing identity with the 4-O-dimethylallyl-l-tyrosine synthase TcpD (32%) (49) and SirD (29%) (50). In addition, this BGC contains genes for DUF6534 domain-containing protein (g1287.t1), phenylalanine ammonia-lyase (g1289.t1), flavin adenine dinucleotide (FAD)-binding domain-containing protein (g1291.t1), (4-O-methyl)-d-glucuronate–lignin esterase (g1292.t1), and major facilitator superfamily (MFS)-type transporter (g1293.t1) (Fig. 5F). This rare BGC inspired us to explore its widespread distribution, and three similar BGCs derived from Basidiomycetes were found in the NCBI genome database using this BGC as a probe. All three clusters contain at least a core gene (tyrosine dimethylallyl transferase gene) and two modifier genes (phenylalanine ammonia-lyase and FAD-binding domain-containing protein genes) (Fig. 5F). This gene cluster, which uses l-tyrosine as the starting substrate, has been reported only for filamentous fungi so far (49, 50).
To further analyze these BGCs, they were subjected to gene cluster family (GCF) analysis with BIG-SCAPE; the results showed that 4 gene clusters were in the GCF network (Fig. S11). Clusters 15, 26, and 27, all three BGCs containing T1PKS, are all in a complex network of PKS and PKS-NRPS. Clusters 26 and 27 are directly connected, and the similarity analysis of these two also proves this association (Fig. S12A). Cluster 9 is in a GCF network composed entirely of terpenoid BGCs, and the similarity analysis of cluster 9 and the three BGCs directly connected to cluster 9 did not reveal the association (Fig. S12B).
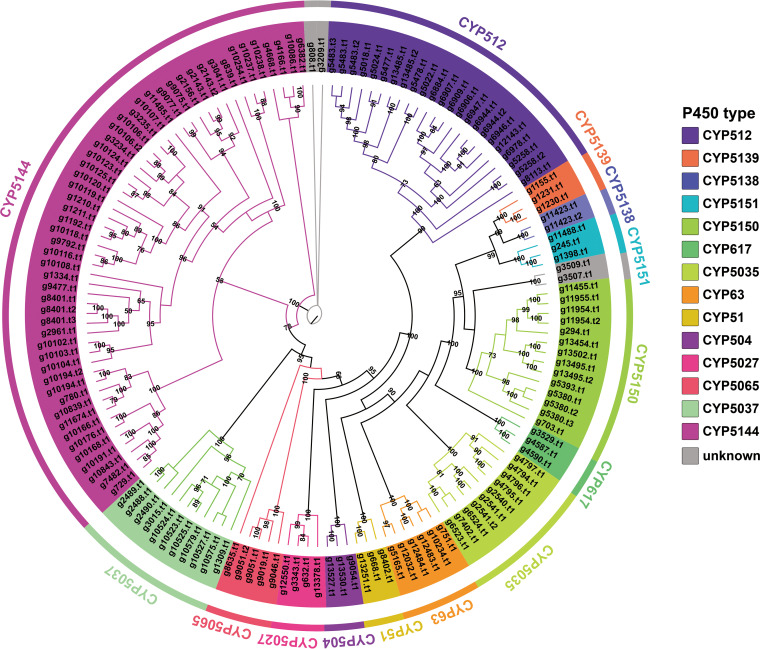
Cytochrome P450 monooxygenase family analysis.
Cytochrome P450 is a ubiquitous superfamily of monooxygenases, which are considered to be essential enzymes involved in fungal primary and secondary metabolic processes, including detoxification, exogenous degradation, and biosynthesis of secondary production (51). According to Pfam prediction based on domain features, a total of 130 P450-coding genes (147 P450 proteins) were screened in the genome of strain NWAFU-1 (Table S18). The cluster analysis revealed a clear classification based on the evolutionary association of these 147 protein sequences with the representative basidiomycete P450 sequences of the Fungal Cytochrome P450 Database (Fig. 6, Fig. S13). There were 126 coding genes (143 proteins) identified and divided into 14 cytochrome P450 families, with the CYP5144 family having the most P450s (52), followed by the CYP512 (31), CYP5141 (14), CYP5037 (11), and CYP5035 (10) families. The remaining nine families all have no more than 10 members, accounting for about 16% in total. The P450 family characteristics in L. sulphureus are basically consistent with those in Phanerochaete carnosa (Thelephorales) (53).
FIG 6.
Maximum likelihood method tree of 147 cytochrome P450s from strain NWAFU-1. Each cytochrome P450 family is shown in a separate color, and the branch reliability value of not less than 50 is marked on the corresponding branch node.
Difference and identification of metabolites from fruiting bodies and mycelium.
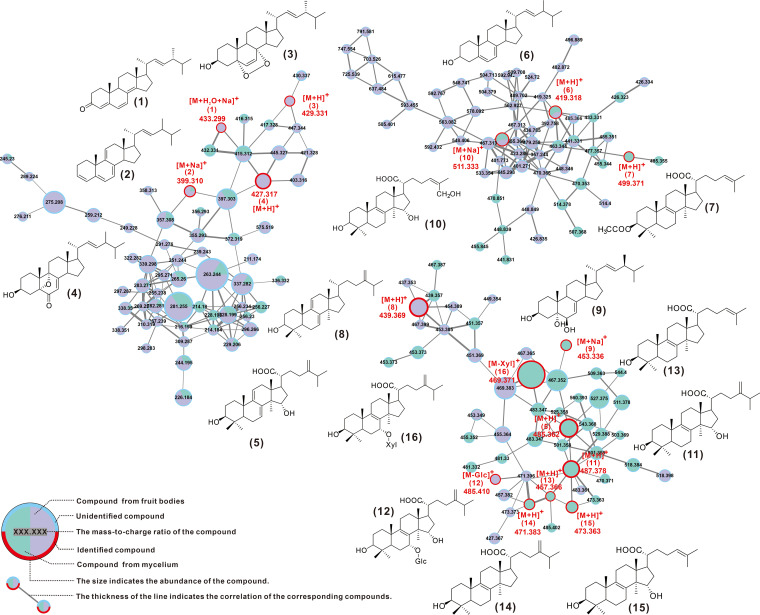
Most current reports on secondary metabolites of L. sulphureus concern the fermentation products of mycelium. In order to investigate whether there are differences in the chemical composition and content of the mycelium and fruiting bodies of L. sulphureus, the metabolites from fruiting bodies and the liquid fermentation of the mycelium were used for quantitative high resolution LCMS analysis and compared with GNPS molecular networks. The visualized molecular network showed that the chemical compositions and the contents of the two are significantly different (Fig. S14).
