ABSTRACT
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is a global public health threat. In this study, we employed whole-genome sequencing (WGS) to determine the genomic epidemiology of a longitudinal collection of clinical CRKP isolates recovered from a large public acute care hospital in Singapore. Phylogenetic analyses, a characterization of resistance and virulence determinants, and plasmid profiling were performed for 575 unique CRKP isolates collected between 2009 and 2020. The phylogenetic analyses identified the presence of global high-risk clones among the CRKP population (clonal group [CG] 14/15, CG17/20, CG147, CG258, and sequence type [ST] 231), and these clones constituted 50% of the isolates. Carbapenemase production was common (n = 497, 86.4%), and KPC was the predominant carbapenemase (n = 235, 40.9%), followed by OXA-48-like (n = 128, 22.3%) and NDM (n = 93, 16.2%). Hypervirulence was detected in 59 (10.3%) isolates and was most common in the ST231 carbapenemase-producing isolates (21/59, 35.6%). Carbapenemase genes were associated with diverse plasmid replicons; however, there was an association of blaOXA-181/232 with ColKP3 plasmids. This study presents the complex and diverse epidemiology of the CRKP strains circulating in Singapore. Our study highlights the utility of WGS-based genomic surveillance in tracking the population dynamics of CRKP.
IMPORTANCE In this study, we characterized carbapenem-resistant Klebsiella pneumoniae clinical isolates collected over a 12-year period in the largest public acute-care hospital in Singapore using whole-genome sequencing. The results of this study demonstrate significant genomic diversity with the presence of well-known epidemic, multidrug-resistant clones amid a diverse pool of nonepidemic lineages. Genomic surveillance involving comprehensive resistance, virulence, and plasmid gene content profiling provided critical information for antimicrobial resistance monitoring and highlighted future surveillance priorities, such as the emergence of ST231 K. pneumoniae strains bearing multidrug resistance, virulence elements, and the potential plasmid-mediated transmission of the blaOXA-48-like gene. The findings here also reinforce the necessity of unique infection control and prevention strategies that take the genomic diversity of local circulating strains into consideration.
KEYWORDS: carbapenem-resistant, Klebsiella pneumoniae, whole-genome sequencing, drug resistance mechanisms, plasmid-mediated resistance, virulence factors
INTRODUCTION
Klebsiella pneumoniae, a key member of the Enterobacterales family, is a medically important pathogen that is implicated in several nosocomial infections, including urinary tract infections and pneumonia. It is also a commonly encountered organism in intensive care units (1). This pathogen is a major public health threat, owing to its ability to acquire multidrug resistance, including carbapenem resistance. The emergence of carbapenem-resistant K. pneumoniae (CRKP) has been singled out as a leading priority by the World Health Organization and the United States Centers for Disease Control and Prevention (2, 3).
Singapore’s strategic geographical location has positioned it as an international hub for travel, trade, and medical tourism. However, overseas travel has also provided the opportunity for the importation of diverse, novel, multidrug resistant (MDR) bacteria into the country (4, 5). Coupled with the endemicity of extended-spectrum β-lactamases (ESBLs) and the corresponding high carbapenem usage, it is not surprising that a dramatic increase in the incidence of carbapenem-resistant Enterobacterales (including K. pneumoniae) was observed (6). Among the Enterobacterales, K. pneumoniae presented with the highest carbapenem resistance rates; the national prevalence of CRKP in 2017 was 7.0 per 100 000 patient days, more than twice that of carbapenem-resistant E. coli (7).
Antimicrobial resistance (AMR) has emerged significantly in certain clones and clonal groups (CGs). The global dissemination of carbapenem resistance is associated with a limited number of successful “high-risk” clones. These problematic clones are defined as global high-risk clones based primarily on the following characteristics: (i) isolation from various international geographical locations; (ii) multidrug resistance; (iii) enhanced fitness, virulence, and pathogenicity; (iv) extended host colonization and persistence; and (v) extensive transmission among hosts (8, 9). The most recognized international clones belong to CG258, which includes sequence type (ST) 258, ST11, and ST512. These strains are commonly implicated in the dissemination of KPC-2/KPC-3 carbapenemases, especially in the United States and Israel. Other common high-risk clones include those belonging to CG14/15, CG17/20, CG43, CG147, and ST101 (10, 11).
Of even greater concern is the genotypic convergence of carbapenem resistance and virulence. Multi-drug resistance and hypervirulence were often deemed to have followed distinct evolutionary directions, with each phenotype occupying its own clonal lineage and possessing a largely nonoverlapping genomic signature (12). AMR often occurs in less virulent classical pathotypes that are encountered more frequently in healthcare settings (13). On the other hand, well-known hypervirulent K. pneumoniae (hvKP) clones (e.g., ST23, ST25, ST65, and ST86) are mostly implicated in severe community-acquired infections and are often highly susceptible to antibiotics (14). However, instances of the acquisition of virulence genes by CRKP and the acquisition of resistance genes by hvKP are increasingly being reported, posing a significant public health challenge (15, 16).
Limited evidence has pointed toward the high dynamicity and diversity of carbapenem-resistant Enterobacterales locally (4, 6, 17). Such diversity can introduce complexities in the management of these resistant organisms. Contemporary surveillance data would thus be beneficial in deciphering AMR trends, informing public health policies and interventions, guiding treatment selection, and assessing the impact of interventions (18). Therefore, we aimed to use whole-genome sequencing (WGS) to understand the population structure and the resistance and virulence determinants of CRKP, the predominant carbapenem-resistant Enterobacterales in Singapore, from samples collected between 2009 and 2020 from a local public health hospital.
RESULTS
CRKP isolates and antibiotic susceptibilities.
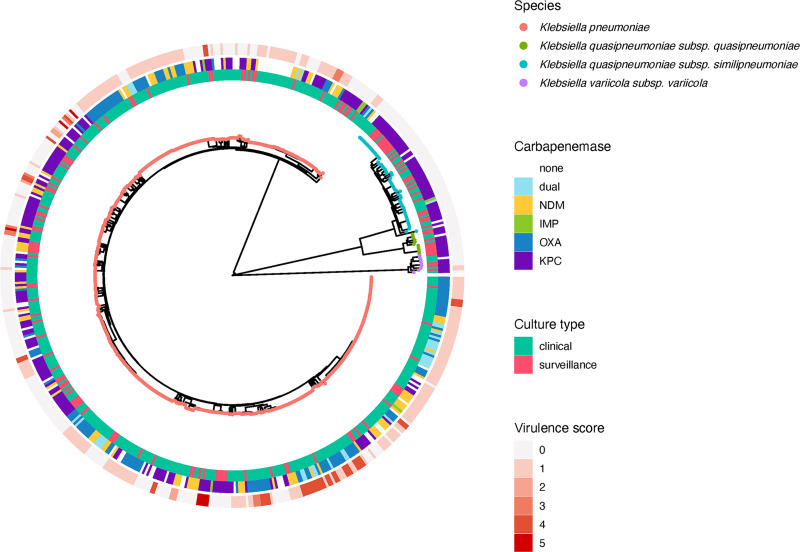
In silico species identification showed that K. pneumoniae sensu stricto (n = 500, 87.0%) accounted for the majority of the isolates, followed by K. quasipneumoniae subsp. similipneumoniae (n = 55, 9.6%), K. quasipneumoniae subsp. quasipneumoniae (n = 11, 1.9%), and K. variicola subsp. variicola (n = 9, 1.6%). Similar to previous population structure studies of K. pneumoniae, we observed three distinct phylogroups corresponding to these three species groups: KpI (K. pneumoniae sensu stricto), KpII (K. quasipneumoniae), and KpIII (K. variicola) (indicated by the different colored nodes in Fig. 1) (10).
FIG 1.
Core SNP phylogenetic tree of 575 carbapenem-resistant K. pneumoniae genomes. Tree tips are color-labeled by K. pneumoniae subspecies. The culture type (innermost ring), carbapenemase type (middle ring), and Kleborate virulence score (outermost ring) are annotated according to the legend.
CRKP was resistant to multiple antibiotic classes, whereas most isolates were resistant to levofloxacin (66.4%) in addition to the various β-lactams. Lower resistance rates were observed for amikacin (25.2%), polymyxin B (11.1%), and tigecycline (4.2%) (Table 1). The antibiotic susceptibilities of the KpI and non-KpI isolates were comparable, except for cefepime, levofloxacin, and amikacin; KpI isolates had higher cefepime susceptibility rates (5.8% versus 0%, P = 0.032) but lower amikacin (69.0% versus 97.3%, P < 0.001) and levofloxacin susceptibility rates (17.0% versus 48.0%, P < 0.001).
