ABSTRACT
Bacterial vaginosis (BV) is the most common cause of abnormal vaginal discharge. BV represents a dysbiosis with the acquisition of a diverse community of anaerobic bacteria and a reduction in lactobacilli burden. Our objective was to evaluate the Aptima BV assay kit for the diagnosis of BV. From May to August 2019, we enrolled outpatients and inpatients, including nonpregnant women above 18 with vaginosis symptoms, consulting at Nantes University hospital. The Aptima BV assay measures the loads of Gardnerella vaginalis, Atopobium vaginae, and Lactobacillus species in relation to overall bacterial load. The Aptima BV assay was compared to Nugent scoring (NS). A total of 456 women were enrolled, and 347 patients met the inclusion criteria with data available for the analysis. NS was used to classify the samples and 144 (41.5%) samples were classified as normal (NS = 0–3), 45 (13%) as BV (NS = 7–10), 38 (11%) presented an intermediate vaginal microbiota (3 < NS < 7), 79 (22.7%) had various bacteria (excluding vaginal flora), 29 (8.3%) had insufficient bacterial density, and 12 (3.5%) had a predominance of yeasts. The Aptima BV kit displayed a sensitivity of 91.1% and specificity of 94.4% with a positive predictive value (PPV) of 83.7% and a negative predictive (NPV) value of 97.1%. The results of this monocentric retrospective study show that Aptima BV kit has a good diagnostic correlation compared to standard of care for dysbiotic diagnosis cases.
IMPORTANCE The possibility exists of the involvement of a new molecular test in the routine algorithm of bacterial vaginosis diagnosis in microbiology laboratories. This manuscript reports on our experience, and we propose an organization combining Nugent scoring and molecular testing, especially for intermediate Nugent scores.
KEYWORDS: Nugent scoring, molecular test, Lactobacillus, Gardnerella vaginalis, Atopobium vaginae, bacterial vaginosis
INTRODUCTION
Bacterial vaginosis (BV) corresponds to a vaginal dysbiosis, the main cause of vaginal discharge in women, affecting 29% of the overall population (1–3). In 1955, Gardner and Dukes first defined and described BV. Despite its benign character for nonpregnant women, in pregnant women, BV cause spontaneous abortions, premature births, premature membranes ruptures, or chorioamnionitis (4–6).
BV is characterized by a shift in vaginal flora from the dominant Lactobacillus species (which normally represent 95% of the total bacteria of the vaginal flora [7–9]) to a polymicrobial anaerobe-dominated microbiota, including especially Gardnerella vaginalis, Atopobium vaginae, Mobiluncus sp., Prevotella, Bacteroides sp., and Peptostreptococcus sp. (3). This modification within the vaginal microbiota is also associated with biochemical and cytological changes, most commonly pathognomonic of BV (10).
In routine clinical practice, BV diagnosis is based on a set of criteria rather than on the detection of a specific causative microorganism. Indeed, BV can be diagnosed clinically by using the Amsel’s criteria (10) and/or by using Nugent scoring (NS) after Gram staining (11). BV is defined by the Amsel’s criteria if three out of the four of the following criteria are met: presence of thin watery homogenous discharge; elevated vaginal pH (>4.5); “fishy” smell either spontaneously or after the addition of 10% potassium hydroxide to vaginal secretions (“whiff test”); and direct microscopic examination revealing vaginal clue cells (exocervical cells lined with small Gram-positive or-negative bacilli) (10, 12, 13). Exploring the cervicovaginal microbiome can also allow the characterization of normal and healthy vaginal ecosystems or modified ones based on their composition. Thus, it has been revealed in 2011 that the vaginal microbiome contains different bacterial communities clustered into five groups: four were dominated by Lactobacillus iners, L. crispatus, L. gasseri, or L. jensenii, whereas the fifth had lower proportions of lactic acid bacteria and higher proportions of strictly anaerobic organisms (14). However, this innovative method remains costly and not available in all laboratories.
Therefore, the NS is the most widely used microbiological method for BV diagnosis in routine clinical practice. After Gram staining, microscopic observation allows for evaluation the presence of lactobacilli, as well as some anaerobic microorganisms such as Gardnerella vaginalis or Mobiluncus sp.