Further, a total of 16 compounds were identified by comparing their mass spectrometry (MS) and/or nuclear magnetic resonance (NMR) data with reported literature values, including dehydrosulfurenic acid (5), sulfurenic acid (11), tsugaric acid A (12), eburicoic acid (14), laetiposide E (16), trametenolic acid (13) and its derivatives acetyl trametenolic acid (7) and 15α-hydroxytrametenolic acid (15) and other lanostanoids, as well as ergosta-4,6,8(14),22-tetraen-3-one (1), ergost-3,5,7,9(11),22-pentaen (2), ergost-5,7,22-trien-3-ol (6), ergosta-7,22-diene-3,5,6-triol (9), ergosterol peroxide (10), and other ergosteroids (Fig. 7, Table 3, Fig. S15 and S16, and Table S19). Two of the steroids, ergosta-4,6,8(14),22-tetraen-3-one (1) and 3β-hydroxy-5,9-epoxy-(22E,24R)-ergosta-7,22-dien-6-one (4), are reported for the first time for the genus Laetiporus. Except for compound 2, which was present in both mycelium and fruiting body, the other compounds were only distributed in one of the two tissues, which reflected the tissue specificity of the distribution of basidiomycete-derived compounds.
FIG 7.
GNPS-based molecular network identification of metabolites.
TABLE 3.
Identified metabolites from strain NWAFU-1
| No. | Source | Putative metabolite | Molecular formula | Adduct | m/z | Reference |
|---|---|---|---|---|---|---|
| 1 | Fruiting body | Ergosta-4,6,8(14),22-tetraen-3-one | C28H40O | [M+Na+H2O]+ | 415.2977 | 52 |
| 2 | Fruiting body and mycelium | Ergost-3,5,7,9(11),22-pentaen | C28H40 | [M+Na]+ | 377.3130 | 62 |
| 3 | Fruiting body | 5α,8α-epidioxy-(22E,24R)-ergosta-6,22-dien-3b-ol | C28H44O3 | [M+H]+ | 429.3290 | 63 |
| 4 | Fruiting body | 3β-Hydroxy-5,9-epoxy-(22E,24R)-ergosta-7,22-dien-6-one | C28H42O3 | [M+H]+ | 427.3207 | 64 |
| 5 | Fruiting body | Dehydrosulfurenic acid | C31H48O4 | [M+H]+ | 485.3625 | 65 |
| 6 | Mycelium | Ergost-5,7,22-trien-3-ol | C30H48O | [M+H]+ | 425.3778 | 62 |
| 7 | Mycelium | Acetyl trametenolic acid | C32H50O4 | [M+H]+ | 499.3782 | 66 |
| 8 | Fruiting body | 24-methylenelanost-7,9-dien-3-ol | C31H50O | [M+H]+ | 439.3934 | 62 |
| 9 | Fruiting body | Ergosta-7,22-dien-3,5,6-triol | C28H46O3 | [M+Na]+ | 431.3520 | 62 |
| 10 | Mycelium | Ergosterol peroxide | C28H46O3 | [M+Na]+ | 453.3339 | 62 |
| 11 | Mycelium | Sulfurenic acid | C31H50O4 | [M+Na]+ | 509.3601 | 67 |
| 12 | Mycelium | Tsugaric acid A | C31H50O4 | [M+H]+ | 487.3782 | 66 |
| 13 | Mycelium | Trametenolic acid | C30H48O3 | [M+H]+ | 457.3676 | 67 |
| 14 | Mycelium | Eburicoic acid | C31H50O3 | [M+H]+ | 471.3833 | 67 |
| 15 | Mycelium | 15α-Hydroxytrametenolic acid | C30H4804 | [M+H]+ | 473.3625 | 67 |
| 16 | Fruiting body | Laetiposide E | C36H58O8 | [M+H]+ | 619.4310 | 67 |
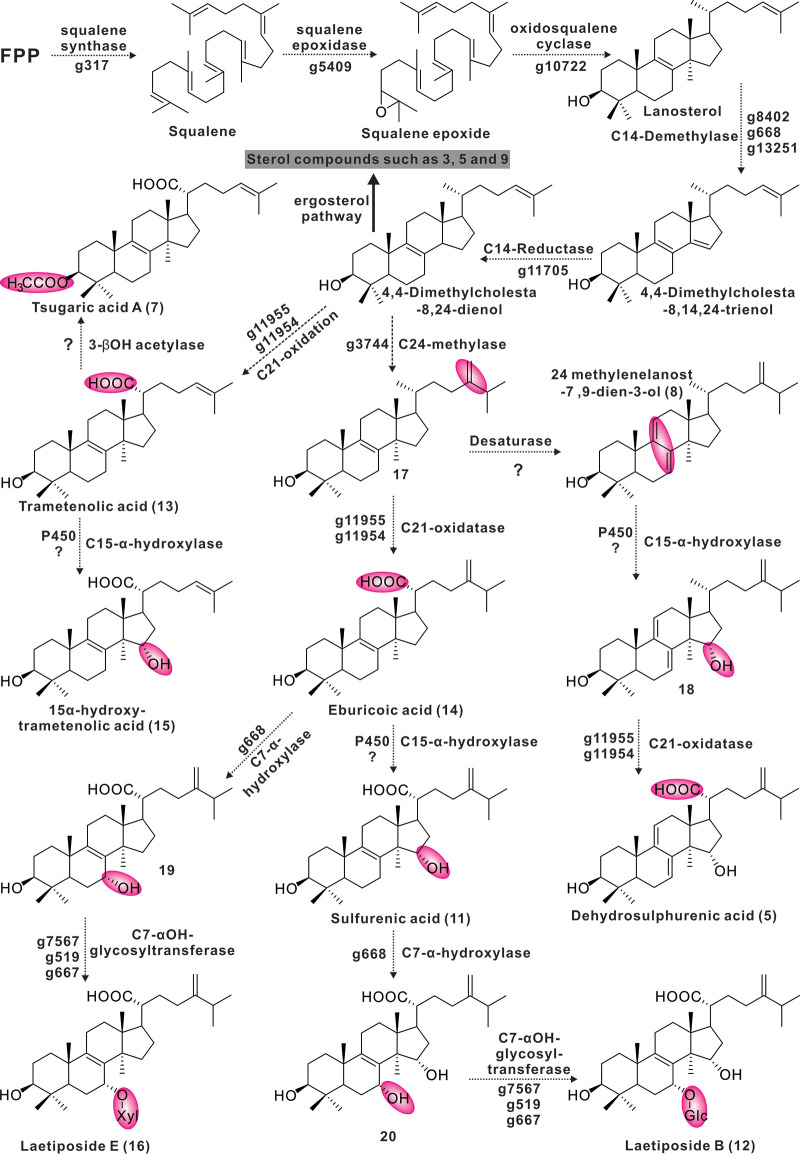
Speculation on the biosynthesis of lanostanoids derived from L. sulphureus.