TABLE 1.
Antibiotic susceptibility patterns of 575 carbapenem-resistant K. pneumoniaea
| Antibiotic | % S | % I/SDD | % R | MIC data (mg/L) |
||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||||
| Ertapenem | 0.0 | 1.6 | 98.4 | ≥32 | ≥32 | ≤1 to ≥32 |
| Doripenem | 7.0 | 5.9 | 87.1 | 16 | ≥32 | ≤0.5 to ≥32 |
| Imipenem | 5.6 | 4.0 | 90.4 | 16 | ≥32 | ≤0.5 to ≥32 |
| Meropenem | 6.6 | 5.6 | 87.8 | ≥32 | ≥32 | ≤0.5 to ≥32 |
| Aztreonam | 4.9 | 1.4 | 93.7 | ≥64 | ≥64 | ≤2 to ≥64 |
| Cefepime | 5.0 | 11.0 | 84.0 | ≥64 | ≥64 | ≤1 to ≥64 |
| Piperacillin-tazobactam | 0.4 | 3.1 | 96.5 | ≥128/4 | ≥128/4 | 8/4 to ≥128/4 |
| Levofloxacin | 21.1 | 12.5 | 66.4 | 16 | ≥64 | ≤0.25 to ≥64 |
| Amikacin | 72.7 | 2.1 | 25.2 | 8 | ≥128 | ≤4 to ≥128 |
| Polymyxin B | - | 88.9 | 11.1 | 0.5 | 4 | ≤0.25 to ≥16 |
| Tigecycline | 88.3 | 7.5 | 4.2 | 1 | 4 | ≤0.25 to ≥16 |
The categorical susceptibilities were determined using CLSI breakpoints, except for tigecycline (FDA). S, susceptible; I, intermediate; SDD, susceptible dose-dependent; R, resistant; MIC, minimum inhibitory concentration.
Clonal diversity and phylogenetic analyses.
There was significant diversity observed among the K. pneumoniae species. 151 distinct STs were identified in this study, including 22 novel STs which have been submitted to the Institute Pasteur K. pneumoniae MLST database. The prevalence for each ST ranged from a single isolate to 54 isolates (ST14, 9.4%). Other predominant STs include: ST147 (n = 41, 7.1%), ST16 (n = 39, 6.8%), and ST231 (n = 33, 5.7%).
Based on the goeBURST analyses using the criteria where all members assigned to the same group are single locus variants of each other, 52 different STs were grouped into 20 CGs. The remaining 99 STs were singletons (single STs that do not correspond to any CG). The largest CGs were CG14/15, which consisted of ST14, ST15, ST709, and ST2096 (n = 79, 13.7%), and CG17/20, which consisted of ST16, ST17, ST20, and ST3247 (n = 78, 13.6%). Other noteworthy groups include CG147 (ST147, ST273, ST392, ST885, ST4843, ST5612, and ST5613) (n = 56, 9.7%), CG258 (ST11, ST258, ST340, and ST437) (n = 39, 6.8%), and ST231 (n = 33, 5.7%). Together, these CGs represented only 49.6% of the study population. The high diversity is emphasized by the large numbers of STs which were represented by only one isolate (n = 93/151, 61.6%). The high-risk MDR global clones ST15, ST20, ST37, ST101, ST147, ST258, and ST307, as well as the known hvKP clones ST23, ST25, ST65, and ST86 were detected in our population (19).
While CG17/20 was equally prevalent in both the clinical and the surveillance isolates, the other dominant CG/STs in clinical isolates were less frequently observed in the surveillance isolates (CG14/15: 6.8% versus 15.8%, P = 0.009; CG258: 1.5% versus 8.4%, P = 0.005; CG147: 4.5% versus 11.3%, P = 0.022; ST231: 2.3% versus 6.8%, P = 0.055). Instead, ST477 was observed frequently (8.3% versus 0.9%, P < 0.001) in the surveillance isolates, in view of the overrepresentation of K. quasipneumoniae in this cohort.
Carbapenemase production.
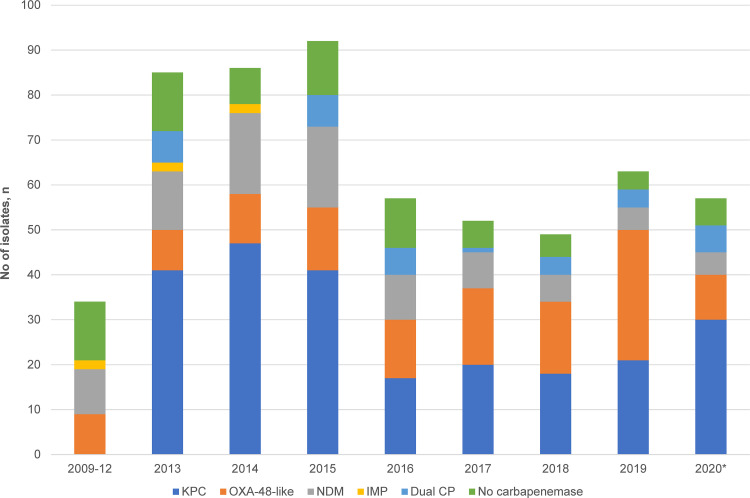
Overall, carbapenemase production was detected in 497 isolates (86.4%). KPC was the predominant carbapenemase (n = 235, 40.9%), followed by OXA-48-like (n = 128, 22.3%) and NDM enzymes (n = 93, 16.2%). A small proportion (n = 35, 6.1%) harbored two carbapenemases concurrently. Of these, most were carrying OXA-48-like enzymes in combination with NDM.
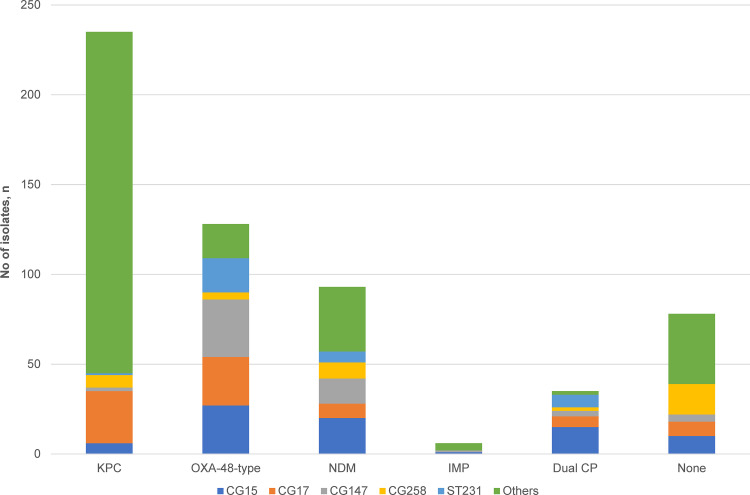
Carbapenemases were carried by diverse clones. KPC carbapenemases were disseminated by an array of clones (n = 103). Among the KPC-producers (all harbored KPC-2), a significant proportion was borne by non-KpI isolates (67/235, 28.5%; P < 0.001) (Fig. 1). The non-KpI species, K. quasipneumoniae and K. variicola, were predominantly KPC-producers (67/75, 89.3%). The NDM-producing (NDM-1, NDM-4, NDM-5, NDM-7, and NDM-9) isolates were distributed across 31 STs and displayed less diversity than the KPC-producers (distributed across 103 STs), although the predominant STs (e.g., ST14, ST15, and ST147) accounted for 61% of the NDM-carrying isolates (Fig. 2). The diversity of strains carrying OXA-48-like carbapenemases and those harboring two carbapenemases concurrently were even lower (distributed across 23 STs); isolates belonging to CG14/15, CG17/20, CG147, and ST231 accounted for 84% of the OXA-48-like population (Fig. 2). Among these OXA-48-like-carriers, the frequency of blaOXA-232 was the highest (67/161, 41.6%), followed by blaOXA-181 (51/161, 31.7%) and blaOXA-48 (43/161. 26.7%).
FIG 2.
Distribution of carbapenemases among the major clonal groups in carbapenem-resistant K. pneumoniae. The carriage of OXA-48-type enzymes was associated with CG14/15 (P ≤ 0.001), CG17/20 (P = 0.002), CG147 (P ≤ 0.001), and ST231 (P ≤ 0.001). The carriage of NDM enzymes was associated with CG14/15 (P ≤ 0.001) and ST231 (P = 0.015). KPC carriage was not associated with any of the clonal groups. The P values indicated here are determined by chi-square tests for two-group comparisons of each CG/ST versus all other CG/STs. Abbreviations: CG, clonal group; CP, carbapenemase; ST, sequence type.