With an experienced technologist, Gram staining is more sensitive, whereas the clinical Amsel criteria are more specific. Overall, a concordance of 80% to 90% has been reported (15). In routine clinical practice, the NS is currently the reference test available in around 3 h for BV diagnosis. As a comparison, Amsel’s criteria are based on nonquantifiable, nonreproducible clinical symptoms only (16–19).
Although Gram staining has been widely used for almost 3 decades and is considered the “reference test” of BV diagnosis, this method is not without limitations. For example, this method is often subject to interobserver variability depending on the observer’s skills and experience (20, 21); this technique is also proven to be limited in detecting some species such as A. vaginae, Ureaplasma spp., and Mycoplasma spp.
Recent studies suggest that molecular diagnostic tools would be beneficial to improve BV diagnosis efficiency and that nucleic acid amplification, targeting several BV-associated bacteria, could be performed (22). The Aptima BV assay is an in vitro nucleic acid amplification test that uses a real-time transcription-mediated amplification (TMA) for detection and quantitation of rRNA from bacteria associated with BV, including Lactobacillus (L. gasseri, L. crispatus, and L. jensenii), Gardnerella vaginalis, and Atopobium vaginae. The assay reports a qualitative result for BV, according to a specific algorithm based on the quantification and ratio of these microorganisms detected or not. However, it does not report results for each microorganism. The assay is intended to help in BV diagnosis using the automated Panther system with dedicated clinician- or patient-collected vaginal swab samples from females with a clinical suspicion or presenting profiles consistent with vaginosis.
This study aimed to assess the performance of the Aptima BV kit on the Panther automation system for the detection of BV through a prospective analysis of a routine clinical practice use of the technique. The experiment was conducted using vaginal samples (VS) taken from nonpregnant women over 18 years of age.
RESULTS
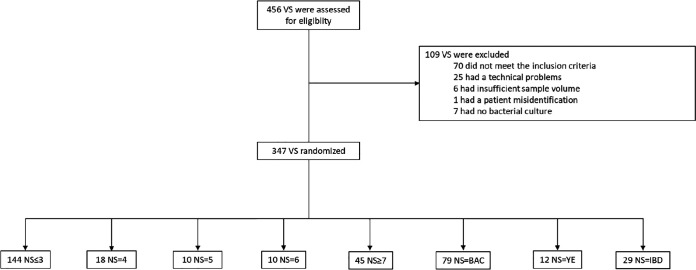
Only specimens meeting the study inclusion criteria were included in the analysis (Fig. 1).
FIG 1.
Study flow chart of the vaginal specimens included. NS, Nugent scoring; VS, vaginal samples; BAC, vaginal samples with various bacteria (excluding vaginal flora); YE, vaginal samples with predominance of yeast; IBD, vaginal samples with insufficient bacteria density.
Characteristics of women enrolled in the study.
A total of 456 women were enrolled in this study; 347 patients met the inclusion criteria with all data available for the analysis. The demographic characteristics associated with these 347 specimens prospectively analyzed in this study are presented in Table 1. Most of the samples (61.7%) were collected from the gynecological emergency ward, from the family planning department, and from hospitalized and outpatients, represented by 6.3%, 7.5%, and 24.5% of the total study population, respectively. According to NS interpretation (positive, negative, or intermediate) after an expert reading by the technician, among the 347 women selected for vaginosis, there were 144 patients (41.5%) with a normal vaginal microbiota (low NS). There were 45 patients (13%) with a positive NS consistent with BV and 38 patients (11%) presenting with an intermediate vaginal microbiota. Finally, 79 patients (22.7%) had various bacteria (excluding vaginal flora), 29 (8.3%) had insufficient bacterial density, and 12 (3.5%) had predominance of yeasts according to NS (Table 1).
TABLE 1.