Structurally, the 16 identified compounds can be divided into two classes, lanostanoids and ergosteroids. Based on what is known, it is assumed that these compounds are all derivatives of the lanosterol-to-ergosterol pathway, with 4,4-dimethylcholestta-8,14,24-trienol acting as a point of divergence (Fig. 8). Enzymes related to the identified ganoderic acid biosynthesis enzymes responsible for the pathway of farnesyl pyrophosphate to lanosterol (54) and the corresponding homologous genes were highly similar. Three candidate genes (g8402, g668, and g13251) responsible for the conversion of lanosterol to 4,4-dimethylcholestta-8,14,24-trienol were identified, and the corresponding three cytochrome P450s are members of the CYP51 family (Fig. 6). The protein encoded by g11705 is responsible for the C-14 reduction of 4,4-dimethylcholestta-8,14,24-trienol, which is very similar to the sterol C-14 reductase from Phellinus igniarius (Table S20). Trametenolic acid (13) is formed by the sequential oxidation of C-21 of 4,4-dimethylcholestta-8,24-dienol, which is presumably accomplished by g11955 or g11954. Its protein is highly like CYP5150L8, an enzyme responsible for a similar process in the formation of ganoderic acid 3-hydroxy-lanosta-8,24-dien-26-oic acid (54). Compound 15 is formed by α-hydroxylation of C-15 on compound 13, which is presumably also carried out by cytochrome P450. Tsugaric acid A (12) is produced by methylating the C-4 of compound 15. The 3-hydroxyl group of compound 13 is acetylated to form compound 8. The presumed intermediate compound 17 is produced by C-24 methylation of 4,4-dimethylcholestta-8,24-dienol, which opens another pathway. Methylation of C-24 is thought to be the responsibility of the homolog of ERG6, g3744, and the two proteins catalyze very similar reactions. C-7 of eburicoic acid (14) undergoes α-hydroxylation and then glycosylation to form Laetipides E (16). The methylation of the C-7 position of this tetracyclic backbone (sterol) is rarely reported, such as in the cases of Homo sapiens (55) and Rattus norvegicus (56), and the results of genome scanning with the corresponding proteins showed that g668 was the best-matching hit (Fig. S17). It is speculated that g668 is responsible for the former step, and there are three candidate genes (g7567, g519, and g667) for the latter step. α-Methylation of C-15 of compound 14 resulted in a shunting metabolism to form eburicoic acid (11) (Fig. 8). The network in which eburicoic acid (16) is found contains all three putative intermediate compounds 17 to 19 (Fig. S18 and Table S20).
FIG 8.
The proposed biosynthetic pathway of tetracyclic triterpenes from L. sulphureus.
DISCUSSION
The fungi of the family Polyporaceae, represented by G. lucidum and Inonotus obliquus, are significant complementary and alternative medicines as well as ancient traditional medicines that have been proven to have significant health-promoting effects. Ganoderic acids, lanosterol tetracyclic triterpenoids from G. lucidum, have been proven by modern medicine to have medicinal value, and their biosynthesis has been extensively analyzed (57). Eburicoic acid, sulfurenic acid, and their derivatives are lanosterol tetracyclic triterpenoids derived from L. sulphureus and have also been reported for other Polyporaceae fungi (11, 16). Such functional molecules have been shown to have antitumor, analgesic, anti-inflammatory, and hepatoprotective effects (11, 16, 58, 59). The present work provides detailed speculation on the biosynthesis of these metabolites and identifies the candidate genes responsible for transformation processes. This investigation provides a clear baseline for subsequent experiments to validate these processes.
The active ingredients of many ancient medicinal macrofungi are commonly derived from the fruiting bodies, and prescriptions in medical classics also emphasize that the medicinal parts are fruiting bodies. In fact, it is not easy for medicinal fungi to form seeds in their natural environment, which explains the rarity of large medicinal fungi. The lack of available sources of fruiting bodies is a major limitation to the large-scale medicinal application of macrofungi. A viable solution to this limitation is artificial cultivation (2). Although there has been a report claiming to have achieved artificial cultivation of L. sulphureus, the mating type of L. sulphureus has not yet been determined. The present work investigated the mating type of L. sulphureus based on genomic information and found that the MAT-A locus on chromosome 13, far from the MAT-B locus on ctg17, was unrelated, and it is reasonable to conclude that L. sulphureus has a tetrapolar mating system. This conclusion will provide valuable clues for the efficient large-scale cultivation of L. sulphureus.
The genome sequencing, assembly, and functional annotation of macrofungi provide valuable information for understanding the genetic background and studying gene function and can provide a theoretical basis for the construction of genetic manipulation systems, molecular breeding, and high-yield cultivation. In this study, the whole-genome sequencing and analysis of L. sulphureus NWAFU-1 showed that this strain was rich in CAZymes and had the potential to synthesize various secondary metabolites, especially in terms of the highly enriched polysaccharide and terpenoid biosynthesis genes. Comparative genomic analysis showed that strain NWAFU-1, unlike other sequenced subspecies, underwent a larger genome duplication event. Based on the molecular network analysis of metabolites and genome scanning, it was speculated that the active triterpenoids originated from the lanosterol pathway, and several potential genes of the transformation process were pointed out. These findings provide important insights into the biological characteristics of the medicinal edible fungus L. sulphureus, including growth characteristics and biosynthetic pathways of active compounds. In addition, this study is expected to provide basic information for the development of mushroom genomes and germplasm resources, to provide a reference for elucidating the biosynthetic mechanisms of various secondary metabolites in medicinal edible fungi, and possibly to promote synthetic biology research in mushrooms.