In addition, the distribution of carbapenemases differed between the clinical and the surveillance isolates. While the carbapenemase types were more evenly distributed among the 443 clinical isolates (OXA-48-like: 33.9%, KPC: 33.6%, NDM: 21.0%), there was an overrepresentation of KPCs (88, 66.7%) in the 132 surveillance isolates. The NDM and OXA-48 prevalence in the surveillance isolates were 26.5% and 8.3%, respectively.
Other AMR determinants.
Acquired resistance genes to the various classes were identified in the majority of the isolates. The median number of resistance genes identified was 12 (range: 1 to 27), encompassing a median of nine antibiotic classes (range: 1 to 12).
ESBL-encoding genes were detected in 392 (68.1%) isolates, and blaCTX-M-15 was the most prevalent (328/392, 83.7%). We observed multiple ESBL genes in 16 isolates. Plasmidic ampC genes (including blaDHA and blaCMY) were far less common, occurring only in 66 (11.5%) isolates. Aside from carbapenemases, carbapenem resistance is also mediated by the combination of ESBLs and porin loss in 231 (40.2%) of the isolates, consisting of 176/497 (35.4%) of the carbapenemase-producing isolates and 55/78 (70.5%) of the non-carbapenemase producing isolates.
The prevalence of acquired genes mediating resistance to aminoglycosides (n = 467, 81.2%) and fluoroquinolones (n = 374, 65.0%) were equally high. Of note, the prevalence of 16S rRNA methyltransferases (16S RMTases), enzymes capable of mediating broad-spectrum aminoglycoside resistance, was relatively high (143/575, 24.9%). The distribution of the 16S RMTase genes were as follows: armA (72/143, 50.3%), rmtF (54/143, 37.8%), rmtB (24/143, 16.8%), and rmtC (5/143, 3.5%). 16S RMTases were primarily associated with isolates carrying OXA-48-like (71/143, 49.7%), NDM (32/143, 22.4%), or both carbapenemases (33/143, 23.1%), and they were overrepresented in ST43 (6/7, 85.7%; P < 0.001), ST231 (27/33, 81.8%; P < 0.001), and ST14 (38/54, 70.4%; P < 0.001). The prevalence of 16S RMTases was lower in the ST147 isolates (16/41, 39.0%), although this sequence type has been associated with the cocarriage of NDM and 16S RMTases in both Spain and the United States (20, 21).
Acquired AMR determinants mediating resistance to last-line agents polymyxin, tigecycline, and fosfomycin appeared to be rare. Polymyxin resistance-mediating mcr genes (mcr1.1, mcr8.2, mcr9.1, mcr9.2, and mcr10.1) were detected in 11 (1.9%) isolates. We noted that phenotypic resistance (polymyxin B minimum inhibitory concentration [MIC] > 2 mg/L) was only detected in the three mcr1.1 isolates, whereas the isolates with the other mcr variants had low MICs (0.5 to 2 mg/L). Tigecycline resistance-mediating tet(X4) genes were detected in only two (0.4%) isolates. This is much lower than the fecal carriage rate (10%) reported in another local study involving healthy volunteers (22). All isolates harbored chromosomal fosA, but plasmid-mediated fos genes (fosA3 and fosA4) were detected in only 12 (2.1%) isolates. High fosfomycin MICs were observed for all 12 isolates (11 isolates, MIC ≥ 2048mg/L; 1 isolate, MIC = 16mg/L).
Virulence-associated genetic determinants.
We detected 83 different known K loci encoding the core capsule biosynthesis machinery, with KL51 (n = 70, 12.2%), KL2 (n = 46, 8.0%), KL64 (n = 45, 7.8%), KL15 (n = 30, 5.2%), and KL10 (n = 29, 5.0%) representing the most common loci. K1, the capsular serotype associated with K. pneumoniae abscesses, was rare and was found exclusively in the five ST23 hvKP isolates observed in this study. Of these, three of the isolates have been extensively profiled previously (23). We did not observe any strong association of K serotypes with phylogenetic lineages, which paralleled previous observations, in which K serotypes may be associated with several clones and vice versa (24). There were far fewer distinct O loci observed (n = 11), with O1 (n = 213, 37.0%), O2 (106, 18.4%), and O3b (84, 14.6%) representing the most common loci in concordance with previous reports (25).
The prevalence of the other virulence factors is displayed in Table 2. The gene encoding yersiniabactin (ybt), which was associated with 9 different chromosomally integrated mobile elements (ICEKp) and two plasmids, was detected in 202 (41.6%) isolates. ICEKp3, ICEKp4, and ICEKp5 were the most common among these mobile elements. The gene encoding aerobactin (iuc), which was used to define hypervirulence in this study, was detected in 59 (10.3%) isolates. All 59 isolates harbored carbapenemase-encoding genes, denoting the convergence of hypervirulence and multidrug resistance. Aside from the known hvKP lineages (ST23, ST65, and ST86), the presence of iuc was observed primarily in the ST231-K51/O1 strains (21/59, 35.6%), all of which harbored blaOXA-181/232 and/or blaNDM. The remaining virulence genes (iro, clb, rmpA, and rmpA2), were far less prevalent, occurring in only 2.4 to 4.5% of the isolates.
TABLE 2.
Virulence characteristics of 575 carbapenem-resistant K. pneumoniae isolates
| Isolate type | No. of isolates | % prevalence |
Kleborate virulence score |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ybt | clb | iuc | iro | rmp/rmp2 | Mean | Median | Range | ||
| All isolates | 575 | 46.4 | 2.4 | 10.3 | 3.7 | 4.5 | 0.8 | 0 | 0 to 5 |
| KPC | 235 | 18.7 | 4.7 | 6.8 | 6.8 | 6.4 | 0.4 | 0 | 0 to 5 |
| OXA | 128 | 88.3 | 0 | 23.4 | 0 | 3.9 | 1.6 | 1 | 0 to 4 |
| NDM | 93 | 58.1 | 3.2 | 6.5 | 4.3 | 5.4 | 0.8 | 1 | 0 to 4 |
| IMP | 6 | 16.7 | 0 | 0 | 0 | 0 | 0.2 | 0 | 0 to 1 |
| Dual carbapenemase | 35 | 74.3 | 0 | 14.3 | 0 | 0 | 1.2 | 1 | 0 to 4 |
| No carbapenemase | 78 | 37.2 | 0 | 2.6 | 1.3 | 1.3 | 0.5 | 0 | 0 to 4 |
| Clinical isolates | 443 | 54.0 | 2.7 | 12.4 | 3.8 | 5.0 | 0.9 | 1 | 0 to 5 |
| Surveillance isolates | 132 | 21.2 | 1.5 | 3.0 | 3.0 | 3.0 | 0.3 | 0 | 0 to 5 |
Virulence factors were absent in all non-KpI isolates, except for two ybt+ K. variicola isolates. This was also reflected by the generally low virulence scores in this group (Fig. 1), which suggested a limited virulence potential in these species and was in concordance with earlier studies that reported low virulence frequencies in these species (26, 27). Similarly, the surveillance isolates generally possessed fewer virulence factors and corresponding lower virulence scores (Table 2).
Plasmid analyses.
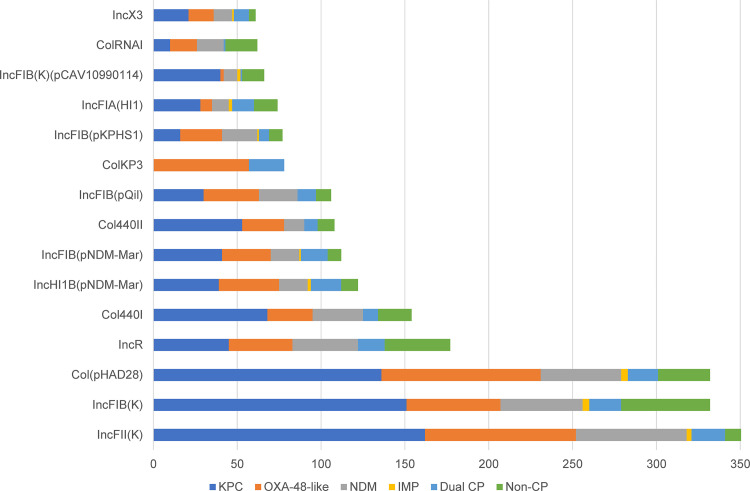
The plasmid analyses identified highly diverse incompatibility (Inc) groups (n = 53) within the population surveyed. The frequencies of the various replicon types are shown in Fig. 3. The number of replicons detected in each isolate ranged from 1 to 11 (median: 6), suggesting that some of our isolates might carry multiple plasmids, therefore resulting in the high prevalence of acquired resistance genes from different classes observed in this study.