The Aptima BV assay validationa
| Patient sample characteristics | Nugent score (0–3) n = 144 | Nugent score (4–6) n = 38 | Nugent score (7–10) n = 45 | BAC n = 79 | YE n = 12 | IBD n = 29 |
|---|---|---|---|---|---|---|
| Median age in yrs; (IQR) | 31 (25–38) | 33.5 (25.5–37.75) | 31 (25–38) | 33 (27–38) | 29.5 (27–32.25) | 35 (32–44) |
| Hospital type: | ||||||
| Gynecological emergency | 82 (57%) | 28 (73.7%) | 25 (55.6%) | 56 (70.9%) | 7 (58.3 %) | 16 (64%) |
| Hospitalised patient | 4 (2.8%) | 3 (7.9%) | 2 (4.4%) | 9 (11.4%) | 0 (0 %) | 8 (32%) |
| Outpatient | 48 (33.3%) | 6 (15.8%) | 12 (26.7%) | 12 (15.2%) | 3 (25 %) | 4 (16%) |
| Family planning services | 10 (6.9%) | 1 (2.6%) | 6 (13.3%) | 2 (2.5%) | 2 (16.7 %) | 1 (4%) |
| Presence of bacterial vaginosis with the vaginosis Aptima BV kit | 8 (5.6%) | 15 (39%) | 41 (91.1%) | 23 (29.1 %) | 5 (41.7 %) | 2 (6.9 %) |
BAC, vaginal samples with various bacteria (excluding vaginal flora); YE, vaginal samples with predominance of yeasts; IBD, vaginal sample with insufficient bacterial density; IQR, interquartile range.
The Aptima BV assay assessment.
For BV diagnosis, sensitivity, specificity, PPV, and NPV of the Aptima BV test were calculated using all samples (n = 189) for which both NS and Aptima BV results were conclusive (either positive or negative) for diagnosis of BV, excluding then the intermediate NS. The Aptima BV test was highly sensitive (91.1%) and specific (94.4%) for diagnosis of BV, with a PPV of 83.7% and a NPV of 97.1% for BV (Table 2).
TABLE 2.
Test characteristics of the Aptima bacterial vaginosis assay for BV diagnosis
| Aptima bacterial vaginosis assay performance on Panther system |
||||
|---|---|---|---|---|
| Reference method: Nugent score (NS) | True positive samples | True negative samples | False positive samples | False negative samples |
| NS ≤ 3 | 0 | 136 | 8 | 0 |
| NS ≥ 7 | 41 | 0 | 0 | 4 |
A total of 91.1% of NS up to 7 were considered a BV with the Aptima BV test, whereas only 5.6% were also considered positive in the NS less than 3. It is important to note that 39% of intermediate NS was considered BV at the molecular level, which could send a signal in case of doubt (Table 1). It is interesting to note that the higher the NS, the more positive the Aptima BV test is. Indeed, for the 10 NS = 6, 10 NS = 5, and the 18 NS = 4, 70%, 50%, and 16.7%,respectively, were positive with the Aptima BV test.
According to our routine clinical practice organization, the NS was not evaluable for 120 samples. However, when the presence of many bacteria was revealed on direct examination, 29.1% were positive with the Aptima BV test. Coinfection with candidiasis and bacterial dysbiosis was found in almost 42% when yeasts were reported after direct examination. Finally, the Aptima BV test revealed BV in only 6.9% when an insufficient bacterial density on direct examination was observed.
DISCUSSION
The diagnosis of BV remains a challenge in a microbiology laboratory. Most laboratories still use labor-intensive tests. The reference test (23, 24) still relies on a labor-intensive and time-consuming test performed by experienced technicians (24, 25). This results in consistent difficulties in Gram staining reading. Moreover, despite the formation and the need of a second reading sometimes, the NS may yield inter and intravariability results (26). However, the categorical cutoff value definition allows us to segregate among three groups with an intermediary score between the negative one (score less than 3 = normal flora) and positive one (up to 7 = high probability of bacterial vaginosis). Culture is not always performed routinely. In several cases, the availability of a rapid molecular test may help microbiologists with the diagnosis and clinicians with the management of patients.
Although traditional methods that diagnose BV have relied much more on methods such as the microscopic assessment of bacterial morphotypes (NS) or some combination of patient examination and vaginal discharge (Amsel’s criteria [10]), there exists limited data on molecular detection and BV diagnosis in routine clinical practice and in real life. Although this investigational test is an FDA-approved nucleic acid amplification test for detection of the major causes of bacterial vaginosis, there are limited data on the accuracy of this method (i) to compare with this study, (ii) to assess the relevance of the three-bacteria group detection, and (iii) to implement the routine into clinical practice for a well-defined population.
However, the results of this monocentric retrospective study demonstrate that the Aptima BV assay provides a BV diagnosis with a good correlation and a negative predictive value of 97.1% compared to the NS determination. This new test presents a higher sensitivity compared to previous molecular tests evaluated (24). Indeed, the Aptima BV test assessed during this study presents a 91.1% sensitivity and a 94.4% specificity, whereas the Aptima IVD, BD Affirm, and Hologic ASR revealed 84.4%, 86.7%, and 75.6% sensitivity, and 86.3%, 60.6%, and 81.8% specificity, respectively (24).