MATERIALS AND METHODS
Microbial strains and culture conditions.
L. sulphureus NWAFU-1 was isolated from the bark of lacquer tree (Toxicodendron vernicifluum) in the Qinling Mountains. The strain was stored in Shaanxi Province Key Laboratory of Chemical Biology & Natural Products. Solid cultures of the strains were carried out on potato dextrose agar medium at 25°C, and liquid cultures of the strains were accomplished in shake flasks containing potato dextrose broth medium at 200 rpm and 25°C.
Genome sequencing and assembly.
The genomic DNA was extracted from mycelia with a Covaris ultrasonic crusher. The sample then underwent end repair, addition of an A tail, addition of sequencing connectors, purification, and PCR amplification. High-quality bulk DNA was collected to build the library after checking purity, concentration, and integrity. Qubit 2.0 was used for preliminary quantification to detect whether the inserted fragment of the library met expectation before the concentration of the library was accurately quantified by quantitative PCR to ensure the quality of the library. Then, the prepared library was pooled to flow cell, and Illumina NovaSeq was used for sequencing after clustering. The Oxford Nanopore PromethION sequenator was supported by the software Guppy to automatically distinguish between pass and fail data. After removal of reads with an N base content exceeding 5%, reads with 50% of bases in low quality (mass value less than or equal to 5), reads contaminated with adapters, and the repeat sequence caused by PCR amplification, a total of 79,622,624 clean reads and 5,582,739,300 bases were generated. The genome size of 47.7 Mb was estimated by the k-mer method using sequencing data from the DNA library. NECAT software (https://github.com/xiaochuanle/NECAT) was used to perform genome error correction, and splicing was performed to obtain the initial splicing result; then, Racon (https://github.com/isovic/racon) 1.4.11 software was used to perform two rounds of error correction on the splicing result based on the third-generation sequencing data, and, finally, two rounds of Pilon were performed (v.1.23). Error correction was performed, and after removal of heterozygosity, the final assembly result was obtained. BUSCO 4.1.4 software (https://github.com/RoyNexus/busco) evaluated 93.3% of the predicted genes integrity based on the fungal database (fungi_odb10) (v.4.0.6), showing an excellent assembly level. The mitochondrial genome reads were extracted from the whole-genome data by bowtie, and the mitochondrial genome was assembled using GeSeq, which performed annotation using MITOS and GeSeq, based on genetic code 4. Genome collinearity analysis and visualization were achieved by McscanX (https://opensourcelibs.com/lib/mcscanx).
Gene prediction and annotation.
Gene prediction was performed mainly using BRAKER software (v.2.1.4); first, GeneMark-EX was used to train the model, and then, AUGUSTUS was called for prediction. INFERNAL (v.1.1.2) was used to predict and classify ncRNA based on the Rfam database. RepeatModeler software (v. 1.0.4) was used to build its own repeat library, and RepeatMasker (v.4.0.5) was used to annotate the repeated sequence of the genome after merging the repbase library. Functions of the gene products were annotated based on BLAST searches of nonredundant protein sequences from the NCBI, Swiss-Prot, COG, and KEGG databases. GC-Profile characterizes changes of GC content in genomic sequences and predicts genomic islands, which were generated on a web-based tool. Nr annotation showed that strain NWAFU-1 shared 95% genes with L. sulphureus 93-53.
Phylogenomic and comparative genomic analyses.
Homologous gene identification and phylogenetic analysis were performed with strain NWAFU-1 and another a total of 21 representative strains in Russulales, Agaricales, and Hymenochaetales. Single-copy homologous genes were identified using OrthoFinder (v.2.5.4) (60) with the parameters as “-S diamond -M msa -T raxml-ng.” MCMCtree (http://abacus.gene.ucl.ac.uk/software/paml.html) was utilized to predict divergence time with a total of 520 single-copy ortholog sequences of 24 strains. Several groups of recent ancestor divergence times were queried as calibrated points in TimeTree (http://www.timetree.org/) (H. irregulare versus S. hirsutum, 71.3 to 125.1 MYA; Phanerochaete sordida versus Phlebiopsis gigantea, 65.3 to 103.7 MYA; and G. sinense versus Dichomitus squalens, 18.2 to 34.7 MYA).
Comparative genomic analysis within L. sulphureus subspecies was performed through single-copy genes and visualized by jVenn (http://jvenn.toulouse.inra.fr/app/index.html). Ks calculation was performed among three L. sulphureus subspecies. The homolog protein sequence identifier (ID) lists were transformed to CDS lists by ParaAT 2.0 (https://github.com/wonaya/ParaAT). Homologous sequence pairs among the three subspecies were calculated by KaKs Calculator 3.0 (https://ngdc.cncb.ac.cn/biocode/tools/BT000001) and finally visualized by Rstudio (v. 4.2.0).
CAZy family analysis.
The database CAZy (http://bcb.unl.edu/dbCAN2/) was used to annotate and class the genes encoding CAZymes from NWAFU-1 and other brown-rot Basidiomycota, with HMMER (v.3.2.1; filter parameter E value < 1e−5; coverage > 0.35).
Prediction and phylogenetic analyses of gene clusters involved in secondary metabolites.
The antibiotic and secondary-metabolite production gene clusters were examined using antiSMASH 6.1 software and FramePlot with default parameters. To verify the predicted results, the obtained gene clusters were manually checked. BLASTP analysis and gene annotation were performed using the NCBI genome portal software platform. We searched all hypothetical gene models in the database using blastP and tblastN algorithms. BIG-SCAPE was performed to build a BGC network between BGCS predicted by antiSMASH (https://antismash.secondarymetabolites.org/) and confirmed BGCs in the MiBIG1.4 database with a cutoff of 0.75, and the BGC network was visualized by Cytoscape3.9.1. Farnesyl diphosphate synthase (FPPs) were selected from terpene synthases predicted by antiSMASH by 2ndFind for further analysis. The FPPs phylogenetic tree showed a clear classification. Prediction for PKS domains was analyzed by PKS/NRPS analysis program online (http://nrps.igs.umaryland.edu/tutorial.html). For PKS BGCs, the KS domains of L. sulphureus and related species were collected from the NCBI and JGI database to perform phylogenetic analysis to generate a maximum likelihood tree using OrthoFinder software to describe its evolutionary status. For NRPS-like BGCs, all the domains of L. sulphureus and other species that had been identified in UniProt (https://www.uniprot.org) were collected to generate a basic phylogenetic tree to understand the polygenetic status. Comparison of indole-type BGCs with similar BGCs was achieved by clinker (61).