FIG 3.
Frequency of plasmid replicon types in carbapenem-resistant K. pneumoniae.
Unfortunately, due to the limitations of short-read sequencing, we were unable to conduct detailed plasmid investigations. Based on our analyses, we were unable to detect the replicon types for all of the blaKPC-containing contigs using PlasmidFinder, which may be related to the inability of the current typing scheme to detect all known plasmid types or the contiguous rearrangement/mutations in the regions used for plasmid typing. Here, the majority of the blaKPC appeared to be driven by non-Tn4401 elements; most KPC sequences (234/237, 98.7%) mapped to the non-Tn4401 prototype sequence pFP10-1 (GenBank: HQ651092.1), concurring with previous observations (17, 28). Additionally, we observed that 216/237 (91.1%) isolates also had sequence reads that mapped to that of a previously identified local blaKPC plasmid, pKPC2_sg2 (GenBank: MN542378.1) (23).
Similarly, we were unable to identify the replicon types of the blaNDM-containing contigs for many samples (89/128, 69.5%). Among the replicons identified, the broad-host range IncN2 replicons were the most frequently detected (31/128, 24.2%). All 31 samples (17 different STs) had reads that mapped to pNDM-ECS01 (GenBank accession: KJ413946.1), which was previously described in NDM-producing E. coli and K. pneumoniae in Singapore (29). Other minority replicons identified included IncC, IncFIB, IncFII, IncHI1B, and IncX3. Only one isolate (ST147) mapped to the pSg1-NDM plasmid (GenBank accession: CP011839.1), which was associated with an interinstitutional outbreak in Singapore (29).
On the other hand, most of the contigs containing blaOXA-181 or blaOXA-232 were associated with ColKP3 plasmid replicons (78/118, 66.1%), while a smaller subset carrying blaOXA-181 were associated with IncC replicons (19/118, 16.1%). The strong association between the ColKP3 plasmid and blaOXA-232 has been confirmed previously (30). We could not identify the replicons within the remaining five blaOXA-181 contigs or the replicons within all 43 blaOXA-48 contigs with PlasmidFinder.
Read-mapping to the pKP3-A reference plasmid (GenBank accession: JN205800), the plasmid bearing blaOXA-181 isolated from K. pneumoniae and recovered from a patient in Oman (31) revealed its presence in 62/67 (92.5%) of the blaOXA-232-carrying samples and in 7/51 (13.7%) of the blaOXA-181-carrying samples, accounting for 42.9% (69/161) of all of the blaOXA-48-like samples. pKP3-A was not detected in any of the blaOXA-48 samples; instead, read-mapping identified the presence of the pOXA-48a IncL/M plasmid (GenBank accession: JN626286), albeit in only 20/43 (46.5%) of our blaOXA-48 isolates.
Analyses of the major clones.
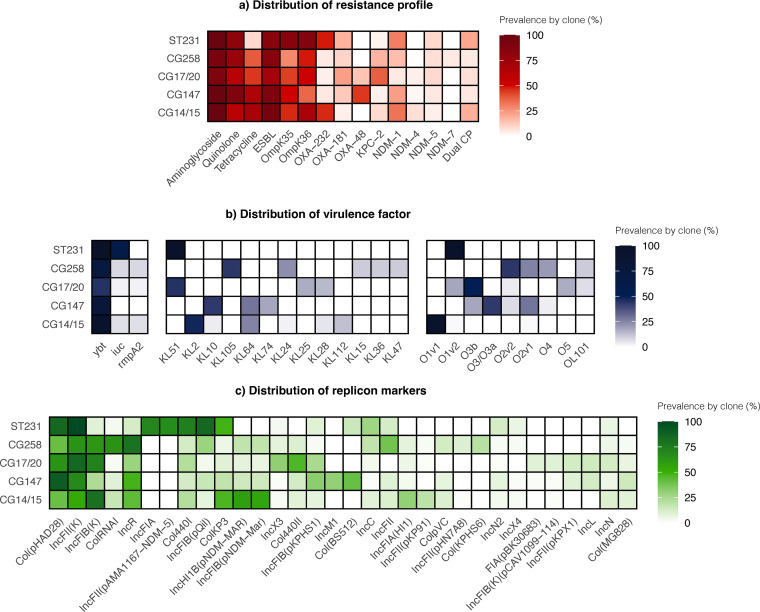
We also sought to explore the major clones in our study. The prevalence of the major classes of AMR determinants, virulence factors, and plasmid replicons within the various CGs is depicted in Fig. 4. Generally, we did not observe any strong overall pattern of association of genomic characteristics with the individual lineages; the various resistance, virulence and plasmid markers were distributed among the major lineages.
FIG 4.
Clonal group heatmaps of carbapenem-resistant K. pneumoniae displaying distributions of (a) key resistance mechanisms. The aminoglycoside, quinolone, and tetracycline groups refer to acquired genes conferring resistance to the antibiotic group. (b) Virulence factors, K locus, and O locus. (c) Plasmid replicons. The values represent the proportions of isolates within each clonal group bearing the individual genetic elements displayed on the horizontal axis. Factors/elements with overall prevalence <10% are not displayed in this figure. Abbreviations: CG, clonal group; CP, carbapenemase-producing; ESBL, extended-spectrum β-lactamase; ST, sequence type.
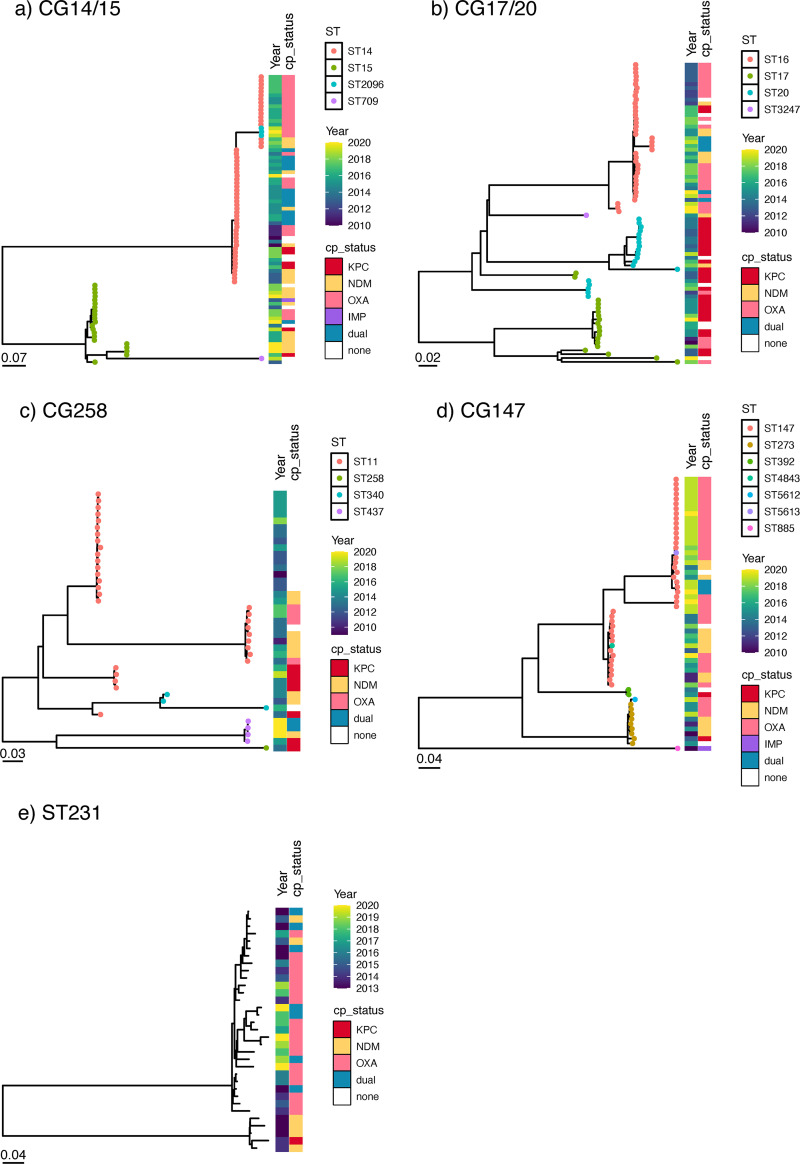
Genomic diversity within each ST is also demonstrated as indicated by the different clades on the individual CG/ST phylogenetic trees (Fig. 5). There were subsets of strains with pairwise SNP differences of ≤25 (Fig. S3A–E) (32). These clusters may represent putative clonal transmission events and should be further confirmed with clinical epidemiological findings.