Recent research by Frederick et al. has indicated more complex process implicating the role of bacterial pathogens in the etiology of BV (27–29). Many of these organisms, such as Atopobium vaginae, Prevotella species, and others, can be detected only by amplification tests (18). Indeed, despite the increasing use of MALDI-TOF technology in microbiology laboratories (with large databases), the identification of these fastidious microorganisms remains challenging, necessitating anaerobic prolonged cultures, not compatible with the delay of results (turnaround time) in the routine clinical practice of BV diagnosis (30). The growth time of these microorganisms is long and some of them are uncultivable (30). Therefore, a molecular assay based on the presence of lactobacilli and the absence of deleterious organisms represents an opportunity to improve the diagnosis of BV in routine clinical practice (28, 31, 32).
Although this new assay performed well, a fuller understanding of its true performance is constrained by the known limitations of the reference methods. Indeed, by using Gram staining, a challenge exists to clearly distinguish Lactobacillus iners (type-III cervicovaginal microbiota) from G. vaginalis (33). However, the performance of tests for BV diagnosis could be relevant in the case of specimens with an intermediate NS. In the same way, to limit BV recurrence and optimize the safe and efficacious vaginal probiotic treatment, which reduces the negative effect of antibiotic treatment on other microbiomes or the emergence of antimicrobial resistance (34), this test allows for faster analysis times (3 h on average).
As BV has been significantly associated with preterm labor, premature membrane rupture, preterm delivery, miscarriage, birth asphyxia, low birth weight, and neonatal intensive care unit admission, this rapid molecular detection test may represent an option in early BV screening to prevent adverse maternal and fetal outcomes (35).
This study has limitations. First, as already highlighted in previous studies, the reference test based on the NS with the uncertainty of microscopy reading (24, 25), despite well-trained technicians for BV diagnosis, may explain discrepancies and constitutes a limitation. Second, future studies are needed and should be designed to analyze specific populations of women in order to determine the correlation between the treatment and clinical outcomes, especially in pregnant women. Third, molecular methods are highly specific, which can potentially impact the sensitivity to vaginosis due to minor strains or depending on the initial cervical microbiota. Indeed, this system also does not evaluate the presence of other bacterial strains such as Prevotella species, Megasphera, Mobiluncus species, Ureaplasma spp., and Mycoplasma spp. yet associated with BV.
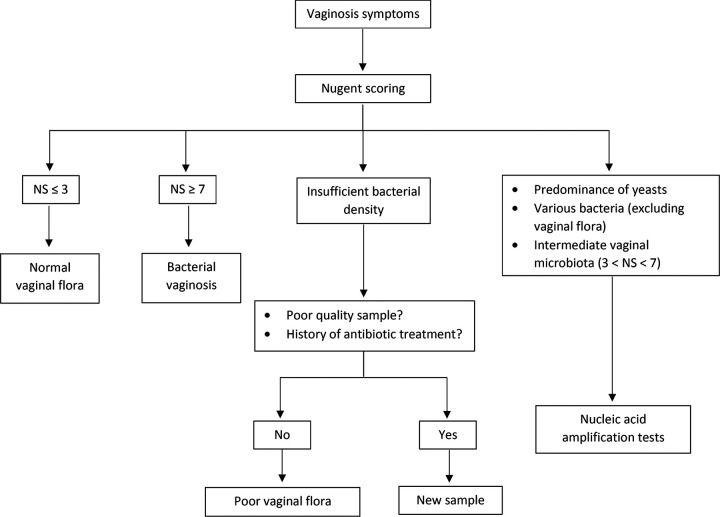
To conclude, the availability of a highly sensitive molecular test like the Aptima BV test may improve accurate diagnosis of BV and reduce the delay. In the future, the validation of such multiplexed sample-to-answer tests may change the way patients are managed, and we propose here an algorithm (Fig. 2). Research will be required to demonstrate performance and outcomes in various populations, including pregnant women, asymptomatic women, or women with intermediate NS. In summary, this test appears to be a promising molecular tool for BV routine clinical practice.
FIG 2.