Prediction and analysis of cytochrome P450s.
The package Hmmer was used to predict cytochrome P450s with Diamond (version > 2.9.0; E value > e−5) and annotate the target protein sequence. The reference CYP sequences for cluster analyses were downloaded from the Fungal Cytochrome P450 Database (http://p450.riceblast.snu.ac.kr/index.php?a=view). A total of 147 cytochrome P450 proteins of strain NWAFU-1 and several other similar species selected from the fungal cytochrome P450 database were clustered to perform phylogenetic tree analysis with clear classification. A maximum likelihood tree was built by IQ-tree 2.2.3 with parameters “-m MFP -bb 1000 -alrt 1000 -abayes -nt AUTO.”
Metabolite analysis and structural evaluation.
To analyze the metabolites of strain NWAFU-1, liquid fermentation and solid fermentation were performed; the solid fermentation was performed on PDA medium at 25°C for 14 days. Liquid fermentations were grown in shake flasks with PDB at 200 rpm and 25°C for 10 days. After fermentation was complete, the cultures were harvested and extracted 3 times with an equal volume of ethyl acetate to obtain as many secondary metabolites as possible. The bioactive metabolite extracts and their fractions were analyzed using high-resolution electrospray ionization mass spectrometry (HRESIMS) was carried out using AB Sciex TripleTOF 6600 mass spectrometer. The electrospray mass spectra were recorded on a time-of-flight mass spectrometer in both positive-ion and negative-ion modes. One-dimensional nuclear magnetic resonance (1D-NMR) data were obtained with a Bruker (Germany) ADVANCE IIIHD 400-MHz NMR spectrometer including 1H NMR and 13C NMR. The structures of the isolated compounds were established based on spectroscopic analyses and comparisons to previously reported data sets. Molecular network analysis of high-performance liquid chromatography (HPLC)–HRESIMS data of the crude extract was performed by GNPS with default parameters. The network file based on negative-ion-mode MS data can be found and accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=ec420f568ec64f7d82470a7cf507b35f. The molecular network was visualized by Cytoscape 3.9.1. The compounds identified by NMR and HRMS are in the same network and show a good correlation with analogs with similar structure.
Data availability.
The ITS sequence of strain L. sulphureus NWAFU-1 has been deposited in the NCBI GenBank under accession number ON797482. The final genome assembly results and related data of L. sulphureus NWAFU-1 have been submitted to the National Center for Biotechnology Information (NCBI) under the BioProject JAMXVQ000000000 and BioSample SAMN29145082, respectively. For details, see Data Set S1 (gene distribution of 22 brown-rot fungi based on the six major modules of CAZymes), Data Set S2 (the protein sequences of the predicted genes for Laetiporus sulphureus NWAFU-1), Data Set S3 (the DNA sequences of the predicted genes for Laetiporus sulphureus NWAFU-1), and Data Set S4 (the chromosome where the predicted gene is located and its specific location on the chromosome).
ACKNOWLEDGMENTS
We thank André Damasio and Fabio Squina from LEBIMO—University of Campinas for providing the protein sequences, CDS, and GFF files of Laetiporus sulphureus ATCC 52600.
J.Q., W.-G.D., and Z.-X.W. designed the project. W.-G.D., Z.-X.W., X.-L.F., and R.-Q.Z. carried out experiments and analyzed the primary data. D.-Y.S. and S.D. provided and identified the strains. J.Q., X.-L.F., and W.-G.D. conducted bioinformatic analyses. X.-L.F. and R.-Q.Z. analyzed NMR data and identified structures. J.Q. and W.-G.D. finished the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31800031), the China Postdoctoral Science Foundation (2019M653760), and the Natural Science Basic Research Plan in Shaanxi Province of China (2019JQ-046).
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Jianzhao Qi, Email: qjz@nwafu.edu.cn.
Matthew Zack Anderson, The Ohio State University.
REFERENCES
- 1.Sandargo B, Chepkirui C, Cheng T, Chaverra-Muñoz L, Thongbai B, Stadler M, Hüttel S. 2019. Biological and chemical diversity go hand in hand: Basidiomycota as source of new pharmaceuticals and agrochemicals. Biotechnol Adv 37:107344. doi: 10.1016/j.biotechadv.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Lou H, Hu J, Liu Z, Chen Q. 2020. Macrofungi: a review of cultivation strategies, bioactivity, and application of mushrooms. Compr Rev Food Sci Food Saf 19:2333–2356. doi: 10.1111/1541-4337.12602. [DOI] [PubMed] [Google Scholar]
- 3.Niazi AR, Ghafoor A. 2021. Different ways to exploit mushrooms: a review. All Life 14:450–460. doi: 10.1080/26895293.2021.1919570. [DOI] [Google Scholar]
- 4.Murrill WA. 1904. The Polyporaceae of North America-IX. Inonotus, Sesia and monotypic genera. Bull Torrey Bot Club 31:593–610. doi: 10.2307/2478612. [DOI] [Google Scholar]
- 5.Kirk PM, Canon PF, Stalpers JA. 2008. Dictionary of the fungi, 10th ed. CABI Publishing, Wallingford, Oxfordshire, United Kingdom. [Google Scholar]
- 6.Song J, Cui B-K. 2017. Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evol Biol 17:102. doi: 10.1186/s12862-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ota Y, Hattori T, Banik MT, Hagedorn G, Sotome K, Tokuda S, Abe Y. 2009. The genus Laetiporus (Basidiomycota, Polyporales) in East Asia. Mycol Res 113:1283–1300. doi: 10.1016/j.mycres.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Pires RM, Motato-Vasquez V, Gugliotta AD. 2016. A new species of Laetiporus (Basidiomycota) and occurrence of L. gilbertsonii Burds. in Brazil. Nova_Hedwigia 102:477–490. doi: 10.1127/nova_hedwigia/2016/0320. [DOI] [Google Scholar]