FIG 5.
Core SNP phylogenetic trees of major carbapenem-resistant K. pneumoniae lineages. (a) A maximum likelihood tree of CG 14/15 was inferred from the mapping of 79 genomes to reference EC1155. (b) A maximum likelihood tree of CG 17/20 was inferred from the mapping of 78 genomes to reference EC0720. (c) A maximum likelihood tree of CG258 was inferred from the mapping of 37 genomes to reference EC3204. (d) A maximum-likelihood tree of CG147 was inferred from the mapping of 56 genomes to reference EC0325. (e) A maximum-likelihood tree of ST231 was inferred from the mapping of 33 genomes to reference EC2886. Abbreviations: CG, clonal group; CP, carbapenemase producer; SNP, single nucleotide polymorphism; ST, sequence type.
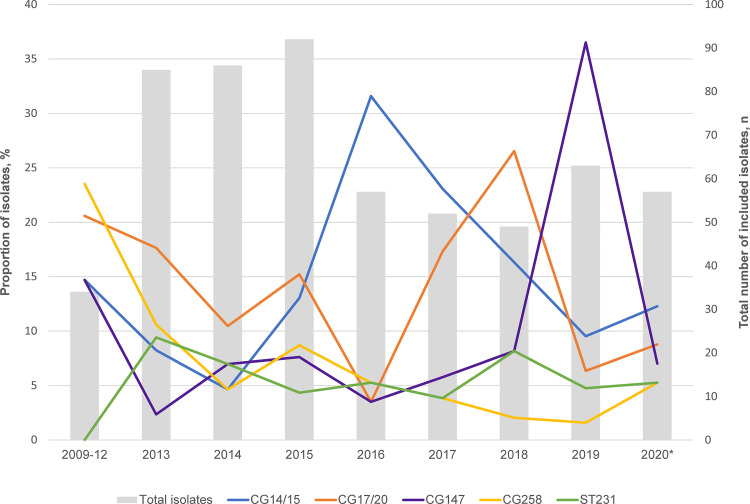
However, it was striking that there was a convergence of resistance and hypervirulence elements in ST231 K. pneumoniae, a lineage more commonly associated with multidrug resistance (Fig. 4). ST231 was first recorded in our collection in 2013, and its prevalence remained stable through the study period (Fig. 6). More than half of the ST231 isolates (21/33, 63.6%) were assigned virulence scores of 4. Additionally, these ST231 isolates exhibited conservation of the K-locus and O-locus (KL51:O1V2), in contrast to the diversity of serotypes observed with the other lineages (Fig. 4B). Furthermore, many of these ST231 isolates carried blaOXA-181 or blaOXA-232 on similar ColKP3 plasmid backbones, although the possibility of a clonal expansion was not supported by the phylogenetic analyses, as most pairs had SNP distances of >25 (Mean pairwise SNP distance: 286, range: 6 to 958) (Fig. 5e; Fig. S3E).
FIG 6.
Temporal distribution of the major circulating carbapenem-resistant K. pneumoniae lineages. The primary axis (line graph) denotes the proportion of each CG/ST of the 575 isolates which was included for sequencing. The secondary axis (bar graph) denotes the number of included isolates per year. Isolates from 2009 to 2012 were pooled due to the low number of isolates collected in that time period.
Another noteworthy finding was the spike in isolates belonging to CG147 in 2019, primarily driven by ST147 (Fig. 6). This may also have contributed to the increasing trend in blaOXA-48-like in the recent years, since more than half of ST147 CRKP (30/41, 73.2%) harbored the gene with/without NDM (Fig. 7; Fig. 5D). The phylogenetic analyses suggested the possibility of an undetected transmission cluster, although this requires verification with clinical epidemiological evidence (Mean pairwise SNP difference: 13, range: 9 to 16) (Fig. 5D; Fig. S3D).
FIG 7.
Temporal distribution of carbapenemases in carbapenem-resistant K. pneumoniae. Isolates from 2009 to 2012 were grouped together, as cases were few and collection was sporadic. *Does not include data for the full year.
DISCUSSION
In this study, we utilized WGS concurrently with phenotypic surveillance to characterize CRKP isolates collected from a large tertiary public health hospital in Singapore. To our knowledge, this is the largest WGS-based survey of CRKP in Singapore. This study serves to complement and update the growing body of epidemiological findings regarding this pathogen in Singapore.
Circulating CRKP in the institution are highly diverse, composing a polyclonal population structure represented by several well-known international, high-risk clones (ST231, CG147, CG258, CG14/15, and CG17/20) (24). Our findings indicate the limited role of clonal transmission, as we observed only 2,448 of the 330,625 pairs (0.7%) possessing SNP differences below the threshold of 25. It appears that infection prevention measures, such as targeted surveillance, isolation precautions, and strict hand hygiene, were successful in controlling CRKP transmissions between patients residing in the hospital wards, resulting in the stabilization of CRKP incidence rates after 2013. Nevertheless, the acquisition of carbapenem resistance could have also occurred through indirect ward/hospital contact, which was not as easily detected. This hypothesis is supported by the findings from another local paper, which identified an independent association of indirect contact with putative clonal transmissions (33). Furthermore, the larger contribution of plasmid transmission toward carbapenem resistance dissemination may be related to non-human reservoirs, such as the hospital environment, which has been identified in previous studies as an important source of MDR organisms. Future control efforts should be directed to environmental surveillance that includes high-risk areas, such as sinks and toilets, to identify hidden CRKP reservoirs (34, 35).
In a previous study of acquired carbapenemases in the institution from 1996 to 2012, ST14 K. pneumoniae was the most common strain that contributed to carbapenem resistance in the institution (36). blaOXA-181-producing ST14 strains have also been detected in other public hospitals in Singapore (37). In our study, CG14 (comprising largely of ST14 and ST15) was observed to be one of the predominant carbapenem-resistant clonal groups over the study period. CG14 represents the most common third-generation, cephalosporin-resistant clones in various regions, particularly on the Asian continent (19, 38). This might explain its high prevalence (13.7%) in this study, considering the endemicity of ESBL-producing Enterobacterales in our region. Carbapenem resistance has emerged in this clonal complex and has been described in isolates producing the OXA-48-type with or without NDM carbapenemases in Tanzania, South Korea, the United Arab Emirates, and Turkey (19, 39–42). While some of the earliest cases of ST14 carbapenemase producers (observed in 2001) were reported to have been isolated from patients with travel history to Bangladesh, where OXA-181-producing Enterobacterales are endemic (43), it is likely that circulating CG14 strains that have established a successful foothold in the local setting have also acquired different carbapenemases (including KPC, IMP, OXA-48, and and NDM) over multiple independent events, as exemplified by the large pairwise SNP differences observed with the ST14 isolates.
Likewise, the CG258 subclone ST11 is historically one of the most common carbapenemase-carrying clones in the local population (36). ST11 K. pneumoniae is the dominant KPC clone in China, and it is the single-locus variant of ST258, the widely circulating KPC-2/3 clone in the United States and parts of Europe (Italy and Greece) (44). ST11 constituted the majority of the CG258 strains in our study; there was only one occurrence of the ST258 strain which was a KPC-2 carrier, while the ST512 strain was absent. A 2011 study involving four KPC-producing isolates from various Singapore hospitals identified that all strains belonged to ST11, suggesting that the early dissemination of KPC locally may be attributed to ST11 (28).
However, we found that KPC enzymes were later disseminated via diverse clones over the study period. KPC carriage was not independently associated with any of the prevalent clones in this study, and it often occurred in non-CG258 strains. CG258 accounted only for 7 (2.9%) of the 235 KPC-carrying isolates. Instead, the CG258 strains were either non-carbapenemase producing or carriers of NDM/OXA-48 enzymes. This is in concordance with the findings by the CaPES group, which attributed KPC-2 prevalence in Singapore public health hospitals primarily to non-CG258 isolates (17). Furthermore, the findings here point toward the likelihood of local KPC dissemination being mediated by a few successful plasmids transmitted across multiple different lineages, as evidenced by majority of the KPC-containing contigs mapping to a local blaKPC plasmid. This observation is also supported by various other local studies and may explain the predominance and endemicity of KPC enzymes locally (17, 23).