Proposal of an algorithm for routine BV diagnosis. NS, Nugent’s scoring.
MATERIALS AND METHODS
Study design.
This study was designed as a monocentric, retrospective, observational study of adult women. From May to August 2019, we enrolled outpatients and inpatients, including nonpregnant women over18 years of age with vaginosis symptoms consulting at Nantes University hospital.
Ethics approval.
According to the French and European legislation, the use of data in this monocentric study does not need approval from the ethics committee. This study was recorded at Nantes University hospital by the local data privacy officer under the reference: TS005.BIO.AP.2019_14.
Samples for this study, results, and data had been recorded during normal medical care of patients by professionals who are following them. All the data collected for this new study from patient medical folders has been filed in a board under an anonymized code which cannot be tied to the patient.
Clinical samples.
Vaginal samples (VS) from the patients meeting the inclusion criteria were selected for the study. For the routine clinical practice diagnosis, the vaginal samples (collected using the Copan ESwab in Amies transport medium) for Gram staining and culture, if necessary, was received by the microbiology laboratory. These VS with sufficient volume for Gram staining and standard bacterial culture were also used for the assessment of the Aptima bacterial vaginosis kit on the Panther system. In the laboratory, 200 μL of Copan ESwab collection and transport system was transferred in a tube containing specimen transport media (STM) (Hologic Inc., San Diego, CA, USA) within 24 h of collection and tested immediately on the Panther platform.
Vaginal sample comparator testing.
Vaginal samples were tested for BV according to the standard laboratory protocol: NS was performed blind to the molecular test. Gram staining was read by experienced technicians (in case of doubt, a new independent reading was performed). According to Gram staining, (i) a large predominance of Gram-positive bacilli suggesting the presence of Lactobacillus spp. from different strains was consistent with a normal vaginal flora (NS between 0–3), (ii) a large predominance of Gram-variable bacilli suggesting the presence of G. vaginalis (presence of clue cells), Mobiluncus sp., or other anaerobe bacteria with a lack or total absence of Lactobacillus spp. was consistent with BV (NS between 7–10), and (iii) an intermediate NS (NS between 4–6) corresponded to potential transition from a normal vaginal flora to a dysbiosis. At Nantes University Hospital, in the microbiology laboratory, a bacterial culture was only performed in four cases: intermediate NS, presence of yeasts on direct examination (YE), insufficient bacterial density on direct examination (IBD) and presence of numerous bacteria on direct examination that did not belong to the vaginal flora (BAC). All media blood (Thermo Fisher, Basingstoke, United Kingdom) and chocolate (Becton, Dickinson, Heidelberg, Germany) agar plates, were incubated at 35 to 37°C in ambient air for 48 h.
Aptima BV assay.
Each tube containing STM (Hologic Inc., San Diego, CA, USA) was inserted directly into the Panther platform. The Panther system detects and discriminates between four fluorescent signals corresponding to the Lactobacillus group, Atopobium vaginae, Gardnerella vaginalis, and internal control (IC) amplification products. The Panther system software compares signal emergence times for each target microorganism to calibration information to determine either positive BV status, negative BV status, or invalid test of each sample.
An IC is added to each sample. During processing, IC acceptance criteria are automatically verified by the Panther system software. If an IC result is invalid, the sample result is invalidated. Every sample with an invalid IC result must be retested to obtain a valid result.
Results and discrepant analysis.
Performance of the Aptima BV test was assessed routinely by comparing the results to real life NS scores 0 to 3 (normal vaginal flora) and 7 to 10 (bacterial vaginosis). An Aptima BV result was considered a true positive (TP) or a true negative (TN) only when it matched the result from the comparator method: NS for bacterial culture.
Calculations and statistical analysis.
Sensitivity was calculated as 100× (no. of TP/[no. of TP + no. of FN]), and specificity was calculated as 100× (no. of TN/[no. of TN + no. of FP]). Positive predictive value was calculated as 100× (no. of TP/[no. of TP + no. of FP]), and negative predictive value was calculated as 100× (no. of TN/[no. of TN + no. of FN]).
ACKNOWLEDGMENTS
We thank all the lab technicians for their daily involvement in the NS reading. We also thank the HOLOGIC company for providing laboratory diagnostic reagents and technical help. They did not participate in data collection, analysis, the decision to publish or the preparation of this manuscript.
This research project was carried out as part of our routine work and received no external funding.