- 9.Sinclair WA, Lyon H, Johnson WT. 1987. Diseases of trees and shrubs, vol 25. Cornell University Press, New York, NY. [Google Scholar]
- 10.de Figueiredo FL, de Oliveira ACP, Terrasan CRF, Gonçalves TA, Gerhardt JA, Tomazetto G, Persinoti GF, Rubio MV, Peña JAT, Araújo MF, de Carvalho Silvello MA, Franco TT, Rabelo SC, Goldbeck R, Squina FM, Damasio A. 2021. Multi-omics analysis provides insights into lignocellulosic biomass degradation by Laetiporus sulphureus ATCC 52600. Biotechnol Biofuels 14:96. doi: 10.1186/s13068-021-01945-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grienke U, Zöll M, Peintner U, Rollinger JM. 2014. European medicinal polypores—a modern view on traditional uses. J Ethnopharmacol 154:564–583. doi: 10.1016/j.jep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Petrović J, Glamočlija J, Stojković DS, Ćirić A, Nikolić M, Bukvički D, Guerzoni ME, Soković MD. 2013. Laetiporus sulphureus, edible mushroom from Serbia: investigation on volatile compounds, in vitro antimicrobial activity and in situ control of Aspergillus flavus in tomato paste. Food Chem Toxicol 59:297–302. doi: 10.1016/j.fct.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Hassan K, Matio KB, Stadler M. 2021. Two new triterpenes from Basidiomata of the medicinal and edible mushroom, Laetiporus sulphureus. Molecules 26:7090. doi: 10.3390/molecules26237090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He JB, Tao J, Miao XS, Bu W, Zhang S, Dong ZJ, Li ZH, Feng T, Liu JK. 2015. Seven new drimane-type sesquiterpenoids from cultures of fungus Laetiporus sulphureus. Fitoterapia 102:1–6. doi: 10.1016/j.fitote.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 15.He JB, Tao J, Miao XS, Feng YP, Bu W, Dong ZJ, Li ZH, Feng T, Liu JK. 2015. Two new illudin type sesquiterpenoids from cultures of Phellinus tuberculosus and Laetiporus sulphureus. J Asian Nat Prod Res 17:1054–1058. doi: 10.1080/10286020.2015.1040774. [DOI] [PubMed] [Google Scholar]
- 16.Zjawiony JK. 2004. Biologically active compounds from Aphyllophorales (polypore) fungi. J Nat Prod 67:300–310. doi: 10.1021/np030372w. [DOI] [PubMed] [Google Scholar]
- 17.Turkoglu A, Duru ME, Mercan N, Kivrak I, Gezer K. 2007. Antioxidant and antimicrobial activities of Laetiporus sulphureus (Bull.) Murrill. Food Chem 101:267–273. doi: 10.1016/j.foodchem.2006.01.025. [DOI] [Google Scholar]
- 18.Karaman M, Jovin E, Malbaša R, Matavuly M, Popović M. 2010. Medicinal and edible lignicolous fungi as natural sources of antioxidative and antibacterial agents. Phytother Res 24:1473–1481. doi: 10.1002/ptr.2969. [DOI] [PubMed] [Google Scholar]
- 19.Jonathan G, Chikwem J, Hull A, Aina D, Asemoloye M. 2021. Antimicrobial activities of Laetiporus conifericola (Agaricomycetes) Burds & Banik. Collected from Pennsylvania, USA. Int J Med Mushrooms 23:69–77. doi: 10.1615/IntJMedMushrooms.2021038407. [DOI] [PubMed] [Google Scholar]
- 20.Seibold PS, Lenz C, Gressler M, Hoffmeister D. 2020. The Laetiporus polyketide synthase LpaA produces a series of antifungal polyenes. J Antibiot 73:711–720. doi: 10.1038/s41429-020-00362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber RWS, Mucci A, Davoli P. 2004. Laetiporic acid, a new polyene pigment from the wood-rotting basidiomycete Laetiporus sulphureus (Polyporales, Fungi). Tetrahedron Lett 45:1075–1078. doi: 10.1016/j.tetlet.2003.11.073. [DOI] [Google Scholar]
- 22.Davoli P, Mucci A, Schenetti L, Weber RWS. 2005. Laetiporic acids, a family of non-carotenoid polyene pigments from fruit-bodies and liquid cultures of Laetiporus sulphureus (Polyporales, Fungi). Phytochemistry 66:817–823. doi: 10.1016/j.phytochem.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Hwang HS, Lee SH, Yu MB, Sang WK, Yong KJ, Yun JW. 2008. Production of extracellular polysaccharides by submerged mycelial culture of Laetiporus sulphureus var. miniatus and their insulinotropic properties. Appl Microbiol Biotechnol 78:419–429. doi: 10.1007/s00253-007-1329-6. [DOI] [PubMed] [Google Scholar]
- 24.Pleszczyńska M, Wiater A, Siwulski M, Szczodrak J. 2013. Successful large-scale production of fruiting bodies of Laetiporus sulphureus (Bull.: Fr.) Murrill on an artificial substrate. World J Microbiol Biotechnol 29:753–758. doi: 10.1007/s11274-012-1230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, Luo H, Li Y, Song J, Henrissat B, Levasseur A, Qian J, Li J, Luo X, Shi L, He L, Xiang L, Xu X, Niu Y, Li Q, Han MV, Yan H, Zhang J, Chen H, Lv A, Wang Z, Liu M, Schwartz DC, Sun C. 2012. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun 3:913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang N, Hu S, Peng B, Li Z, Yuan X, Xiao S, Fu Y. 2021. Genome of Ganoderma species provides insights into the evolution, conifers substrate utilization, and terpene synthesis for Ganoderma tsugae. Front Microbiol 12:724451. doi: 10.3389/fmicb.2021.724451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan Y, Han H, Qi J, Gao J-m, Xu Z, Wang P, Zhang J, Liu C. 2022. Genome sequencing of Inonotus obliquus reveals insights into candidate genes involved in secondary metabolite biosynthesis. BMC Genomics 23:314. doi: 10.1186/s12864-022-08511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong W, Wang Y, Xie C, Zhou Y, Zhu Z, Peng Y. 2020. Whole genome sequence of an edible and medicinal mushroom, Hericium erinaceus (Basidiomycota, Fungi). Genomics 112:2393–2399. doi: 10.1016/j.ygeno.