Our study also highlights the emergence of several notable high-risk lineages. ST231 is an emerging CRKP epidemic clone which has been reported in several countries, including India, Thailand, Brunei, France, and Switzerland (45, 46). In India, there were several reports describing this lineage as one of the predominant STs in the region (47, 48). Our isolates share common genomic characteristics with these previously described ST231 strains; they were MDR and mostly carried blaOXA-181/232, blaCTX-M, 16S RMTases (armA/rmtB/rmtF), porin mutations, and the virulence genes, ybt and iuc. Cocarriage of blaNDM was also frequently observed, conferring high levels of resistance. The convergence of virulence and resistance elements (including 16S RMTases with the ability to hydrolyze new aminoglycosides, such as plazomicin) in this emerging CRKP high-risk clone requires high vigilance, as this may represent an important reservoir of genetic elements which may be horizontally transferred to other lineages.
We also observed ST147, which is an emerging epidemic clone capable of disseminating various resistance elements. This clone likely emerged in Europe in the early 1990s, where it acquired gyrA and parC mutations following the introduction of ciprofloxacin, and it is associated with blaCTX-M-15. In the late 2000s, the clone came to be associated with the various carbapenemases, according to the geographical regions in which specific carbapenemases prevailed (e.g., NDM and OXA-181 in India and KPC in Greece), allowing the clones to establish themselves as endemic pathogens in these locales (49). The ST147 strains in our study were most frequently associated with OXA-48-like enzymes. Additionally, the genetic relatedness of the strains suggests local clonal expansion, which may explain the increasing trend of ST147 and the corresponding increasing prevalence of OXA-48-like enzymes from 2018 to 2020. Interestingly, this clone has been implicated in a local interinstitutional transmission of blaNDM on a novel plasmid, pSG1-NDM, previously, although we only detected the plasmid in one sample within our collection (29).
There is evidence of recent OXA-48-like outbreaks that involved multiple Enterobacterales within the hospital (50). While the clonal transmission of ST147 may have contributed to the displacement of the other carbapenemases by OXA-48-like enzymes, horizontal transmission of OXA-48-plasmids likely played an important role in the increasing prevalence of OXA-48 in the institution, as it can even spread to/via other species. Previous studies have shown that blaOXA-48 dissemination in certain regions, such as the Mediterranean and Western Europe, was a consequence of the successful spread of single plasmids. Different clones from different geographical areas, and even different Enterobacterales species, carried blaOXA-48 on plasmids that shared similar genetic features/plasmid backbones. These were typically self-conjugative plasmids of 60 to 70 kb in size (51–54). Similar observations exist for the blaOXA-232 plasmids. There were several reports documenting the carriage of blaOXA-232 on ~6kb plasmids with ColE/KP3-like backbones in isolates from India, the United States, and China, and these plasmids often showed high similarity to the blaOXA181-bearing pKP3-A plasmid from Oman, which was also found in high frequency in our study (31, 55, 56).
This study is not without limitations. Sample collection and sequencing were not exhaustive, due to resource constraints. As surveillance was not largely mandated or standardized, we were reliant on the sampling criteria and procedures of the institution. Surveillance isolates were tested for carbapenamase production using GeneXpert Xpert Carba-R, and they were not consistently cultured out after 2015, resulting in the uneven distribution of surveillance isolates throughout the study period. The lack of systematic sampling could potentially have introduced unintentional sampling bias. Moreover, the exclusion of antibiotic-susceptible strains and the lack of such existing local info impeded our analysis of the evolution of the AMR strains. As the study was conducted in a single center, the results here may not be generalizable to the local population at large. In addition, the lack of clinical epidemiological information prohibited us from establishing conclusive evidence regarding transmission clusters, as epidemiologically unrelated samples could also display little genetic variability at the SNP level. Lastly, we relied on short-read sequencing, which is limited in its resolution, in the investigation of the mobile genetic elements.
Conclusions.
Through this study, we have demonstrated the utility of WGS in improving the understanding of AMR transmission. We established that the local population structure of CRKP encompasses well-known epidemic, MDR clones amid a diverse pool of nonepidemic lineages. The multiple phylogenetic clusters of the major circulating clones here primarily demonstrated multiple independent acquisition events, as opposed to clonal expansion/transmission. However, subsets of strains found to be clustering together may point toward outbreaks/reservoirs (e.g., ST147) which otherwise would not have been resolved via conventional typing methods. Furthermore, the plasmid analyses illustrated a divergence in the transmission dynamics of the three carbapenemases. Further exhaustive analyses could potentially uncover the local evolutionary perspectives of these carbapenemases.
The comprehensive resistance, virulence, and plasmid gene content profiling provided critical information for use in AMR monitoring and highlighted future surveillance priorities. While problematic multidrug resistance elements, such as mcr and tet(X), remained rare, the emergence of ST231 K. pneumoniae bearing multidrug resistance genes (including those that confer resistance to novel agents) and virulence elements concurrently warrants closer attention. The seemingly increasing blaOXA-48-like trend is also concerning and deserves ongoing surveillance. The establishment of this detailed hospital-specific repository will not only enable more precise and rapid analyses of future isolates to allow for the timelier identification of areas of concerns and corresponding interventions, but also serves to inform future treatment guidelines and research priorities.
Lastly, the findings here reinforce the necessity of unique infection control and prevention strategies that take into consideration the genomic diversity of local circulating strains. We should be mindful that strategies developed or employed in geographical areas with a limited number of circulating clones may not necessarily be adopted here effectively.
MATERIALS AND METHODS
Study setting.
This study was conducted at the Singapore General Hospital, the largest public acute care tertiary hospital in Singapore. The hospital has approximately 1,800 beds and accounts for approximately 25% of the total acute hospital beds in the public sector and 20% of the acute beds nationwide. A wide range of medical and surgical specialties are offered by the hospital, and it is the national and regional referral center for services such as plastic surgery, burns, renal medicine, nuclear medicine, pathology, and hematology.
Specific interventions targeted at carbapenemase-producing Enterobacterales control included the active surveillance of carbapenemase carriage in high-risk patients (hospitalization in the preceding year; admission to intensive care, renal, hematology, or oncology units; and a duration of hospital stay of ≥2 weeks) and contact precautions for colonized/infected patients (isolation in single/cohort rooms). The targeted surveillance of carbapenemase-producing organisms only commenced in the hospital after 2013 (50, 57). The incidence rates of CRKP in the hospital over the study period are displayed in Fig. S1.
Bacterial isolates.
A total of 575 CRKP isolates from 547 adult inpatients collected between 2009 and 2020 were available for WGS and were included in the analyses. Only one isolate was included per patient per year, unless the isolates displayed different genotypic characteristics (e.g., different carbapenemases). The sequenced isolates represented approximately 75% and 14% of the clinical and surveillance CRKP isolates collected over the study period, respectively (Fig. S2). Carbapenem resistance was defined as nonsusceptibility to at least one carbapenem (ertapenem, doripenem, meropenem, or imipenem). These isolates were collected at the institution’s Microbiology Laboratory and tested at the Pharmacy Research Laboratory as part of an informal surveillance study of carbapenem nonsusceptible Gram-negative organisms. Isolates received at the Pharmacy Research Laboratory for antibiotic combination testing were also included. The collection included both routine surveillance (n = 132, 23.0%) and clinical isolates (n = 443, 77.0%) from various sources (urinary, blood, gastrointestinal/abdominal, wound, respiratory, and bone). Generally, invasive isolates from sterile sites were prioritized for WGS.
All isolates were identified using Vitek GNI+ cards with the Vitek 2 instrument (bioMérieux, Hazelwood, MO, USA) and/or a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) system (Bruker Daltonik, Germany) as part of the routine workflow of the institution’s microbiology laboratory. Only isolates which were eventually identified in silico as K. pneumoniae sensu lato were included in the analyses.
Antibiotic susceptibilities.
MICs were obtained using customized 96-well broth microdilution panels (TREK Diagnostics, East Grinstead, UK) in accordance with the manufacturer’s recommendations and were interpreted according to the Clinical & Laboratory Standards Institute (CLSI) breakpoints (58), except for tigecycline, which was interpreted according to the US FDA criteria. Fosfomycin MICs were obtained using gradient MIC test strips (bioMérieux, Marcy l’Etoile, France). Escherichia coli ATCC 25922 was used as the quality control strain.
DNA preparation and whole-genome sequencing.