We declare no conflict of interest.
Contributor Information
S. Corvec, Email: stephane.corvec@chu-nantes.fr.
Salika M. Shakir, University of Utah and ARUP Laboratories
REFERENCES
- 1.Morris M, Nicoll A, Simms I, Wilson J, Catchpole M. 2001. Bacterial vaginosis: a public health review. BJOG 108:439–450. doi: 10.1111/j.1471-0528.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- 2.van de Wijgert JHHM, Borgdorff H, Verhelst R, Crucitti T, Francis S, Verstraelen H, Jespers V. 2014. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One 9:e105998. doi: 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnall P, Rizzolo D. 2017. Bacterial vaginosis: a practical review. JAAPA 30:15–21. doi: 10.1097/01.JAA.0000526770.60197.fa. [DOI] [PubMed] [Google Scholar]
- 4.Taylor BD, Darville T, Haggerty CL. 2013. Does bacterial vaginosis cause pelvic inflammatory disease? Sex Transm Dis 40:117–122. doi: 10.1097/OLQ.0b013e31827c5a5b. [DOI] [PubMed] [Google Scholar]
- 5.Sobel R, Sobel JD. 2015. Metronidazole for the treatment of vaginal infections. Expert Opin Pharmacother 16:1109–1115. doi: 10.1517/14656566.2015.1035255. [DOI] [PubMed] [Google Scholar]
- 6.Laxmi U, Agrawal S, Raghunandan C, Randhawa VS, Saili A. 2012. Association of bacterial vaginosis with adverse fetomaternal outcome in women with spontaneous preterm labor: a prospective cohort study. J Matern-Fetal Neonatal Med off J Eur Assoc Perinat Med Fed Asia Ocean Perinat Soc Int Soc Perinat Obstet 25:64–67. [DOI] [PubMed] [Google Scholar]
- 7.Spiegel CA, Amsel R, Eschenbach D, Schoenknecht F, Holmes KK. 1980. Anaerobic bacteria in nonspecific vaginitis. N Engl J Med 303:601–607. doi: 10.1056/NEJM198009113031102. [DOI] [PubMed] [Google Scholar]
- 8.Eschenbach DA. 1989. Bacterial vaginosis: emphasis on upper genital tract complications. Obstet Gynecol Clin North Am 16:593–610. doi: 10.1016/S0889-8545(21)00410-1. [DOI] [PubMed] [Google Scholar]
- 9.France M, Alizadeh M, Brown S, Ma B, Ravel J. 2022. Towards a deeper understanding of the vaginal microbiota. Nat Microbiol 7:367–378. doi: 10.1038/s41564-022-01083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. 1983. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med 74:14–22. doi: 10.1016/0002-9343(83)91112-9. [DOI] [PubMed] [Google Scholar]
- 11.Nugent RP, Krohn MA, Hillier SL. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol 29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eschenbach DA, Hillier S, Critchlow C, Stevens C, DeRouen T, Holmes KK. 1988. Diagnosis and clinical manifestations of bacterial vaginosis. Am J Obstet Gynecol 158:819–828. doi: 10.1016/0002-9378(88)90078-6. [DOI] [PubMed] [Google Scholar]
- 13.Mohammadzadeh F, Dolatian M, Jorjani M, Alavi Majd H. 2014. Diagnostic value of Amsel’s clinical criteria for diagnosis of bacterial vaginosis. Glob J Health Sci:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livengood CH. 2009. Bacterial Vaginosis: an Overview for 2009. Rev Obstet Gynecol 2:28–37. [PMC free article] [PubMed] [Google Scholar]
- 16.Chawla R, Bhalla P, Chadha S, Grover S, Garg S. 2013. Comparison of Hay’s criteria with Nugent’s scoring system for diagnosis of bacterial vaginosis. Biomed Res Int 2013:365194. doi: 10.1155/2013/365194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amegashie CP, Gilbert NM, Peipert JF, Allsworth JE, Lewis WG, Lewis AL. 2017. Relationship between nugent score and vaginal epithelial exfoliation. PLoS One 12:e0177797. doi: 10.1371/journal.pone.0177797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman JS, Gaydos CA. 2018. Molecular diagnosis of bacterial vaginosis: an update. J Clin Microbiol 56. doi: 10.1128/JCM.00342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonucci F, Mirandola W, Fontana C. 2017. Comparison between Nugent’s and Hay/Ison scoring criteria for the diagnosis of bacterial vaginosis in WASP prepared vaginal samples. Clin Invest 7(3). [Google Scholar]