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Sun T, Zhang Y, Jiang H, Yang K, Wang S, Wang R, Li S, Lei P, Xu H, Qiu Y, Sun D. 2021. Whole genome sequencing and annotation of Naematelia aurantialba (Basidiomycota, edible-medicinal fungi). J Fungi 8:6. doi: 10.3390/jof8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu MY, Fan WL, Wang WF, Chen T, Tang YC, Chu FH, Chang TT, Wang SY, Li MY, Chen YH, Lin ZS, Yang KJ, Chen SM, Teng YC, Lin YL, Shaw JF, Wang TF, Li WH. 2014. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc Natl Acad Sci USA 111:E4743–E4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J-H, Wu S-H, Zhou L-W. 2021. The first whole genome sequencing of Sanghuangporus sanghuang provides insights into its medicinal application and evolution. J Fungi 7:787. doi: 10.3390/jof7100787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Wu S, Ma X, Chen W, Zhang J, Duan S, Gao Y, Kui L, Huang W, Wu P, Shi R, Li Y, Wang Y, Li J, Guo X, Luo X, Li Q, Xiong C, Liu H, Gui M, Sheng J, Dong Y. 2018. The genome sequences of 90 mushrooms. Sci Rep 8:9982. doi: 10.1038/s41598-018-28303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy LG, Riley R, Tritt A, Adam C, Daum C, Floudas D, Sun H, Yadav JS, Pangilinan J, Larsson K-H, Matsuura K, Barry K, Labutti K, Kuo R, Ohm RA, Bhattacharya SS, Shirouzu T, Yoshinaga Y, Martin FM, Grigoriev IV, Hibbett DS. 2016. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol Biol Evol 33:959–970. doi: 10.1093/molbev/msv337. [DOI] [PubMed] [Google Scholar]
- 34.Wright R, Woof K, Douglas B, Gaya E, Consortium DToL . 2022. The genome sequence of the chicken of the woods fungus, Laetiporus sulphureus (Bull.) Murrill, 1920. Wellcome Open Res 7:83. doi: 10.12688/wellcomeopenres.17750.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown AJ, Casselton LA. 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet 17:393–400. doi: 10.1016/s0168-9525(01)02343-5. [DOI] [PubMed] [Google Scholar]
- 36.Kües U, James TY, Heitman J. 2011. Mating type in basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality, p 97–160. In Pöggeler S, Wöstemeyer J (ed), Evolution of fungi and fungal-like organisms. Springer, Berlin, Germany. doi: 10.1007/978-3-642-19974-5_6. [DOI] [Google Scholar]
- 37.Bistis GN, Raper JR. 1967. Genetics of sexuality in higher fungi. Mycologia 59:384. doi: 10.2307/3756816. [DOI] [Google Scholar]
- 38.Alquini G, Carbonero ER, Rosado FR, Cosentino C, Iacomini M. 2004. Polysaccharides from the fruit bodies of the basidiomycete Laetiporus sulphureus (Bull.: Fr.) Murr. FEMS Microbiol Lett 230:47–52. doi: 10.1016/S0378-1097(03)00853-X. [DOI] [PubMed] [Google Scholar]
- 39.Lung MY. 2012. Antioxidant properties of polysaccharides from Laetiporus sulphureus in submerged cultures. Afr J Biotechnol 11:6350–6358. [Google Scholar]
- 40.Gong M, Zhang H, Wu D, Zhang Z, Zhang J, Bao D, Yang Y. 2021. Key metabolism pathways and regulatory mechanisms of high polysaccharide yielding in Hericium erinaceus. BMC Genomics 22:160. doi: 10.1186/s12864-021-07480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N, Tang Z, Zhang J, Li X, Yang Z, Yang C, Zhang Z, Huang Z. 2019. Comparative transcriptome analysis reveals the genetic basis underlying the biosynthesis of polysaccharides in Hericium erinaceus. Bot Stud 60:15. doi: 10.1186/s40529-019-0263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng L, Qiao S, Xu Z, Guan F, Ding Z, Gu Z, Zhang L, Shi G. 2015. Effects of culture conditions on monosaccharide composition of Ganoderma lucidum exopolysaccharide and on activities of related enzymes. Carbohydr Polym 133:104–109. doi: 10.1016/j.carbpol.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Wawrzyn GT, Quin MB, Choudhary S, López-Gallego F, Schmidt-Dannert C. 2012. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem Biol 19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagamine S, Liu C, Nishishita J, Kozaki T, Sogahata K, Sato Y, Minami A, Ozaki T, Schmidt-Dannert C, Maruyama J-i, Oikawa H, Master ER. 2019. Ascomycete Aspergillus oryzae is an efficient expression host for production of basidiomycete terpenes by using genomic DNA sequences. Appl Environ Microbiol 85:e00409-19. doi: 10.1128/AEM.00409-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agger S, Lopez-Gallego F, Schmidt-Dannert C. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichinose H, Kitaoka T. 2018. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb Biotechnol 11:952–965. doi: 10.1111/1751-7915.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engels B, Heinig U, Grothe T, Stadler M, Jennewein S. 2011. Cloning and characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. J Biol Chem 286:6871–6878. doi: 10.1074/jbc.M110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nofiani R, de Mattos-Shipley K, Lebe KE, Han L-C, Iqbal Z, Bailey AM, Willis CL, Simpson TJ, Cox RJ. 2018. Strobilurin biosynthesis in basidiomycete fungi. Nat Commun 9:3940. doi: 10.1038/s41467-018-06202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dopstadt J, Neubauer L, Tudzynski P, Humpf H-U. 2016. The epipolythiodiketopiperazine gene cluster in Claviceps purpurea: dysfunctional cytochrome P450 enzyme prevents formation of the previously unknown clapurines. PLoS One 11:e0158945. doi: 10.1371/journal.pone.0158945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MSC, Howlett BJ. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol Microbiol 53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 51.Črešnar B, Petrič Š. 2011. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta 1814:29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 52.Xu G-H, Choo S-J, Kim Y-H, Ryoo I-J, Seok S-J, Ahn J-S, Yoo I-D. 2010. Secondary metabolites of Volvariella bombycina and their inhibitory effects on melanogenesis. J Microbiol Biotechnol 20:78–81. doi: 10.4014/jmb.0906.06017. [DOI] [PubMed] [Google Scholar]