Overnight bacterial cultures in cation-adjusted Muller-Hinton were prepared and used for genomic DNA extraction using the DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. The genomic DNA was then sent for paired-end WGS using MiSeq/HiSeq systems (Illumina Inc., CA, USA), with a resultant sequencing depth of at least 50-fold. Additionally, some of the raw sequences were retrieved from the multicenter study (Carbapenemase-Producing Enterobacteriaceae Study [CaPES]), which involved major public health hospitals in Singapore (BioProject PRJNA342893) (6). Raw sequences were assessed for quality using FastQC (v0.11.3, Babraham Institute), followed by the removal of adaptors and poor-quality bases and sequences using Trimmomatic (59, 60). The trimmed sequences were then assembled de novo using the SPAdes software (61).
Antimicrobial resistance profiling, serotyping, and virulence gene characterization.
Relevant resistance genes were identified using the NCBI-AMRFinderPlus database (v3.10.16) and the Kleborate tool (v.2.0.4) (https://github.com/katholt/Kleborate) (62). The Kleborate tool was also used to identify the ICEKp-associated and plasmid-associated virulence loci (i.e., genes encoding yersiniabactin [ybt], aerobactin [iuc], salmochelin [iro], the colibactin toxin [clb], and the mucoid phenotype regulators [rmpA, rmpA2]). Kleborate assigns a virulence score to each of the samples. Virulence scores are ranked from 0 to 5 based on the presence of ybt, clb, and iuc as follows: 0 = none present, 1 = ybt only, 2 = clb with/without ybt, 3 = iuc only, 4 = iuc and ybt, and 5 = ybt, clb, and iuc. “Convergent” K. pneumoniae are defined as strains which are both hypervirulent (carrying the iuc aerobactin locus) and MDR (carrying ESBL or carbapenemase genes) (39).
The Kaptive tool was also used to perform capsular typing, in which capsule polysaccharide (K) and lipopolysaccharide (O) loci were determined (63). Plasmid replicons were identified with PlasmidFinder (64).
In silico multilocus sequence typing and phylogenetic analyses.
STs were identified by BLAST, using the scheme in the Pasteur database (https://bigsdb.pasteur.fr/klebsiella/). Core genome alignment of the assembled K. pneumoniae draft genomes was performed using Parsnp from the Harvest suite of phylogenetic tools (65). The alignment was run with EC0295, which was randomly chosen by Parsnp as the reference genome for the main tree. For the individual CG/ST trees, the reference genomes were EC1155, EC0720, EC3204, EC0325, and EC2886 for the CG14/15, CG17/20, CG147, CG258, and ST231 phylotrees, respectively. The resultant phylogenetic trees were visualized using the R ggtree package (66). The transmission inference threshold used to identify genomically closely related isolates was a SNP difference of ≤25 (32).
Statistical methods.
Comparisons involving dichotomous variables were tested using a Chi-square test or Fisher’s exact test, as appropriate. No adjustment was made for multiple comparisons, due to the exploratory nature of this study. All statistical analyses were conducted with IBM SPSS Statistics for Windows, Version 26.0 (IBM Corp., Armonk, NY). A two-tailed value of P < 0.05 was considered to be indicative of statistical significance.
Ethics statement.
This study is exempt from review by the Singhealth Centralised Institutional Review Board, as it is a retrospective study involving archival bacterial isolates, which does not fall under the Human Biomedical Research Act. No identifiable data were collected.
Data availability.
The whole-genome sequences of CRKP used in this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA577535 (https://www.ncbi.nlm.nih.gov/bioproject/).
ACKNOWLEDGMENTS
We thank the laboratory staff at the Singapore General Hospital Microbiology Laboratory and Pharmacy Research Laboratory for assisting in the collection of the isolates.
This study was supported by funding from the National Medical Research Council, Singapore (NMRC/CG/C005/2017, NMRC/CG/M011/2017, NMRC/MOH-000018-00, NMRC/CG21APR2005, NMRC/CG21APR1011) and the Saw Swee Hock School of Public Health Infectious Diseases Program (SSHSPH ID-PRG/SeedFund/2018/05). Funding for Cheng Yee Tang and Rick Twee-Hee Ong was provided by the NUS Start-Up Grant, which was awarded to Rick Ong. The funders had no role in the study design, data collection, data interpretation, or decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Rick Twee-Hee Ong, Email: ephoth@nus.edu.sg.
Florence Claude Doucet-Populaire, University Paris-Saclay, AP-HP Hôpital Antoine Béclère, Service de Microbiologie, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS.
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2019. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC. [Google Scholar]
- 3.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Teo JQ, Cai Y, Lim TP, Tan TT, Kwa AL. 2016. Carbapenem resistance in Gram-negative bacteria: the not-so-little problem in the little red dot. Microorganisms 4:13. doi: 10.3390/microorganisms4010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Bij AK, Pitout JD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 6.Marimuthu K, Venkatachalam I, Khong WX, Koh TH, Cherng BPZ, Van La M, De PP, Krishnan PU, Tan TY, Choon RFK, Pada SK, Lam CW, Ooi ST, Deepak RN, Smitasin N, Tan EL, Lee JJ, Kurup A, Young B, Sim NTW, Thoon KC, Fisher D, Ling ML, Peng BAS, Teo YY, Hsu LY, Lin RTP, Ong RT, Teo J, Ng OT, Carbapenemase-Producing Enterobacteriaceae in Singapore Study Group . 2017. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae among adult inpatients in Singapore. Clin Infect Dis 64:S68–S75. doi: 10.1093/cid/cix113. [DOI] [PubMed] [Google Scholar]
- 7.One Health Antimicrobial Resistance Working Group. 2017. One Health report on antimicrobial utilisation and resistance. Singapore: One Health Antimicrobial Resistance Working Group. [Google Scholar]
- 8.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baquero F, Tedim AP, Coque TM. 2013. Antibiotic resistance shaping multi-level population biology of bacteria. Front Microbiol 4:15. doi: 10.3389/fmicb.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 12.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decre D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marr CM, Russo TA. 2019. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther 17:71–73. doi: 10.1080/14787210.2019.1555470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Kreiswirth BN. 2018. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis 18:2–3. doi: 10.1016/S1473-3099(17)30517-0. [DOI] [PubMed] [Google Scholar]
- 17.Octavia S, Kalisvar M, Venkatachalam I, Ng OT, Xu W, Sridatta PSR, Ong YF, Wang L, Chua A, Cheng B, Lin RTP, Teo JWP. 2019. Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J Antimicrob Chemother 74:3205–3210. doi: 10.1093/jac/dkz332. [DOI] [PubMed] [Google Scholar]
- 18.Prestinaci F, Pezzotti P, Pantosti A. 2015. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 20.Arca-Suarez J, Rodino-Janeiro BK, Perez A, Guijarro-Sanchez P, Vazquez-Ucha JC, Cruz F, Gomez-Garrido J, Alioto TS, Alvarez-Tejado M, Gut M, Gut I, Oviano M, Beceiro A, Bou G, Group G-SRES . 2022. Emergence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacterales in Spain studied by whole-genome sequencing. Int J Antimicrob Agents 59:106456. doi: 10.1016/j.ijantimicag.2021.106456. [DOI] [PubMed] [Google Scholar]
- 21.Lee C-S, Vasoo S, Hu F, Patel R, Doi Y. 2014. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J Clin Microbiol 52:4109–4110. doi: 10.1128/JCM.01404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y, Saw WY, Tan LWL, Moong DKN, Nagarajan N, Teo YY, Seedorf H. 2020. Emergence of tigecycline- and eravacycline-resistant Tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J Antimicrob Chemother 75:3480–3484. doi: 10.1093/jac/dkaa372. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, Prakki SRS, Xu W, Tan YH, Nguyen LC, Koh TH, Ng OT, Gan YH. 2020. Acquisition of plasmid with carbapenem-resistance gene blaKPC2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis 26:549–559. doi: 10.3201/eid2603.191230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Follador R, Heinz E, Wyres KL, Ellington MJ, Kowarik M, Holt KE, Thomson NR. 2016. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom 2:e000073. doi: 10.1099/mgen.0.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4:e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chew KL, Octavia S, Lai D, Lin RTP, Teo JWP. 2021. Genomic characterization of Klebsiella quasipneumoniae from clinical specimens in Singapore. Antimicrob Agents Chemother 65:e0041221. doi: 10.1128/AAC.00412-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balm MN, Ngan G, Jureen R, Lin RT, Teo J. 2012. Molecular characterization of newly emerged blaKPC-2-producing Klebsiella pneumoniae in Singapore. J Clin Microbiol 50:475–476. doi: 10.1128/JCM.05914-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khong WX, Marimuthu K, Teo J, Ding Y, Xia E, Lee JJ, Ong RT, Venkatachalam I, Cherng B, Pada SK, Choong WL, Smitasin N, Ooi ST, Deepak RN, Kurup A, Fong R, Van La M, Tan TY, Koh TH, Lin RT, Tan EL, Krishnan PU, Singh S, Pitout JD, Teo YY, Yang L, Ng OT, Carbapenemase-Producing Enterobacteriaceae in Singapore Study G . 2016. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother 71:3081–3089. doi: 10.1093/jac/dkw277. [DOI] [PubMed] [Google Scholar]