- 20.Klebanoff MA, Schwebke JR, Zhang J, Nansel TR, Yu KF, Andrews WW. 2004. Vulvovaginal symptoms in women with bacterial vaginosis. Obstet Gynecol 104:267–272. doi: 10.1097/01.AOG.0000134783.98382.b0. [DOI] [PubMed] [Google Scholar]
- 21.Modak T, Arora P, Agnes C, Ray R, Goswami S, Ghosh P, Das NK. 2011. Diagnosis of bacterial vaginosis in cases of abnormal vaginal discharge: comparison of clinical and microbiological criteria. J Infect Dev Ctries 5:353–360. doi: 10.3855/jidc.1153. [DOI] [PubMed] [Google Scholar]
- 22.Plummer EL, Garland SM, Bradshaw CS, Law MG, Vodstrcil LA, Hocking JS, Fairley CK, Tabrizi SN. 2017. Molecular diagnosis of bacterial vaginosis: does adjustment for total bacterial load or human cellular content improve diagnostic performance? J Microbiol Methods 133:66–68. doi: 10.1016/j.mimet.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 23.Forsum U, Jakobsson T, Larsson PG, Schmidt H, Beverly A, Bjørnerem A, Carlsson B, Csango P, Donders G, Hay P, Ison C, Keane F, McDonald H, Moi H, Platz-Christensen J-J, Schwebke J. 2002. An international study of the interobserver variation between interpretations of vaginal smear criteria of bacterial vaginosis. APMIS 110:811–818. doi: 10.1034/j.1600-0463.2002.1101107.x. [DOI] [PubMed] [Google Scholar]
- 24.Richter SS, Otiso J, Goje OJ, Vogel S, Aebly J, Keller G, Van Heule H, Wehn D, Stephens AL, Zanotti S, Johnson T, Leal SM, Procop GW. 2019. Prospective evaluation of molecular assays for diagnosis of vaginitis. J Clin Microbiol 58:e01264-19. doi: 10.1128/JCM.01264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Veer C, van Houdt R, van Dam A, de Vries H, Bruisten S. 2018. Accuracy of a commercial multiplex PCR for the diagnosis of bacterial vaginosis. J Med Microbiol 67:1265–1270. doi: 10.1099/jmm.0.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dols JAM, Molenaar D, van der Helm JJ, Caspers MPM, de Kat Angelino-Bart A, Schuren FHJ, Speksnijder AGCL, Westerhoff HV, Richardus JH, Boon ME, Reid G, de Vries HJC, Kort R. 2016. Molecular assessment of bacterial vaginosis by Lactobacillus abundance and species diversity. BMC Infect Dis 16:180. doi: 10.1186/s12879-016-1513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 28.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. 2007. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol 45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onderdonk AB, Delaney ML, Fichorova RN. 2016. The Human Microbiome during Bacterial Vaginosis. Clin Microbiol Rev 29:223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Hanlon DE, Moench TR, Cone RA. 2013. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One 8:e80074. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shipitsyna E, Roos A, Datcu R, Hallén A, Fredlund H, Jensen JS, Engstrand L, Unemo M. 2013. Composition of the vaginal microbiota in women of reproductive age–sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS One 8:e60670. doi: 10.1371/journal.pone.0060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrova MI, Reid G, Vaneechoutte M, Lebeer S. 2017. Lactobacillus iners: friend or Foe? Trends Microbiol 25:182–191. doi: 10.1016/j.tim.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 34.van de Wijgert JHHM, Verwijs MC, Agaba SK, Bronowski C, Mwambarangwe L, Uwineza M, Lievens E, Nivoliez A, Ravel J, Darby AC. 2020. Intermittent Lactobacilli-containing vaginal probiotic or metronidazole use to prevent bacterial vaginosis recurrence: a pilot study incorporating microscopy and sequencing. Sci Rep 10:3884. doi: 10.1038/s41598-020-60671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhakta V, Aslam S, Aljaghwani A. 2021. Bacterial vaginosis in pregnancy: prevalence and outcomes in a tertiary care hospital. Afr J Reprod Health 25:49–55. [DOI] [PubMed] [Google Scholar]