- 53.Syed K, Shale K, Pagadala NS, Tuszynski J. 2014. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS One 9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W-F, Xiao H, Zhong J-J. 2018. Biosynthesis of a ganoderic acid in Saccharomyces cerevisiae by expressing a cytochrome P450 gene from Ganoderma lucidum. Biotechnol Bioeng 115:1842–1854. doi: 10.1002/bit.26583. [DOI] [PubMed] [Google Scholar]
- 55.Sezer E, Can Demirdöğen B, Demirkaya Ş, Bulut G, Akkulak M, Evin E, Adalı O. 2022. Association of cholesterol 7α-hydroxylase (CYP7A1) promoter polymorphism (rs3808607) and cholesterol 24S-hydroxylase (CYP46A1) intron 2 polymorphism (rs754203) with serum lipids, vitamin D levels, and multiple sclerosis risk in the Turkish population. Neurol Sci 43:2611–2620. doi: 10.1007/s10072-021-05597-1. [DOI] [PubMed] [Google Scholar]
- 56.Wicher G, Norlin M. 2015. Estrogen-mediated regulation of steroid metabolism in rat glial cells; effects on neurosteroid levels via regulation of CYP7B1-mediated catalysis. J Steroid Biochem Mol Biol 145:21–27. doi: 10.1016/j.jsbmb.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 57.Angulo-Sanchez LT, López-Peña D, Torres-Moreno H, Gutiérrez A, Gaitán-Hernández R, Esqueda M. 2022. Biosynthesis, gene expression, and pharmacological properties of triterpenoids of Ganoderma species (Agaricomycetes): a review. Int J Med Mushrooms 24:1–17. doi: 10.1615/IntJMedMushrooms.2022044016. [DOI] [PubMed] [Google Scholar]
- 58.Deng J-S, Huang S-S, Lin T-H, Lee M-M, Kuo C-C, Sung P-J, Hou W-C, Huang G-J, Kuo Y-H. 2013. Analgesic and anti-inflammatory bioactivities of eburicoic acid and dehydroeburicoic acid isolated from Antrodia camphorata on the inflammatory mediator expression in mice. J Agric Food Chem 61:5064–5071. doi: 10.1021/jf303820k. [DOI] [PubMed] [Google Scholar]
- 59.Huang GJ, Deng JS, Huang SS, Lee CY, Hou WC, Wang SY, Sung PJ, Kuo YH. 2013. Hepatoprotective effects of eburicoic acid and dehydroeburicoic acid from Antrodia camphorata in a mouse model of acute hepatic injury. Food Chem 141:3020–3027. doi: 10.1016/j.foodchem.2013.03.061. [DOI] [PubMed] [Google Scholar]
- 60.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilchrist CLM, Chooi YH. 2021. Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 62.Ericsson D, Ivonne J. 2009. Sterol composition of the macromycete fungus Laetiporus sulphureus. Chem Nat Compd 45:193–196. doi: 10.1007/s10600-009-9301-6. [DOI] [Google Scholar]
- 63.Kobori M, Yoshida M, Ohnishi-Kameyama M, Takei T, Shinmoto H. 2006. 5α,8α-epidioxy-22E-ergosta-6,9(11),22-trien-3β-ol from an edible mushroom suppresses growth of HL60 leukemia and HT29 colon adenocarcinoma cells. Biol Pharm Bull 29:755–759. doi: 10.1248/bpb.29.755. [DOI] [PubMed] [Google Scholar]
- 64.Yu FX, Li Z, Chen Y, Yang YH, Li GH, Zhao PJ. 2017. Four new steroids from the endophytic fungus Chaetomium sp. M453 derived of Chinese herbal medicine Huperzia serrata. Fitoterapia 117:41–46. doi: 10.1016/j.fitote.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Chepkirui C, Matasyoh JC, Decock C, Stadler M. 2017. Two cytotoxic triterpenes from cultures of a Kenyan Laetiporus sp. (Basidiomycota). Phytochem Lett 20:106–110. doi: 10.1016/j.phytol.2017.04.009. [DOI] [Google Scholar]
- 66.León F, Quintana J, Rivera A, Estévez F, Bermejo J. 2004. Lanostanoid triterpenes from Laetiporus sulphureus and apoptosis induction on HL-60 human myeloid leukemia cells. J Nat Prod 67:2008–2011. doi: 10.1021/np049762o. [DOI] [PubMed] [Google Scholar]
- 67.Yoshikawa K, Matsumoto K, Mine C, Bando S, Arihara S. 2000. Five lanostane triterpenoids and three saponins from the fruit body of Laetiporus versisporus. Chem Pharm Bull (Tokyo) 48:1418–1421. doi: 10.1248/cpb.48.1418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S18 and Tables S1 to S20. Download spectrum.02439-22-s0001.pdf, PDF file, 6.2 MB (6.3MB, pdf)
Data Set S1. Download spectrum.02439-22-s0002.xlsx, XLSX file, 0.1 MB (93.2KB, xlsx)
Data Set S2. Download spectrum.02439-22-s0003.csv, CSV file, 6.2 MB (6.3MB, csv)
Data Set S3. Download spectrum.02439-22-s0004.csv, CSV file, 18.3 MB (18.7MB, csv)
Data Set S4.1. Download spectrum.02439-22-s0005.csv, CSV file, 13.4 MB (13.7MB, csv)
Data Set S4.2. Download spectrum.02439-22-s0006.csv, CSV file, 14.8 MB (15.1MB, csv)
Data Availability Statement
The ITS sequence of strain L. sulphureus NWAFU-1 has been deposited in the NCBI GenBank under accession number ON797482. The final genome assembly results and related data of L. sulphureus NWAFU-1 have been submitted to the National Center for Biotechnology Information (NCBI) under the BioProject JAMXVQ000000000 and BioSample SAMN29145082, respectively. For details, see Data Set S1 (gene distribution of 22 brown-rot fungi based on the six major modules of CAZymes), Data Set S2 (the protein sequences of the predicted genes for Laetiporus sulphureus NWAFU-1), Data Set S3 (the DNA sequences of the predicted genes for Laetiporus sulphureus NWAFU-1), and Data Set S4 (the chromosome where the predicted gene is located and its specific location on the chromosome).