- 30.Argimon S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, Abudahab K, Yeats CA, Goater R, Taylor B, Harste H, Muddyman D, Feil EJ, Brisse S, Holt K, Donado-Godoy P, Ravikumar KL, Okeke IN, Carlos C, Aanensen DM, Resistance NGHRUoGSoA . 2021. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis 73:S325–S335. doi: 10.1093/cid/ciab784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorrie CL, Da Silva AG, Ingle DJ, Higgs C, Seemann T, Stinear TP, Williamson DA, Kwong JC, Grayson ML, Sherry NL, Howden BP. 2021. Key parameters for genomics-based real-time detection and tracking of multidrug-resistant bacteria: a systematic analysis. Lancet Microbe 2:e575–e583. doi: 10.1016/S2666-5247(21)00149-X. [DOI] [PubMed] [Google Scholar]
- 33.Marimuthu K, Venkatachalam I, Koh V, Harbarth S, Perencevich E, Cherng BPZ, Fong RKC, Pada SK, Ooi ST, Smitasin N, Thoon KC, Tambyah PA, Hsu LY, Koh TH, De PP, Tan TY, Chan D, Deepak RN, Tee NWS, Kwa A, Cai Y, Teo Y-Y, Thevasagayam NM, Prakki SRS, Xu W, Khong WX, Henderson D, Stoesser N, Eyre DW, Crook D, Ang M, Lin RTP, Chow A, Cook AR, Teo J, Ng OT, Marimuthu K, Venkatachalam I, Cherng BPZ, Fong RKC, Pada SK, Ooi ST, Smitasin N, Thoon KC, Hsu LY, Koh TH, De PP, Tan TY, Chan D, Deepak RN, Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group, et al. 2022. Whole genome sequencing reveals hidden transmission of carbapenemase-producing Enterobacterales. Nat Commun 13. doi: 10.1038/s41467-022-30637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chia PY, Sengupta S, Kukreja A, Ponnampalavanar SSL, Ng OT, Marimuthu K. 2020. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob Resist Infect Control 9:29. doi: 10.1186/s13756-020-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, Program NCS, Henderson DK, Palmore TN, Segre JA, Frank KM, NISC Comparative Sequencing Program . 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9 doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh TH. 2013. Acquired carbapenemases in Gram-negative bacilli in Singapore. Doctoral dissertation. National University of Singapore, Singapore. [Google Scholar]
- 37.Balm MN, Ngan G, Jureen R, Lin RT, Teo JW. 2013. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect Dis 13:58. doi: 10.1186/1471-2334-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C. 2013. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis 13:466. doi: 10.1186/1471-2334-13-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, Ip M, Karkey A, Ling CL, Miliya T, Newton PN, Lan NPH, Sengduangphachanh A, Turner P, Veeraraghavan B, Vinh PV, Vongsouvath M, Thomson NR, Baker S, Holt KE. 2020. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med 12:11. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YJ, Kim S, Kim J, Bae S. 2020. Tracking short-term changes in the genetic diversity and antimicrobial resistance of OXA-232-producing Klebsiella pneumoniae ST14 in clinical settings. Clin Microbiol Infect 26:78–86. doi: 10.1016/j.cmi.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Moubareck CA, Mouftah SF, Pal T, Ghazawi A, Halat DH, Nabi A, AlSharhan MA, AlDeesi ZO, Peters CC, Celiloglu H, Sannegowda M, Sarkis DK, Sonnevend A. 2018. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents 52:90–95. doi: 10.1016/j.ijantimicag.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Hosbul T, Guney-Kaya K, Guney M, Sakarya S, Bozdogan B, Oryasin E. 2021. Carbapenem and colistin resistant Klebsiella pneumoniae ST14 and ST2096 dominated in two hospitals in Turkey. Clin Lab 67. [DOI] [PubMed] [Google Scholar]
- 43.Koh TH, Cao DY, Chan KS, Wijaya L, Low SB, Lam MS, Ooi EE, Hsu LY. 2012. bla(OXA-181)-positive Klebsiella pneumoniae, Singapore. Emerg Infect Dis 18:1524–1525. doi: 10.3201/eid1809.111727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 45.Boonyasiri A, Jauneikaite E, Brinkac LM, Greco C, Lerdlamyong K, Tangkoskul T, Nguyen K, Thamlikitkul V, Fouts DE. 2021. Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC Infect Dis 21:142. doi: 10.1186/s12879-021-05790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mancini S, Poirel L, Tritten ML, Lienhard R, Bassi C, Nordmann P. 2018. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J Antimicrob Chemother 73:821–823. doi: 10.1093/jac/dkx428. [DOI] [PubMed] [Google Scholar]
- 47.Nagaraj G, Shamanna V, Govindan V, Rose S, Sravani D, Akshata KP, Shincy MR, Venkatesha VT, Abrudan M, Argimon S, Kekre M, Underwood A, Aanensen DM, Ravikumar KL, Resistance NGHRUoGSoA . 2021. High-resolution genomic profiling of carbapenem-resistant Klebsiella pneumoniae isolates: a multicentric retrospective Indian study. Clin Infect Dis 73:S300–S307. doi: 10.1093/cid/ciab767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shankar C, Mathur P, Venkatesan M, Pragasam AK, Anandan S, Khurana S, Veeraraghavan B. 2019. Rapidly disseminating blaOXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol 19:137. doi: 10.1186/s12866-019-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liapis E, Pantel A, Robert J, Nicolas-Chanoine MH, Cavalie L, van der Mee-Marquet N, de Champs C, Aissa N, Eloy C, Blanc V, Guyeux C, Hocquet D, Lavigne JP, Bertrand X, Onerba . 2014. Molecular epidemiology of OXA-48-producing Klebsiella pneumoniae in France. Clin Microbiol Infect 20:O1121–3. doi: 10.1111/1469-0691.12727. [DOI] [PubMed] [Google Scholar]
- 50.Venkatachalam I, How MKB, Ko KKK, Abdul Rahman NB, Conceicao EP, Aung MK, Aung MO, Yang Y, Tan KY, Sim JXY, Lee LC, Ling ML. 2022. Healthcare-associated multispecies outbreaks of OXA-48-positive carbapenemase-producing Enterobacteriaceae in a Singapore tertiary-care hospital. Infect Control Hosp Epidemiol: 1–9. doi: 10.1017/ice.2022.28. [DOI] [PubMed] [Google Scholar]
- 51.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrer A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brehony C, McGrath E, Brennan W, Tuohy A, Whyte T, Brisse S, Maiden M, Jolley K, Morris D, Cormican M. 2019. An MLST approach to support tracking of plasmids carrying OXA-48-like carbapenemase. J Antimicrob Chemother 74:1856–1862. doi: 10.1093/jac/dkz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Lutgring JD, Zhu W, de Man TJB, Avillan JJ, Anderson KF, Lonsway DR, Rowe LA, Batra D, Rasheed JK, Limbago BM. 2018. Phenotypic and genotypic characterization of Enterobacteriaceae producing oxacillinase-48-like carbapenemases, United States. Emerg Infect Dis 24:700–709. doi: 10.3201/eid2404.171377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ministry of Health. 2013. Guidelines for control and prevention of multi-drug resistant organisms (MDROs) in healthcare facilities. Singapore: MOH. [Google Scholar]
- 58.Clinical and Laboratory Standards Institute. 2021. Performance standards for antimicrobial susceptibility testing. 31st ed CLSI supplement M100, Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 59.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. doi: 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wick RR, Heinz E, Holt KE, Wyres KL. 2018. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol 56:e00197-18. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY, McInerny G. 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3. Download spectrum.00957-22-s0001.pdf, PDF file, 3.2 MB (3.3MB, pdf)
Data Availability Statement
The whole-genome sequences of CRKP used in this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA577535 (https://www.ncbi.nlm.nih.gov/bioproject/).